Abstract
Nitric oxide (NO) and reactive oxygen intermediates (ROIs) play key roles in the activation of disease resistance mechanisms both in animals and plants. In animals NO cooperates with ROIs to kill tumor cells and for macrophage killing of bacteria. Such cytotoxic events occur because unregulated NO levels drive a diffusion-limited reaction with O2− to generate peroxynitrite (ONOO−), a mediator of cellular injury in many biological systems. Here we show that in soybean cells unregulated NO production at the onset of a pathogen-induced hypersensitive response (HR) is not sufficient to activate hypersensitive cell death. The HR is triggered only by balanced production of NO and ROIs. Moreover, hypersensitive cell death is activated after interaction of NO not with O2− but with H2O2 generated from O2− by superoxide dismutase. Increasing the level of O2− reduces NO-mediated toxicity, and ONOO− is not a mediator of hypersensitive cell death. During the HR, superoxide dismutase accelerates O2− dismutation to H2O2 to minimize the loss of NO by reaction with O2− and to trigger hypersensitive cell death through NO/H2O2 cooperation. However, O2− rather than H2O2 is the primary ROI signal for pathogen induction of glutathione S-transferase, and the rates of production and dismutation of O2− generated during the oxidative burst play a crucial role in the modulation and integration of NO/H2O2 signaling in the HR. Thus although plants and animals use a similar repertoire of signals in disease resistance, ROIs and NO are deployed in strikingly different ways to trigger host cell death.
Attempted infection of plants by an avirulent pathogen elicits a battery of defenses often accompanied by the collapse of challenged host cells. This hypersensitive cell death results in a restricted lesion delimited from surrounding healthy tissue and is thought to contribute to pathogen restriction. An early event in this hypersensitive response (HR) is the generation of superoxide (O2−) and accumulation of hydrogen peroxide (H2O2) in an oxidative burst reminiscent of that producing such reactive oxygen intermediates (ROIs) in activated macrophages (1).
Activation of the oxidative burst in the plant HR is part of a highly amplified and integrated signal system that also involves salicylic acid and perturbations of cytosolic Ca2+ to trigger defense mechanisms (2) and to mediate the establishment of systemic immunity (3). The oxidative burst is necessary but not sufficient to trigger host cell death, and recent data indicate that nitric oxide (NO) cooperates with ROIs in the activation of hypersensitive cell death (4).
NO and ROIs also interact in the mammalian native immune system where macrophage killing of pathogens and tumor cells involves the diffusion-limited reaction of NO and O2 to generate ONOO−, a long lived and highly reactive oxidant species that freely crosses membranes (5), which may modulate NO signal functions (6). ONOO− induces apoptosis in some human tumor cells (7), and it is also directly cytotoxic, e.g. by causing protein-tyrosine nitration and oxidative tissue damage (8, 9). Although the effects of NO depend on many factors including rates of production and diffusion, levels of ROIs, and the activities of ROI scavengers such as superoxide dismutase (SOD) and catalase (10), unregulated NO production causes cell death through oxidative stress, disrupted energy metabolism, DNA damage, activation of poly(ADP-ribose) polymerase, or dysregulation of cytosolic Ca2+ (11).
Here we demonstrate that in soybean cell suspensions, the efficient induction of hypersensitive cell death requires a balance between ROIs and NO production such that high levels of NO are ineffective in the absence of a correspondingly strong oxidative burst. Moreover unlike animal cells, ONOO−, formed by the reaction between O2− and NO, is not an effective inducer of hypersensitive cell death. Although O2− seems to be the ROI involved in the induction of glutathione S-transferase (GST) in a cellular protectant response, H2O2, formed by the SOD-catalyzed dismutation of O2−, functions with NO in the triggering of plant hypersensitive cell death. The rates of production and dismutation of O2− generated during the pathogen-induced oxidative burst seem to play a crucial role in modulating and integrating the binary NO/H2O2 trigger. We conclude that the functional interactions between NO and ROIs in the plant HR are strikingly different from those observed previously in the vertebrate native immune system.
Materials and Methods
Plant Material.
Physiological experiments were performed with soybean (Glycine max cv. Williams 82) cell suspensions 3 days after subculture (12). Cells were incubated in 12-well tissue culture plates (1 ml per well) agitated at the indicated speed. Pseudomonas syringae pv. glycinea race 4 with the plasmid pLAFR1 carrying the avrA avirulence gene (13) was grown as described (14). Except where noted, reagents were added to cells simultaneously with bacteria.
Cell Death.
Cell death was assayed 24 h after the indicated treatments by incubating the soybean cell suspensions for 15 min with 0.05% Evan's blue (Sigma). Unbound dye was removed by extensive washing, and dye bound to dead cells was solubilized in 50% (vol/vol) methanol/1% SDS for 30 min at 50°C and quantified by absorbance at 600 nm (12). The data are expressed as a percentage of total killing calibrated by Evan's blue staining of equivalent cells treated with ethanol (14).
ROIs.
H2O2 accumulation was assayed by incubating cell suspensions for 5 min with scopoletin (Sigma) and measuring the loss of fluorescence at 460 nm after excitation at 350 nm (12). O2− accumulation was assayed by monitoring cytochrome c or sodium,3′-[1-(phenylamino-carbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene-sulfonic acid hydrate (XTT) reduction. Then 100 μM cytochrome c (Sigma) was added to suspension cells, and the shift in absorbance of the medium from 540 to 550 nm was recorded after 10 min (15). XTT (0.5 mM, Diagnostic Chemicals, Charlottetown, PE, Canada) was prepared as described (16), and reduction of the tetrazolium dye was monitored by recording the absorbance of the medium at 470 nm after 24 h.
NO.
NO accumulation was assayed by monitoring the conversion of HbO2 to metHb as described previously (4). Sodium nitroprusside (SNP) was added to the soybean cell suspensions 5 min before the addition of HbO2 to a final concentration of 10 μM. After 2 min, the changes in absorbance of the medium at 421 and 401 nm were measured, and the NO levels were calculated by using an extinction coefficient of 77 mM−1⋅cm−1 [A401 (metHb) − A421 (HbO2)]. Under conditions of a strong oxidative burst NO was measured by a NO electrode (17) calibrated under nonburst conditions to the metHb assay.
SOD.
Total SOD activity in the low speed supernatant of whole-cell extracts was assayed by a kit (Calbiochem). Cu,Zn-SOD activity was measured after ethanol/chloroform extraction, which inactivates Mn-SOD and Fe-SOD (18).
RNA Blot Hybridization.
Total RNA was isolated by using Trizol reagent (Life Technologies, Inc., Rockville, MD). RNA blot hybridization (12) was performed with the following probes: Gmhsp-26 gst cDNA (19) and a cDNA encoding Cu,Zn-SOD from soybean (20).
Results
NO/ROI Regulation of Cell Death.
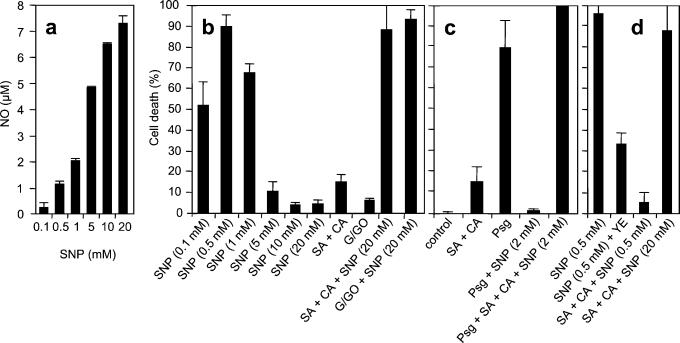
Treatment of soybean cells with 20 mM SNP delivers >7 μM NO as measured by the metHb assay (Fig. 1a), but in the absence of ROIs even such high concentration did not affect cell viability (data not shown). In contrast, when cells were agitated rapidly (100 rpm) to trigger a mechanically induced oxidative burst (21) giving a steady-state H2O2 concentration of ≈1 μM (4), SNP at concentrations between 0.1 and 1.0 mM, generating 0.25–2.0 μM NO, induced cell death with the optimal effect at 0.5 mM SNP, equivalent to 1.2 μM NO (Fig. 1 a and b). However, the addition of higher concentrations of the NO donor reversed the response causing a dramatic reduction in ROI-dependent cell death, and in the presence of 10 mM SNP (generating 6.5 μM NO) cell death was abolished completely (Fig. 1b). Thus, in rapidly shaking cells a NO/H2O2 ratio between 0.25 and 2 was effective in inducing cell death.
Figure 1.
Efficient induction of cell death in soybean cells requires a balance between ROI and NO production. (a) Generation of NO by SNP. (b–d) Cell death in cell suspensions in the presence of different concentrations of NO and ROI. In b and d, cells were agitated at 100 rpm; in c, cells were agitated slowly (60 rpm) and challenged with 108 P. syringae pv. glycinea carrying avrA (Psg). The final concentrations of indicated reagents were: 50 μM salicylic acid (SA), 4 μM cantharidin (CA), 100 μg of yeast elicitor (YE), 500 μM glucose + 0.5 units⋅ml−1 glucose oxidase (G/GO). Each datum point is the mean and standard deviation of three replicates. Experiments were repeated three times with similar results. Suppression of cell death at either supraoptimal SNP or ROI concentrations as well as its reactivation by balancing the binary signal system were all significant over appropriate controls by the Student's t test (P < 0.001).
The inhibition of ROI-induced cell death at high concentrations of NO suggests that an appropriate balance between ROI and NO production is required. We therefore examined the effect of increasing the levels of endogenous ROIs in the presence of high levels of NO. To do this we exploited the fact that the signal transduction pathway leading to the oxidative burst is regulated by a balance between phosphorylation and dephosphorylation events (12). Salicylic acid synergistically enhances H2O2 accumulation in response to the protein phosphatase type 2A inhibitor cantharidin by switching this regulatory balance to pathway activation (14), generating H2O2 at a steady-state concentration of ≈30 μM (data not shown). Although salicylic acid + cantharidin in the absence of NO caused only a modest induction of cell death, this massive enhancement of the oxidative burst strongly reactivated the cell death program in the presence of high levels of SNP (Fig. 1b). The same reversion could be observed by the addition of the H2O2-generating system glucose/glucose oxidase (G/GO) at levels generating ≈30 μM H2O2 (G/GO; Fig. 1b). Salicylic acid or cantharidin alone do not induce a strong oxidative burst (14), and as expected 20 mM SNP in the presence of either salicylic acid or cantharidin separately did not induce cell death (data not shown).
The metHb assay cannot be used to measure NO levels under conditions of a strong oxidative burst because of interference by high levels of ROI. However, we used a NO electrode to show that under these conditions high levels of SNP generate the same concentration of NO as in the absence of an oxidative burst. Thus, although 7 μM NO is ineffective in conjunction with a weak oxidative burst generated by rapid shaking of cells, this high concentration of NO becomes effective with a strong oxidative burst, generating 30 μM H2O2 (NO/H2O2 ≈ 4).
We next investigated whether a balance between NO and ROI was required also for hypersensitive cell death triggered by the recognition of an avirulent pathogen. P. syringae pv. glycinea carrying the avirulence gene avrA is recognized by soybean cv. Williams 82, which possesses the corresponding Rpg2 resistance gene (13), leading to rapid ROI- and NO-dependent hypersensitive cell death. The NO/H2O2 ratio is ≈0.3 during the pathogen-induced response (4) and hence falls within the effective range established by the SNP dose-response experiment. Perturbation of this ratio by the addition of high concentrations of SNP blocked hypersensitive cell death in cells challenged with avirulent Pseudomonas (Fig. 1c). Moreover, this effect could be reversed by the addition of salicylic acid + cantharidin to supplement the pathogen-induced oxidative burst (Fig. 1c).
Further evidence that the binary NO/ROI signal system must be balanced for an optimal response was obtained from analysis of the effects of increased ROI levels on NO-induced cell death. Thus, the potentiation of the oxidative burst in cell suspensions agitated at 100 rpm by the addition of either a yeast elicitor or salicylic acid + cantharidin dramatically reduced the cell death induced by 0.5 mM SNP, and the effect could be reversed by increasing the concentration of SNP up to 20 mM (Fig. 1d).
Peroxynitrite.
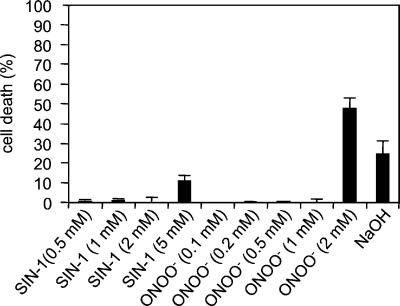
The cytotoxic effects of NO and ROI often derive from the reaction of NO with O2 to form ONOO− (1). To determine whether a similar mechanism is involved in NO-mediated cell death in plants, we analyzed the effect of exogenous ONOO− on soybean cells. Exposure of animal cells to ONOO− in the range of 1–1,000 μM causes concentration-dependent cell death (7, 22). However, exposure of soybean cells to ONOO− did not cause cell death at concentrations up to 1 mM (Fig. 2) despite driving extensive tyrosyl nitration of cellular proteins (J.Z. and C.L., unpublished data). At higher concentrations ONOO− killed soybean cells, but this likely reflects the high concentration of NaOH (1 M) in which ONOO− is dissolved (Fig. 2). We also investigated the effects of ONOO− when cell suspensions were exposed for a prolonged period. SIN-1 gradually decomposes to yield equimolar amounts of NO and O2−. This reaction continues for ≈24 h (23), and at pH 7.4 100 μM SIN-1 releases 1.24 μM NO/min and 1.12 μM O2−/min (24) for continuous steady-state generation of ONOO−. The addition of 0.1–5 mM SIN-1 to soybean suspensions did not cause any cell death (Fig. 2) and failed also to protect against cell death triggered by challenge with avirulent P. syringae (data not shown).
Figure 2.
The effect of ONOO− and 3-morpholinosydnonimine N-ethylcarbamide (SIN-1) on cell death in soybean cells. NaOH indicates cells receiving an amount of NaOH equal to cells treated with 2 mM ONOO−. Each datum point is the mean and standard deviation of three replicates. Experiments were repeated two times each at slow (60 rpm) and rapid (100 rpm) agitation with similar results.
SOD Function.
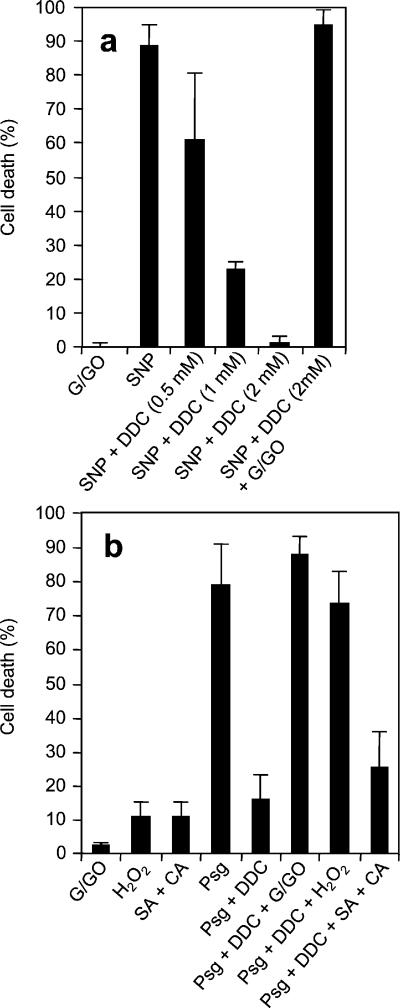
O2 is an important initial product of the pathogen-induced oxidative burst (2). H2O2 then can be formed nonenzymatically by dismutation of O2− (25) or enzymatically by the action of SOD (26). To distinguish between the contributions of O2− and H2O2 we analyzed the effects of sodium diethyldithiocarbamate (DDC), an inhibitor of Cu,Zn-SOD (27). DDC blocks H2O2 accumulation in harpin-induced Arabidopsis cells, leading to increased O2− and inhibition of hypersensitive cell death (28). The addition of DDC to soybean cells agitated at 100 rpm blocked NO-induced ROI-dependent cell death, and this effect could be abrogated by the simultaneous addition of G/GO (Fig. 3a). Likewise, DDC abolished both H2O2 accumulation (data not shown) and cell death (Fig. 3b) in cells challenged with avirulent Pseudomonas. The concomitant addition of sublethal concentrations of either H2O2 or G/GO restored cell death, indicating that DDC was not affecting either pathogen recognition or early downstream events. Consistent with the lack of synergism between O2− and NO, the potentiation of the endogenous oxidative burst with salicylic acid + cantharidin had a very limited effect (Fig. 3b). Thus, H2O2 from the pathogen-induced oxidative burst derives from the SOD-catalyzed dismutation of O2− and that H2O2 not O2− is the ROI effector in hypersensitive cell death. In contrast, O2− seems to be the primary signal in ROI-mediated induction of GST transcripts, because DDC dramatically enhanced the accumulation of GST transcripts in soybean cells inoculated with avirulent Pseudomonas (Fig. 4).
Figure 3.
Cell death in soybean cell suspensions. (A) Cells were agitated at 100 rpm. (B) Cells were agitated slowly (60 rpm) and challenged with P. syringae pv. glycinea carrying avrA (Psg). DDC was added 15 min in advance, and all other reagents were added simultaneously to the following final concentrations: 200 μM G + 0.5 units⋅ml−1 GO, 0.5 mM SNP, 2 mM H2O2, 50 μM salicylic acid (SA), and 3 μM cantharidin (CA). The final concentration of DDC was 2 mM unless indicated. Each datum point is the mean and standard deviation of three replicates. Experiments were repeated three times with similar results. Suppression of cell death by the addition of 2 mM DDC as well as its reactivation by balancing the binary signal system with exogenous H2O2 were all significant over appropriate controls by the Student's t test (P < 0.001)
Figure 4.
The accumulation of transcripts encoding GST and Cu,Zn-SOD was monitored by gel blot hybridization analysis of total cellular RNA isolated at indicated times after treatment of soybean cells with P. syringae pv. glycinea carrying avrA (Psg) or P. syringae pv. glycinea carrying avrA plus 2 mM DDC (Psg + DDC). RNA loading was checked by gel staining with ethidium bromide (RNA).
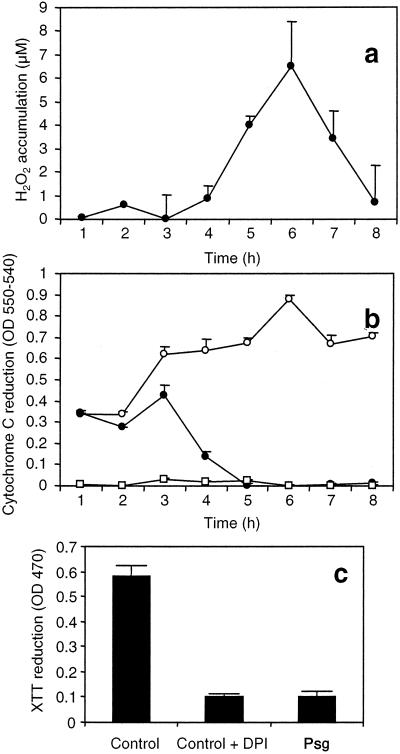
In cells agitated at high speed, the accumulation of H2O2 in response to avirulent Pseudomonas (Fig. 5a) is accompanied by a dramatic reduction in the steady-state level of O2− (Fig. 5 b and c). These data suggest a stimulation of O2− dismutation to H2O2 during the oxidative burst. Cu,Zn-SOD transcripts showed a marked DDC-insensitive accumulation within 1 h of the addition of the avirulent pathogen (Fig. 4). Soybean cells undergoing the HR possess high levels of SOD (Fig. 6). A partial purification, which caused the inactivation of Fe- and Mn-SOD isoforms, revealed that most of this activity could be accounted for by Cu,Zn-SOD, and the dramatic reduction of total SOD activity in DDC-treated cells reflected the complete inhibition of this form of SOD (Fig. 6).
Figure 5.
ROI accumulation in soybean cells agitated at 90 rpm. (A) Kinetics of H2O2 accumulation in response to P. syringae pv. glycinea carrying avrA. (B) Kinetics of O2− accumulation measured by cytochrome c reduction in response to P. syringae pv. glycinea carrying avrA (●), unchallenged cells (○), and unchallenged cells + 10 μM diphenylene iodonium (□). (c) Cumulative O2− accumulation 24 h postinoculation with P. syringae pv. glycinea carrying avrA (Psg) measured by XTT reduction. Each datum point is the mean and standard deviation of three replicates. Experiments were repeated three times with similar results.
Figure 6.
SOD activity in soybean cells agitated at 60 rpm 4 h postinoculation with P. syringae pv. glycinea carrying avrA: black bars, total SOD; white bars, Cu,Zn-SOD. DDC was added 15 min in advance to a final concentration of 2 mM. Each datum point is the mean and standard deviation of three replicates. Experiments were repeated twice with similar results.
Discussion
Inflammatory cells such as macrophages and neutrophils produce large amounts of NO and O2−, which in turn rapidly form ONOO− (6). Although ONOO can damage normal tissue, at the whole organism level its reactive chemistry can be considered beneficial because of ONOO− cytotoxicity to invading pathogens (1) and tumor cells (7). In plants ROIs and NO produced during the onset of a pathogen-induced HR cooperate to trigger hypersensitive cell death (4). We therefore investigated whether ROIs modulate NO signaling to trigger hypersensitive cell death through similar mechanisms. Surprisingly, we found that in soybean cells ONOO− is not an essential intermediate. Moreover, whereas treatment of various cultured animal cells with high levels of NO leads to their death (11), treatment of soybean cells with high levels of NO did not affect cell viability unless compensated by high levels of ROIs. Likewise, additional NO was required to compensate for increased levels of ROIs. This series of experiments, which was based on perturbation of NO and ROI production by several different stimuli followed by counterbalancing treatments within the same experimental system and confirmation of key points using more than one treatment, demonstrated that efficient activation of hypersensitive cell death required a balance between NO and ROI production.
The lack of a direct interaction between NO and O2− in triggering hypersensitive cell death prompted us to investigate the role of H2O2 by monitoring NO killing in the absence of H2O2 formation. The SOD inhibitor DDC abolished hypersensitive cell death induced by NO in soybean cells undergoing an oxidative burst. Cell death could be rescued by the addition of sublethal amounts of H2O2, whereas enhancement of O2− production by potentiation of the endogenous oxidative burst had a very limited effect, consistent with the ineffectiveness of O2− in triggering NO-induced cell death. We therefore conclude that during the HR, SOD-mediated dismutation of O2− to H2O2 is required to activate cell death, which depends on synergistic interactions between NO and H2O2. The SNP dose response with rapidly agitated cells generating a weak oxidative burst indicates that a NO/H2O2 ratio in the range of 0.25–2.0 gives effective induction of cell death. The NO/H2O2 ratios during pathogen induction of the HR and for induction of cell death by exogenous NO in association with a strong oxidative burst also fall within this range. Although we cannot exclude the possibility that other physiological conditions distort the effective window, it is clear that independent perturbations in either NO or H2O2 levels can be compensated by appropriate changes in the other component to re-establish an effective balance in the binary signal system.
The relative rates of O2− dismutation to H2O2 and reaction with NO to generate ONOO− are critical in the integration of the signal system to deliver the NO and H2O2 required as coactivators of hypersensitive cell death. The rate of NO reaction with O2− is approximately three times faster than the reaction of O2− with SOD (8). However, NO likely is generated in the cytosol, whereas O2− from the pathogen-induced oxidative burst seems to be generated in the apoplast, likely mediated at least in part by a plasma membrane NADPH oxidase. Cu,Zn-SOD is among the key cellular enzymes by which animal cells protect against NO-mediated damage (29). In macrophages undergoing apoptotic cell death, Cu,Zn-SOD is down-regulated (30). Down-regulation of Cu,Zn-SOD in PC12 cells leads to their death via the ONOO− pathway (29), whereas its overexpression protects RAW 264.7 macrophages against NO cytotoxicity (30). In contrast we observed pathogen induction of Cu,Zn-SOD transcripts. SOD activity increases in tobacco infected with tobacco mosaic virus during the expression of the HR (31), and in Phaseolus vulgaris Cu,Zn-SOD activity increases in the HR of resistant leaves (32). Our observation that H2O2 but not O2− is the key ROI coeffector of plant hypersensitive cell death and that ONOO−, formed by an O2− reaction with NO, is not a plant cell death signal is consistent with the striking inverse patterns of SOD regulation during the expression of disease resistance mechanisms in plants and animals.
Although in animals the reaction of NO with H2O2 does not seem to be generally (and directly) involved in killing, NO cooperates with H2O2 to induce DNA fragmentation and cell lysis in murine lymphoma, hepatoma, and endothelial cells (23, 33). In vitro studies suggest that reaction of NO gas with H2O2 produces singlet oxygen or hydroxyl radicals (34). Alternatively, the toxicity of NO/H2O2 may be caused by the production of a potent oxidant formed via a trace metal-, H2O2-, and NO-dependent process (23). The iron liberated from ferritin by NO indeed may promote oxidative stress caused by H2O2 (11, 35). Iron homeostasis can be regulated by NO through the activation of iron regulatory proteins (IRPs) by reducing the translation of mRNA encoding proteins that use or sequester iron such as σ-aminolevulinate synthase (36) and ferritin (37). One of these IRPs is identical in primary amino acid sequence to cytosolic aconitase, and the protein functions either as an RNA-binding protein or a functional enzyme depending on cytosolic iron levels and NO and ROI modulation (38, 39). NO has been shown to inhibit a tobacco cytosolic aconitase that shares a strong homology with the human IRP-1 and possesses the conserved mRNA binding domain for modulation of key proteins involved in intracellular iron homeostasis (40). Although tobacco aconitase has not been shown to be a functional IRP, NO generation in vivo could also increase free iron by mobilization from ferritin (35) and destruction of the iron-sulfur clusters of aconitase (40).
The relative rates of production of NO and O2− are critical in determining whether NO acts as a pro- or antioxidant (41). NO also can protect against oxidative damage by intercepting reactive species, such as the hydroxyl radical (11). NO protects potato against oxidative damage caused by methylviologen herbicides (42), and because ONOO− is not toxic to soybean cells, it is likely that NO can serve a protective function by diverting O2− from reactions causing cellular damage.
We propose a model (Fig. 7) in which NO is scavenged before it can react with H2O2 if the balance between NO and O2− production is in favor of O2−, whereas if the balance is in favor of NO, O2− is scavenged before it can dismutate to H2O2. Scavenging NO with O2− or scavenging O2− with NO leads to the formation of ONOO−, which is not an essential intermediate of NO-mediated cell death. The ability of NO to scavenge O2− may help reconcile our conclusion that H2O2 is the key ROI effector of pathogen-induced hypersensitive cell death and the observations that O2− is necessary and sufficient for propagation of cell death in the Arabidopsis lsd1 mutant (43). Thus, LSD1, which functions as a negative regulator of cell death by monitoring an O2−-dependent signal (43), regulates salicylic acid induction of Cu,Zn-SOD (44), and the spreading lesion phenotype of lsd1 mutants is correlated with a failure to up-regulate Cu,Zn-SOD (44). Likewise, our data indicate that O2− rather than H2O2 is the primary ROI signal for induction of GST. Failure to activate such cellular protectant genes in lsd1 mutants may allow constitutive generation of O2− in the absence of NO to drive the accumulation of O2− to toxic levels, whereas in a pathogen-induced HR, O2− from the oxidative burst is channeled toward H2O2 production. This model identifies fluctuations in O2− levels as the key indicator of redox stress in uninfected plants and the sensor integrating NO and H2O2 coactivation of pathogen-induced hypersensitive cell death.
Figure 7.
Balance model for NO and ROI interactions. (a) Although the NO/H2O2 cooperation triggers the HR cell death, the NO/O2− reaction leads to the formation of ONOO−, which is not an essential intermediate of NO-mediated cell death. (b) SOD activity is required for H2O2 accumulation during the HR. (c) When the NO/O2− balance is in favor of O2−, there is no NO left for interaction with H2O2. (d) When the NO/O2− balance is in favor of NO, there is no O2− left for SOD-mediated dismutation to H2O2.
Acknowledgments
J.Z. was a Fellow of the Alexander von Humboldt Foundation. This work was supported by a grant to M.D. from the Ministero dell'Università e della Ricerca Scientifica e Tecnologica and by the United Kingdom Biotechnology and Biological Sciences Research Council.
Abbreviations
- HR
hypersensitive response
- ROI
reactive oxygen intermediate
- SOD
superoxide dismutase
- GST
glutathione S-transferase
- XTT
sodium,3′-[1-[phenylamino-carbonyl]-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene-sulfonic acid hydrate
- SNP
sodium nitroprusside
- G/GO
glucose/glucose oxidase
- SIN-1
3-morpholinosydnonimine N-ethylcarbamide
- DDC
sodium diethyldithiocarbamate
References
- 1.Fang F C. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb C, Dixon R A. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:251–275. doi: 10.1146/annurev.arplant.48.1.251. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez M E, Pennell R I, Meijer P J, Ishikawa A, Dixon R A, Lamb C. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 4.Delledonne M, Xia Y, Dixon R A, Lamb C. Nature (London) 1998;394:585–588. doi: 10.1038/29087. [DOI] [PubMed] [Google Scholar]
- 5.Marla S S, Lee J, Groves J T. Proc Natl Acad Sci USA. 1997;94:14243–14248. doi: 10.1073/pnas.94.26.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beckman J S, Beckman T W, Chen J, Marshall P A, Freeman B A. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin K T, Xue J Y, Nomen M, Spur B, Wong P Y. J Biol Chem. 1995;270:16487–16490. doi: 10.1074/jbc.270.28.16487. [DOI] [PubMed] [Google Scholar]
- 8.Ischiropoulos H, al-Mehdi A B. FEBS Lett. 1995;364:279–282. doi: 10.1016/0014-5793(95)00307-u. [DOI] [PubMed] [Google Scholar]
- 9.Szabo C, Zingarelli B, O'Connor M, Salzman A L. Proc Natl Acad Sci USA. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamir S, Lewis R S, de Rojas Walker T, Deen W M, Wishnok J S, Tannenbaum S R. Chem Res Toxicol. 1993;6:895–899. doi: 10.1021/tx00036a021. [DOI] [PubMed] [Google Scholar]
- 11.Murphy M P. Biochim Biophys Acta. 1999;1411:401–414. doi: 10.1016/s0005-2728(99)00029-8. [DOI] [PubMed] [Google Scholar]
- 12.Levine A, Tenhaken R, Dixon R A, Lamb C. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 13.Keen N T, Buzzell R I. Theor Appl Genet. 1991;81:133–138. doi: 10.1007/BF00226123. [DOI] [PubMed] [Google Scholar]
- 14.Shirasu K, Nakajima H, Rajasekhar V K, Dixon R A, Lamb C. Plant Cell. 1997;9:261–270. doi: 10.1105/tpc.9.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCord J M, Fridovich I. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 16.Able A J, Guest D I, Sutherland M W. Plant Physiol. 1998;117:491–499. doi: 10.1104/pp.117.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipton S A, Choi Y B, Pan Z H, Lei S Z, Chen H S, Sucher N J, Loscalzo J, Singel D J, Stamler J S. Nature (London) 1993;364:626–632. doi: 10.1038/364626a0. [DOI] [PubMed] [Google Scholar]
- 18.Beauchamp C O, Fridovich I. Biochim Biophys Acta. 1973;317:50–64. doi: 10.1016/0005-2795(73)90198-0. [DOI] [PubMed] [Google Scholar]
- 19.Czarnecka E, Nagao R T, Key J L, Gurley W B. Mol Cell Biol. 1988;8:1113–1122. doi: 10.1128/mcb.8.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arahira M, Nong V H, Kadokura K, Kimura K, Udaka K, Fukazawa C. Biosci Biotechnol Biochem. 1998;62:1018–1021. doi: 10.1271/bbb.62.1018. [DOI] [PubMed] [Google Scholar]
- 21.Yahraus T, Chandra S, Legendre L, Low P S. Plant Physiol. 1995;109:1259–1266. doi: 10.1104/pp.109.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foresti R, Sarathchandra P, Clark J E, Green C J, Motterlini R. Biochem J. 1999;339:729–736. [PMC free article] [PubMed] [Google Scholar]
- 23.Farias-Eisner R, Chaudhuri G, Aeberhard E, Fukuto J M. J Biol Chem. 1996;271:6144–6151. doi: 10.1074/jbc.271.11.6144. [DOI] [PubMed] [Google Scholar]
- 24.Kelm M, Dahmann R, Wink D, Feelisch M. J Biol Chem. 1997;272:9922–9932. doi: 10.1074/jbc.272.15.9922. [DOI] [PubMed] [Google Scholar]
- 25.Fridovich I. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 26.Bowler C, Van Camp W, Van Montagu M, Inze D. Crit Rev Plant Sci. 1994;13:199–218. [Google Scholar]
- 27.Auh C K, Murphy T M. Plant Physiol. 1995;107:1241–1247. doi: 10.1104/pp.107.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desikan R, Hancock J T, Coffey M J, Neill S J. FEBS Lett. 1996;382:213–217. doi: 10.1016/0014-5793(96)00177-9. [DOI] [PubMed] [Google Scholar]
- 29.Troy C M, Derossi D, Prochiantz A, Greene L A, Shelanski M L. J Neurosci. 1996;16:253–261. doi: 10.1523/JNEUROSCI.16-01-00253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockhaus F, Brune B. Biochem J. 1999;338:295–303. [PMC free article] [PubMed] [Google Scholar]
- 31.Montalbini P, Buonaurio R. Plant Sci Lett. 1986;47:135–143. [Google Scholar]
- 32.Buonaurio R, Torre G d, Montalbini P. Physiol Mol Plant Pathol. 1987;31:173–184. [Google Scholar]
- 33.Filep J G, Lapierre C, Lachance S, Chan J S. Biochem J. 1997;321:897–901. doi: 10.1042/bj3210897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noronha-Dutra A A, Epperlein M M, Woolf N. FEBS Lett. 1993;321:59–62. doi: 10.1016/0014-5793(93)80621-z. [DOI] [PubMed] [Google Scholar]
- 35.Reif D W, Simmons R D. Arch Biochem Biophys. 1990;283:537–541. doi: 10.1016/0003-9861(90)90680-w. [DOI] [PubMed] [Google Scholar]
- 36.Dandekar T, Stripecke R, Gray N K, Goossen B, Constable A, Johansson H E, Hentze M W. EMBO J. 1991;10:1903–1909. doi: 10.1002/j.1460-2075.1991.tb07716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aziz N, Munro H N. Proc Natl Acad Sci USA. 1987;84:8478–8482. doi: 10.1073/pnas.84.23.8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Philpott C C, Klausner R D, Rouault T A. Proc Natl Acad Sci USA. 1994;91:7321–7325. doi: 10.1073/pnas.91.15.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy M C, Antholine W E, Beinert H. J Biol Chem. 1997;272:20340–20347. doi: 10.1074/jbc.272.33.20340. [DOI] [PubMed] [Google Scholar]
- 40.Navarre D A, Wendehenne D, Durner J, Noad R, Klessig D F. Plant Physiol. 2000;122:573–582. doi: 10.1104/pp.122.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Darley-Usmar V, Wiseman H, Halliwell B. FEBS Lett. 1995;369:131–135. doi: 10.1016/0014-5793(95)00764-z. [DOI] [PubMed] [Google Scholar]
- 42.Beligni M V, Lamattina L. Planta. 1999;208:337–344. [Google Scholar]
- 43.Jabs T, Dietrich R A, Dangl J L. Science. 1996;273:1853–1856. doi: 10.1126/science.273.5283.1853. [DOI] [PubMed] [Google Scholar]
- 44.Kliebenstein D J, Dietrich R A, Martin A C, Last R L, Dangl J L. Mol Plant–Microbe Interact. 1999;12:1022–1026. doi: 10.1094/MPMI.1999.12.11.1022. [DOI] [PubMed] [Google Scholar]