Abstract
Introduction
Enteroaggregative Escherichia coli (EAEC) has been implicated as an emerging cause of traveler’s diarrhea, persistent diarrhea among children, and immunocompromised patients. The present study aimed to investigate the prevalence of antibiotic resistance, extendedspectrum β-lactamase (ESBL) production, and virulence factors of EAEC isolates obtained from Iranian children suffered from diarrhea.
Materials and methods
In this cross-sectional study, from March 2015 to February 2016, 32 EAEC isolates were collected from fecal samples of children aged <12 years with diarrhea in southwest of Iran. All EAEC isolates identified using phenotypic and molecular methods and the cell line adhesion assay. Antimicrobial susceptibility testing was determined using disk diffusion method. The presence of virulence factors and ESBL resistance genes were determined by polymerase chain reaction.
Results
Overall, 28.1% (9/32) of the isolates were positive for at least one of virulence genes. The most frequent gene was aap with a frequency of 96.9%. Neither aafA nor aggA gene was detected among all of the EAEC isolates. Antimicrobial susceptibility testing revealed the highest resistance rate to ampicillin (100%) and co-trimoxazole (100%), followed by ceftriaxone (81.3%). Further analysis revealed that the rate of ESBLs-producing isolates was 71.9% (23/32). Polymerase chain reaction screening revealed that 87.5% and 65.5% of EAEC isolates were positive for blaTEM and blaCTX-M genes, respectively, and 17 (53.1%) of isolates contained both blaTEM and blaCTX-M genes.
Conclusion
The high detection rate of ESBL-producing EAEC isolates accompanied with virulence genes highlights a need to restrict infection control policies in order to prevent further dissemination of the resistant and virulent EAEC strains.
Keywords: enteroaggregative Escherichia coli, diarrhea, adherence, antibiotic resistance, ESBLs, Iran
Introduction
Acute diarrheal diseases are an important health problem among children and are among the commonest causes of death among infants and children in developing countries.1 About 70% of cases of acute diarrheal illness occurs in the first 5 years of life. Pathogenic bacteria and viruses are responsible for ~20% of the episodes of acute gastroenteritis in children.2,3
Among the bacterial pathogens, diarrheagenic Escherichia coli (DEC) is a common cause of acute infectious diarrhea.4 DECs are classified into six groups based on clinical associations, phenotypic assays, and virulence factors: enteroaggregative E. coli (EAEC), diffusely adherent E. coli, enteroinvasive E. coli, enteropathogenic E. coli, enterohemorrhagic E. coli, and enterotoxigenic E. coli.4 EAEC has been implicated as an emerging cause of traveler’s diarrhea and persistent diarrhea among children and immunocompromised patients in both developing and developed countries.5 EAEC has also been associated with chronic intestinal inflammation, leading to malnutrition and growth retardation in infants.6
The pathogenesis of EAEC infection involves the adherence of the bacterium to the intestinal mucosa, forming a mucoid biofilm, and induces toxic effects on the intestinal mucosa, which result in diarrhea.7 The identification of EAEC depends on the HEp-2 adherence test, in which EAEC strains exhibit a “stacked-brick” appearance in a characteristic aggregative adherence (AA) pattern.7 The majority of EAEC strains carry a large (100-kb) plasmid, which encodes most putative EAEC virulence factors, including fimbrial adhesins, designated AA fimbria I (AAF/I, encoded by aggA gene), and AA fimbria II (AAF/II; encoded by aafA gene), which are responsible for the AA phenotype.8 The other plasmid-borne virulence factors include the enteroaggregative heat stable toxin (EAST; encoded by astA gene), dispersin secretory protein (encoded by aap gene), and plasmid-encoded toxin (Pet).8
In general, the first-choice agents for the treatment of EAEC infections are β-lactams, sulfonamides, and quinolones.9 However, in the recent years, therapeutic options are limited due to the emergence of E. coli strains resistant to third-generation cephalosporins, associated with the production of extended-spectrum β-lactamases (ESBLs).10 Bacterial strains producing ESBLs enzymes (TEM-1, SHV- 1, and CTX-M-type) inactivate the drugs by hydrolyzing the b-lactam ring.10 The ESBL encoding genes are located on large plasmids, which can carry the genes for resistance to numerous other groups of antimicrobials. Thus, worldwide dissemination of plasmid-borne ESBLs among E. coli isolates is a global problem.10 The objectives of the present study was to investigate the prevalence of EAEC strains, virulence factors, antibiotic resistance, and ESBL production in children suffering from diarrhea in Ahvaz, southwest Iran.
Materials and methods
Ethics
The study was approved by the Research Ethics Committee of the Ahvaz Jundishapur University of Medical Sciences (No: IR.AJUMS.REC.1395.462), Ahvaz, Iran. Written informed consent was obtained from all the children’s parents.
Study design and bacterial samples
In this cross-sectional study, from March 2015 to February 2016, 255 fecal samples were collected from children aged <12 years with diarrhea attending two teaching hospitals Golestan and Abuzar Children’s Hospital, affiliated to Ahvaz Jundishapur University of Medical Sciences, southwest of Iran. The specimens were cultured on MacConkey agar and incubated at 37°C for 24 h. Subsequently, E. coli isolates were identified using standard microbiologic methods including Gram-staining, colony characteristics and reaction on Triple Sugar Iron agar, Simmons’ citrate agar, Christensen’s urea agar, Indole test, Methyl red, and Voges-Proskauer tests. The strains that confirmed as E. coli were stored in tripticase soy broth with 15% glycerol at −70°C for long preservation. E. coli strains were then screened for EAEC identification using molecular method and cell line adhesion assay.
DNA extraction and molecular assay
Genomic DNA was extracted from all E. coli isolates by boiling method as described previously11 and subjected to polymerase chain reaction (PCR) after evaluating concentration and quality by measuring the absorbance of A260 and A280 nm with spectrophotometer and agarose gel electrophoresis, respectively. E. coli isolates were confirmed as EAEC by the amplification of aggR gene as previously described.12 Subsequently, the presence of five virulence factors were determined by assessing the presence of pCVD, aggA, ast, aap, and aafA genes.13 Moreover, the presence of ESBL resistance genes, blaTEM, blaPER, blaCTX-M, and blaSHV were determined by specific primers.14 Gene control strains were prepared from National E. coli Reference Laboratory, Pasteur Inestitute of Iran. The targeted genes and nucleotide sequences of the oligonucleotide primers used in this study were chosen as described in Table 1. PCR amplifications of the study genes were carried out in the following condition: initial denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 60 s, annealing for 45 s (temperature was depending on the sequence of primers), extension at 72°C for 50 s and final extension at 72°C for 5 min. PCR amplifications for studied genes were carried out on a thermal cycler 5530 (Eppendorf master, Germany). The amplicons were separated on 1.5% agarose gel prepared in 1× TAE (Tris/Acetate/EDTA) buffer and visualized using ultraviolet light after staining with ethidium bromide (CinnaGen Co., Tehran, Iran).
Table 1.
List of used primers in the present study
| Primer | Oligonucleotide sequence (5′ to 3′) | Gene | Product size | Annealing | Reference |
|---|---|---|---|---|---|
| pCVD-F | CTGGCGAAAGACTGTATCAT | pCVD432 | 630 | 57 | 13 |
| pCVD-R | AATGTATAGAAATCCGCTGTT | ||||
| aggR-F | GTATACACAAAAGAAGGAAGC | aggR | 254 | 57 | 13 |
| aggR-R | ACAGAATCGTCAGCATCAGC | ||||
| aggA-F | TTAGTCTTCTATCTAGGG | aggA | 457 | 49 | 13 |
| aggA-R | AAATTAATTCCGGCATGG | ||||
| aafA-F | TGCGATTGCTACTTTATTAT | aafA | 242 | 56 | 13 |
| aafA-R | ATTGACCGTGATTGGCTTCC | ||||
| aap-F | CTTGGGTATCAGCCTGAATG | aap | 310 | 58 | 13 |
| aap-R | AACCCATTCGGTTAGAGCAC | ||||
| astA-F | CCATCAACACAGTATATCCGA | astA | 111 | 58 | 13 |
| astA-R | GGTCGCGAGTGACGGCTTTGT | ||||
| TEM-F | GAGTATTCAACATTTCCGTGTC | blaTEM | 800 | 60 | 14 |
| TEM-R | TAATCAGTGAGGCACCTATCTC | ||||
| PER-F | AATTTGGGCTTAGGGCAGAA | BlaPER | 925 | 48 | 14 |
| PER-R | ATGAATGTCATTATAAAAGC | ||||
| CTX-M-F | CGCTTTGCGATGTGCAG | BlaCTX-M | 550 | 60 | 14 |
| CTX-M-R | ACCGCGATATCGTTGGT | ||||
| SHV-F | CGCCTGTGTATTATCTCCCTGTTAGCC | BlaSHV | 843 | 62 | 14 |
| SHV-R | TTGCCAGTGCTCGATCAGCG |
Adhesion to HEp-2 cells
All of the E. coli isolates positive for the presence of aggR gene were confirmed as EAEC by aggregative adhesion to HEp-2 cells by a method described previously.15 Briefly, an overnight culture of E. coli was prepared, and then, a concentration of 107 bacteria was incubated with monolayers of HEp-2 cells grown to 50% confluence on circular cover slips in wells of 24-well tissue culture plates. After 0.5-1 h of incubation at 37°C in 5% CO2, the wells were gently washed three times with phosphate-buffered saline, and then, 200 µL of Dulbecco’s minimum essential medium was added to each well, and the cultures were incubated at 37°C for 3 h in 5% CO2. Fixation was done by 70% ethanol and stained with Giemsa stain. The aggregative adhesion was examined under the oil immersion lens of a light microscope. The HEp-2 cell lines were purchased from Razi Vaccine and Serum Research Institute, Karaj, Iran.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was carried out on all EAEC isolates to 14 antibiotics by standard disk diffusion method on Mueller-Hinton agar medium (EMD Millipore, Billerica, MA, USA) as described by the Clinical and Laboratory Standards Institute (CLSI) guidelines.16 The antimicrobial agents used were gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), cefoxitin (30 µg), ceftazidime (30 µg), cefotaxime (30 µg), cefpodoxime (10 µg), ampicillin (10 µg), meropenem (10 µg), imipenem (10 µg), tetracycline (30 µg), cefteriaxone (30 µg), trimethoprim/sulfamethoxazole (25 µg), and azithromycin (15 µg). E. coli ATCC 25922 strain was used for quality control purposes. Multiple-drug-resistant (MDR) isolates (resistant to three or more of antimicrobials) were estimated according to previously described definitions.17 All isolates were tested for ESBL production using the combined-disk test using ceftazidime (30 µg) and cefotaxime (30 µg) disks and combination with clavulanic acid (10 µg) disk as described by CLSI guidelines.16 E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used as negative and positive control strains for ESBL production, respectively.
Statistical analysis
The analysis was performed by using SPSS™ software, version 21.0 (IBM Corporation, Armonk, NY, USA). The results are presented as descriptive statistics in terms of relative frequency. Values are expressed as the percentages of the group (categorical variables). Chi-square or Fisher’s exact tests were used to determine the significance of differences. A difference was considered statistically significant if the p-value was <0.05.
Results
Virulance gene patterns and HEp-2 cell assay
Thirty-two EAEC isolated from 17 (53.1%) males and 15 (46.9%) females aged from 5 months to 11 years showed an overall prevalence of EAEC of 12.5% (32/255) in our region. All isolates were positive for aggR gene and adhered in a HEp-2 cell adherence assay in the AA pattern (Figure 1). The frequency of the detected virulence genes among EAEC isolates is shown in Table 2. The data revealed that 21 (65.6%), 7 (21.9%), and 31 (96.9%) of strains were positive for the pCVD, astA, and aap genes, respectively. The most frequent gene was aap with a frequency of 96.9%. Neither aafA nor aggA genes were detected among all of the EAEC isolates. Regarding to the coexistence of the virulence genes, our isolates showed three distinct virulence patterns (Table 2). The most prevalent combination was pCVD-aap, found in 21 (65.6%) strains. In 32 strains analyzed, six (18.7%) isolates were positive for astA-aap and four (12.5%) isolates were positive for pCVD-astA-aap genes.
Figure 1.
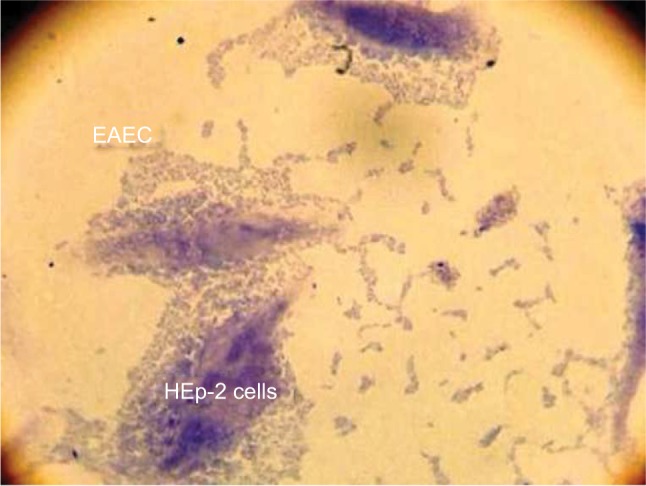
The AA pattern of EAEC to HEp-2 cells after 3 h of incubation.
Abbreviations: EAEC, enteroaggregative Escherichia coli; AA, aggregative adherence.
Table 2.
The distribution of virulence and extended-spectrum β-lactamase (ESBL) genes
| Virulence genes | Positive, N (%) | Negative, N (%) |
|---|---|---|
| pCVD | 21 (65.6) | 11 (34.4) |
| aggA | 0 | 32 (100) |
| astA | 7 (21.9) | 25 (78.1) |
| aap | 31 (96.9) | 1 (3.1) |
| aafA | 0 | 32 (100) |
|
| ||
| Coexistence of virulence genes | N (%) | |
|
| ||
| astA-aap | 6 (18.7) | |
| pCVD-aap | 21 (65.6) | |
| pCVD-astA-aap | 4 (12.5) | |
|
| ||
| ESBL genes | Positive, N (%) | Negative, N (%) |
|
| ||
| blaTEM | 28 (87.5) | 4 (12.5) |
| blaCTX-M | 21 (65.6) | 11 (34.4) |
| blaPER | 0 | 32 (100) |
| blaSHV | 0 | 32 (100) |
| blaTEM,blaCTX-M | 17 (53.1) | 15 (46.9) |
Antibiotic resistance patterns and ESBL genes
The results of antimicrobial susceptibility testing of the 32 EAEC isolates to 14 antibiotics are summarized in Table 3.
Table 3.
The antibiotic susceptibility testing results of 32 EAEC isolates
| Antibiotic | Total EAEC, N (%)
|
ESBL producing EAEC, N (%)
|
||||
|---|---|---|---|---|---|---|
| R | I | S | R | I | S | |
| Gentamicin | 10 (31.2) | 0 | 22 (68.8) | 6 (26.1) | 0 | 17 (73.9) |
| Amikacin | 4 (12.5) | 1 (3.1) | 27 (84.4) | 3 (13) | 1 (4.3) | 19 (82.6) |
| Ciprofloxacin | 6 (18.8) | 1 (3.1) | 25 (78.1) | 5 (21.7) | 1 (4.3) | 17 (73.9) |
| Cefoxitin | 5 (15.6) | 0 | 27 (84.4) | 0 | 0 | 23 (100) |
| Ceftazidime | 17 (53.1) | 4 (12.5) | 11 (34.4) | 12 (52.2) | 3 (13) | 8 (34.8) |
| Cefotaxime | 26 (81.3) | 0 | 6 (18.8) | 20 (87) | 0 | 3 (13) |
| Cefpodoxime | 26 (81.3) | 0 | 6 (18.8) | 20 (87) | 0 | 3 (13) |
| Cefteriaxon | 26 (81.3) | 1 (3.1) | 5 (15.6) | 21 (91.3) | 0 | 2 (8.7) |
| Ampicillin | 32 (100) | 0 | 0 | 23 (100) | 0 | 0 |
| Meropenem | 3 (9.4) | 3 (9.4) | 26 (81.3) | 1 (4.3) | 2 (8.7) | 20 (87) |
| Imipenem | 0 | 0 | 32 (100) | 0 | 0 | 23 (100) |
| Tetracycline | 15 (46.9) | 0 | 17 (53.1) | 7 (30.4) | 0 | 16 (69.6) |
| Trimethoprim/sulfamethoxazole | 32 (100) | 0 | 0 | 23 (100) | 0 | 0 |
| Azithromycin | 25 (78.1) | 0 | 7 (21.9) | 18 (78.3) | 0 | 5 (21.7) |
Abbreviations: EAEC, enteroaggregative Escherichia coli, ESBL, extended-spectrum β-lactamase.
From 32 confirmed EAEC isolates, all of them (100%) were resistant to ampicillin and trimethoprim/sulfamethoxazole, followed by 26 (81.3%) to cefteriaxon, cefotaxime, and cefpodoxime, 25 (78.1%) to azithromycin, 17 (53.1%) to ceftazidime, 15 (46.9%) to tetracycline, 10 (31.2%) to gentamicin, 6 (18.8%) to ciprofloxacin, 5 (15.6%) to cefoxitin, 4 (12.5%) to amikacin, and 3 (9.4%) to meropenem. The results disclosed that the most effective antibiotic against EAEC isolates was imipenem with 100% susceptibility.
MDR profiles
According to the antimicrobial susceptibility testing, all 32 EAEC isolates were resistant to at least two antibiotics, and the majority of isolates (n=31, 96.9%) were MDR with 21 different patterns (Table 4). The most prevalent resistance profile was XII (12.5%) (cefotaxime-cefpodoxime-ampicillin-ceftriaxone-trimethoprim/sulfamethoxazole-azithromycin).
Table 4.
Antibiotic resistance phenotypic patterns of EAEC isolates
| Resistance pattern | Phenotypic resistance | Number of resistant EAEC isolates (%) |
|---|---|---|
| I | AMP-SXT | 1 (3.1%) |
| II | AMP-TET-SXT | 2 (6.2%) |
| III | AMP-TET-SXT-AZM | 1 (3.1%) |
| IV | GEN-CTX-CPDX-AMP-CRO-SXT-AZM | 1 (3.1%) |
| V | GEN-CTX-CPDX-AMP-TET-SXT-AZM | 1 (3.1%) |
| VI | GEN-AN-CIP-AMP-CRO-SXT-AZM | 1 (3.1%) |
| VII | GEN-CAZ-CTX-CPDX-AMP-CRO-SXT-AZM | 3 (9.3%) |
| VIII | GEN-CIP-CAZ-CTX-CPDX-AMP-CRO-SXT-AZM | 1 (3.1%) |
| IX | GEN-FOX-CAZ-CTX-CPDX-AMP-CRO-TET-SXT-AZM | 1 (3.1%) |
| X | GEN-FOX-CAZ-CTX-CPDX-MEM-AMP-CRO-TET-SXT-AZM | 2 (6.2%) |
| XI | CTX-CPDX-AMP-CRO-SXT | 1 (3.1%) |
| XII | CTX-CPDX-AMP-CRO-SXT-AZM | 4 (12.5%) |
| XIII | CTX-CPDX-AMP-CRO-TET-SXT-AZM | 2 (6.2%) |
| XIV | AN-AMP-TET-SXT-AZM | 1 (3.1%) |
| XV | AN-CAZ-CTX-CPDX-MEM-AMP-CRO-SXT | 1 (3.1%) |
| XVI | AN-CAZ-CTX-CPDX-AMP-CRO-SXT-AZM | 1 (3.1%) |
| XVII | CIP-CAZ-CTX-CPDX-AMP-CRO-SXT-AZM | 3 (9.3%) |
| XVIII | CIP-FOX-CAZ-CTX-CPDX-AMP-CRO-TET-SXT-AZM | 1 (3.1%) |
| XIX | CAZ-CTX-CPDX-AMP-CRO-TET-SXT | 2 (6.2%) |
| XX | CAZ-CTX-CPDX-AMP-CRO-TET-SXT-AZM | 1 (3.1%) |
| XXI | FOX-CAZ-CTX-CPDX-AMP-CRO-TET-SXT-AZM | 1 (3.1%) |
Abbreviations: EAEC, enteroaggregative Escherichia coli; AMP, ampicillin; AN, amikacin; AZM, azithromycin; CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime; CPDX, cefpodoxime; CIP, ciprofloxacin; FOX, cefoxitin; GEN, gentamicin; MEM, meropenem; SXT, trimethoprimsulfamethoxazole; TET, tetracycline.
Phenotypic results for ESBLs
The overall occurrence of ESBL-producing isolates was 71.9% (23/32) of EAEC. All isolates that were tested positive for ESBLs were also MDR. The results of antimicrobial susceptibility testing of the ESBL-producing EAEC isolates are summarized in Table 3.
Molecular assay of ESBL genes
PCR screening for the presence of ESBL genes showed that 28 (87.5%) and 21 (65.5%) of EAEC isolates were positive for blaTEM and blaCTX-M genes, respectively, and 17 (53.1%) of isolates contained both blaTEM and blaCTX- M genes. Neither blaSHV nor blaPER genes were detected among all of the EAEC isolates (Table 2). The detailed characteristics of all 32 studied EAEC isolates including virulence profile, MDR pattern, and ESBL production are summarized in Table 5.
Table 5.
The detailed results of virulence genes, MDR, and ESBL gene patterns in the 32 EAEC isolates
| EAEC No. | Virulance gene profile | MDR pattern | ESBL phenotype | ESBL gene patter |
|---|---|---|---|---|
| 1 | pCVD-astA-aap | II | POS | TEM, CTX-M |
| 2 | pCVD-aap | IV | POS | TEM, CTX-M |
| 3 | pCVD-aap | II | NEG | TEM, CTX-M |
| 4 | astA | IX | NEG | TEM |
| 5 | pCVD-aap | XII | POS | CTX-M |
| 6 | pCVD-aap | XIV | NEG | TEM |
| 7 | pCVD-aap | XI | POS | TEM, CTX-M |
| 8 | pCVD-astA-aap | XV | POS | TEM, CTX-M |
| 9 | astA -aap | V | NEG | TEM |
| 10 | pCVD-aap | XVII | POS | CTX-M |
| 11 | pCVD-aap | I (not MDR) | NEG | TEM, CTX-M |
| 12 | pCVD-aap | XIX | POS | TEM, CTX-M |
| 13 | pCVD-aap | XVII | POS | TEM, CTX-M |
| 14 | pCVD-aap | III | POS | TEM |
| 15 | aap | X | NEG | TEM |
| 16 | pCVD-aap | XX | POS | TEM, CTX-M |
| 17 | pCVD-aap | VI | POS | TEM, CTX-M |
| 18 | pCVD-aap | VII | POS | TEM, CTX-M |
| 19 | pCVD-aap | VIII | POS | TEM |
| 20 | aap | XVIII | NEG | CTX-M |
| 21 | pCVD-aap | XVII | POS | TEM |
| 22 | pCVD-astA-aap | XIII | POS | TEM |
| 23 | aap | XXI | NEG | TEM, CTX-M |
| 24 | aap | XIX | POS | TEM, CTX-M |
| 25 | aap | X | NEG | TEM |
| 26 | aap | XII | POS | CTX-M |
| 27 | astA -aap | XII | POS | TEM, CTX-M |
| 28 | aap | XII | POS | TEM, CTX-M |
| 29 | pCVD-astA-aap | XIII | POS | TEM |
| 30 | aap | VII | POS | TEM, CTX-M |
| 31 | pCVD-aap | VII | POS | TEM |
| 32 | pCVD-aap | XVI | POS | TEM,CTX-M |
Abbreviations: EAEC, enteroaggregative Escherichia coli; ESBL, extended-spectrum β-lactamase; MDR, multiple-drug resistant.
Discussion
During the past decade, EAEC E. coli have been shown to cause persistent diarrhea and have received increasing attention globally.13 In addition to persistent diarrhea, EAEC have been isolated from acute sporadic cases and outbreaks worldwide, affecting children and adults.13,18 Previous studies have shown that EAEC strains are one of the most important agents of diarrhea in Iranian children.13,19 Our study also revealed EAEC as a cause of sporadic diarrhea.
In this study, a total of 32 EAEC were isolated from 255 fecal specimens. The PCR results for the aggR gene disclosed good agreement with the HEp-2 cell adhesion assay, as 100% of aggR-positive isolates were confirmed as EAEC. Nowadays, different genes are used to recognize EAEC in molecular studies. One of them that is commonly used to detect EAEC by PCR includes aggR.20 In our research, the aggR PCR method compared to the HEp-2 cell culture assay indicated 100% sensitivity and 100% specificity.
In the present study, three diverse combinations of the virulence genes were found among the EAEC isolates and 31 (96.9%) of them were positive for app gene (Table 2). Neither aafA nor aggA genes were found in our study. In this regard, Aslani et al in the west of Iran reported 11 different patterns of the virulence markers in EAEC isolates.13 However, in contrast to our results, Aslani et al showed the prevalence of aggA and aafA in EAEC isolates. 13 This might be due to epidemiologic differences between studied regions. A limitation of our study is that we only tried to search for the most common types of fimbriae. Our PCR targeted only AAFI and AAFII, but all isolates in this study were negative for these two fimbrial types indicating that the EAEC isolates in our region have fimbrial adhesins belonging to the other three established types or have novel types. In a study by Bouzari et al in the north of Iran, the prevalence of astA, aggA, and aafA among EAEC isolates obtained from children were reported as 8%, 38.8%, and 25%, respectively.19 Bafandeh et al in the north-west of Iran showed the prevalence of aap (88.6%), astA (83.5%), aggR (79.4%), aafA (46.4%), and aggA (5.1%) virulence determinants in EAEC isolates obtained from adult patients with diarrhea.21 As a general concept from Iranian results and reports from other parts of the world, EAEC are heterogeneous, and no virulence factor has been identified as common to all EAEC strains.8,13,19–25
Antimicrobial agents belonging to β-lactams family, particularly ampicillin and cephalosporins, and sulfonamides are widely used for the treatment of severe or persistent diarrhea in developing countries.9 However, the frequent use of these antimicrobial agents and the emergence of resistant strains have become a serious public health concern.25 In our results, similar to previous reports from developing countries, the majority of isolates were resistant to ampicillin, cephalosporins, and co-trimoxazole.3,13,26,27 In Iran, cephalosporins are widely used due to their low degree of side effects. The high incidence of resistance to these agents may be due to the inappropriate and widespread use of antibiotics. Hopefully, based on our results, carbapenems, aminoglycosides, and fluoroquinolones can be used as an alternative for the treatment of EAEC-associated diarrhea in our area.
The emergence of MDR strains, particularly ESBL-producing Enterobacteriaceae is a global challenge for clinicians.28 In the present study, we observed a high frequency of MDR (96.9%) and ESBL (71.9%) EAEC isolates which were resistance to the most tested antimicrobial agents. This high level of resistance is justified by the availability of medications without doctors’ prescription from pharmacies in developing countries. Aslani et al closest to our findings showed the high rate of MDR EACE (71.4%) in Iranian children.13 Reports from other parts of the country showed the prevalence of ESBLs producing clinical isolates of E. coli ranging from 22% to 74%. The ESBL production in EAEC strains from two Asian countries China and Bangladesh was reported as 50% and 49.1%, respectively.23,29 The differences in the prevalence of ESBLs producing isolates can be due to dissimilarities in geographical distribution, sample types, studied population, and hospital or community origin of isolates.
One of the major concern is the spread of ESBL-positive bacteria, which may mainly be due to the transfer of resistance genes via mobile genetic elements.10 ESBLs are enzymes most commonly derived from blaTEM or blaSHV, but the prevalence of blaCTX-M types has risen recently.30 In our results, the genotype TEM was predominant with the prevalence of 87.5% followed by CTX-M type with 65.5%. The mechanisms of ESBL resistance in EAEC in Iran are poorly understood, and no similar study can be found. However, in agreement with our findings, several authors in our region showed the prevalence of blaCTX-M, blaSHV, and blaTEM as the main mechanisms responsible for ESBL production in clinical isolates of E. coli among Iranian patients.31–34 Meanwhile, some authors showed the global spread of blaCTX-M, blaSHV, and blaTEM harboring DEC strains same as Iran.24,27,35,36
Conclusion
In this study, the high detection rate of MDR and ESBL producing EAEC isolates accompanied with virulence genes highlights a need to restrict infection control policies to prevent further dissemination of the resistant and virulent EAEC strains. Hopefully, several locally available antibiotics still have promising effects against MDR isolates in our region. These findings provide experimental evidence for safe and effective management of EAEC associated infections.
Acknowledgments
This research was financially supported by the Infectious and Tropical Disease Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran (Grant No: OG-95124). The authors also wish to thank Dr Masoud Alebouyeh (Research Center for Gastroenterology and Liver Diseases) for providing the control strains.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Koletzko S, Osterrieder S. Acute infectious diarrhea in children. Dtsch Arztebl Int. 2009;106(33):539–547. doi: 10.3238/arztebl.2009.0539. quiz 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciccarelli S, Stolfi I, Caramia G. Management strategies in the treatment of neonatal and pediatric gastroenteritis. Infect Drug Resist. 2013;6:133–161. doi: 10.2147/IDR.S12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vu Nguyen T, Le Van P, Le Huy C, Nguyen Gia K, Weintraub A. Etiology and epidemiology of diarrhea in children in Hanoi, Vietnam. Int J Infect Dis. 2006;10(4):298–308. doi: 10.1016/j.ijid.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Hien BT, Scheutz F, Cam PD, et al. Diarrheagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. J Clin Microbiol. 2008;46(3):996–1004. doi: 10.1128/JCM.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomes TA, Elias WP, Scaletsky IC, et al. Diarrheagenic Escherichia coli. Braz J Microbiol. 2016;47(Suppl 1):3–30. doi: 10.1016/j.bjm.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaur P, Chakraborti A, Asea A. Enteroaggregative Escherichia coli: an emerging enteric food borne pathogen. Interdiscip Perspect Infect Dis. 2010;2010:254159. doi: 10.1155/2010/254159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang DB, Okhuysen PC, Jiang ZD, DuPont HL. Enteroaggregative Escherichia coli: an emerging enteric pathogen. Am J Gastroenterol. 2004;99(2):383–389. doi: 10.1111/j.1572-0241.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 8.Kahali S, Sarkar B, Rajendran K, et al. Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2004;42(9):4111–4120. doi: 10.1128/JCM.42.9.4111-4120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen TV, Le PV Le CH, Weintraub A. Antibiotic resistance in diarrheagenic Escherichia coli and Shigella strains isolated from children in Hanoi, Vietnam. Antimicrob Agents Chemother. 2005;49(2):816–819. doi: 10.1128/AAC.49.2.816-819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Antibiotic resistance and extended spectrum beta-lactamases: types, epidemiology and treatment. Saudi J Biol Sci. 2015;22(1):90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hosseinzadeh Z, Sedigh Ebrahim-Saraie H, Sarvari J, et al. Emerge of blaNDM-1 and blaOXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J Chin Med Assoc. 2018;81(6):536–540. doi: 10.1016/j.jcma.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Kubomura A, Misaki T, Homma S, Matsuo C, Okabe N. Phenotypic and molecular characterization of enteroaggregative Escherichia coli isolated in Kawasaki, Japan. Jpn J Infect Dis. 2017;70(5):507–512. doi: 10.7883/yoken.JJID.2016.387. [DOI] [PubMed] [Google Scholar]
- 13.Aslani MM, Alikhani MY, Zavari A, Yousefi R, Zamani AR. Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int J Infect Dis. 2011;15(2):e136–9. doi: 10.1016/j.ijid.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Kjerulf A, Hansen DS, Sandvang D, Hansen F, Frimodt-Moller N. The prevalence of ESBL-producing E. coli and Klebsiella strains in the Copenhagen area of Denmark. Apmis. 2008;116(2):118–124. doi: 10.1111/j.1600-0463.2008.00777.x. [DOI] [PubMed] [Google Scholar]
- 15.Iwanaga M, Song T, Higa N, Kakinohana S, Toma C, Nakasone N. Enteroaggregative Escherichia coli: incidence in Japan and usefulness of the clump-formation test. J Infect Chemother. 2002;8(4):345–348. doi: 10.1007/s10156-002-0199-1. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PE: Clinical and Laboratory Standards Institute; 2016. [Accessed July 30, 2018]. (Twenty-sixth Informational Supplement. CLSI document M100-S26). Available from: www.facm.ucl.ac.be/intranet/CLSI/CLSI-2016-M100-S26.pdf. [Google Scholar]
- 17.Rzewuska M, Czopowicz M, Kizerwetter-Świda M, Chrobak D, Błaszczak B, Binek M. Multidrug resistance in Escherichia coli strains isolated from infections in dogs and cats in Poland (2007-2013) Sci World J. 2015;2015:408205. doi: 10.1155/2015/408205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dedeic-Ljubovic A, Hukic M, Bekic D, Zvizdic A. Frequency and distribution of diarrhoeagenic Escherichia coli strains isolated from pediatric patients with diarrhoea in Bosnia and Herzegovina. Bosn J Basic Med Sci. 2009;9(2):148–155. doi: 10.17305/bjbms.2009.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouzari S, Jafari A, Zarepour M. Distribution of virulence related genes among enteroaggregative Escherichia coli isolates: using multiplex PCR and hybridization. Infect Genet Evol. 2005;5(1):79–83. doi: 10.1016/j.meegid.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Jensen BH, Olsen KEP, Struve C, Krogfelt KA, Petersena AM. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev. 2014;27:614–630. doi: 10.1128/CMR.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bafandeh S, Haghi F, Zeighami H. Prevalence and virulence characteristics of enteroaggregative Escherichia coli in a case-control study among patients from Iran. J Med Microbiol. 2015;64(Pt 5):519–524. doi: 10.1099/jmm.0.000055. [DOI] [PubMed] [Google Scholar]
- 22.Lima IF, Boisen N, Quetz Jda S, et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J Med Microbiol. 2013;62(Pt 5):683–693. doi: 10.1099/jmm.0.054262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chattaway MA, Day M, Mtwale J, et al. Clonality, virulence and antimicrobial resistance of enteroaggregative Escherichia coli from Mirzapur, Bangladesh. J Med Microbiol. 2017;66(10):1429–1435. doi: 10.1099/jmm.0.000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonnevend A, Al Dhaheri K, Mag T, et al. CTX-M-15-producing multidrug-resistant enteroaggregative Escherichia coli in the United Arab Emirates. Clin Microbiol Infect. 2006;12(6):582–585. doi: 10.1111/j.1469-0691.2006.01413.x. [DOI] [PubMed] [Google Scholar]
- 25.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi E, Sultan Z, Shimada S, et al. Studies on diarrheagenic Escherichia coli isolated from children with diarrhea in Myanmar. Microbiol Immunol. 2008;52(1):2–8. doi: 10.1111/j.1348-0421.2008.00001.x. [DOI] [PubMed] [Google Scholar]
- 27.Albert MJ, Rotimi VO, Dhar R, et al. Diarrhoeagenic Escherichia coli are not a significant cause of diarrhoea in hospitalised children in Kuwait. BMC Microbiol. 2009;9:62. doi: 10.1186/1471-2180-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaikh S, Fatima J, Shakil S, Rizvi SM, Kamal MA. Risk factors for acquisition of extended spectrum beta lactamase producing Escherichia coli and Klebsiella pneumoniae in North-Indian hospitals. Saudi J Biol Sci. 2015;22(1):37–41. doi: 10.1016/j.sjbs.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Gu DX, Huang YL, Chan EW, Chen GX, Chen S. Comparative genetic characterization of enteroaggregative Escherichia coli strains recovered from clinical and non-clinical settings. Sci Rep. 2016;6:24321. doi: 10.1038/srep24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandramohan L, Revell PA. Prevalence and molecular characterization of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in a pediatric patient population. Antimicrob Agents Chemother. 2012;56(9):4765–4770. doi: 10.1128/AAC.00666-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohajeri P, Rostami Z, Farahani A, Norozi B. Distribution of ESBL producing uropathogenic Escherichia coli and carriage of selected ß-lactamase genes in hospital and community isolates in west of Iran. Ann Trop Med Public Health. 2014;7(5):219–222. [Google Scholar]
- 32.Haghighatpanah M, Mozaffari Nejad AS, Mojtahedi A, Amirmozafari N, Zeighami H. Detection of extended-spectrum beta-lactamase (ESBL) and plasmid-borne blaCTX-M and blaTEM genes among clinical strains of Escherichia coli isolated from patients in the north of Iran. J Glob Antimicrob Resist. 2016;7:110–113. doi: 10.1016/j.jgar.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Memariani M, Najar Peerayeh S, Zahraei Salehi T, Shokouhi Mostafavi SK. Occurrence of SHV TEM and CTX-M beta-lactamase genes among enteropathogenic Escherichia coli strains isolated from children with diarrhea. Jundishapur J Microbiol. 2015;8(4):e15620. doi: 10.5812/jjm.8(4)2015.15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghorbani-Dalini S, Kargar M, Doosti A, Abbasi P, Sarshar M. Molecular epidemiology of ESBL genes and multi-drug resistance in diarrheagenic Escherichia coli strains isolated from adults in Iran. Iran J Pharm Res. 2015;14(4):1257–1262. [PMC free article] [PubMed] [Google Scholar]
- 35.Guiral E, Mendez-Arancibia E, Soto SM, et al. CTX-M-15-producing enteroaggregative Escherichia coli as cause of travelers’ diarrhea. Emerg Infect Dis. 2011;17(10):1950–1953. doi: 10.3201/eid1710.110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JS, Kim J, Kim SJ, et al. Characterization of CTX-M-type extended-spectrum beta-lactamase-producing diarrheagenic Escherichia coli isolates in the Republic of Korea during 2008-2011. J Microbiol Biotechnol. 2014;24(3):421–426. doi: 10.4014/jmb.1401.01023. [DOI] [PubMed] [Google Scholar]


