Summary
While the canonical assembly of a GABAA receptor contains two α subunits, two β subunits, and a fifth subunit, it is unclear which variants of each subunit are necessary for native receptors. We used CRISPR/Cas9 to dissect the role of the GABAA receptor β subunits in inhibitory transmission onto hippocampal CA1 pyramidal cells and found that deletion of all β subunits 1, 2, and 3 completely eliminated inhibitory responses. In addition, only knockout of β3, alone or in combination with another β subunit, impaired inhibitory synaptic transmission. We found β3 knockout impairs inhibitory input from PV but not SOM expressing interneurons. Furthermore, expression of β3 alone on the background of the β1-3 subunit knockout was sufficient to restore synaptic and extrasynaptic inhibitory transmission. These findings reveal a crucial role for the β3 subunit in inhibitory transmission and identify a synapse specific role of the β3 subunit in GABAergic synaptic transmission.
ETOC BLURB
Nguyen and Nicoll use CRISPR/Cas9 to dissect the role of the β subunits in functional GABAA receptors, uncovering synapse-specific requirements governing GABAA receptor subunit composition.
Introduction
GABAA receptors (GABAARs) are heteropentameric ionotropic receptors which mediate the majority of fast inhibitory neurotransmission in the brain (Olsen and Sieghart, 2008; Sieghart, 2006). These receptors are comprised of a multitude of different subunit families each with their own number of isoforms, including α1-6, β1-3, γ1-3, δ, ε, θ, π, and ρ1-3 (Simon et al., 2004). The vast diversity of subunits makes studying this particular receptor especially daunting. However, previous work has established that the canonical assembly of the GABAAR contains the presence of two α subunits, two β subunits, and a fifth subunit (Chang et al., 1996; Sieghart and Sperk, 2002).
Much of the work on assembly and function of GABAARs has been done using heterologous expression systems to observe functional assembly of discrete sets of overexpressed subunits. A key criticism of this approach is the potential assembly of subunit combinations that normally would not occur in the native mammalian system (Olsen and Sieghart, 2008). In addition, the different GABAAR subunits vary in their expression within the brain, suggesting that particular receptor subunit profiles may exist within specific brain regions and in particular neuronal circuits (Fritschy and Panzanelli, 2014; Laurie et al., 1992). This insight however has not translated into much progress in understanding the role of native receptor subunits since knockouts do not exist for all known subunits of the GABAA receptor (Rudolph and Mohler, 2004).
CRISPR/Cas9 technology enables efficient gene editing without the time and expense necessary to generate entire knockout mice. This approach has previously been shown to enable complete deletion of synaptic proteins in post-mitotic neurons (Incontro et al., 2014; Straub et al., 2014). We utilize this technology to probe the function of the β subunits of the GABAA receptor in inhibitory synaptic transmission onto hippocampal CA1 pyramidal cells. We find that, indeed, the β subunit is required for assembly of functional GABAARs and that expression of the β3 subunit in particular is important for proper inhibitory transmission. We show that knockout of β3 affects inputs from parvalbumin (PV) but not somatostatin (SOM) expressing interneurons onto pyramidal cells. Furthermore, we show that expression of β3 alone is sufficient to rescue inhibitory currents in the context of a β1-3 subunit knockout. Our findings identify a key requirement for the β subunit and highlight the unique importance of the β3 subunit in regulating GABAAR function, showcasing an approach that can be used for the study of native receptor composition and function at inhibitory synapses.
Results
Functional GABAA receptors require the β subunit
To determine the importance of the β subunits to inhibitory transmission, we utilized CRISPR-Cas9 technology to develop a knockout construct containing individual single guide RNA (sgRNA) sequences for β1, β2, and β3 chained together (Figure 1A). Lentiviral-mediated expression of these guides along with Cas9 in dissociated rat hippocampal cultures resulted in a dramatic reduction in protein levels of all three subunits (Figure 1B). For all remaining experiments we biolistically transfected rat organotypic hippocampal slices with these constructs to enable sparse transfection (Figure 1C). The organotypic slice prep enables high-throughput screening of our manipulations while preserving the endogenous circuitry (Di Cristo et al., 2004). Since our constructs co-express a fluorophore, we are able to specifically record from cells expressing both constructs. We measured inhibitory currents induced with a stimulus electrode placed in area CA1. Simultaneous recording of a transfected and untransfected neighboring CA1 pyramidal cell revealed complete loss of evoked inhibitory currents (IPSCs) in transfected cells, showing that functional GABAARs require inclusion of the β subunit (Figure 1D-F). Regardless of the size of the evoked inhibitory current elicited in the control cell, we consistently failed to see a significant response in the transfected cell (Figure 1E). However, evoked excitatory responses (EPSCs) were unchanged (Figure S1A). We also failed to see any spontaneous inhibitory responses (Figure S1B). Time course for the full effect is about 3 weeks and therefore all recordings were made after 3 weeks.
Figure 1. GABAA β1-3 subunits are necessary for inhibitory transmission.
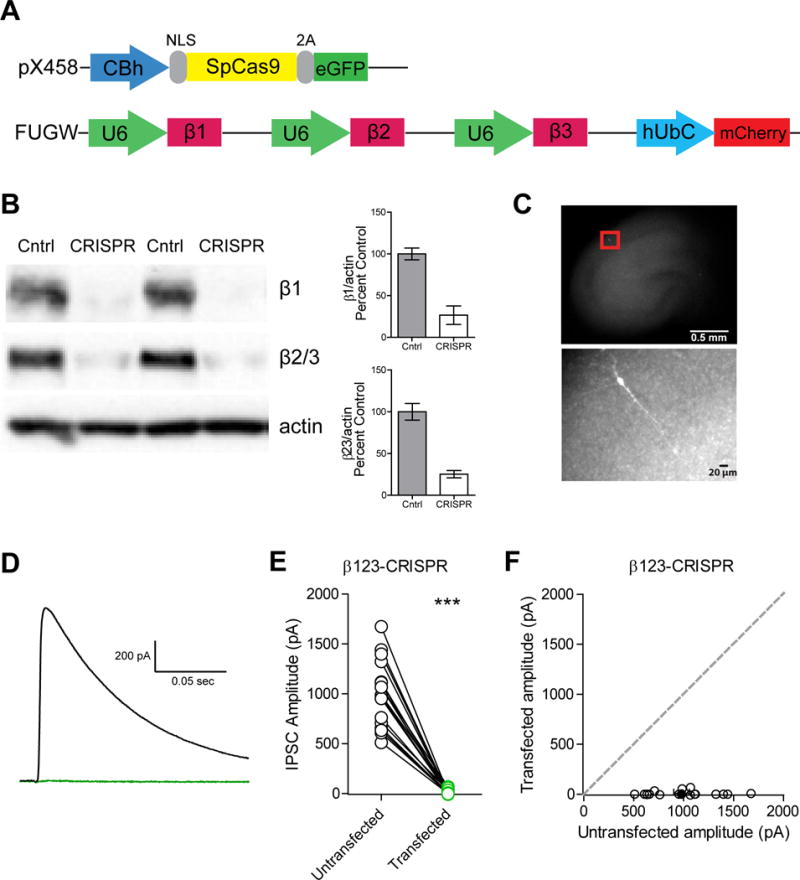
A) Diagram of the vector constructs used to express the chained sgRNAs (in pink) for β1, β2, and β3 and Cas9 (in yellow). CBh: promoter; NLS: nuclear localization sequence; 2A: cleavage peptide; U6: promoter; hUbC: promoter. B) Western blot of rat dissociated hippocampal cultures infected with lentiviruses for both Cas9 and sgRNA (CRISPR lanes) or cultures transfected with lentivirus only for Cas9 (Control lanes). Lysates were probed for expression of β1, β2/3, and actin (n=4 replicates). C) Representative image of our organotypic hippocampal slice culture preparation with a zoomed in image of a transfected cell. D) Representative evoked IPSC traces of an untransfected cell (in black) and cell transfected with the sgRNAs for β1-3 and Cas9 (in green). E) Varying absolute amplitudes were observed in control untransfected cells while transfected cells displayed no inhibitory current (***p = 0.0002, n = 18) F) Scatter plot showing no significant IPSCs observed in β1-3 CRISPR transfected neurons compared to controls (***p = 0.0002, n = 18). Open circles are individual pairs, filled circle is mean ± s.e.m. See also Figure S1.
The β3 subunit is most important for inhibitory transmission
While transgenic knockout animals exist for the β2 and β3 subunit, there is no knockout for β1. Previous work looking at the effects of β2 knockout on synaptic transmission in dentate gyrus granule cells found no effect on miniature IPSCs (Herd et al., 2008). β3 knockout mice display deficits in inhibitory synaptic transmission in granule cells but not mitral cells of the olfactory bulb (Nusser et al., 2001). In order to determine the relative contribution of each β subunit to inhibitory synaptic transmission in the hippocampus, we expressed individual sgRNAs for the β subunits (Figure 2A). Knockout of neither β1 nor β2 had a significant effect on inhibitory transmission (Figure 2B-C and 2E). This is consistent with the low expression of β2 in the hippocampus (Laurie et al., 1992; Sperk et al., 1997). Knockout of β3 resulted in a significant impairment of inhibitory currents (Figure 2D and 2E), indicating its importance for proper inhibitory transmission.
Figure 2. GABAA β3 subunit is important for inhibitory transmission.
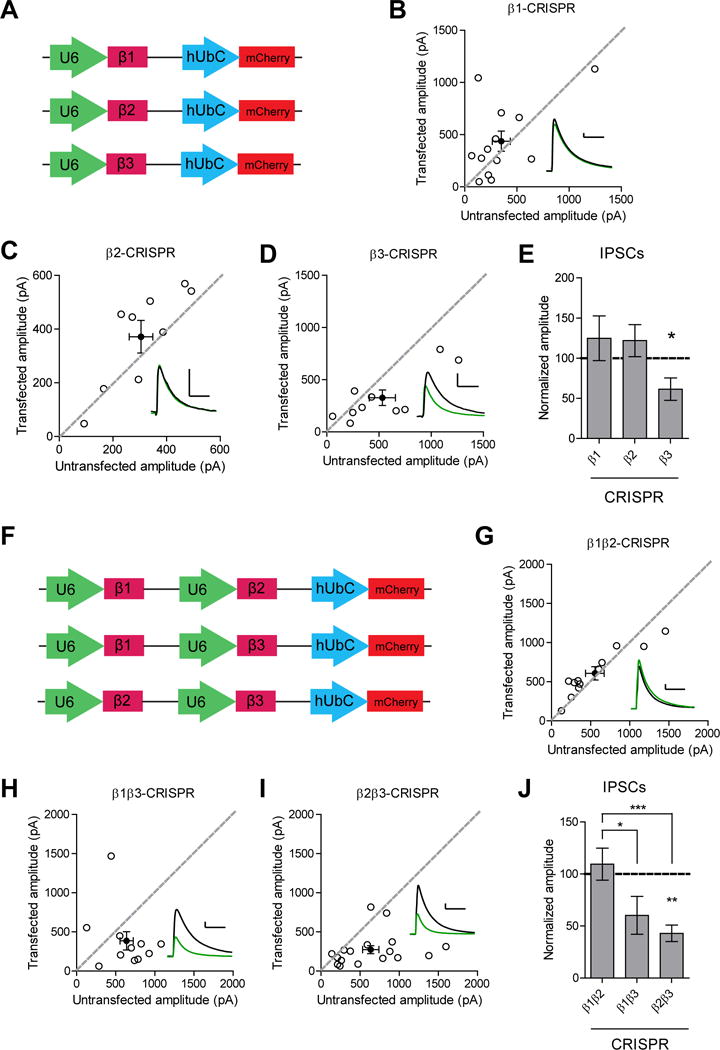
A) Diagram of the vector constructs used to express the individual sgRNAs for β1, β2, and β3. B) Scatter plot showing no reduction in IPSCs in β1 CRISPR transfected neurons compared to controls (p = 0.4, n = 13). C) Scatter plot showing no reduction in IPSCs in β2 CRISPR transfected neurons compared to controls (p = 0.1, n = 9). D) Scatter plot showing reduction in IPSCs in β3 CRISPR transfected neurons compared to controls (*p = 0.037, n = 10). E) Summary graph of B-D. F) Diagram of the vector constructs used to express two of the sgRNAs for β1, β2, and β3 at once. G) Scatter plot showing no reduction in IPSCs in β1 and β2 CRISPR transfected neurons compared to controls (p = 0.2, n = 12). H) Scatter plot showing inhibitory currents in β1 and β3 CRISPR transfected neurons were not significantly different compared to controls (p = 0.1, n = 11). I) Scatter plot showing reduction in IPSCs in β2 and β3 CRISPR transfected neurons compared to controls (**p = 0.0024, n = 17). J) Summary graph of G-I. Knockout of both β1 and β3 or both β2 and β3 resulted in currents that were significantly reduced compared to those seen in knockout of both β1 and β2 (*p = 0.015, ***p = 0.0006). For panels B-D, G-I open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 msec. For panels E and J summary graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure S2 and S3.
We then proceeded to knockout two different subunits at once to determine the properties of the endogenous subunit that was left (Figure 2F). Knockout of both β1 and β2 did not result in a significant change to inhibitory currents, suggesting that the endogenous β3 subunit remaining is fully able to maintain proper inhibitory synaptic transmission (Figure 2G and 2J). While the currents observed after knockout of β1 and β3 were not significantly different than control, the effect was significantly different than that seen with knockout of both β1 and β2, suggesting that the presence of β2 alone may not be sufficient to maintain inhibitory transmission (Figure 2H and 2J). Knockout of β2 and β3 resulted in a significant depression of inhibitory currents, suggesting that the presence of β1 alone is not sufficient to maintain inhibitory transmission (Figure 2I and 2J). Overall, these results show that β3 is necessary for maintaining proper inhibitory transmission and is the most able to compensate for the loss of the other β subunits.
Knockout of β3 preferentially affects PV but not SOM inputs
It was interesting that neither our single nor double knockout manipulations fully recapitulated the dramatic elimination of all inhibitory current observed with the triple knockout. Since CA1 pyramidal cells receive inhibitory inputs from a multitude of interneuron subtypes, we wondered whether there are subtype specific effects in our knockout manipulations which could underlie the partial reduction observed with single and double knockouts. In particular, we wanted to parse the inputs from PV positive interneurons, which target somatic and proximal dendritic regions, from SOM positive interneurons, which target distal dendritic regions of pyramidal cells (Rudy et al., 2011; Xu et al., 2010). We wanted to know if these anatomical differences in the inhibitory projections translated to molecular differences in the requirements for GABAAR composition at these synapses. We focused on knockout of β3 since it was the only single knockout manipulation that had exhibited a significant reduction in inhibitory currents. In addition, the inhibitory currents remaining after knockout of either β3 or both β2 and β3 displayed faster decay kinetics both in evoked (Figure S2C and S2D) and spontaneous inhibitory currents (Figure S3), suggesting that the inhibitory synapses remaining are electrophysiologically distinct.
We utilized transgenic mice expressing Cre either under the PV or SOM promoter along with Cre-dependent expression of Channelrhodopsin (ChR2). Experiments were performed wherein electrical stimulation was given to sample the population of inhibitory inputs followed by a brief pulse of blue light to stimulate the particular inputs from either PV or SOM cells. Using this approach, we found that β3 knockout significantly impaired responses mediated by PV interneurons while inputs from SOM interneurons were unaffected, suggesting that loss of β3 preferentially affects PV but not SOM inputs (Figure 3B-C and 3E-F). Importantly, electrical stimulation of slices from both PV-ChR2 and SOM-ChR2 mice still showed a deficit in inhibitory currents due to loss of β3, consistent with what was observed in rat slices (Figure 3A and 3C, 3D and 3F).
Figure 3. Knockout of GABAA β3 subunit preferentially affects PV not SOM inputs.
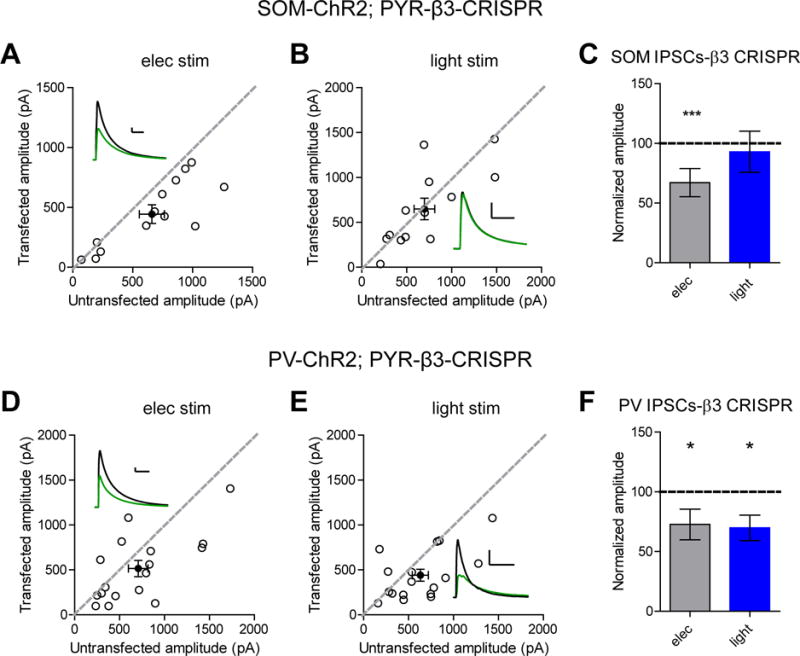
A) Scatter plot showing electrical stimulation of hippocampal organotypic slices from SOM-ChR2 transgenic mice still displayed a deficit in inhibitory transmission in pyramidal cells transfected with β3 CRISPR compared to controls (***p = 0.0005, n = 13). B) Scatter plot showing no reduction in IPSCs in β3 CRISPR transfected pyramidal cells when SOM currents are specifically elicited with blue light (p = 0.3, n = 13). C) Summary graph of A-B. D) Scatter plot showing electrical stimulation of hippocampal organotypic slices from PV-ChR2 transgenic mice still displayed a deficit in inhibitory transmission in pyramidal cells transfected with β3 CRISPR compared to controls (*p = 0.04, n = 17). E) Scatter plot showing reduction in IPSCs in β3 CRISPR transfected pyramidal cells when PV currents are specifically elicited with blue light (*p = 0.01, n = 17). F) Summary graph of D-E. For panels A-B and D-E, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 msec. For panels C and F summary graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. Significance above each column represents pairwise comparison between transfected and untransfected cells. See also Figure S4 and S6.
To determine whether these synapse-specific effects of β3 knockout are due to differences in the endogenous levels of β3 containing GABAARs at these two types of inhibitory synapses, we washed on etomidate, a positive allosteric modulator of β2/β3-containing GABAARs in untransfected slices from PV-ChR2 and SOM-ChR2 mice. Prior work with this drug has shown that it significantly slows the decay kinetics of IPSCs (Drexler et al., 2009). Since our data suggests minimal contribution of the β2 subunit in hippocampal GABAAR function, we expect that any effect from washing on etomidate would be mostly due to the presence of the β3 subunit. Bath application of etomidate significantly slowed the decay kinetics of light-evoked IPSCs in PV-ChR2 slices but not in SOM-ChR2 slices, suggesting enrichment of β2/β3-containing GABAARs at synapses formed by PV versus SOM interneurons (Figure S4A-C).
Expression of β3 alone is sufficient to restore inhibitory transmission
To follow up on our result that knockout of both β1 and β2 did not change inhibitory synaptic transmission, which suggested that the presence of β3 alone was sufficient to maintain proper inhibitory transmission, we expressed a human ortholog of β3 that would not be recognized by our sgRNA in combination with the β1-3 subunit knockout. Inhibitory currents recorded from these cells did not show a significant difference compared to control, indicating that expression of β3 alone is able to restore inhibitory synaptic currents (Figure 4A-B). Converse to what we observed with the β3 knockout, these rescued currents displayed slower kinetics (Figure S2E). We also looked to see if expression of the β3 subunit alone is able to restore extrasynaptic currents, since we see no response after applying a 100μM puff of GABA, which samples both synaptic and extrasynaptic GABAARs (Song et al., 2011), onto cells expressing the β1-3 subunit knockout (Figure 4C). Indeed, expression of β3 on the background of the β1-3 subunit knockout is able to fully restore responses to control levels (Figure 4D).
Figure 4. GABAA β3 subunit is sufficient to restore inhibitory transmission.
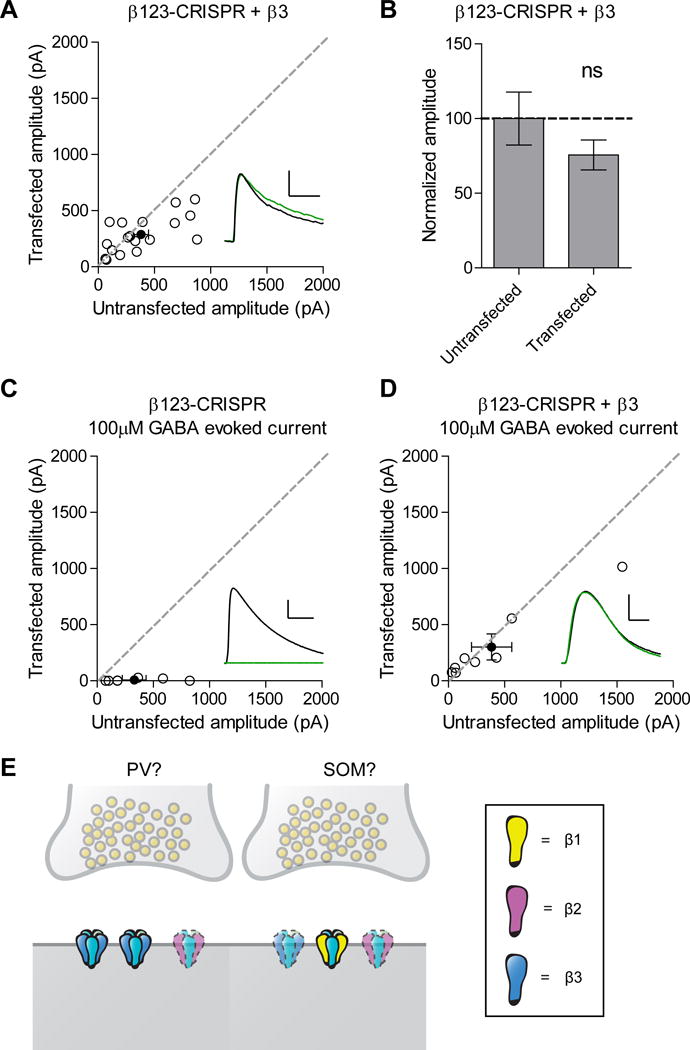
A) Scatter plot showing expression of β3 on the background of the triple β subunit CRISPR is able to restore inhibitory synaptic currents (p = 0.1071, n = 18). B) Summary graph of A. C) Scatter plot showing no responses to a 100μM puff of GABA in cells transfected with the triple β subunit CRISPR compared to controls (*p = 0.0156, n = 7). D) Scatter plot showing rescue of responses to a 100μM puff of GABA in cells transfected with the triple β subunit CRISPR and β3 (p = 0.5, n = 8). E) Our model proposes the existence of two different types of inhibitory synapses, corresponding to PV and SOM-mediated inputs. One has GABAARs with β3 and possibly β2 subunits and one has receptors comprised of all three β isoforms. Receptors containing β2 are more opaque and outlined with a dashed line at both types of synapses and receptors containing β3 are more opaque and outlined with a dashed line at the SOM synapse to highlight the low expression levels of these subunits. For panels A and C-D, open circles are individual pairs, filled circle is mean ± s.e.m. Black sample traces are control, green are transfected. Scale bars represent 100 pA and 50 msec for A and 200 pA and 1 sec for C-D. For panel B summary graph plots mean transfected amplitude ± s.e.m, expressed as a percentage of control amplitude. See also Figure S2 and S5.
To determine whether this property is unique to β3 or whether β1 or β2 could also restore GABAAR function, we expressed human orthologs of β1 or β2 that would not be recognized by our sgRNAs in combination with the β1-3 subunit knockout. This also allowed us to determine whether the lack of effect we observed with our knockout manipulations involving β2, which is the most abundant β subunit in the brain outside of the hippocampus, is due to its unique low expression in the hippocampus or a lack of function of the subunit in this region of the brain (Laurie et al., 1992). We found that rescue with β1 was unable to fully restore inhibitory synaptic transmission while rescue with β2 is able to restore currents similar to β3, suggesting that β1 has a limited role and β2 can indeed function at hippocampal synapses if expressed at higher levels (Figure S5A-D).
Discussion
Dissection of the molecular and circuit mechanisms of inhibitory neurotransmission is complicated by the diversity in subunit composition of the GABAARs in addition to the diversity in population of the inhibitory interneurons themselves. The emergence of new tools to hone in on the precise workings of native receptors within defined neuronal circuits hold much promise toward accelerating progress in understanding inhibition in the brain.
Here we utilize both CRISPR/Cas9 as well as optogenetic approaches to dissect the involvement of the β subunits of the GABAAR in inhibitory transmission. We find that the presence of the β subunit is absolutely required for the assembly of functional GABAARs. In addition, we find that only knockout of β3 impairs inhibitory transmission and this manipulation preferentially affects PV but not SOM-mediated inhibitory synapses onto CA1 pyramidal cells due to the enrichment of β2/β3-containing GABAARs at these synapses. Finally, we show that expression of β3 alone is sufficient to rescue the complete loss of inhibitory currents observed in the triple β subunit knockout. Together these results highlight the crucial role of the β subunit, especially β3, of the GABAAR in inhibitory transmission both at the level of individual circuits and in overall transmission.
Efficacy of CRISPR/Cas9
CRISPR/Cas9 works with a remarkable efficiency in eliminating the expression of synaptic proteins in post-mitotic neurons (Incontro et al., 2014; Straub et al., 2014). A key question in further applying this technology for genetic manipulations concerns whether this approach can be used to target multiple different gene targets at once while maintaining the same efficacy and efficiency as seen with single targets. In our biolistic approach, we were able to observe >90% co-expression and functionality of Cas9 and all three sgRNAs after about 3 weeks. It is possible that by using post-mitotic cells we maintain high expression of the Cas9 and sgRNAs, enabling multiple rounds of gene editing until expression of the target is effectively eliminated.
Our knockout of all three β subunits eliminated all inhibitory current, both synaptic and extrasynaptic. This manipulation can be used in future studies as a way to eliminate all inhibition postsynaptically. It is possible that other cell types in other brain regions have different requirements for GABAAR functional assembly. Our approach provides a guide and framework in which to address this possibility and other remaining questions such as the precise nature of GABAAR composition and the identity of specific interneuron populations affected by loss of the β3 subunit in particular.
The critical requirement of the β subunit
We were able to show that native assembly of a functional GABAAR requires inclusion of the β subunit. This is consistent with observations in heterologous systems that GABAARs are composed of two α subunits, two β subunits, and one other subunit (Chang et al., 1996; Sieghart and Sperk, 2002). Furthermore, our results show that expression of just one β subunit isoform, in this case β2 or β3, is able to restore the deficits seen when all three are eliminated, suggesting that within the synaptic receptor complex of 2 α, 2 β, and a γ subunit the two β subunits can be the same isoform. While biochemical studies have suggested the existence of two different β subunit isoforms in the same GABAAR, our results show that this criteria does not prevent functional assembly (Jechlinger et al., 1998; Li and De Blas, 1997).
β3 is critical for proper inhibitory transmission and the limited role of β2 in the hippocampus
We found in both our single and double knockout manipulations that it was only when β3 is knocked out, either alone or in combination with another β isoform, that inhibitory currents are depressed, suggesting that out of the three β subunits, β3 is most important for proper inhibitory transmission in the hippocampus. The role of β3 was observed to be even more critical in the absence of β1 and β2 or all endogenous β subunits where expression of β3 alone is sufficient to maintain or restore inhibitory currents respectively.
The decay kinetics of the GABAAR depend on the identity of the particular α subunit isoform expressed, with α1 containing receptors having faster kinetics than α2/3 containing receptors (Gingrich et al., 1995). Previous characterization of neurons from β3 knockout mice found faster mIPSC decay kinetics due to the reduction in expression of α2/3 subunits (Ramadan et al., 2003). Our results showing faster kinetics in the β3 knockout and slower kinetics in the β3 rescue with the triple β subunit knockout corroborate this finding and suggest that the β3 subunit preferentially associates with α2/3 subunits to mediate slower IPSC decay kinetics and therefore longer-lasting inhibition (Figure S2 and S3). This difference in kinetics can have profound effects on network processes such as temporal coding (Xie and Manis, 2013).
In contrast to β3, the role of β2 in the hippocampus is less crucial, most likely due to minimal expression of the β2 subunit in the hippocampus compared to most other regions of the brain (Laurie et al., 1992; Sperk et al., 1997). This low level of expression explains why we did not see a significant effect with knockout of β2 whereas previous work characterizing β2 knockout mice found a significant decrease in GABAergic currents in Purkinje neurons of the cerebellum where expression levels are high (Sur et al., 2001). A key question of this observation is whether this reduced expression underlies a fundamental inability of the β2 subunit to function at inhibitory synapses in this particular region. Our results showing that overexpression of the β2 subunit alone on the background of a triple β subunit knockout is able to rescue inhibitory synaptic currents suggests that indeed the β2 subunit can function at hippocampal synapses if expressed at higher levels. It remains to be determined why the hippocampus in particular has such low expression of this subunit and how subunit expression is regulated. While this manuscript was in preparation a paper appeared showing the effect of etomidate on decay kinetics at SOM but not PV synapses onto L2/3 pyramidal cells in prefrontal cortex (Chiu et al., 2018). This could be due to different expression of the β subunits between the two brain regions in adult rodents, with cortex having greater expression of β2 versus β3 subunits, opposite of what is observed in hippocampus, and is consistent with their conclusion that β2 subunit containing GABAARs are enriched at SOM synapses (Laurie et al., 1992). These results thus complement our findings and demonstrate how different brain regions can use different subunits to regulate inhibition.
Towards a model
From our results, we can assemble a possible model for GABAAR β subunit localization to explain our observations (Figure 4E). Interestingly, none of our single and double knockout manipulations fully recapitulated the dramatic elimination of inhibitory responses seen in the triple knockout. To illustrate these findings, our model proposes the existence of two different types of inhibitory synapses: one that has β3 and possibly β2 containing GABAARs and one that has β1, β2, and β3 containing GABAARs. In manipulations where either β1 or β2 or both are knocked out, β3 is present to maintain proper inhibitory transmission. It is only in instances where β3 is knocked out do we see a deficit owing to the fact that the presence of either β1 or β2 or both is not enough to compensate for the loss of β3. For instance, when β1 and β3 are both knocked out the low level of endogenous expression of β2 is not enough to fully compensate for the loss of these receptor populations. This model also explains why we only see a partial deficit when β3 is knocked out since β1 and/or β2 containing receptors are able to compensate at synapses where these isoforms are present.
Our optogenetic results suggest that these two types of inhibitory synapses correlate with PV- and SOM-mediated inputs. Knockout of β3 affects inhibitory transmission from PV but not SOM inputs because there is preferential enrichment of GABAARs containing this subunit at these synapses. While compensation by β2 could explain why we did not see a complete loss of PV responses in our β3 knockout manipulation, unfortunately we are unable to determine the effect of knocking out both β2 and β3 on PV synapses as our β2 sgRNA is only able to target the rat ortholog of the gene. A PV-Cre transgenic rat line has recently been developed but currently is not commercially available (Oh et al., 2017). Compensation could also explain why we did not see a deficit in SOM inputs after loss of β1 (Figure S6), suggesting that normally the presence of this subunit prevents receptors with β2 or β3 from getting to these synapses. In addition, while PV interneurons are a major subset of interneurons and account for 26% of GABAergic neurons in the CA1, they themselves can be subdivided into different classes of interneurons including PV-positive basket cells, bistratified cells, and axo-axonic (chandelier) cells (Kosaka et al., 1987; Somogyi and Klausberger, 2005). It is therefore possible that the effect we see on PV inputs is due to changes from one particular type of PV to pyramidal cell connection and future experiments can refine our findings even further to hone in on this possibility. Additionally, there are many other classes of interneurons besides PV and SOM cells and it remains to be determined whether there are differential effects among other inhibitory inputs as well (Bezaire and Soltesz, 2013).
Our work highlights the critical role of the β subunits in inhibitory transmission and identifies the β3 subunit as an important subunit regulating GABAAR function at both the molecular and circuit level. These findings and approach will provide multiple avenues for future study in elucidating mechanisms of GABAAR function and how its dysfunction leads to disease.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Roger Nicoll (roger.nicoll@ucsf.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse genetics
All experiments were performed in accordance with established protocols approved by the University of California San Francisco Institutional Animal Care and Use Committee (UCSF IACUC). All animals were housed according to the UCSF IACUC guidelines. Animals (postnatal day 6-8) of either sex were used in all experiments. Rat hippocampal slices were obtained from Sprague Dawley rats. To obtain channelrhodopsin (ChR2) expression in PV positive interneurons, PV-ires-Cre mice were bred with Ai32 mice. To obtain ChR2 expression in SOM positive interneurons, Sst-ires-Cre mice were bred with Ai32 mice. Mice that were either heterozygous or homozygous for each gene were used for preparation of organotypic slice cultures. PV-ires-Cre and Sst-ires-Cre mice were generously donated by Dr. V.S. Sohal, while Ai32 mice were generously donated by Dr. Z.A. Knight.
METHOD DETAILS
Experimental constructs
All constructs used for biolistic transfection and lentiviral production co-expressed either a GFP or mCherry fluorophore for visualization. The human codon-optimized Cas9 and chimeric sgRNA expression plasmid (pX458) and lentiviral plasmid for expression of Cas9 (lentiCRISPRv.2) were developed by the Zhang lab and obtained from Addgene (plasmid #48138 and #52961) (Ran et al., 2013; Sanjana et al., 2014). LentiCRISPRv.2 was modified by insertion of an EGFP sequence after a p2a promoter to detect expression after infection. Design of sgRNAs for CRISPR/Cas9 was followed as previously described (Incontro et al., 2014). Briefly, to ensure that sgRNAs targeted both rat and mouse genomic sequences, we isolated conserved genomic regions of rat and mouse DNA for Gabrb1, Gabrb2, and Gabrb3. The conserved region in rat and mouse for Gabrb2 was too short to use as an input sequence in our sgRNA design algorithms (we used both http://crispr.mit.edu/ and Benchling design tools) and therefore we used the rat genomic sequence. We chose the top two scoring guide candidates across the two different sgRNA design platforms and performed electrophysiological and biochemical assays to test their efficacy, identifying the best sgRNA candidates to use for our experiments.
Primers used to generate sgRNA oligos were: β1 FOR 5′ - CACC GTTGATCCAAAACGACACCC - 3′ and REV 5′ - AAAC GGGTGTCGTTTTGGATCAAC -3′; β2 FOR 5′ - CACC GGATGAACAAAACTGCACGT - 3′ and REV 5′ - AAAC ACGTGCAGTTTTGTTCATCC - 3′; β3 FOR 5′ - CACC GTAAAATTCAATGTCATCCG - 3′ and REV 5′ - AAAC CGGATGACATTGAATTTTAC -3′. sgRNA oligos were initially cloned into pX458 and then subcloned into pFUGW-mCherry. For subcloning and generation of chained sgRNAs the following primers were used: insertion of first sgRNA cassette FOR 5′ -TTAATCGTACGAATTCGAGGGCCTATTTCCC - 3′ and REV 5′ - GGGTTAATTAATTCGAATGGCGTTACTATTGA - 3′; insertion of second and third sgRNA cassette FOR 5′ - TAGTAACGCCATTGCAAGAGGGCCTATTTCCC - 3′ and REV 5′ - GGGTTAATTAATTCGAATGGCGTTACTATTGA - 3′. The first sgRNA cassette was inserted into pFUGW-mCherry digested with BstBI and EcoRI HF. The resulting oligo was cut with BstBI for each subsequent cassette insertion. sgRNAs for β1 and β3 were optimized to recognize both rat and mouse genomic sequences whereas the sgRNA for β2 only recognizes the rat genomic sequence due to high disparity between the rat and mouse genome for this gene.
For rescue experiments cDNA was obtained from GE Dharmacon for GABRB1 (CloneId: 4797401), GABRB2 (CloneId: 40006779), GABRB3 (CloneId: 3871111) and cloned into NheI and XmaI sites of pCAGGS-ires-GFP using the following primers: β1 FOR 5′ – ATTCGCGGCCGCTAGCGCCACC ATGTGGACAGTACAAAATCG - 3′ and REV 5′ – AGGGGCGGATCCCGGG TCAGTGTACATAGTAAAGCCAATAGACGACATTAAAAAGAGA; β2 FOR 5′ – ATTCGCGGCCGCTAGCGCCACC ATGTGGAGAGTGCGGAAAAG - 3′ and REV 5′ – AGGGGCGGATCCCGGG TTAGTTCACATAATAAAGCCAATAGACGATGTTGAAGAAGGA; β3 FOR 5′ -ATTCGCGGCCGCTAGCGCCACC ATGTGGGGCCTTGCGGGAGG - 3′ and REV 5′ -AGGGGCGGATCCCGGG TCAGTTAACATAGTACAGCCAGTAAACTAAGTTGAAAAGAGA - 3′. Editing to prevent recognition of the cDNA by the CRISPR sgRNA was done using the following primers: β1 FOR 5′ - GTCCT GGGTCTCTTTCTGGATTAAT TATGA - 3′ and REV 5′ - TCATAATTAATCCAGAAAGAGACCCAGGAC - 3′; β2 FOR 5′ - CCACT GGACGAGCAGAATTGTACCT TGGAA - 3′ and REV 5′ - TTCCAAGGTACAATTCTGCTCGTCCAGTGG - 3′; β3 FOR 5′ - CACCA CGGACGATATCGAGTTCTAT TGGCG - 3′ and REV 5′ - CGCCAATAGAACTCGATATCGTCCGTGGTG - 3′. Editing was done on the GABRB1 coding sequence to make it compatible with the human reference genome (GenBank: BC022449.1) at residues: 429 FOR 5′ – AATAGGGTAGCTGACCAACTCTG G GTACCAGACACCTACTTTCTGAAT - 3′ and REV 5′ - ATTCAGAAAGTAGGTGTCTGGTACCCAGAGTTGGTCAGCTACCCTATT - 3′; 1354 FOR 5′ - GCA T CCGCAGGCGTGCCTCCCAGCTCAAA - 3′ and REV 5′ - TTTGAGCTGGGAGGCACGCCTGCGGATGC - 3′.
All cloning was done using the In-Fusion HD Cloning Kit (Clontech).
Lentivirus production
HEK293T cells were co-transfected with psPAX2, pVSV-G, and either lentiCRISPRv.2 (for Cas9 expression) or FUGW-β123-CRISPR-sgRNAs using FuGENE HD (Promega). Supernatant was collected 40 hours later, filtered, and concentrated using PEG-it Virus Precipitation Solution (System Biosciences). Resulting pellet was resuspended in Opti-mem, flash-frozen, and stored at −80°C.
Immunoblotting
Primary rat hippocampal dissociated neurons were prepared at E18.5 and infected with lentivirus for lentiCRISPR and sgRNAs at DIV 4-7. Neurons were harvested at DIV 25-28 in Tris-buffered saline (25mM Tris pH 7.4, 150 mM NaCl) plus 0.5% Triton-X and protease inhibitor mix (Roche Applied Sciences, cOmplete Protease Inhibitor Cocktail Tablets). After incubation at 4°C for 30 minutes, cell lysates were centrifuged for 15 minutes at 12000g. Proteins were resolved by SDS-PAGE and analyzed by western blot using antibodies against β1 (1:1000, UC Davis/NIH NeuroMab Facility Cat# 75-137, RRID: AB_2109406), β2/3 (1:500, UC Davis/NIH NeuroMab Facility Cat# 75-363, RRID:AB_2315838), and actin (Millipore Cat# MAB1501, RRID: AB_2223041).
Slice culture and biolistic transfection
Rat and mouse slice cultures were prepared on P6–8 as previously described (Stoppini et al., 1991).
Sparse biolistic transfections of organotypic slice cultures were performed 1 day after culturing as previously described (Schnell et al., 2002). Briefly, 100 g total of mixed plasmid DNA was coated on 1 m-diameter gold particles in 0.5 mM spermidine, precipitated with 0.1 mM CaCl2, and washed four times in pure ethanol. The gold particles were coated onto PVC tubing, dried using ultra-pure N2 gas, and stored at 4°C in desiccant. DNA-coated gold particles were delivered with a Helios Gene Gun (BioRad). When biolistically expressing two plasmids, gold particles were coated with equal amounts of each plasmid and plasmids always expressed different fluorescent markers. Recordings were obtained from cells that clearly had co-expression of both constructs. Slices were maintained at 34 °C with media changes every other day.
Electrophysiological recording
Recordings were performed at DIV 22-36 after 3-5 weeks of expression. Dual whole-cell recordings of CA1 pyramidal neurons were done by simultaneously recording responses from a fluorescent transfected neuron and a neighboring untransfected control neuron. Synaptic responses were evoked by stimulating with a monopolar glass electrode filled with aCSF in stratum radiatum of CA1. Typically each pair of neurons is from a separate slice, whereas on rare occasions two pairs may come from one slice. For all paired recordings, the number of experiments (n) reported in the figure legends refer to the number of pairs. Pyramidal neurons were identified by morphology and location. To ensure stable recording, membrane holding current, input resistance, and pipette series resistance were monitored throughout recording. Series resistance was monitored on-line and recordings in which series increased to >30 MOhm or varied by >50% between neurons were discarded. All recordings were made at 20–25 °C using glass patch electrodes filled with an internal solution consisting of 135 mM CsMeSO4, 8 mM NaCl, 10 mM HEPES, 0.3 mM EGTA, 4 mM Mg-ATP, 0.3 mM Na-GTP, 5 mM QX-314, and 1 mM spermine and an external solution containing 119 mM NaCl, 2.5 mM KCl, 4 mM MgSO4, 4 mM CaCl2, 1 mM NaH2PO4, 26.2 mM NaHCO3 and 11 mM glucose bubbled continuously with 95% O2 and 5% CO2. Recordings of IPSCs were made in the presence of D-APV (100 μM) and NBQX (10 μM) to block NMDA and AMPA-mediated currents respectively. Stimulation was delivered using 8 sec interstimulus intervals. Data were acquired using a Multiclamp 700B amplifier (Axon Instruments) controlled by a Master 8 stimulator (A.M.P.I.).
GABA puff experiments were performed using a Picospritzer II (General Valve Corporation). 100μM final concentration of GABA was dissolved in 140 mM NaCl, 5 mM KCl, 5 mM EGTA, 1.4 mM MgSO4, 1 mM NaH2PO4, 10 mM glucose, and 10 mM Hepes, pH 7.2. Agonist was applied using 100 msec pulses at a pressure of 100-300 kPa.
For optogenetic experiments, a TLED+ transmitted light source (Sutter Instruments) was used to deliver blue light through the 40x objective. Light pulse duration and onset were controlled by the Master 8 stimulator. Duration of light pulses ranged from 0.5-2 msec and intensity ranged from 0.5-2.5 mW/mm2. Optogenetic stimulation was delivered at 15 sec interstimulus intervals. For etomidate experiments, 1μM final concentration of etomidate (Sigma) was bath applied.
QUANTIFICATION AND STATISTICAL ANALYSIS
All paired whole-cell data were analyzed using a two-tailed Wilcoxon matched-pairs signed rank test. For comparisons between different experimental groups, a Mann Whitney test was used on the ratios of the transfected cell to the control cell.
Decay kinetics for evoked currents were analyzed using a two-tailed Wilcoxon matched-pairs signed rank test and decay kinetics for spontaneous currents were analyzed using a Mann Whitney test. Sample traces were fit and normalized as previously described (Cathala et al., 2000; Gray et al., 2011). For each experimental condition, the average trace was scaled such that the peak amplitude corresponded to either 100pA for evoked currents or the maximum amplitude of either the control or transfected cell for spontaneous currents. Normalized traces were peak-aligned to generate the overlaid traces.
Data analysis was carried out in Igor Pro (Wavemetrics), GraphPad Prism (GraphPad Software) and Excel (Microsoft).
DATA AND SOFTWARE AVAILABILITY
Data and custom-written Igor Pro analysis code are available upon request.
Supplementary Material
Highlights.
Functional GABAA receptors require the β subunit
The β3 subunit is necessary and sufficient for hippocampal inhibitory transmission
Loss of the β3 subunit affects PV- but not SOM- mediated inhibition
CRISPR/Cas9 allows for combinatorial dissection of multiple molecular components
Acknowledgments
We thank S. Tomita for discussions on the property of GABAA β subunits, D. Qin for excellent technical assistance and members of the Nicoll lab for helpful feedback and comments on the manuscript including S. Incontro for discussions and assistance regarding the design of CRISPR constructs, M. Horn for assistance with obtaining transgenic mice, and M. Bemben for advice regarding Western blots. This work was funded by grants from the National Institutes of Mental Health to R.A.N. All primary data are archived in the Department of Cellular and Molecular Pharmacology, University of California, San Francisco.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: QAN performed research and analyzed data; QAN and RAN designed experiments; QAN and RAN designed the figures and wrote the paper.
Declaration of Interests: The authors declare no competing interests.
References
- Bezaire MJ, Soltesz I. Quantitative assessment of CA1 local circuits: knowledge base for interneuron-pyramidal cell connectivity. Hippocampus. 2013;23:751–785. doi: 10.1002/hipo.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathala L, Misra C, Cull-Candy S. Developmental profile of the changing properties of NMDA receptors at cerebellar mossy fiber-granule cell synapses. J Neurosci. 2000;20:5899–5905. doi: 10.1523/JNEUROSCI.20-16-05899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Wang R, Barot S, Weiss DS. Stoichiometry of a recombinant GABAA receptor. J Neurosci. 1996;16:5415–5424. doi: 10.1523/JNEUROSCI.16-17-05415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin YC, Zimmerman CA, Essner RA, Knight ZA. Hunger neurons drive feeding through a sustained, positive reinforcement signal. Elife. 2016;5 doi: 10.7554/eLife.18640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CQ, Martenson JS, Yamazaki M, Natsume R, Sakimura K, Tomita S, Tavalin SJ, Higley MJ. Input-Specific NMDAR-Dependent Potentiation of Dendritic GABAergic Inhibition. Neuron. 2018;97:368–377. doi: 10.1016/j.neuron.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristo G, Wu C, Chattopadhyaya B, Ango F, Knott G, Welker E, Svoboda K, Huang ZJ. Subcellular domain-restricted GABAergic innervation in primary visual cortex in the absence of sensory and thalamic inputs. Nat Neurosci. 2004;7:1184–1186. doi: 10.1038/nn1334. [DOI] [PubMed] [Google Scholar]
- Drexler B, Jurd R, Rudolph U, Antkowiak B. Distinct actions of etomidate and propofol at beta3-containing gamma-aminobutyric acid type A receptors. Neuropharmacology. 2009;57:446–455. doi: 10.1016/j.neuropharm.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Fritschy JM, Panzanelli P. GABAA receptors and plasticity of inhibitory neurotransmission in the central nervous system. Eur J Neurosci. 2014;39:1845–1865. doi: 10.1111/ejn.12534. [DOI] [PubMed] [Google Scholar]
- Gingrich KJ, Roberts WA, Kass RS. Dependence of the GABAA receptor gating kinetics on the alpha-subunit isoform: implications for structure-function relations and synaptic transmission. J Physiol. 1995;489(Pt 2):529–543. doi: 10.1113/jphysiol.1995.sp021070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herd MB, Haythornthwaite AR, Rosahl TW, Wafford KA, Homanics GE, Lambert JJ, Belelli D. The expression of GABAA beta subunit isoforms in synaptic and extrasynaptic receptor populations of mouse dentate gyrus granule cells. J Physiol. 2008;586:989–1004. doi: 10.1113/jphysiol.2007.146746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechlinger M, Pelz R, Tretter V, Klausberger T, Sieghart W. Subunit composition and quantitative importance of hetero-oligomeric receptors: GABAA receptors containing alpha6 subunits. J Neurosci. 1998;18:2449–2457. doi: 10.1523/JNEUROSCI.18-07-02449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T, Katsumaru H, Hama K, Wu JY, Heizmann CW. GABAergic neurons containing the Ca2+-binding protein parvalbumin in the rat hippocampus and dentate gyrus. Brain Res. 1987;419:119–130. doi: 10.1016/0006-8993(87)90575-0. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III Embryonic and postnatal development. J Neurosci. 1992;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AT, Gee SM, Vogt D, Patel T, Rubenstein JL, Sohal VS. Pyramidal neurons in prefrontal cortex receive subtype-specific forms of excitation and inhibition. Neuron. 2014;81:61–68. doi: 10.1016/j.neuron.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, De Blas AL. Coexistence of two beta subunit isoforms in the same gamma-aminobutyric acid type A receptor. J Biol Chem. 1997;272:16564–16569. doi: 10.1074/jbc.272.26.16564. [DOI] [PubMed] [Google Scholar]
- Nusser Z, Kay LM, Laurent G, Homanics GE, Mody I. Disruption of GABA(A) receptors on GABAergic interneurons leads to increased oscillatory power in the olfactory bulb network. J Neurophysiol. 2001;86:2823–2833. doi: 10.1152/jn.2001.86.6.2823. [DOI] [PubMed] [Google Scholar]
- Oh YM, Karube F, Takahashi S, Kobayashi K, Takada M, Uchigashima M, Watanabe M, Nishizawa K, Kobayashi K, Fujiyama F. Using a novel PV-Cre rat model to characterize pallidonigral cells and their terminations. Brain Struct Funct. 2017;222:2359–2378. doi: 10.1007/s00429-016-1346-2. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Sieghart W. International Union of Pharmacology. LXX Subtypes of gamma-aminobutyric acid(A) receptors: classification on the basis of subunit composition, pharmacology, and function. Update Pharmacol Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan E, Fu Z, Losi G, Homanics GE, Neale JH, Vicini S. GABA(A) receptor beta3 subunit deletion decreases alpha2/3 subunits and IPSC duration. J Neurophysiol. 2003;89:128–134. doi: 10.1152/jn.00700.2002. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph U, Mohler H. Analysis of GABAA receptor function and dissection of the pharmacology of benzodiazepines and general anesthetics through mouse genetics. Annu Rev Pharmacol Toxicol. 2004;44:475–498. doi: 10.1146/annurev.pharmtox.44.101802.121429. [DOI] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell E, Sizemore M, Karimzadegan S, Chen L, Bredt DS, Nicoll RA. Direct interactions between PSD-95 and stargazin control synaptic AMPA receptor number. Proc Natl Acad Sci U S A. 2002;99:13902–13907. doi: 10.1073/pnas.172511199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieghart W. Structure, pharmacology, and function of GABAA receptor subtypes. Adv Pharmacol. 2006;54:231–263. doi: 10.1016/s1054-3589(06)54010-4. [DOI] [PubMed] [Google Scholar]
- Sieghart W, Sperk G. Subunit composition, distribution and function of GABA(A) receptor subtypes. Curr Top Med Chem. 2002;2:795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- Simon J, Wakimoto H, Fujita N, Lalande M, Barnard EA. Analysis of the set of GABA(A) receptor genes in the human genome. J Biol Chem. 2004;279:41422–41435. doi: 10.1074/jbc.M401354200. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song I, Savtchenko L, Semyanov A. Tonic excitation or inhibition is set by GABA(A) conductance in hippocampal interneurons. Nat Commun. 2011;2:376. doi: 10.1038/ncomms1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperk G, Schwarzer C, Tsunashima K, Fuchs K, Sieghart W. GABA(A) receptor subunits in the rat hippocampus I: immunocytochemical distribution of 13 subunits. Neuroscience. 1997;80:987–1000. doi: 10.1016/s0306-4522(97)00146-2. [DOI] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One. 2014;9:e105584. doi: 10.1371/journal.pone.0105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur C, Wafford KA, Reynolds DS, Hadingham KL, Bromidge F, Macaulay A, Collinson N, O’Meara G, Howell O, Newman R, et al. Loss of the major GABA(A) receptor subtype in the brain is not lethal in mice. J Neurosci. 2001;21:3409–3418. doi: 10.1523/JNEUROSCI.21-10-03409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Manis PB. Target-specific IPSC kinetics promote temporal processing in auditory parallel pathways. J Neurosci. 2013;33:1598–1614. doi: 10.1523/JNEUROSCI.2541-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


