Abstract
The early detection of lung cancer is a major clinical challenge. Long noncoding RNAs (lncRNAs) have important functions in tumorigenesis. Plasma lncRNAs directly released from primary tumors or the circulating cancer cells might provide cell-free cancer biomarkers. The objective of this study was to investigate whether the lncRNAs could be used as plasma biomarkers for early-stage lung cancer. By using droplet digital polymerase chain reaction, we determined the diagnostic performance of 26 lung cancer–associated lncRNAs in plasma of a development cohort of 63 lung cancer patients and 33 cancer-free individuals, and a validation cohort of 39 lung cancer patients and 28 controls. In the development cohort, 7 of the 26 lncRNAs were reliably measured in plasma. Two (SNHG1 and RMRP) displayed a considerably high plasma level in lung cancer patients vs. cancer-free controls (all P < .001). Combined use of the plasma lncRNAs as a biomarker signature produced 84.13% sensitivity and 87.88% specificity for diagnosis of lung cancer, independent of stage and histological type of lung tumor, and patients' age and sex (all P > .05). The diagnostic value of the plasma lncRNA signature for lung cancer early detection was confirmed in the validation cohort. The plasma lncRNA signature may provide a potential blood-based assay for diagnosing lung cancer at the early stage. Nevertheless, a prospective study is warranted to validate its clinical value.
Introduction
Approximately 155,870 Americans will die from lung cancer each year, more than the other 3 leading cancers combined (breast, prostate, and colorectal cancers). Over 85% lung cancers are non–small cell lung cancers (NSCLCs). NSCLC mainly consists of adenocarcinoma (AC) and squamous cell carcinoma (SCC). Tobacco smoking is the major cause of NSCLC. The early detection of lung cancer in a large randomized trial using low-dose CT (LDCT) has revealed a 20% reduction in mortality as compared to chest X-rays [1]. Therefore, LDCT is recently recommended to be used for lung cancer early detection among smokers [2], [3]. However, LDCT is associated with overdiagnosis, excessive cost, and radiation exposure [2], [3]. The development of noninvasive or circulating biomarkers that can accurately and cost-effectively diagnose early-stage lung cancer is required [4].
Long noncoding RNAs (lncRNAs) have minimum transcript length of 200 bp and play vital roles in various biological processes [5], [6]. lncRNAs can regulate different molecular signaling pathways via changing gene expression, and therefore, their dysregulations are implicated in numerous mechanisms of carcinogenesis [7], [8]. Dysregulation of some lncRNAs has been found in relation to oncogenesis and metastasis of lung tumor [9], [10], [11]. Importantly, plasma lncRNAs directly released from primary tumors or the circulating cancer cells might provide biomarkers for human malignancies [12]. To date, several plasma lncRNAs have been identified that show the potential to distinguish lung cancer patients from noncancer subjects [12], [13], [14], [15], [16], [17], [18], [19], [20]. Yet none of them has been accepted in the clinical settings for lung cancer diagnosis, mainly due to the low sensitivity and specificity.
Recent studies have characterized 21 lncRNAs whose aberrations are associated with lung cancer [10], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]. Furthermore, using whole-genomic next-generation sequencing (NGS) to analyze ncRNA profile of primary lung tumor tissues, we recently identified additional five lung cancer–related lncRNAs [47], [48]. These lung tumor–associated lncRNAs may provide a comprehensive list of biomarker candidates for developing circulating lung cancer biomarkers. The objective of this study was to investigate whether the lung cancer–associated lncRNAs could be used as plasma biomarkers for lung cancer.
Materials and Methods
Patients and Clinical Specimens
This study was approved by the Institutional Review Boards of University of Maryland Baltimore and Veterans Affairs Maryland Health Care System. We recruited lung cancer patients and cancer-free smokers by using the inclusion and/or exclusion criteria recommended by the US Preventive Services Task Force for lung cancer screening in heavy smokers [49]. We collected blood in BD Vacutainer spray-coated K2EDTA Tubes (BD, Franklin Lakes, NJ) and prepared plasma using the standard operating protocols developed by The NCI-Early Detection Research Network [50], [51]. The specimens were processed within 2 hours of collection by centrifugation at 1300×g for 10 minutes at 4°C. A total of 102 NSCLC patients and 55 cancer-free smokers were recruited. Among the cancer patients, 24 patients were female and 78 were male. Twenty-three had stage I NSCLC, 18 with stage II, 28 with stage III, 28 with stage IV, and 5 with unknown stage. Of the cancer-free smokers, 14 patients were female and 41 were male. There were no significant differences of age, gender, and smoking status between the NSCLC patients and cancer-free smokers. The cases and controls were randomly grouped into two cohorts: a development cohort and a validation cohort. The development cohort consisted of 63 lung cancer patients and 33 cancer-free smokers, while the validation cohort comprised 39 lung cancer patients and 28 cancer-free smokers. The demographic and clinical variables of the two cohorts are shown in Tables 1 and 2.
Table 1.
Characteristics of NSCLC Patients and Cancer-Free Smokers in a Development Cohort
| NSCLC Cases (n = 63) | Controls (n = 33) | P Value | |
|---|---|---|---|
| Age | 67.93 (SD 9.16) | 63.79 (SD 16.12) | .18 |
| Sex | .36 | ||
| Female | 15 | 8 | |
| Male | 48 | 25 | |
| Smoking pack-years (median) | 32.1 | 31.76 | .19 |
| Stage | |||
| Stage I | 14 | ||
| Stage II | 10 | ||
| Stage III | 17 | ||
| Stage IV | 18 | ||
| Unknown | 4 | ||
| Histological type | |||
| Adenocarcinoma | 32 | ||
| Squamous cell carcinoma | 31 |
Table 2.
Characteristics of NSCLC Patients and Cancer-Free Smokers in a Validation Cohort
| NSCLC Cases (n = 39) | Controls (n = 28) | P Value | |
|---|---|---|---|
| Age | 66.58 (SD 9.93) | 63.68 (SD 13.27) | .25 |
| Sex | .45 | ||
| Female | 9 | 6 | |
| Male | 30 | 22 | |
| Smoking pack-years (median) | 33.39 | 29.64 | .26 |
| Stage | |||
| Stage I | 9 | ||
| Stage II | 8 | ||
| Stage III | 11 | ||
| Stage IV | 10 | ||
| Unknown | 1 | ||
| Histological type | |||
| Adenocarcinoma | 22 | ||
| Squamous cell carcinoma | 17 |
RNA Isolation and Quantitative Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
RNA was extracted from the specimens by using Trizol LS reagent (Invitrogen, Carlsbad, CA) and RNeasy Mini Kit (Qiagen, Hilden, Germany) [52], [53], [54]. RT was carried out to generate cDNA by using an RT Kit (Applied Biosystems, Foster City, CA) as described in our published works [52], [53], [54]. PCR was performed to measure expressions of target genes by using a PCR kit (Applied Biosystems) on a Bio-Rad IQ5 Multicolor RT-PCR Detection System (Bio-Red, Hercules, CA). Expression levels of the genes were determined using comparative cycle threshold (CT) method with miR-1228 as an internal control. The targeted genes with CT values >35 were considered to be below the detection level of qRT-PCR [55].
Droplet Digital PCR (ddPCR)
ddPCR for analysis of the genes was performed as described in our previous work. Briefly, TaqMan reaction mix (Applied Biosystems) containing sample cDNA was partitioned into aqueous droplets in oil via the QX100 Droplet Generator (Bio-Rad) and then transferred to a 96-well PCR plate. A two-step thermocycling protocol (95°C × 10 minutes; 40 cycles of 94°C × 30 seconds and 60°C × 60 seconds, and 98°C × 10 minutes) was undertaken in a Bio-Rad C1000 (Bio-Rad). The PCR plate was loaded on Droplet Reader (Bio-Rad), by which copy number of each gene per μl PCR reaction was directly determined. We used QuantaSoft 1.7.4 analysis software (Bio-Rad) and Poisson statistics to compute droplet concentrations (copies/μl). Only genes that had at least 10,000 droplets were considered to be robustly detectable by ddPCR in plasma and subsequently underwent further analysis [56], [57]. All assays were done in triplicates, and one no-template control and two interplate controls were carried along in each experiment.
Statistical Analysis
Pearson correlation analysis was applied to assess relationship between gene expressions and demographic and clinical characteristics of the lung cancer patients and control individuals. The area under receiver operating characteristic curve (AUC) analyses were used to determine sensitivity, specificity, and corresponding cutoff value of each gene [58]. All P values shown were two sided, and a P value of < .05 was considered statistically significant.
Results
Developing a Plasma lncRNA Signature for Lung Cancer Early Detection
We first measured expression levels of the 26 lncRNAs (Supplementary Table 1) in plasma by using qRT-PCR in a discovery cohort of 63 cases and 33 controls. The lncRNAs had a CT value of ≥35 in 75% plasma samples. However, the internal control gene, miR-1228, stably displayed a CT value of 20-22 across the plasma samples. The results suggested that the amplification curves for the lncRNAs were not reliably generated, and their expression levels in plasma were too low to be detectable by qRT-PCR. We have proven that ddPCR is a direct method for absolutely and quantitatively measuring ncRNAs with a higher sensitivity compared with qRT-PCR [56], [57], [59]. Therefore, we used ddPCR to determine expression level of the lncRNAs in the plasma samples. Seven (26.9%) of the 26 lncRNAs could generated at least 10,000 droplets in each well of the plasma samples. Therefore, the seven lncRNAs could be successfully “read” by ddPCR for the absolute quantification in the plasma samples. The seven genes are SNHG1, MALAT1, HOTAIR, H19, MEG3, MEG8, and RMRP.
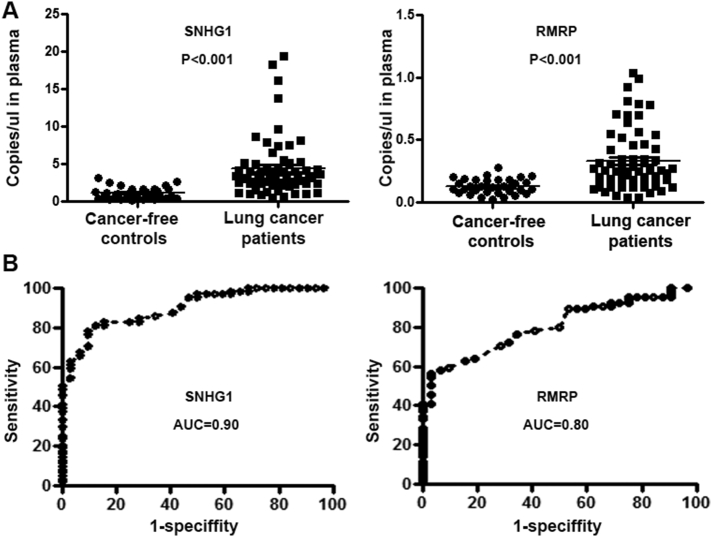
Among the seven genes, SNHG1 and RMRP had a higher expression level in plasma of lung cancer patients versus cancer-free controls (all P ≤ .05) (Figure 1A). Conversely, other five lncRNAs did not display a different plasma level in lung cancer cases versus cancer-free controls (all P ≥ .05). Furthermore, the plasma expression levels of the SNHG1 and RMRP were independent of stages and histological types of lung cancer. In addition, SNHG1 and RMRP exhibited AUC values of 0.90 and 0.80, respectively, in distinguishing NSCLC patients from healthy individuals (Figure 1B). Using Youden's index [60], we set up optimal cutoff for the two genes at 1.11 and 0.12, respectively. As a result, the use of the individual genes alone produced 61.00%-78.78% sensitivities and 87.88%-90.91% (Table 3). Combined use of the two genes based on at least one positive result in either SNHG1 or RMRP produced the highest classification accuracy (85.42%) compared to any one used alone (all P < .05) (Table 3). The two genes used in combination produced a sensitivity of 84.13% and a specificity of 87.88% for diagnosis of lung cancer, thus considerably improving the detection rate by a single gene with only a 2% decline in specificity (Table 3). Furthermore, the estimated correlation determined by Pearson correlation analysis among levels of the two lncRNAs was very low (R2 = 0.011, P = .53), further supporting that the combined analysis of the two genes outperformed a single one. In addition, combined analysis of the two plasma biomarkers did not show special association with stage and histological type of lung cancer and patients' age, gender, and smoking status (all P > .05).
Figure 1.
Expression levels of SNHG1 and RMRP in plasma samples of 63 lung cancer patients and 33 cancer-free controls. (A) SNHG1 and RMRP displayed a higher plasma level in lung cancer patients vs. cancer-free controls (all P < .001). (B) The receiver operating characteristic curves of SNHG1 and RMRP produced an AUC of 0.90 and 0.80, respectively, in diagnosis of lung cancer.
Table 3.
Diagnostic Performance of One-Gene vs. a Plasma lncRNA Signature for Lung Cancer Diagnosis in a Development Cohort
| Accuracy | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|
| SNHG1 | 81.25% (72.00%-88.49%) | 77.78% (65.54%-87.28%) | 87.88% (71.80%-96.60%) |
| RMRP | 71.88% (61.78%-80.58%) | 61.90% (48.80%-73.85%) | 90.91% (75.67%-98.08%) |
| A plasma lncRNA signature | 85.42% (76.74%-91.79%) | 84.13% (72.74%-92.12%) | 87.50% (71.80%-96.60%) |
Abbreviation: CI, confidence interval.
Validating the Plasma lncRNA Marker Signature in an Independent Set of Lung Cancer Patients and Controls
To evaluate the diagnostic performance of the biomarker signature, the two lncRNAs (SNHG1 and RMRP) were assessed by using ddPCR in plasma of additional 39 NSCLC patients and 28 healthy controls. The two genes used in combination could differentiate the NSCLC patients from healthy controls with 82.05% sensitivity and 83.36% specificity (Table 4). Furthermore, no statistically significant difference was found in the sensitivity and specificity of the biomarker signature for stages and histological types of NSCLC (all P > .05). Moreover, there was no association of expressions of the two genes with the age, gender, or smoking status of the lung cancer patients and normal individuals (all P > .05). Taken together, the results confirm the potential of combined use of the two lncRNAs as a plasma biomarker signature for the early detection of lung cancer.
Table 4.
Diagnostic Performance of One-Gene vs. a Plasma lncRNA Signature for Lung Cancer Diagnosis in a Validation Set
| Accuracy | Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|---|
| SNHG1 | 80.33% (68.16%-89.40%) | 76.92% (60.67%-88.87) | 86.36% (65.09%-97.09%) |
| RMRP | 72.13% (59.17%-82.85%) | 61.54% (44.62%-76.64%) | 90.91% (70.84%-98.88%) |
| A plasma lncRNA signature | 83.62% (71.91%-91.85%) | 82.05% (66.47%-92.46%) | 86.36% (65.09%-97.09%) |
Discussion
Circulating cell-free lncRNAs biomarkers show promise as biomarkers for cancer diagnosis. However, unlike other ncRNA (e.g., microRNAs), lncRNAs have the lowest levels in plasma among several different RNA species [61], presenting a major challenge in the development of cell-free lncRNAs as cancer biomarkers. For instance, Schlosser et al. recently demonstrated that expressions of lncRNAs were robustly detectable in tissues yet undetectable or sporadically measurable in the matched plasma by using qRT-PCR, a routine platform used for nucleic acid detection [61]. Regular qPCR has some limitations in determining expression of ncRNAs: i) It is an indirect and labor-consuming approach. ii) It requires an internal control gene for normalization. Yet none of the investigated genes has been accepted as a standard control. iii) Its sensitivity for a low copy number of genes is very low. Our current observations are consistent with Schlosser's finding [61]: of the 26 lung cancer–associated lncRNAs, none is reliably measurable in plasma using qRT-PCR. Therefore, conventional qPCR might not be an appropriate tool for the development of lncRNAs as circulating biomarkers given that circulating lncRNAs in body fluids are present in low abundance. We have shown that ddPCR is a direct method for sensitively measuring ncRNAs [56], [59] since it depends on limiting partition of the PCR volume, where a positive result of a large number of microreactions indicates the presence of a single molecule in a given reaction [62]. The number of positive reactions, together with Poisson's distribution, produces a straight and high-confidence measurement of the original target concentration. Importantly, ddPCR does not require a reliance on rate-based measurements (CT values), endogenous controls, and calibration curves and therefore overcome the obstacles linked to the regular qPCR in quantification of genes in plasma. Here we demonstrate that 7 of the 26 lung cancer–associated lncRNAs that are not detectable by qRT-PCR are robustly measurable by ddPCR in plasma. Therefore, ddPCR may address the limitations of the qPCR in quantification of lncRNAs in plasma and hence help develop the genes as cell-free cancer biomarkers.
The previous plasma lncRNA-based assays were mostly developed from the limited number of lung cancer–associated lncRNAs and only consisted of a single lncRNA gene [12], [13], [14], [15], [16], [17], [18], [19], [20]. Since lung tumor is a heterogeneous group of neoplasms and develops from a multitude of molecular changes, a single lncRNA-based assay may not achieve the performance required to move forward for clinically detecting lung cancer. The development of a panel of multiple biomarkers by integrating analysis of multifaceted and diverse lncRNAs would provide a synergistic test for lung cancer diagnosis. By searching published data, we found 21 lncRNAs whose malfunction was well characterized in lung tumorigenesis [10], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46]. Furthermore, by systematically and comprehensively defining ncRNA changes of NSCLC in surgical lung tumor tissues using whole-transcriptome NGS [47], [48], we recently identified additional five lung cancer–associated lncRNAs [47]. Both the published and our NGS-defined lncRNAs of lung tumors may provide a comprehensive set of biomarker candidates for lung cancer. From the 26 lncRNAs, our present study identified and optimized a plasma signature consisting of two lncRNAs that created a higher diagnostic value for lung cancer detection than did individual lncRNAs used alone [12], [13], [14], [15], [16], [17], [18], [19], [20]. In addition, the diagnostic performance of the biomarkers was further blindly validated in a different cohort, suggesting that the plasma signature might be a robust assay for lung cancer diagnosis. Moreover, the performance of this plasma lncRNA signature for lung cancer diagnosis was independent of tumor stage and histology. This might be an important characteristic if the plasma lncRNA signature is employed for identifying early-stage lung cancer.
The two lncRNAs (SNHG1 and RMRP) have diverse and important functions in lung tumorigenesis through regulating different molecular pathways. Elevated expression of SNHG1 was frequently observed in lung cancer tissues and significantly correlated with larger tumor size, advanced stage, lymph node metastasis, and poor overall survival of the patients [63]. Furthermore, SNHG1 could promote NSCLC progression of lung cancer via miR-101-3p/SOX9/Wnt/β-catenin regulatory network and miR-145-5p/ MTDH axis [63], [64]. In addition, SNHG1 plays an oncogenic role in lung squamous cell carcinoma through ZEB1 signaling pathway by inhibiting TAp63 [65]. RMRP is best known for being a component of the nuclear RNase MRP complex, which participates in the processing of ribosomal RNA to generate the short mature 5.8S rRNA [66] and cleaves B-cyclin mRNA, lowering B-cyclin levels during mitosis [67]. In addition, RMRP interacts with telomerase to form a complex with RNA-dependent RNA polymerase activity capable of synthesizing dsRNA precursors processed by DICER1 into siRNAs [68]. Moreover, RMRP is important for mitochondrial DNA replication and RNA processing [69]. Upregulation of RMRP is found in lung adenocarcinoma tissues [70]. RMRP might act as an oncogenic lncRNA to promote the expression of KRAS, FMNL2, and SOX9 by inhibiting miR-206 expression in lung cancer [70]. Our current study extends the previous findings by developing them as a biomarker signature that might be clinically useful in the early detection of lung cancer.
There are some limitations in this present study. 1) The size of the cohorts is small. Furthermore, the plasma samples were obtained from the hospital-based patients with clinical diagnosis. The participants might not be representative of high-risk populations (e.g., heavy smokers) in screening setting for lung cancer. We will perform a prospective and multisite lung cancer screening trial to validate the diagnostic value of the plasma lncRNA signature. 2) The early diagnosis of lung cancer by using LDCT could reduce the mortality [3]. However, LDCT has a low specificity for the early detection of lung cancer, presenting a major clinical challenge [71]. We are evaluating whether the plasma lncRNA signature could improve the specificity of LDCT for the early detection of NSCLC by specifically distinguishing malignant from benign pulmonary growths. 3) The cell-free circulating tumor DNA, microRNAs, or DNA methylation provides other important types of biomarkers for lung cancer [54], [72], [73], [74], [75], [76], [77], [78]. Our ongoing efforts are to investigate whether integrating the lncRNA signature with other types of biomarkers might improve the early detection of lung cancer. 4) In the current study, SNHG1 or RMRP expression level in plasma is independent of histological types of lung cancer. Furthermore, the combined use of the two genes is also not associated with histological types of lung cancer. The findings in plasma samples are inconsistent with the two previous studies in tissue specimens that showed that SNHG1 or RMRP was associated with SCC or AC, respectively [65], [70]. A new study using paired plasma and tumor tissue specimens from the same lung cancer patients is needed to validate the discrepancy and further understand the mechanism underlying the divergence.
Conclusions
A plasma lncRNA signature was developed that could differentiate early-stage NSCLC patients from cancer-free smokers. Nevertheless, undertaking a prospective study to further validate this plasma biomarker signature in a large cohort is required.
The following are the supplementary data related to this article.
Lung Cancer–Associated lncRNAs
Declarations
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
The Institutional Review Boards of University of Maryland Baltimore and Veterans Affairs Maryland Health Care System approved this study. Reference number is H40666.
Competing Interests
The authors declare no conflict of interest.
Funding
Grant support: This work was supported in part by NCI R21CA205746, VA Merit Award I01 CX000512, Award from the Geaton and JoAnn DeCesaris Family Foundation, UMD-UMB Research and Innovation Seed Grant, DoD-Idea Development Award, and Maryland Innovation Initiative (MII) Commercialization Program-Phase 1/2 Grant (F.J.)
Authors' Contributions
Y. L., Q. L., M. Z., and F. J. conducted the experiments and participated in study design, coordination, data interpretation, and preparing the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials
Not applicable.
Disclosure of Potential Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, Fagerstrom RM, Gareen IF, Gatsonis C, Marcus PM, Sicks JD. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patz EF, Jr., Pinsky P, Gatsonis C, Sicks JD, Kramer BS, Tammemagi MC, Chiles C, Black WC, Aberle DR. Overdiagnosis in low-dose computed tomography screening for lung cancer. JAMA Intern Med. 2014;174(2):269–274. doi: 10.1001/jamainternmed.2013.12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aberle DR, Berg CD, Black WC, Church TR, Fagerstrom RM, Galen B, Gareen IF, Gatsonis C, Goldin J, Gohagan JK. The National Lung Screening Trial: overview and study design. Radiology. 2011;258:243–253. doi: 10.1148/radiol.10091808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubers AJ, Prinsen CF, Sozzi G, Witte BI, Thunnissen E. Molecular sputum analysis for the diagnosis of lung cancer. Br J Cancer. 2013;109(3):530–537. doi: 10.1038/bjc.2013.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10:925–933. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. doi: 10.1155/2015/320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou M, Guo M, He D, Wang X, Cui Y, Yang H, Hao D, Sun J. A potential signature of eight long non-coding RNAs predicts survival in patients with non–small cell lung cancer. J Transl Med. 2015;13:231. doi: 10.1186/s12967-015-0556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Qiu M, Xu Y, Mao Q, Wang J, Dong G, Xia W, Yin R, Xu L. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumour Biol. 2015;36:9969–9978. doi: 10.1007/s13277-015-3714-6. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A. The long noncoding MALAT-1 RNA indicates a poor prognosis in non–small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 12.Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng J, Yang W, Yin J, Song Y. Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine (Baltimore) 2016;95(37) doi: 10.1097/MD.0000000000004608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non–small cell lung cancer. Int J Clin Exp Pathol. 2015;8:7887–7895. [PMC free article] [PubMed] [Google Scholar]
- 14.Li N, Feng XB, Tan Q, Luo P, Jing W, Zhu M, Liang C, Tu J, Ning Y. Identification of circulating long noncoding RNA Linc00152 as a novel biomarker for diagnosis and monitoring of non–small-cell lung cancer. Dis Markers. 2017;17:743–749. doi: 10.1155/2017/7439698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M, Tu J. Identification of circulating long noncoding RNA HOTAIR as a novel biomarker for diagnosis and monitoring of non–small cell lung cancer. Technol Cancer Res Treat. 2017;17:1533–1539. doi: 10.1177/1533034617723754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan L, Zhang L, Fan K, Wang JJ. Diagnostic significance of circulating long noncoding RNA PCAT6 in patients with non–small cell lung cancer. Onco Targets Ther. 2017;10:5695–5702. doi: 10.2147/OTT.S149314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan Q, Zuo J, Qiu S, Yu Y, Zhou H, Li N, Wang H, Liang C, Yu M, Tu J. Identification of circulating long non-coding RNA GAS5 as a potential biomarker for non–small cell lung cancer diagnosis, long non-coding RNA, plasma, GAS5, biomarker. Int J Oncol. 2017;50:1729–1738. doi: 10.3892/ijo.2017.3925. [DOI] [PubMed] [Google Scholar]
- 18.Zhu Q, Lv T, Wu Y, Shi X, Liu H, Song Y. Long non-coding RNA 00312 regulated by HOXA5 inhibits tumour proliferation and promotes apoptosis in non–small cell lung cancer. J Cell Mol Med. 2017;21:2184–2198. doi: 10.1111/jcmm.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, Zhang L, Yan S, Li W, Cui J, Zhu M, Xia N, Yang Y, Yuan J, Chen X. LncRNA16 is a potential biomarker for diagnosis of early-stage lung cancer that promotes cell proliferation by regulating the cell cycle. Oncotarget. 2017;8:7867–7877. doi: 10.18632/oncotarget.13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HM, Lu JH, Chen WY, Gu AQ. Upregulated lncRNA-UCA1 contributes to progression of lung cancer and is closely related to clinical diagnosis as a predictive biomarker in plasma. Int J Clin Exp Med. 2015;8:11824–11830. [PMC free article] [PubMed] [Google Scholar]
- 21.Wei MM, Zhou YC, Wen ZS, Zhou B, Huang YC, Wang GZ, Zhao XC, Pan HL, Qu LW, Zhang J. Long non-coding RNA stabilizes the Y-box-binding protein 1 and regulates the epidermal growth factor receptor to promote lung carcinogenesis. Oncotarget. 2016;7:59556–59571. doi: 10.18632/oncotarget.10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen L, Chen L, Wang Y, Jiang X, Xia H, Zhuang Z. Long noncoding RNA MALAT1 promotes brain metastasis by inducing epithelial-mesenchymal transition in lung cancer. J Neurooncol. 2015;121:101–108. doi: 10.1007/s11060-014-1613-0. [DOI] [PubMed] [Google Scholar]
- 23.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90–96. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li P, Li J, Yang R, Zhang F, Wang H, Chu H, Lu Y, Dun S, Wang Y, Zang W. Study on expression of lncRNA RGMB-AS1 and repulsive guidance molecule b in non–small cell lung cancer. Diagn Pathol. 2015;10:63–68. doi: 10.1186/s13000-015-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS, Feng XJ. Increased expression of the lncRNA PVT1 promotes tumorigenesis in non–small cell lung cancer. Int J Clin Exp Pathol. 2014;7:6929–6935. [PMC free article] [PubMed] [Google Scholar]
- 26.Whiteside EJ, Seim I, Pauli JP, O'Keeffe AJ, Thomas PB, Carter SL, Walpole CM, Fung JN, Josh P, Herington AC. Identification of a long non-coding RNA gene, growth hormone secretagogue receptor opposite strand, which stimulates cell migration in non–small cell lung cancer cell lines. Int J Oncol. 2013;43:566–574. doi: 10.3892/ijo.2013.1969. [DOI] [PubMed] [Google Scholar]
- 27.Hu T, Lu YR. BCYRN1, a c-MYC-activated long non-coding RNA, regulates cell metastasis of non–small-cell lung cancer. Cancer Cell Int. 2015;15:36–40. doi: 10.1186/s12935-015-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Li P, Zhao W, Yang R, Chen S, Bai Y, Dun S, Chen X, Du Y, Wang Y. Expression of long non-coding RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 2015;15:48–53. doi: 10.1186/s12935-015-0201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Z, Bo H, Gong Z, Lian Y, Li X, Zhang W, Deng H, Zhou M, Peng S, Li G. AFAP1-AS1, a long noncoding RNA upregulated in lung cancer and promotes invasion and metastasis. Tumour Biol. 2016;37:729–737. doi: 10.1007/s13277-015-3860-x. [DOI] [PubMed] [Google Scholar]
- 30.Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C, Chang C, Bi H, Zou J, Yao X. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol. 2014;53:380–388. doi: 10.1016/j.biocel.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Luo J, Tang L, Zhang J, Ni J, Zhang HP, Zhang L, Xu JF, Zheng D. Long non-coding RNA CARLo-5 is a negative prognostic factor and exhibits tumor pro-oncogenic activity in non–small cell lung cancer. Tumour Biol. 2014;35:11541–11549. doi: 10.1007/s13277-014-2442-7. [DOI] [PubMed] [Google Scholar]
- 32.Luo H, Sun Y, Wei G, Luo J, Yang X, Liu W, Guo M, Chen R. Functional characterization of long noncoding RNA Lnc_bc060912 in human lung carcinoma cells. Biochemistry. 2015;54:2895–2902. doi: 10.1021/acs.biochem.5b00259. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, Yin R. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non–small cell lung cancer. Tumour Biol. 2014;35:5375–5380. doi: 10.1007/s13277-014-1700-z. [DOI] [PubMed] [Google Scholar]
- 35.Qiu M, Xu Y, Wang J, Zhang E, Sun M, Zheng Y, Li M, Xia W, Feng D, Yin R. A novel lncRNA, LUADT1, promotes lung adenocarcinoma proliferation via the epigenetic suppression of p27. Cell Death Dis. 2015;6:18–22. doi: 10.1038/cddis.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L, Zhou XF, Pan GF, Zhao JP. Enhanced expression of long non-coding RNA ZXF1 promoted the invasion and metastasis in lung adenocarcinoma. Biomed Pharmacother. 2014;68:401–407. doi: 10.1016/j.biopha.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 37.Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH. Long noncoding RNA ANRIL promotes non–small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther. 2015;14:268–277. doi: 10.1158/1535-7163.MCT-14-0492. [DOI] [PubMed] [Google Scholar]
- 38.Yang R, Li P, Zhang G, Lu C, Wang H, Zhao G. Long non-coding RNA XLOC_008466 functions as an oncogene in human non–small cell lung cancer by targeting miR-874. Cell Physiol Biochem. 2017;42:126–136. doi: 10.1159/000477121. [DOI] [PubMed] [Google Scholar]
- 39.Sang H, Liu H, Xiong P, Zhu M. Long non-coding RNA functions in lung cancer. Tumour Biol. 2015;36:4027–4037. doi: 10.1007/s13277-015-3449-4. [DOI] [PubMed] [Google Scholar]
- 40.Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F, Song Y. A critical role for the long non-coding RNA GAS5 in proliferation and apoptosis in non–small-cell lung cancer. Mol Carcinog. 2015;54:1–12. doi: 10.1002/mc.22120. [DOI] [PubMed] [Google Scholar]
- 41.Han L, Kong R, Yin DD, Zhang EB, Xu TP, De W, Shu YQ. Low expression of long noncoding RNA GAS6-AS1 predicts a poor prognosis in patients with NSCLC. Med Oncol. 2013;30:694–699. doi: 10.1007/s12032-013-0694-5. [DOI] [PubMed] [Google Scholar]
- 42.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non–small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:16–22. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu FQ, Hu R, Wang MS. LncRNA HMlincRNA717 is down-regulated in non–small cell lung cancer and associated with poor prognosis. Int J Clin Exp Pathol. 2014;7:8881–8886. [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Wan L, Lu K, Sun M, Pan X, Zhang P, Lu B, Liu G, Wang Z. The long noncoding RNA MEG3 contributes to cisplatin resistance of human lung adenocarcinoma. PLoS One. 2015;10:11–17. doi: 10.1371/journal.pone.0114586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non–small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68–72. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non–small-cell lung cancer cell. PLoS One. 2013;8:65-60. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma J, Mannoor K, Gao L, Tan A, Guarnera MA, Zhan M, Shetty A, Stass SA, Xing L, Jiang F. Characterization of microRNA transcriptome in lung cancer by next-generation deep sequencing. Mol Oncol. 2014;8:1208–1219. doi: 10.1016/j.molonc.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao L, Ma J, Mannoor K, Guarnera MA, Shetty A, Zhan M, Xing L, Stass SA, Jiang F. Genome-wide small nucleolar RNA expression analysis of lung cancer by next-generation deep sequencing. Int J Cancer. 2015;136:623–629. doi: 10.1002/ijc.29169. [DOI] [PubMed] [Google Scholar]
- 49.Humphrey LL, Deffebach M, Pappas M, Baumann C, Artis K, Mitchell JP, Zakher B, Fu R, Slatore CG. Screening for lung cancer with low-dose computed tomography: a systematic review to update the US Preventive services task force recommendation. Ann Intern Med. 2013;159:411–420. doi: 10.7326/0003-4819-159-6-201309170-00690. [DOI] [PubMed] [Google Scholar]
- 50.Marks JR, Anderson KS, Engstrom P, Godwin AK, Esserman LJ, Longton G, Iversen ES, Mathew A, Patriotis C, Pepe MS. Construction and analysis of the NCI-EDRN breast cancer reference set for circulating markers of disease. Cancer Epidemiol Biomarkers Prev. 2015;24:435–441. doi: 10.1158/1055-9965.EPI-14-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuck MK, Chan DW, Chia D, Godwin AK, Grizzle WE, Krueger KE, Rom W, Sanda M, Sorbara L, Stass S. Standard operating procedures for serum and plasma collection: early detection research network consensus statement standard operating procedure integration working group. J Proteome Res. 2009;8:113–117. doi: 10.1021/pr800545q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma J, Jemal A, Smith R. Reply to lung cancer deaths averted by screening should be considered in the context of tobacco control policies. Cancer. 2013;119:3420–3421. doi: 10.1002/cncr.28192. [DOI] [PubMed] [Google Scholar]
- 53.Shen J, Liu Z, Todd NW, Zhang H, Liao J, Yu L, Guarnera MA, Li R, Cai L, Zhan M. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374–379. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, Guarnera M, Liao J, Chou A, Lu CL. Plasma microRNAs as potential biomarkers for non–small-cell lung cancer. Lab Invest. 2011;91(4):579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J Clin Microbiol. 2008;46:3798–3799. doi: 10.1128/JCM.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li N, Ma J, Guarnera MA, Fang H, Cai L, Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol. 2014;140:145–150. doi: 10.1007/s00432-013-1555-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma J, Li N, Guarnera M, Jiang F. Quantification of plasma miRNAs by digital PCR for cancer diagnosis. Biomark Insights. 2013;8:127–136. doi: 10.4137/BMI.S13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dodd LE, Pepe MS. Partial AUC estimation and regression. Biometrics. 2003;59:614–623. doi: 10.1111/1541-0420.00071. [DOI] [PubMed] [Google Scholar]
- 59.Li H, Jiang Z, Leng Q, Bai F, Wang J, Ding X, Li Y, Zhang X, Fang H, Yfantis HG. A prediction model for distinguishing lung squamous cell carcinoma from adenocarcinoma. Oncotarget. 2017;8:50704–50714. doi: 10.18632/oncotarget.17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schisterman EF, Perkins NJ, Liu A, Bondell H. Optimal cut-point and its corresponding Youden Index to discriminate individuals using pooled blood samples. Epidemiology. 2005;16:73–81. doi: 10.1097/01.ede.0000147512.81966.ba. [DOI] [PubMed] [Google Scholar]
- 61.Schlosser K, Hanson J, Villeneuve PJ, Dimitroulakos J, McIntyre L, Pilote L, Stewart DJ. Assessment of circulating LncRNAs under physiologic and pathologic conditions in humans reveals potential limitations as biomarkers. Sci Rep. 2016;6:36–39. doi: 10.1038/srep36596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Day E, Dear PH, McCaughan F. Digital PCR strategies in the development and analysis of molecular biomarkers for personalized medicine. Methods. 2013;59:101–107. doi: 10.1016/j.ymeth.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 63.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non–small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non–small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;20:17–23. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 65.Zhang HY, Yang W, Zheng FS, Wang YB, Lu JB. Long non-coding RNA SNHG1 regulates zinc finger E-box binding homeobox 1 expression by interacting with TAp63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–658. doi: 10.1016/j.biopha.2017.03.104. [DOI] [PubMed] [Google Scholar]
- 66.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, Kim J, Curtis J, Moad CA, Wohler CM. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016;30:1224–1239. doi: 10.1101/gad.276022.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang DD, Clayton DA. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science. 1987;235:1178–1184. doi: 10.1126/science.2434997. [DOI] [PubMed] [Google Scholar]
- 70.Meng Q, Ren M, Li Y, Song X. LncRNA-RMRP acts as an oncogene in lung cancer. PLoS One. 2016;11:164–184. doi: 10.1371/journal.pone.0164845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marcus PM. Lung cancer screening with low dose computed tomography (LDCT): looking back and moving forward. Ann Transl Med. 2015;3:41–46. doi: 10.3978/j.issn.2305-5839.2015.03.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leng Q, Lin Y, Jiang F, Lee CJ, Zhan M, Fang H, Wang Y. A plasma miRNA signature for lung cancer early detection. Oncotarget. 2017;8:111902–111911. doi: 10.18632/oncotarget.22950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin Y, Leng Q, Jiang Z, Guarnera MA, Zhou Y, Chen X, Wang H, Zhou W, Cai L, Fang H. A classifier integrating plasma biomarkers and radiological characteristics for distinguishing malignant from benign pulmonary nodules. Int J Cancer. 2017;141:1240–1248. doi: 10.1002/ijc.30822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma J, Li N, Lin Y, Gupta C, Jiang F. Circulating neutrophil microRNAs as biomarkers for the detection of lung cancer. Biomark Cancer. 2016;8:1–7. doi: 10.4137/BIC.S37333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma J, Lin Y, Zhan M, Mann DL, Stass SA, Jiang F. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest. 2015;95(10):1197–1206. doi: 10.1038/labinvest.2015.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen J, Jiang F. Applications of microRNAs in the diagnosis and prognosis of lung cancer. Expert Opin Med Diagn. 2012;6:197–207. doi: 10.1517/17530059.2012.672970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Normanno N, Denis MG, Thress KS, Ratcliffe M, Reck M. Guide to detecting epidermal growth factor receptor (EGFR) mutations in ctDNA of patients with advanced non–small-cell lung cancer. Oncotarget. 2017;8:12501–12516. doi: 10.18632/oncotarget.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Balgkouranidou I, Chimonidou M, Milaki G, Tsaroucha E, Kakolyris S, Georgoulias V, Lianidou E. SOX17 promoter methylation in plasma circulating tumor DNA of patients with non–small cell lung cancer. Clin Chem Lab Med. 2016;54:1385–1393. doi: 10.1515/cclm-2015-0776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lung Cancer–Associated lncRNAs
Data Availability Statement
Not applicable.