Abstract
Enzymes of the ALDH1A subfamily of aldehyde dehydrogenases are crucial in regulating retinoic acid (RA) signaling and have received attention as potential drug targets. ALDH1A2 is the primary RA-synthesizing enzyme in mammalian spermatogenesis and is therefore considered a viable drug target for male contraceptive development. However, only a small number of ALDH1A2 inhibitors have been reported, and information on the structure of ALDH1A2 was limited to the NAD-liganded enzyme void of substrate or inhibitors. Herein, we describe the mechanism of action of structurally unrelated reversible and irreversible inhibitors of human ALDH1A2 using direct binding studies and X-ray crystallography. All inhibitors bind to the active sites of tetrameric ALDH1A2. Compound WIN18,446 covalently reacts with the side chain of the catalytic residue Cys320, resulting in a chiral adduct in (R) configuration. The covalent adduct directly affects the neighboring NAD molecule, which assumes a contracted conformation suboptimal for the dehydrogenase reaction. The reversible inhibitors interact predominantly through direct hydrogen bonding interactions with residues in the vicinity of Cys320 without affecting NAD. Upon interaction with inhibitors, a large flexible loop assumes regular structure, thereby shielding the active site from solvent. The precise knowledge of the binding modes provides a new framework for the rational design of novel inhibitors of ALDH1A2 with improved potency and selectivity profiles.

Members of the human aldehyde dehydrogenase (ALDH) family of enzymes exert important physiological and detoxifying functions by catalyzing NAD(P)-dependent oxidation of aldehyde substrates to their corresponding carboxylic acids.1 Of the 19 human ALDH isozymes known, ALDH2 has a primary role in aldehyde detoxification during alcohol metabolism, while members of the ALDH1A subfamily synthesize retinoic acid (RA), the active metabolite of vitamin A1 (retinol). RA signaling is critical for the transcriptional control of many genes, and ALDH1A enzymes have received attention as potential drug targets.2–7
RA is crucial for initiation of meiosis,8 and retinoic acid receptor (RAR) antagonists such as BMS-189453 reversibly inhibit spermatogenesis in mice by inhibiting all three RAR isoforms, RARα, -β, and -γ.9 Another inhibitor affecting spermatogenesis through abrogation of RA metabolism is WIN18,446,10 which inhibits the ALDH1A enzymes of which ALDH1A1 and ALDH1A2 are responsible for the majority of RA production in testis.11–13 ALDH1A1 is expressed in sertoli cells while ALDH1A2 is found in spermatogonia, spermatids, and spermatocytes.12,14 ALDH1A2 is regarded as the primary RA-synthesizing enzyme in mammalian spermatogenesis,15 and ALDH1A2 levels are significantly reduced in the testicular tissue of infertile men.16 Therefore, ALDH1A2 is considered as a promising target for the development of nonhormonal male contraceptives.17
ALDH2 and ALDH2*2, a catalytically deficient mutant enzyme prevalent among the Asian population,18 are the structurally most studied ALDH enzymes. Small molecule inhibitors of ALDH2 include daidzin,19 Aldi compounds,20 and 2P3.21 Disulfiram,22 cyanamide,23 nitroglycerin,24 pargyline,25 molinate,26 and Aldi-320 are irreversible inhibitors of ALDH2 by covalently modifying the catalytic Cys302 residue. Less is known about enzymes of the ALDH1A subfamily and inhibitors thereof, in part because these enzymes can utilize several different substrates. Some of the earlier reports examining ALDH1A inhibition used propanal as a substrate but not the natural substrate retinaldehyde, leading to confusing results. However, several crystal structures of ALDH1A1-inhibitor complexes have been reported recently.21,27,28 Indole-2.3-dinones and diethylaminobenzaldehyde (DEAB) are pan-ALDH1A inhibitors reported to covalently attack the catalytic cysteines of ALDH3A1 and ALDH7A1.29–31 Analogues of duocarmycin form covalent bonds with catalytic and noncatalytic cysteine residues in ALDH1A1.28 WIN18,446 irreversibly inhibits ALDH1A enzymes, but the mechanism of action of covalent modification was unknown.32
ALDH1A2 is one of the least characterized and understood ALDH enzymes. Only two crystal structures of ALDH1A2 are known to date, rat ALDH1A2 liganded with NAD33 (PDB entry 1BI9) and human ALDH1A2 liganded with NAD (PDB entry 4X2Q). However, both structures contain only a fragment of NAD presumably due to incomplete ligand occupancy. Structural information about ALDH1A2–inhibitor complexes has not been available. Here, we describe the mechanisms of action of human ALDH1A2 inhibition by WIN18,446 and two novel reversible small molecule inhibitors using X-ray crystallography, direct binding, and enzymatic studies. The results provide a structural framework toward the rational design of potent and selective ALDH1A2 inhibitors.
RESULTS AND DISCUSSION
Biochemical Characterization of ALDH1A2 Inhibitors.
Human ALDH1A2 was expressed in E. coli and purified to high homogeneity. The interaction of ALDH1A2 with WIN18,446 and two recently developed reversible inhibitors, compounds 6–118 and CM121, was characterized by binding studies and enzyme kinetics (Figure 1). Differential scanning fluorimetry (DSF) was employed to assess the binding potential of each compound toward ALDH1A2. The melting temperature (Tm) of ALDH1A2 is about 63 °C, and the presence of 1 mM NAD+ resulted in a ΔTm value of 2.6 °C (Supporting Information Figure S1). The ΔTm values of ALDH1A2 in the presence of 200 μM compound ranged from 5 to 7.6 °C in the absence of NAD (Table 1). The presence of 1 mM NAD increased the ΔTm values for all compounds by approximately 3 °C, suggesting that the interaction of these inhibitors with ALDH1A2 is NAD-independent. The compound with the highest binding potential for ALDH1A2 was WIN18,446 followed by CM121 and 6–118. Direct binding studies by isothermal titration calorimetry (ITC) yielded apparent Kd values of 12 μM for NAD, 0.26 μM for 6–118, and 1.1 μM for CM121 (Figure 1B). The thermodynamic signatures of 6–118 and CM121 showed enthalpy driven interactions with negligible changes in entropy (Figure 1C).
Figure 1.
Binding potential and activity of ALDH1A2 inhibitors. (A) Chemical structures of the inhibitors studied in this work. (B) Direct binding studies by ITC. The top panels show the raw titration data; the bottom panels show the binding isotherms. (C) Thermodynamic signature of ALDH1A2 interaction with NAD and inhibitors. (D) Dose-dependent inhibition of the ALDH1A2 reaction with retinal and NAD as substrate and cofactor. The dotted lines show the activity of WIN18,446 upon different preincubation times with the enzyme prior to starting the reaction with retinal. The Kd and IC50 values are listed in Table 1.
Table 1.
Binding Potential and Inhibitory Activity of ALDH1A2 Inhibitors
| DSFa |
ITCb |
inhibitionc |
|||
|---|---|---|---|---|---|
| compound | ΔTm (°C); (−) NAD | ΔTm (°C); (+) NAD | Kd (μM) | stoichiometry (N) | IC50 (μM) |
| NAD | n/a | 2.6 ± 0.04 | 11.5 ± 3.9 | 0.54 | n/a |
| WIN18,446 | 7.6 ± 0.17 | 10.3 ± 0.07 | 0.028 ± 0.022d | 1.17 | 0.19 ± 0.05e |
| 6−118 | 5.0 ± 0.03 | 8.8 ± 0.09 | 0.26 ± 0.04 | 0.63 | 0.91 ± 0.33 |
| CM121 | 5.4 ± 0.15 | 8.6 ± 0.07 | 1.1 ± 0.28 | 1.04 | 0.54 ± 0.15 |
Single experiment in quadruplicate.
Single experiment.
Average of two experiments each in triplicate.
Apparent Kd = Ki* assuming a two-step mechanism of inactivation.
Value upon 22 min preincubation of enzyme with inhibitor.
WIN18,446 irreversibly inhibits ALDH1A2 in a two-step reaction, in which the formation of a rapidly reversible complex (Ki = 0.42 μM) is followed by a rate-limiting covalent modification step (kinact = 23 h−1).11,32 In such cases, the calorimetric signals are the result of noncovalent and covalent bond formation, as previously reported for irreversible inhibitors of glutathione S-transferase P1–1 and monoamine oxidase B.34,35 The signature of ALDH1A2 interaction with WIN18,446 showed an unfavorable change in entropy counteracted by a large increase in enthalpy, probably reflecting the covalent reaction with the enzyme (Figure 1C). The apparent Kd value of 0.03 μM indicates tight complex formation compared to the reversible inhibitors. Inhibition of the enzymatic activity of ALDH1A2 yielded IC50 values of 0.91 and 0.54 μM for 6–118 and CM121, respectively (Figure 1D). As expected for a covalent inhibitor, WIN18,446 inactivated ALDH1A2 in a time-dependent manner with an apparent IC50 value of 0.19 μM upon 22 min of preincubation.
WIN18,446 Inactivates ALDH1A2 through Covalent Modification of the Catalytic Residue Cys320.
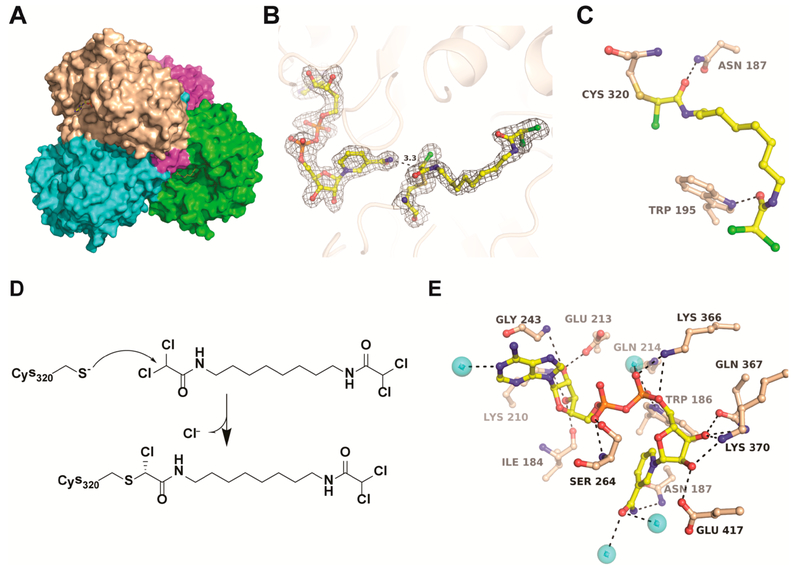
A cocrystal structure of ALDH1A2 liganded with NAD and WIN18,446 was determined at 1.9 Å resolution (Figure 2, Supporting Information Table S1). The asymmetric unit consists of a single copy of tetrameric ALDH1A2 (Figure 2A), each monomer clearly showing the inhibitor bound adjacent to NAD (Figure 2B). Continuous electron density between the inhibitor and the catalytic residue Cys320 indicated a covalent bond, and the resulting refined model was consistent with a Cys320-inhibitor covalent adduct in (R) configuration (Figure 2C). The rest of the inhibitor is stabilized through additional H bonds with Asn187 and Trp195 and multiple hydrophobic interactions with nonpolar residues. The covalent adduct is present in all four active sites of the ALDH1A2 tetramer (Supporting Information Figure S2), suggesting an SN2 reaction mechanism that involves removal of a chloride from WIN18,446 upon nucleophilic attack by the Cys320 thiol group (Figure 2D). NAD is stabilized through multiple H-bonding interactions with residues of the cofactor binding site (Figure 2E).
Figure 2.
Crystal structure of the ALDH1A2-WIN18,446 complex. (A) Surface presentation of the ALDH1A2–WIN18,446 tetramer with each polypeptide chain colored differently. (B) 2Fo–Fc electron density around NAD and WIN18,446 contoured at 1σ. Electron density maps of inhibitor in all four polypeptide chains are shown in Supporting Information Figure S2. (C) The covalent adduct between Cys320 and WIN18,446 along with H-bonding interactions in the active site. A stereo presentation of all binding interactions is shown in Supporting Information Figure S3. (D) Proposed mechanism for the covalent reaction of WIN18,446 with Cys320. (E) Hydrogen bonding interactions of NAD with residues of the cofactor site. Black dotted lines indicate H-bonding interactions.
Compounds 6–118 and CM121 Are Active Site-Directed Reversible Inhibitors of ALDH1A2.
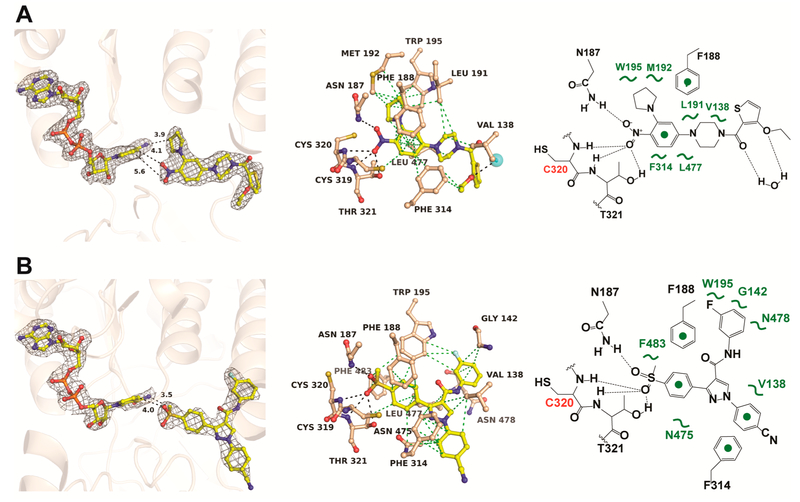
Compound 6–118 was cocrystallized with ALDH1A2 and NAD, and the structure was determined at 2.2 Å resolution (Figure 3A, Supporting Information Table S1). All four monomers showed inhibitor noncovalently bound in the active site adjacent to NAD (Supporting Information Figure S2). The nitro group is within hydrogen bonding distance to the main chain amides of Cys320 and Thr321 as well as the side chains of Thr321 and Asn187. The side chain of Phe188 interacts with the inhibitor through π–π stacking interactions, and several hydrophobic interactions stabilize the inhibitor in the active site. The 3-ethoxythiophene moiety is solvent exposed. Residues Cys320, Asn187, and Met192 are at the interface of the substrate-NAD binding pockets and thus interact with both NAD and compound 6–118.
Figure 3.
Crystal structures of ALDH1A2 in complex with reversible inhibitors. (A) ALDH1A2 in complex with NAD and compound 6–118. (B) ALDH1A2 in complex with NAD and compound CM121. The left panels show the 2Fo–Fc electron density map contoured at 1σ around NAD and inhibitor. The middle panels show potential H-bonding (black dotted lines) and VDW interactions (green dotted lines) of the inhibitors in the active site. The right panels show a schematic drawing of the inhibitor interactions. Electron density maps of inhibitor in all four polypeptide chains are shown in Supporting Information Figure S2. Stereo presentations of the binding interactions are shown in Supporting Information Figure S3.
Co-crystal structures of ALDH1A2 liganded with compound CM121 were determined in the absence and presence of NAD at 2.6 and 2.3 Å resolution, respectively. The two structures differed only slightly with root-mean-square deviations (RMSD) of 0.25 Å over all Cα atoms, the inhibitor assuming identical binding poses with or without NAD. Similar to the nitro group of 6–118, the methylsulfonyl oxygens of CM121 directly interact with the main chain amides of Cys320 and Thr321 as well as with the side chains of Thr321 and Asn187 (Figure 3B). Thus, the principal H-bonding interactions between inhibitor and enzyme are identical for the nitro group of 6–118 and the sulfonyl group of CM121. The side chains of Phe188 and Phe314 establish π–π stacking interactions with the methylsulfonylbenzene and the benzonitrile rings of CM121, respectively. Multiple hydrophobic interactions stabilize the inhibitor in the active site.
Structural Consequences of Covalent vs Reversible Inhibition of ALDH1A2.
The three structures of ALDH1A2 liganded with NAD and different inhibitors are highly similar with overall RMSD values between 0.24 and 0.29 Å (Figure 4A). Only residues Gly263 and Gly288, which are located in the NAD binding site, shift positions significantly. Superposition revealed that all inhibitors occupy a narrow approximately 9-Å-long path extending from Cys320 deep inside the catalytic site toward the surface (Figure 4B). The solvent exposed area of the inhibitor binding site includes a hydrophobic subpocket that accommodates the halogen containing groups of CM121 and WIN18,446. A major difference between the inhibitor complexes is a conformational change of NAD in the dead-end complex with WIN18,446. Upon reaction of Cys320 with WIN18,446, the (R)-chloroethyl group of the resulting covalent adduct protrudes into the adjacent NAD site (Figure 2B). As a consequence, the nicotinamide group gives way, and NAD assumes a contracted conformation (Figure 4C). This conformation is similar to that previously observed in crystal structures of ALDH2 thought to represent reduced NADH before exiting the cofactor site upon catalysis (Figure 4D).36,37 By contrast, the interaction of ALDH1A2 with both reversible inhibitors allows NAD to bind in an extended conformation deemed ideal for hydride transfer (Figures 3, 4E).36 Thus, the interaction with WIN18,446 not only irreversibly modifies a key catalytic residue of the active site but also renders the NAD molecule in a conformation suboptimal for the dehydrogenase reaction.
Figure 4.
Structural consequence of ALDH1A2 interaction with reversible and irreversible inhibitors. (A) Root mean square deviation (RMSD) of the Cα atoms of ALDH1A2 (chain A) liganded with compound 6–118 (blue) and CM121 (red) superimposed onto the ALDH1A2–WIN18,446 complex. (B) Superimposed inhibitors WIN18,446 (green), 6–118 (yellow), and CM121 (magenta) upon alignment of the respective ALDH1A2 cocrystal structures. The inhibitor binding site extends ~9 Å from Cys320 in the interior of the protein toward the solvent exposed area. A hydrophobic subpocket accommodates halogen-containing moieties of WIN18,446 and CM121. (C) Formation of the covalent adduct of Cys320 with WIN18,446 results in a steric clash with the neighboring nicotinamide group of NAD in its extended conformation (yellow). As a consequence, NAD assumes a contracted conformation (green). (D) The contracted conformation of NAD in the ALDH1A2–WIN18,446 complex (green) is similar to that of NADH in ALDH2 (PDB entry 1NZW, cyan). (E) Extended conformation of NAD as observed in the structures of ALDH1A2 with 6–118 (yellow) and CM121 (magenta).
Previously reported crystal structures of uninhibited rat and human ALDH1A2 are characterized by a large disorderd region spanning residues 475–495, which flanks the active site (PDB: 4X2Q, 1BI9).33 In the inhibited enzyme, particularly with WIN18,446, this loop assumes a well-defined regular structure throughout (Figure 5A,B). Few residues of the loop interact with the inhibitors through hydrophic interactions, thereby shielding the active site from solvent. It appears that occupation of the active site by small molecule inhibitors facilitates intermolecular interactions of the 475–495 loop with residues 162–166 of a neighboring monomer in the tetramer (Figure 5C).
Figure 5.
A large flexible loop in ALDH1A2 assumes regular structure upon inhibitor binding. (A) Monomeric human ALDH1A2 with partially occupied NAD (PDB 4X2Q, cyan) superimposed on the ALDH1A2–Win18,446 complex (beige). A 21 residue loop flanking the substrate binding site is flexible in the uninhibited enzyme but rigidifies in the dead-end complex (magenta). (B) 2Fo–Fc electron density of the loop residues contoured at 1σ; the corresponding amino acid sequence is also shown. (C) Intermolecular contacts of tetrameric ALDH1A2 between the 475–495 loop of one monomer (magenta) with the 162–166 loop of a neighboring monomer (green). WIN18,446 is shown in yellow.
Comparison with Other Aldehyde Dehydrogenases.
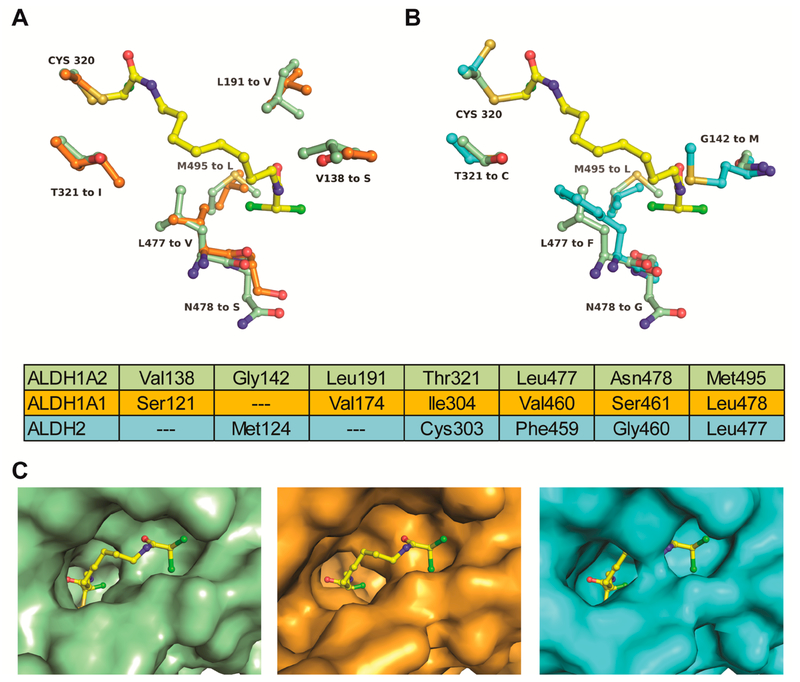
In order to obtain structural insights for future drug design, the active site of ALDH1A2 was compared with those of the structurally similar enzymes human ALDH1A1 (PDB entry 4WPN27) and human mitochondrial ALDH2 (PDB entry 2VLE;19 Figure 6). The amino acid sequences of ALDH1A2 and ALDH1A1 share 69% identity and 83% similarity, and the respective substrate binding sites differ in six residues (Figure 6A). The amino acid substitutions are similar in size and therefore only slightly change the active site architecture. However, the change in polarity from Thr321 to Ile is likely to have a negative impact on the binding potential of inhibitors such as 6–118 and CM121, which directly interact with the Thr321 hydroxyl group (Figure 3). Because of its close proximity to the catalytic Cys320 side chain, the Thr321Ile substitution may explain the 20-fold reduced catalytic efficacy reported for ALDH1A1.12 The change in polarity caused by the Val138Ser substitution at the entrance of the binding site may confer additional differential sensitivity for small molecule inhibitors.
Figure 6.
Comparison of the inhibitor binding sites of aldehyde dehydrogenases. (A) Superimposition of the ALDH1A2-WIN18,446 complex (green) with ALDH1A1 (PDB entry 4WPN, orange) shows six residues of the inhibitor binding site that differ between the two enzymes WIN18,446 is shown in yellow. (B) Superimposition with ALDH2 (PDB entry 2VLE, cyan) shows five amino acid substitutions, Gly142Met and Leu477Phe narrowing the binding site considerably. Aligned residues comprising the respective active sites are tabulated. (C) Surface presentations illustrating the effect of amino acid substitutions on the active site architecture of the respective enzymes. WIN18,446 was overlaid onto the ALDH1A1 and ALDH2 structures.
The amino acid sequences of ALDH1A2 and ALDH2 share 63% identity and 78% similarity, and the active sites differ in five residues (Figure 6B). The introduction of two bulky residues, Gly142Met and Leu477Phe, has a profound effect on the active site architecture of ALDH2, rendering the site considerably narrower than in ALDH1A2 or ALDH1A1 (Figure 6C). This is also consistent with the preference of ALDH2 for the small acetaldehyde molecule as a preferred substrate. While the conformational flexibility of WIN18,446 enables binding to the narrow active site of ALDH2, superposition of ALDH2 on ALDH1A2 predicts considerable steric hindrance for the binding of 6–118 and CM121.
CONCLUSIONS
The comparative analysis of irreversible and reversible ALDH1A2–inhibitor complexes demonstrates that the active site is capable of accommodating the binding of chemically and structurally diverse small molecules. The catalytic residue Cys320 is critical for the interaction with both covalent and reversible inhibitors, the thiol group serving as the nucleophile in the reaction with WIN18,446 and the main chain amide providing H-bonding potential for polar groups of reversible inhibitors. Covalent modification of Cys320 also impacts the neighboring cofactor site, rendering NAD in an unproductive conformation for hydride transfer. Considering the resurgent interest in covalent inhibitors as target-selective drugs,38 it appears that the active site architecture of ALDH1A2 is well suited for the development of such inhibitors using appropriate electrophilic warheads. The precise knowledge of the inhibitor binding modes described herein provides a new framework for the rational design of inhibitors with improved selectivity and potency for ALDH1A2, for example, by applying scaffold hopping approaches.39 The information may be particularly useful for the development of nonhormonal male contraceptives designed to alter testicular retinoic acid biosynthesis.
METHODS
General.
Reagents and compounds for biochemical and crystallographic experiments were purchased from Sigma-Aldrich and Hampton Research unless otherwise indicated. WIN18,446 was purchased from Acros Organics. Compounds 6–118 and CM121 were >95% pure by HPLC chromatography (for details including 1H NMR chemical shift values, see Supporting Information). ALDH1A2 concentration was determined by A280 molar absorbance with a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies) using an extinction coefficient of 52 370 M−1cm−1. Only the oxidized form of NAD (NAD+) was used in the experiments.
Cloning, Overexpression, and Protein Purification.
The previously obtained gene of full length human ALDH1A232 was used to generate a truncated version (residues 27–518) by polymerase chain reaction (PCR) and subcloned into a modified pET28a vector providing an N-terminal His6-tag and a PreScission protease cleavage site using restriction enzymes BamHI, NotI (Fermentas), and T4 DNA ligase (Invitrogen) following the manufacturers’ protocols. The plasmid was transformed into a BL21Star (DE3) competent cell line. Protein expression was induced at OD600 0.6–0.7 by the addition of 0.5 mM IPTG, and cell cultures were allowed to grow at 16 °C overnight. Harvested cell pellets were stored at −80 °C.
Protein purification was performed at 4 °C. Cell culture pellets were resuspended in lysis buffer containing 50 mM Hepes (pH 7.5), 200 mM NaCl, 20 mM imidazole, 0.5 mM TCEP with the addition of 0.5 mg mL−1 lysosyme, and 0.01% Triton X-100. After homogenization (Homogenizers, SPX) and centrifugation (25 000g, 1 h), the supernatant was purified by FPLC using a NiNTA affinity column (50 mL, Qiagen) using a linear gradient from 20 to 250 mM imidazole. The eluted ALDH1A2 peak was subjected to buffer exchange by Sephadex G-25 (GE Healthcare) into 50 mM Hepes (pH 7.5), 150 mM NaCl, and 2 mM DTT. His-tag cleavage was performed by mixing ALDH1A2 with GST-tagged PreScission protease at a ratio of 30:1 (w/w) and incubation overnight. Cleaved ALDH1A2 was loaded onto a Superdex 200 (26/600) column equilibrated with 50 mM Hepes (pH 7.5), 150 mM NaCl, and 2 mM DTT. Highly pure ALDH1A2 was eluted as a tetramer in a single peak. Pooled fractions were concentrated to 40 mg mL−1 using an Amicon centrifugal filter device with a 30 kDa cutoff and stored in 30 μL aliquots at −80 °C for crystallization studies.
Crystallization and Structure Determination.
Crystallization of ALDH1A2 was performed at 18 °C. Initial conditions were obtained by screening multiple precipitants using a Mosquito liquid handling robot (TTP Labtech) in 1μL sitting drops, and conditions were optimized in 4 μL hanging drops. High quality crystals grew reproducibly within 10 days by mixing 2 μL of protein with 2μL of precipitant (0.2 M sodium citrate tribasic dehydrate, 20% w/v polyethylene glycol 3350) supplemented with 1.3 mM inhibitor, 4 mM NAD, and 10% (v/v) DMSO. Compounds WIN18,446 and CM121 were cocrystallized in the absence of NAD followed by indiffusion of crystals with 10 mM NAD for 30 min. Single crystals were transferred to mother liquor including cryo-protectant (30% glycerol), mounted in loops, and flash frozen in liquid nitrogen. X-ray diffraction data were collected at SER-CAT beamlines 22-ID and 22-BM and processed with XDS.40 The structures were solved by molecular replacement using ALDH1A2 from the rat (PDB: 1BI9) as the search model. Molecular replacement and structure refinement were performed with PHENIX.41 Model building was performed using Coot.42 Initial models for the small molecule ligands were generated using MarvinSketch (ChemAxon) with ligand restraints from eLBOW. The covalent adduct of WIN18,446 with residue Cys320 was created with JLigand and modeled in Coot.
Differential Scanning Fluorimetry (DSF).
ALDH1A2 (4 μM) and inhibitor (200 μM) were mixed in 50 mM Hepes (pH 7.5), 50 mM NaCl, 2% DMSO, and SYPRO orange (Sigma, S5692) in a 96- well plate in the absence and presence of NAD (1 mM). Fluorescence was measured using the ROX Reporter channel (620 nm) on a StepOnePlus Real-Time PCR machine (Applied Biosystems). Each sample was measured in quadruplicate and the nonprotein control in duplicate. Unliganded ALDH1A2 with 2% DMSO was used as the reference, and data analysis was performed using the Protein Thermal Shift Software (Applied Biosystems).
Isothermal Titration Calorimetry (ITC).
ITC experiments were performed in a MicroCal iTC200 instrument (GE Healthcare) at 30 °C in 50 mM Hepes at pH 7.5, 50 mM NaCl, and 5% DMSO using reverse titration. ALDH1A2 (0.28–0.8 mM) was titrated into the cell containing NAD or inhibitor (25–52 μM). For inhibitor titrations, 1 mM NAD was present in the syringe and the cell. After the initial injection, a total of 15 injections (2.6 μL each) were performed with a spacing of 300 s and a reference power of 5 μcal/s. The stirring speed was kept at a constant 750 rpm. The dilution effect was measured by titrating protein into buffer and was subtracted from the data points. Data fitting was performed with the ORIGIN software using a onesite fitting mode.
Enzyme Inhibition Studies.
Inhibition of the enzymatic activity of ALDH1A2 was determined in 50 mM Na/K phosphate buffer at pH 7.5, 50 mM NaCl, 0.5 mM TCEP, and 2% DMSO. The reaction was monitored by measuring the formation of NADH at 340 nm on a SpectraMax 340PC384 Microplate Reader. ALDH1A2 (0.8 μg) was mixed with 100 μM NAD, 40 μM retinal, and increasing inhibitor concentrations. The reactions were initiated by the addition of enzyme (for reversible inhibitors) or by the addition of substrate upon incubation of the enzyme with WIN18,446. Data were fit to a fourparameter Hill equation using GraphPad Prism.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Moffitt Chemical Biology Core for access to the protein crystallography facility and ITC instrument (National Cancer Institute grant P30-CA076292) and the Southeast Regional Collaborative Access Team (SER-CAT, University of Georgia) for synchrotron data collection. This work was supported by the Eunice Kennedy Shriver National Institute for Child Health & Human Development (NICHD) grants HHSN275201300017C and U54HD04245.
ABBREVIATIONS
- RA
retinoic acid
- ALDH
aldehyde dehydrogenase
- DSF
differential scanning fluorimetry
- ITC
isothermal titration calorimetry
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00685.
Compound properties, X-ray data collection and refinement statistics, electron density maps of inhibitors in all four chains of ALDH1A2, stereo presentations of inhibitor binding interactions (PDF)
Molecular formula strings (XLSX)
Accession Codes
Atomic coordinates and structure factors of the ALDH1A2-inhibitor complexes have been deposited in the Protein Data Bank with entry codes 6ALJ, 6B5G, 6B5H, and 6B5I.
REFERENCES
- (1).Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, and Vasiliou V (2012) Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol. Rev 64, 520–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Clark DW, and Palle K (2016) Aldehyde dehydrogenases in cancer stem cells: potential as therapeutic targets. Ann. Transl Med. 4, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Liu X, Wang L, Cui W, Yuan X, Lin L, Cao Q, Wang N, Li Y, Guo W, Zhang X, Wu C, and Yang J (2016) Targeting ALDH1A1 by disulfiram/copper complex inhibits non-small cell lung cancer recurrence driven by ALDH-positive cancer stem cells. Oncotarget 7, 58516–58530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Perez-Alea M, McGrail K, Sanchez-Redondo S, Ferrer B, Fournet G, Cortes J, Munoz E, Hernandez-Losa J, Tenbaum S, Martin G, Costello R, Ceylan I, Garcia-Patos V, and Recio JA (2017) ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene 36, 5695–5708. [DOI] [PubMed] [Google Scholar]
- (5).Tomita H, Tanaka K, Tanaka T, and Hara A (2016) Aldehyde dehydrogenase 1A1 in stem cells and cancer. Oncotarget 7, 11018–11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang B, Chen X, Wang Z, Xiong W, Xu T, Zhao X, Cao Y, Guo Y, Li L, Chen S, Huang S, Wang X, Fang M, and Shen Z, (2017) Aldehyde dehydrogenase 1A1 increases NADH levels and promotes tumor growth via glutathione/dihydrolipoic acid-dependent NAD(+) reduction. Oncotarget 8, 67043–67055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Yokoyama Y, Zhu H, Lee JH, Kossenkov AV, Wu SY, Wickramasinghe JM, Yin X, Palozola KC, Gardini A, Showe LC, Zaret KS, Liu Q, Speicher D, Conejo-Garcia JR, Bradner JE, Zhang Z, Sood AK, Ordog T, Bitler BG, and Zhang R (2016) BET Inhibitors Suppress ALDH Activity by Targeting ALDH1A1 Super-Enhancer in Ovarian Cancer. Cancer Res. 76, 6320–6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Griswold MD, Hogarth CA, Bowles J, and Koopman P (2012) Initiating meiosis: the case for retinoic acid. Biol. Reprod. 86, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Chung SS, Wang X, Roberts SS, Griffey SM, Reczek PR, and Wolgemuth DJ (2011) Oral administration of a retinoic Acid receptor antagonist reversibly inhibits spermatogenesis in mice. Endocrinology 152, 2492–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Heller CG, Moore DJ, and Paulsen CA (1961) Suppression of spermatogenesis and chronic toxicity in men by a new series of bis(dichloroacetyl) diamines. Toxicol. Appl Pharmacol. 3, 1–11. [DOI] [PubMed] [Google Scholar]
- (11).Arnold SL, Kent T, Hogarth CA, Griswold MD, Amory JK, and Isoherranen N (2015) Pharmacological inhibition of ALDH1A in mice decreases all-trans retinoic acid concentrations in a tissue specific manner. Biochem. Pharmacol. 95, 177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Arnold SL, Kent T, Hogarth CA, Schlatt S, Prasad B, Haenisch M, Walsh T, Muller CH, Griswold MD, Amory JK, and Isoherranen N (2015) Importance of ALDH1A enzymes in determining human testicular retinoic acid concentrations. J. Lipid Res. 56, 342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Amory JK, Muller CH, Shimshoni JA, Isoherranen N, Paik J, Moreb JS, Amory DW Sr., Evanoff R, Goldstein AS, and Griswold MD (2011) Suppression of spermatogenesis by bisdichloroacetyldiamines is mediated by inhibition of testicular retinoic acid biosynthesis. J. Androl. 32, 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang X, Penzes P, and Napoli JL (1996) Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J. Biol. Chem. 271, 16288–16293. [DOI] [PubMed] [Google Scholar]
- (15).Kasimanickam VR (2016) Expression of retinoic acid-metabolizing enzymes, ALDH1A1, ALDH1A2, ALDH1A3, CYP26A1, CYP26B1 and CYP26C1 in canine testis during postnatal development. Reprod Domest Anim 51, 901–909. [DOI] [PubMed] [Google Scholar]
- (16).Amory JK, Arnold S, Lardone MC, Piottante A, Ebensperger M, Isoherranen N, Muller CH, Walsh T, and Castro A (2014) Levels of the retinoic acid synthesizing enzyme aldehyde dehydrogenase-1A2 are lower in testicular tissue from men with infertility. Fertil. Steril 101, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Amory JK (2016) Male contraception. Fertil. Steril. 106, 1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Chang JS, Hsiao JR, and Chen CH (2017) ALDH2 polymorphism and alcohol-related cancers in Asians: a public health perspective. J. Biomed. Sci. 24, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lowe ED, Gao GY, Johnson LN, and Keung WM (2008) Structure of daidzin, a naturally occurring anti-alcohol-addiction agent, in complex with human mitochondrial aldehyde dehydrogenase. J. Med. Chem. 51, 4482–4487. [DOI] [PubMed] [Google Scholar]
- (20).Khanna M, Chen CH, Kimble-Hill A, Parajuli B, Perez-Miller S, Baskaran S, Kim J, Dria K, Vasiliou V, Mochly-Rosen D, and Hurley TD (2011) Discovery of a novel class of covalent inhibitor for aldehyde dehydrogenases. J. Biol. Chem 286, 43486–43494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Buchman CD, and Hurley TD (2017) Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives. J. Med. Chem. 60, 2439–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shen ML, Johnson KL, Mays DC, Lipsky JJ, and Naylor S (2001) Determination of in vivo adducts of disulfiram with mitochondrial aldehyde dehydrogenase. Biochem. Pharmacol. 61, 537–545. [DOI] [PubMed] [Google Scholar]
- (23).DeMaster EG, Redfern B, and Nagasawa HT (1998) Mechanisms of inhibition of aldehyde dehydrogenase by nitroxyl, the active metabolite of the alcohol deterrent agent cyanamide. Biochem. Pharmacol. 55, 2007–2015. [DOI] [PubMed] [Google Scholar]
- (24).Wenzl MV, Beretta M, Griesberger M, Russwurm M, Koesling D, Schmidt K, Mayer B, and Gorren AC (2011) Site-directed mutagenesis of aldehyde dehydrogenase-2 suggests three distinct pathways of nitroglycerin biotransformation. Mol. Pharmacol. 80, 258–266. [DOI] [PubMed] [Google Scholar]
- (25).Moridani MY, Khan S, Chan T, Teng S, Beard K, and O’Brien PJ (2001) Cytochrome P450 2E1 metabolically activates propargyl alcohol: propiolaldehyde-induced hepatocyte cytotoxicity. Chem.-Biol. Interact. 130–132, 931–942. [DOI] [PubMed] [Google Scholar]
- (26).Allen EM, Anderson DG, Florang VR, Khanna M, Hurley TD, and Doorn JA (2010) Relative inhibitory potency of molinate and metabolites with aldehyde dehydrogenase 2: implications for the mechanism of enzyme inhibition. Chem. Res. Toxicol 23, 1843–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Morgan CA, and Hurley TD (2015) Characterization of two distinct structural classes of selective aldehyde dehydrogenase 1A1 inhibitors. J. Med. Chem. 58, 1964–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Koch MF, Harteis S, Blank ID, Pestel G, Tietze LF, Ochsenfeld C, Schneider S, and Sieber SA (2015) Structural, Biochemical, and Computational Studies Reveal the Mechanism of Selective Aldehyde Dehydrogenase 1A1 Inhibition by Cytotoxic Duocarmycin Analogues. Angew. Chem., Int. Ed. 54, 13550–13554. [DOI] [PubMed] [Google Scholar]
- (29).Kimble-Hill AC, Parajuli B, Chen CH, Mochly-Rosen D, and Hurley TD (2014) Development of selective inhibitors for aldehyde dehydrogenases based on substituted indole-2,3-diones. J. Med. Chem. 57, 714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Luo M, Gates KS, Henzl MT, and Tanner JJ (2015) Diethylaminobenzaldehyde Is a Covalent, Irreversible Inactivator of ALDH7A1. ACS Chem. Biol 10, 693–697. [DOI] [PubMed] [Google Scholar]
- (31).Morgan CA, Parajuli B, Buchman CD, Dria K, and Hurley TD (2015) N,N-diethylaminobenzaldehyde (DEAB) as a substrate and mechanism-based inhibitor for human ALDH isoenzymes. Chem.-Biol. Interact 234, 18–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Paik J, Haenisch M, Muller CH, Goldstein AS, Arnold S, Isoherranen N, Brabb T, Treuting PM, and Amory JK (2014) Inhibition of retinoic acid biosynthesis by the bisdichloroacetyldiamine WIN 18,446 markedly suppresses spermatogenesis and alters retinoid metabolism in mice. J. Biol. Chem. 289, 15104–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lamb AL, and Newcomer ME (1999) The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity,. Biochemistry 38, 6003–6011. [DOI] [PubMed] [Google Scholar]
- (34).Quesada-Soriano I, Primavera A, Casas-Solvas JM, Tellez-Sanz R, Baron C, Vargas-Berenguel A, Lo Bello M, and Garcia-Fuentes L (2012) Identifying and characterizing binding sites on the irreversible inhibition of human glutathione S-transferase P1–1 by S-thiocarbamoylation. ChemBioChem 13, 1594–1604. [DOI] [PubMed] [Google Scholar]
- (35).Rojas RJ, Edmondson DE, Almos T, Scott R, and Massari ME (2015) Reversible and irreversible small molecule inhibitors of monoamine oxidase B (MAO-B) investigated by biophysical techniques. Bioorg. Med. Chem 23, 770–778. [DOI] [PubMed] [Google Scholar]
- (36).Perez-Miller SJ, and Hurley TD (2003) Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase. Biochemistry 42, 7100–7109. [DOI] [PubMed] [Google Scholar]
- (37).D’Ambrosio K, Pailot A, Talfournier F, Didierjean C, Benedetti E, Aubry A, Branlant G, and Corbier C (2006) The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis,. Biochemistry 45, 2978–2986. [DOI] [PubMed] [Google Scholar]
- (38).Lagoutte R, Patouret R, and Winssinger N (2017) Covalent inhibitors: an opportunity for rational target selectivity. Curr. Opin. Chem. Biol 39, 54–63. [DOI] [PubMed] [Google Scholar]
- (39).Sun H, Tawa G, and Wallqvist A (2012) Classification of scaffold-hopping approaches. Drug Discovery Today 17, 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kabsch W (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis W, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, and Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.