Abstract
Over 600 million years ago, animals evolved from a unicellular or colonial organism whose cell(s) captured bacteria with a collar complex, a flagellum surrounded by a microvillar collar. Using principles from evolutionary cell biology, we reason that the transition to multicellularity required modification of pre-existing mechanisms for extracellular matrix synthesis and cytokinesis. We discuss two hypotheses for the origin of animal cell types: division of labor from ancient plurifunctional cells and conversion of temporally alternating phenotypes into spatially juxtaposed cell types. Mechanistic studies in diverse animals and their relatives promise to deepen our understanding of animal origins and cell biology.
Keywords: multicellularity, choanoflagellates, Choanozoa, metazoan origins, evolutionary cell biology, evo-devo
ETOC
In this Review, Brunet and King examine the origin of animal multicellularity. The authors suggest that changes in the mechanisms regulating extracellular matrix synthesis and cytokinesis may underlie the transition to multicellularity, and propose two hypotheses to explain the evolution of cell differentiation in the animal lineage.
Introduction
Every aspect of animal life – from morphology to physiology and behavior – requires the cooperation of thousands to billions of cells. In nearly all animals, the multicellular state is established in each generation through serial divisions of a single founding cell, the zygote. Under joint control by the genome and the environment, daughter cells produced by these divisions change shape, migrate, and selectively attach or detach to give rise to the adult body form through a process known as morphogenesis. In parallel, a process of cell differentiation under fine spatiotemporal control delineates the division of labor between the final cell types. The correct execution of this cellular choreography, repeated anew in every generation, is fundamental to the life of every animal on the planet.
Yet, this type of complex development did not always exist. The discontinuous phylogenetic distribution of multicellularity and differences in cellular mechanisms argue that multicellularity evolved independently in at least 16 different eukaryotic lineages, including animals, plants, and fungi (Bonner 1998; King 2004; Rokas 2008; Knoll 2011). Thus, the mechanisms underpinning animal multicellularity and spatially controlled cell differentiation were likely elaborated in the stem lineage of animals, building upon pathways present in their single-celled ancestors (Richter and King 2013).
Despite the centrality of multicellularity and cell differentiation to animal biology, their origins are little understood. What did the single-celled ancestors of animals look like? How and when did multicellularity and cell differentiation evolve, and what were the underlying molecular mechanisms? Did features of the single-celled progenitors of animals facilitate the early evolution of multicellularity? Conversely, did this single-celled ancestry exert constraints upon the form and function assumed by early animal ancestors?
While the gene complements of animal ancestors have been discussed in depth elsewhere and will not be the focus of this review (see e.g. (King 2004; King et al. 2008; Larroux et al. 2008; Richter and King 2013; Suga et al. 2013; de Mendoza et al. 2013; Sebé-Pedrós et al. 2017; Grau-Bové et al. 2017)), it is notable that many genes required for animal multicellularity (e.g. tyrosine kinases (King and Carroll 2001; Sebé-Pedrós et al. 2016b), cadherins (Abedin and King 2008; Nichols et al. 2012), integrins (Sebé-Pedrós et al. 2010; Suga et al. 2013), and extracellular matrix domains (King et al. 2008; Williams et al. 2014)) evolved before animal origins. Building upon these ancient proteins, the stem-animal lineage was marked by an explosive diversification of transcription factor families and signaling molecules, fueled by the emergence of new gene families (e.g. the Antennapedia and Pax families of transcription factors and the signaling proteins Wnt and BMP) and by expansion of existing gene families (Larroux et al. 2008; Srivastava et al. 2010). The cell biology and morphology implemented by these ancestral genomes, however, have been less explored (Richter and King 2013; Arendt et al. 2015; Cavalier-Smith 2017). In this review, we consider how the evolution of cellular phenotype shaped animal origins.
Although the first animals evolved over 600 million years ago, insights into their origin may be gained through comparison of extant lineages. This approach has revealed a number of features that were likely present in the last common ancestor of animals, the “Urmetazoan” (Figure 1). For example, nearly all extant animals have obligate multicellularity (see (Metzger et al. 2015; Chang et al. 2015) for exceptions) with adult stages typically displaying a specialized morphology and at least five morphologically distinguishable cell types (Valentine 2006). This suggests that the Urmetazoan evolved from a lineage with a long prior history of obligate multicellularity. Likewise, multicellularity in animals is almost invariably the result of a complex embryogenesis initiated by sperm/egg fusion, followed by serial cell division. Finally, in every major animal lineage from sponges and ctenophores to bilaterians, the cells of the future feeding cavity move inside the embryo, morphogenesis establishes the adult body shape, and cells differentiate (Arendt 2004; Leys and Ereskovsky 2006; Nielsen 2012). A form of this elaborate developmental process presumably already existed in the Urmetazoan. Rather than evolving in one step in a single-celled ancestor, it more plausibly resulted from a long and gradual evolution. Therefore, to more fully reconstruct the origin of animal development, we must extend our comparisons beyond animals to include their closest living relatives.
Figure 1. Phylogenetic distribution of traits inferred in the Urmetazoan.

The presence of epithelia (Leys et al. 2009), sperm, eggs and multicellularity (Nielsen 2012) and collar complex (see Supplementary Table 1 for details and references) are mapped onto a consensus eukaryotic phylogeny modified from (Struck et al. 2014; Borner et al. 2014; Laumer et al. 2015; Torruella et al. 2015; Telford et al. 2015; Cannon et al. 2016). The collar complex is inferred to have been present in the Urchoanozoan, and to be a choanozoan synapomorphy. The relationships among sponges (Porifera), ctenophores and other animals are depicted as a polytomy to reflect uncertainties regarding their order of divergence (King and Rokas 2017). Species silhouettes are from PhyloPic (http://phylopic.org).
The “sister-group,” or closest living relatives, of animals has unambiguously been shown to be the choanoflagellates (Figure 1; (King et al. 2008; Ruiz-Trillo et al. 2008; Shalchian-Tabrizi et al. 2008; Torruella et al. 2015)), a globally distributed group of marine and freshwater protozoans (Leadbeater 2014) with a highly distinctive morphology (Figure 2A,B,E). Choanoflagellates are characterized by an apical flagellum surrounded by a collar of microvilli, which together form a “collar complex.” The beating of the flagellum generates forces that can both propel the cell through their aquatic environments and produce a flow that allows the choanoflagellate to collect bacterial prey on the outer surface of the collar. The morphological similarity between choanoflagellates and certain animal cells, particularly sponge choanocytes, was evident even to the first choanoflagellate observers (James-Clark 1867) and inspired the hypothesis of a close relationship between choanoflagellates and animals. This hypothesis, first proposed in the 19th century on morphological grounds, remained otherwise untested for more than a century, as both the affinities of choanoflagellates to sponges and of sponges to other animals were the subject of several competing hypotheses (summarized in (Leadbeater 2014)). The issue was settled by molecular phylogenetics, which provided conclusive evidence that sponges belong to the animal kingdom (Wainright et al. 1993; Srivastava et al. 2010; Telford et al. 2015) and that animals and choanoflagellates are sister-groups. Together, choanoflagellates and animals thus form a monophyletic clade (see Glossary), which we refer to as “Choanozoa” (see box). Furthermore, phylogenomic studies of previously enigmatic taxa have revealed the closest living relatives of choanozoans (Ruiz-Trillo et al. 2008; Torruella et al. 2015) to be Filasterea (a group comprising filopodiated amoebae and recently discovered flagellated protozoans (Hehenberger et al. 2017)) and Ichthyosporea (which alternate between large coenocytic spores and individual amoebae). Together, Choanozoa, Ichthyosporea, and Filasterea make up the clade Holozoa, which is the sister-group of Fungi (Figure 1).
Figure 2. Conserved morphology and ultrastructure of choanoflagellates and sponge choanocytes.
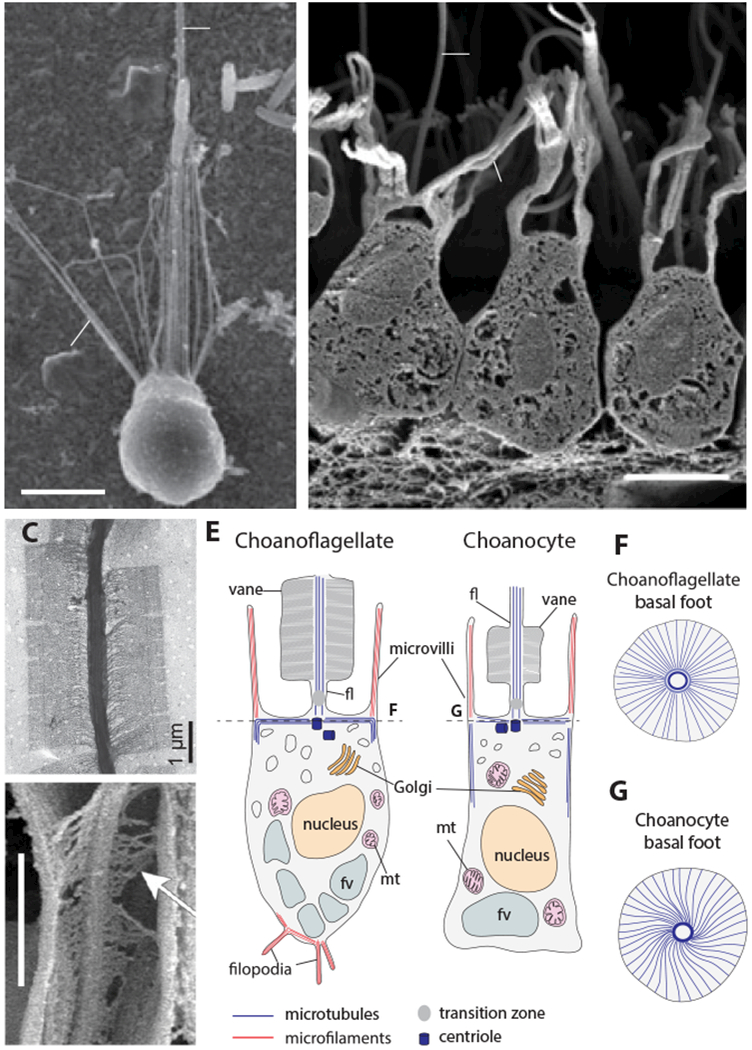
(A, B) The collar complex is conserved in choanoflagellates and sponge collar cells. Both choanoflagellates (A, S. rosetta from (Dayel et al. 2011)) and sponge choanocytes (B, Sycon coactum, from (Leys and Hill 2012)) possess a flagellum (fl), microvilli (mv), a nucleus (nu) and a food vacuole (fv) in the same overall orientation. (C,D) A flagellar vane is present in both choanoflagllates (C, Salpingoeca amphoridium, from (Leadbeater 2014)) and choanocytes (D, Spongilla lacustris, from (Mah et al. 2014); arrow shows the fibrous structure of the vane and lateral contact with the collar). (E) Comparative ultrastructural schematics of a choanoflagellate and a sponge choanocyte, modified from (Maldonado 2004) following (Woollacott and Pinto 1995) for the microtubule cytoskeleton and (Karpov and Leadbeater 1998) for the actin cytoskeleton. (Although filopodia may occasionally be present in choanocytes, as reported in sketches from earlier studies of calcareous sponges (notably Sycon raphanus) (Duboscq and Tuzet 1939; Grassé 1973) and in one scanning electron microscopy study of the demosponge Ephydatia fluviatilis (Weissenfels 1982), they have not been reported so far in transmission electron microscopy or immunofluorescence studies and are thus not indicated here.) (mt): mitochondria. (F,G) basal microtubular foot supporting the flagellum in choanoflagellates and choanocytes, following (Garrone 1969; Woollacott and Pinto 1995; Leadbeater 2014).
Box 1: Choanozoa: the clade composed of choanoflagellates and animals.
We define Choanozoa as the clade containing the most recent common ancestor of animals and choanoflagellates (the Urchoanozoan), along with all of its descendants, including Homo sapiens Linnaeus 1758 (representing animals), and Monosiga brevicollis Ruinen 1938 (representing choanoflagellates). The Greek root “choanē” (or funnel) refers to the collar, which in the current state of knowledge is a synapomorphy (see Glossary) of the clade. Although “Choanozoa” was used previously to refer to an assemblage of protists (Cavalier-Smith et al. 1991) that later proved paraphyletic (see Glossary; (Shalchian-Tabrizi et al. 2008)); this usage was not adopted and the name is more appropriately applied as defined here. The informal term “choanimal” (Fairclough et al. 2013) and the formal term Apoikozoa (Budd and Jensen 2017) have both been previously proposed for the clade containing choanoflagellates and animals, but neither has been formally described nor fully adopted. In particular, the term “Apoikozoa” is less fitting, as the root “apoiko-” refers to colony formation, which is neither universally present in choanozoans, nor exclusive to them.
Choanoflagellates reveal the cellular foundations of animal origins
Because choanoflagellates and animals are each other’s closest relatives, a fundamental question is whether shared cellular features – such as the collar complex – were already present in their last common ancestor, the “Urchoanozoan” (Figure 1). Electron microscopy has revealed that the similarities between choanoflagellates and choanocytes extend beyond morphology to include a shared underlying ultrastructure. In both choanoflagellates and sponge choanocytes, the flagellum is supported by microtubules (Karpov and Leadbeater 1998; Gonobobleva and Maldonado 2009) and often displays a characteristic “vane” – a pair of bilateral wing-like filamentous extensions that are only known in choanoflagellates and choanocytes (Figure 2C,D) (Petersen 1929; Vlk 1938; Hibberd 1975; Mehl and Reiswig 1991; Leadbeater 2006; Mah et al. 2014). Also in both, the ovoid cell body is encased in parallel arrays of sub-membranous microtubules that emerge from the basal body and span the cell from the apical to the basal side. Underneath the flagellum, the basal body is supported by a basal foot surrounded by an elaborate “crown” of transverse microtubules. Like the flagellar vane, this organization appears unique to choanocytes and choanoflagellates (Figure 2F,G) (Garrone 1969; Woollacott and Pinto 1995; Leadbeater 2014). Finally, in both choanoflagellates and choanocytes, the microvilli are supported by bundled actin microfilaments of constant length within a given cell (Karpov and Leadbeater 1998; Rivera et al. 2011). Choanoflagellate genomes encode homologs of most animal microvillar proteins, among which two families appear choanozoan-specific: Ezrin/Radixin/Moesin (ERM) and Whirlin, both of which are involved in controlling microvillar length (Sebé-Pedrós et al. 2013a; Peña et al. 2016). This further supports the notion that the collar is a choanozoan synapomorphy. Interestingly, the protozoan Ministeria vibrans (belonging to Filastera, the sister-group of Choanozoa) sports microvilli-like tentacles that radiate over its entire cell cortex (Cavalier-Smith and Chao 2006), suggesting that microvilli might be more ancient than the collar complex.
Besides its conserved ultrastructure, the idea that the collar complex evolved in choanozan ancestors is further supported by its broad distribution in animals. Beyond sponges, collar complexes featuring a flagellum surrounded by a ring of microvilli are found in most animal phyla (Figure 1, Supplementary Figure 1, Supplementary Table 1) – e.g. in epidermal cells (often sensory), nephridial cells (Supplementary Figure 1B), or as part of diverse inner epithelia (Supplementary Figure 1C; (Nørrevang and Wingstrand 1970; Rieger 1976; Salvini-Plawen 1978)). In modern species, collar cells often function in food absorption: choanoflagellates and sponge choanocytes phagocytose bacteria, and the collar cells lining the gastrodermis of some cnidarians endocytose food particles produced by extracellular digestion (Goldberg and Taylor 1989). In bilaterians and ctenophores, nutrient acquisition through endocytosis is performed by enterocytes lining the midgut that frequently display a motile flagellum and microvilli (packed into a dense brush border rather than forming a ring; (Hernandez-Nicaise 1991; Takashima et al. 2013)), consistent with a possible derivation from ancestral collar cells.
Finally, the homology of the collar complex in animals and choanoflagellates is supported by its restriction to choanozoans. Although morphologically analogous forms exist in a few distantly related species (Mah et al. 2014), in all cases the underlying ultrastructure differs. For example, in the amoebozoan Phalansterium (Cavalier-Smith et al. 2004), the flagellum is surrounded by a “collar” formed by a continuous fold of cytoplasm rather than by independent microvilli (Hibberd 1983). In the pedinellales Actinomonas and Pteridomonas (stramenopiles from the supergroup Bikonta (Cavalier-Smith and Chao 2006)), the flagellum is surrounded by a double ring of tentacles that are supported by microtubule triads rather than actin microfilaments, and are thus not microvilli (Larsen 1985; Patterson and Fenchel 1985). The absence of a true collar complex from all non-choanozoans suggests that it is unlikely to evolve easily through convergence. Although there are subtle differences in the form and function of the collar of choanoflagellates and sponge choanocytes, (Mah et al. 2014) this is unsurprising given that the two lineages diverged more than 600 million years ago and these differences are more likely to be the result of derivation from an ancestral collar cell than the similarities arising through convergent evolution. Therefore, bacterivorous collar cells likely trace their ancestry back to the choanozoan stem lineage, and the study of modern choanoflagellates holds the promise of illuminating the cellular foundations of animal origins.
Division or aggregation? The two paths to multicellularity
How and when did the ancestors of animals become multicellular? Each of the independent transitions to multicellularity (Figure 3) took place through one of two mutually exclusive mechanisms: clonal development, in which multicellularity arises by serial cell division without separation of sister cells, or aggregation, in which separate cells converge and adhere to each other. Clonal and aggregative multicellularity both have scattered distributions in the eukaryote phylogenetic tree, suggesting that they both evolved several times independently (Figure 3A). Interestingly, the two pathways to multicellularity result in different types of multicellular forms, and arguably evolved in response to different selective pressures.
Figure 3. Clonal and aggregative multicellularity.

(A) Phylogenetic distribution of clonal and aggregative multicellularity. Eukaryotic phylogeny is modified from (Keeling et al. 2014). Instances of multicellularity are mapped following (Bonner 1998; King 2004; Raven 2005; Ott et al. 2015; Sebé-Pedrós et al. 2017). (B and C) Examples of aggregative and clonal multicellularity. The organism indicated is shown in bold, and other organisms with similar forms of multicellularity are listed below. (B) Aggregative multicellularity gives rise to spherical masses of spores or cysts, sometimes atop a stalk. From (Brown et al. 2012; Du et al. 2015). (C) Clonal multicellularity gives rise to diverse multicellular forms. From (Bonner 1998; Fairclough et al. 2010).
Independent origins of aggregative multicellularity
Although all extant animals display clonal development, might early animal ancestors have developed by aggregation? Though this hypothesis is formally possible, it appears unlikely in the current state of knowledge. The few cases of aggregation-like processes reported in animals are responses to artificial perturbations: e.g., experimentally dissociated sponge cells can aggregate in vitro (Curtis 1962) but sponge embryonic development is strictly clonal in vivo (Ereskovsky 2010). We argue below that aggregative multicellularity is rarer than clonal multicellularity, and represents a distinct adaptive niche with a more limited evolutionary potential.
Aggregative multicellularity evolved at least seven times in eukaryotes (Figure 3A) as well as in some bacterial lineages such as Myxobacteria (Bonner 1998; Brown et al. 2012; Sebé-Pedrós et al. 2013b). In nearly all well-characterized cases of aggregative multicellularity, cells respond to adverse conditions (e.g. starvation (Souza et al. 1999)) by migrating toward each other and aggregating into a resistant mass of propagules (spores or cysts) – called a sorocarp, sporangium or (if a stalk is present) a fruiting body (Figure 3B). (The few exceptions represent an infrequently observed type of aggregation, sexual agglutination, that has been reported in yeasts (Wickerham 1958), choanoflagellates (Woznica et al. 2017) and Chlamydomonas (Bergman et al. 1975).)
The resulting aggregate is formed of quiescent cells that neither feed nor divide. Motility, if present (as in Dictyostelium slugs), is a transient step toward the formation of the sorocarp. The multicellular form eventually dissociates and the propagules disperse. Aggregation is thus a specialized “emergency response” in which cells transiently assemble under adverse conditions that compromise mitosis, thus preventing clonal multicellularity. The potential advantages of aggregation include resistance to environmental stressors (with the outer cells shielding the inner cells from harmful agents such as UV and toxic chemicals (Stratford 1992)) and, in terrestrial species, formation of a stalk allowing elevation from the substrate and wind dispersal of the propagules (Bonner 1998).
Aggregation presents an impediment to the evolution of division of labor between cell types (Buss 1988): the multicellular mass is composed of cells that are not necessarily genetically related, making it vulnerable to invasion by “cheater” mutants that benefit from their presence in the aggregate without sharing resources or labor with other cells (Strassmann et al. 2000; Santorelli et al. 2008). Indeed, theoretical arguments and experimental data suggest that aggregation can only be evolutionarily stable if it is restricted to closely related individuals (Gilbert et al. 2007; Kuzdzal-Fick et al. 2011). For example, the aggregative multicellularity of dictyostelids has been evolutionarily stable for more than 400 million years (Sucgang et al. 2011) and relies on specific kin recognition mechanisms (Benabentos et al. 2009; Hirose et al. 2011). Perhaps due to the difficulty of overcoming such conflicts, aggregative forms have a transient existence and little division of labor.
Aggregation involves an elaborate series of coordinated steps: mutual cell attraction by chemotaxis, migration, adhesion, and, last, differentiation into propagules (Du et al. 2015). How this cascade originated in each lineage is unknown in detail, but a likely evolutionary prerequisite was the presence of an inducible pathway for the sporulation/encystment of individual cells. Indeed, the cAMP/PKA pathway controlling the switch to multicellularity in slime molds appears to have evolved from an encystment stress response found in single-celled amoebae (Ritchie et al. 2008; Du et al. 2014; Kawabe et al. 2015). As it is contingent on specific prior, permissive evolutionary steps and vulnerable to cheaters, aggregative multicellularity might be both more difficult to evolve and less conducive to the evolution of differentiated cell types than clonal multicellularity.
Independent origins of clonal multicellularity
As clonal multicellularity can in principle result from a failure to complete cell division, it might evolve relatively easily through loss-of-function mutations. All three documented examples of multicellular forms that have evolved de novo in the laboratory developed clonally by incomplete cytokinesis within a small number of generations under selection (100 to 315; (Boraas et al. 1998; Ratcliff et al. 2012, 2013). The relative ease of evolving clonal multicellularity could explain why clonal forms are phylogenetically more widespread (Figure 3A) and morphologically more diverse (Figure 3C) than aggregative forms. Indeed, clonal development underlies all known multicellular forms with active metabolism and proliferation (but also some sorocarps or fruiting bodies). Moreover, clonal multicellularity is likely less vulnerable to cheating than aggregation, as all cells share an identical genome (Buss 1988). These two factors may account for the fact that all five known independently evolved instances of “complex multicellularity” (i.e. obligate multicellularity with different cell types and regulated organismal morphology (Knoll 2011)) involve clonal development (Figure 2A). The selective advantages that favored the evolution and maintenance of clonal multicellularity are unknown, but may include resistance to predators (Boraas et al. 1998), cooperative feeding (Koschwanez et al. 2011; Roper et al. 2013), division of labor (e.g. between flagellated cells and dividing cells) (Margulis 1981; Michod 2007) and formation of a milieu intérieur (i.e. a controlled “internal environment” of stable composition). Finally, one cannot rule out the possibility that the initial evolution of animal multicellularity was a neutral event, that would have first reached fixation by genetic drift.
The origin of animal multicellularity
How ancient is animal multicellularity?
The last common ancestor of animals was clearly multicellular, but might multicellularity extend back to the last common ancestor of choanozoans? Tantalizingly, many choanoflagellates form facultative multicellular forms, including swimming spherical (rosette), linear or flat colonies, and sessile branching colonies (Leadbeater 2014). These forms are all thought to develop clonally (Hibberd 1975; Karpov and Coupe 1998; Fairclough et al. 2010; Dayel et al. 2011). Although the phylogenetic distribution of known instances of multicellularity in choanoflagellates remains patchy, the life cycles of most species are incompletely known, precluding robust parsimony-based inference of the ancestral state. In favor of an ancient origin, the rosette colonies of three distantly related choanoflagellates, Salpingoeca rosetta (Dayel et al. 2011; Fairclough et al. 2013), Codosiga botrytis (Hibberd 1975) and Desmarella Kent (Karpov and Coupe 1998), are composed of cells joined by cytoplasmic bridges with a common ultrastructure featuring two parallel electron-dense plates flanking an electron-dense cytoplasmic core. The last common ancestor of these three species was also the last common ancestor of one of the two main choanoflagellate clades (Carr et al. 2008, 2017), suggesting that clonal multicellularity had already evolved before the first split in the choanoflagellate phylogenetic tree. Characterization of the life cycles and multicellular development of more choanoflagellate species will further test this hypothesis. Whether clonal multicellularity arose once or several times in choanozoans, choanoflagellate rosettes offer a tantalizing proxy for the first stages of animal evolution, due to their close phylogenetic affinity to animals and to similarities in both cell ultrastructure and developmental mode.
Multicellularity might thus be as ancient as stem choanozoans, but could it be even older? The two closest relatives of choanozoans are filastereans and ichthyosporeans (Figure 1). Intriguingly, both form facultative multicellular forms, but in different ways from choanozoans – aggregation in filastereans (Sebé-Pedrós et al. 2013b) and fragmentation of a coenocyte (see Glossary) in ichthyosporeans (Suga and Ruiz-Trillo 2013). If holozoan multicellular forms are homologous to each other, this would imply that interconversions took place between aggregative and clonal multicellularity in stem-filastereans or stem-choanozoans. Perhaps more likely is the possibility that multicellularity evolved independently in the three holozoan clades, although this remains to be tested.
The genetic basis for the origin of animal multicellularity
What molecular mechanisms first supported the evolution of animal multicellularity? This problem cannot be studied solely in animals, as there is no known animal mutant where cleavage of the zygote produces separate free-living cells rather than an embryo, thus reverting to unicellularity. To answer this question, it is therefore necessary to study phylogenetically relevant groups with facultative multicellularity. Thus far, a dozen genes have been found to be either necessary or sufficient for multicellularity in the four groups investigated: green algae, fungi, slime molds and choanoflagellates. All known multicellularity genes encode proteins that belong to one of two major functional categories: extracellular matrix (ECM) proteins and, in the case of clonal multicellularity, cytokinesis regulators (Table 1). This suggests that the initial evolution of multicellularity on different branches of the tree of life repeatedly converged on similar mechanisms (Abedin and King 2010).
Table 1. Cell cycle and ECM genes from diverse eukaryotes regulate the unicellularity/multicellularity switch.
| Species | Type of multicellularity | Gene required for the unicellular/multicellular switch* | Gene function |
|---|---|---|---|
| Gonium pectoral (green alga) | Clonal | retinoblastoma | Cell cycle (transcriptional repressor) (2) |
| Saccharomyces cerevisiae (fungus) | Aggregative (flocculation) | flo1, flo5, flo8, flo9, flo10, flo11, sta1 | ECM (flo1, flo5, flo9, flo10, flo11: lectins, sta1: endoamylase, flo8: transcriptional activator of the formers) (3–7) |
| Saccharomyces cerevisiae (fungus) | Clonal (chain and snowflake mutants) |
cts1 ACE2 |
ECM (chitinase) (8) Cell cycle (transcription factor) (9,10) |
| Dictyostelium discoideum (slime mold) | Aggregative | cbp-26 | ECM (lectin) (12,13) |
| Salpingoeca rosetta (choanoflagellate) | Clonal | rosetteless | ECM (lectin) (11) |
Some pleiotropic genes are necessary for multicellularity as an indirect effect of them being involved in a more general cell function, such as transcription or cell motility. For example, across the 123 Dictyostelium mutants with aberrant or abolished aggregation (1), most are deficient in transcription, cell movement or cAMP synthesis. These genes are not discussed here as their role is indirect. References: 1. (Glöckner et al. 2016) 2. (Hanschen et al. 2016); 3. (Douglas et al. 2007); 4. (Lo and Dranginis 1996); 5. (Soares 2011); 6. (Stratford 1992); 7. (Fidalgo et al. 2006); 8. (Kuranda and Robbins 1991); 9. (Oud et al. 2013); 10. (Ratcliff et al. 2015); 11. (Shinnick and Lerner 1980); 12. (Ray et al. 1979); 13. (Levin et al. 2014)
Among the taxa studied, choanoflagellates occupy a privileged position as the sister-group of animals. The recent establishment of genetics in the model choanoflagellate S. rosetta has revealed the first gene known to be required for multicellular development, named rosetteless for its mutant phenotype (Levin et al. 2014). While wild-type S. rosetta reliably develops into spherical colonies (called “rosettes”) upon induction with bacterial signals (Dayel et al. 2011; Alegado et al. 2012; Woznica et al. 2016), rosetteless mutants are unable to form rosettes under all studied conditions (although the ability to develop into another clonal multicellular form, linear chains, is unaffected). The rosetteless gene encodes a C-type lectin that is secreted into the core of the rosette as an ECM component (Figure 4A). In S. rosetta, the integrity of the colonies is thus likely ensured by the basal extracellular matrix, to which cells appear to anchor by filopodia (Dayel et al. 2011). Both ECM and filopodia also contribute to the cohesion of animal blastulae. Blastomeres are held together by an abundant ECM rich in lectins (Fraser and Zalik 1977; Roberson and Barondes 1983; Harris and Zalik 1985; Outenreath et al. 1988; Lee et al. 1997) and appear linked by filopodia in sponges (Ereskovsky 2010), cnidarians (Benayahu et al. 1989), echinoderms (Vacquier 1968), amphioxus (Hirakow and Kajita 1994), and mice (Salas-Vidal and Lomelı 2004). In mice, laser ablation of the filopodia results in loss of blastomere cohesion (Fierro-González et al. 2013). Thus, the multicellular states of choanoflagellate rosettes and animal embryos are established and maintained by comparable mechanisms.
Figure 4. Morphogenesis in choanoflagellate rosettes, calcareous sponge embryos and volvocale embryos.

(A) Morphogenesis during rosette formation in the choanoflagellate S. rosetta, following (Fairclough et al. 2010). (B) Early embryonic development of the calcareous sponge Sycon ciliatum, including amphiblastula inversion, from (Franzen 1988). (C) Early embryonic development of the volvocale Pleodorina californica (Höhn and Hallmann 2016). In other volvocales such as Volvox, an additional developmental stage is intercalated, in which the embryo first forms a sphere with flagella pointing inward, which later opens up into a continuously bending sheet that finally closes into a sphere with flagella pointing outward.
Choanoflagellate multicellularity and the origin of animal embryogenesis
The similarity of choanoflagellate rosettes to the blastula stage of animal development is consistent with a modern version of Haeckel’s Blastaea hypothesis – which proposes that early animal ancestors were motile spheres of flagellated cells that formed clonally (Haeckel 1874, 1892; Nielsen 2008; Arendt et al. 2015) (possibly alternating with a sessile benthic stage; (Adamska 2016)). Indeed, the first developmental stage of marine invertebrates is often a ciliated free-swimming blastula that resembles choanoflagellate rosettes (in some cases, down to the presence of a collar complex on every cell (Crawford and Campbell 1993)) and, given its widespread taxonomic distribution, was plausibly part of the development of the Urmetazoan (Nielsen 2012).
How did serial cell divisions give rise to these early multicellular forms? In most eukaryotes, including choanoflagellates and animals, cell division has to accommodate a constraint: the two microtubule organizing centers (MTOCs) that form the flagellar basal body also organize the mitotic spindle during division. Moreover, in choanoflagellates, microvilli are directly transmitted to daughter cells. As a consequence, the plane of cell division in choanoflagellates must traverse the apical pole, where MTOCs and microvilli are located, and symmetric cell divisions necessarily take place along the apico-basal axis (Figure 4A). This constraint on the direction of cell division seems universal in choanoflagellates (Leadbeater 2014). As a consequence of this fixed division orientation, in the absence of cell reorientation and/or rearrangements, cell proliferation alone can only produce cells linked together in linear chains (e.g. in S. rosetta (Dayel et al. 2011)) or planar sheets (e.g. in Choanoeca perplexa – formerly Proterospongia choanojuncta (Leadbeater 1983)). Any more complex shape – such as a spherical rosette – must be achieved by cell rearrangements to allow bending and, ultimately, “closure” of the sheet at the point where non-sister cells meet.
These cell rearrangements are apparent during rosette formation in S. rosetta (Fairclough et al. 2010) and the final “closure point” can be identified in mature rosettes as an ECM-free spot in immunostainings for the Rosetteless protein (Woznica et al. 2016). This constraint seems to apply to any spherical colony formed by clonal division of motile flagellated cells: in flagellated volvocale green algae, a similar folding process takes place (Höhn and Hallmann 2016). It plausibly applied to the first animal embryos as well: indeed, a similar series of steps takes place during the early development of calcareous sponges, which first form as concave sheets and secondarily fold into spherical embryos called “amphiblastulae” (Figure 4B) (Franzen 1988; Ereskovsky 2010; Arendt et al. 2015). In both volvocales and calcareous sponges, the hole resorbed at the closure point is called the phialopore. In starfish embryos, the ability to fold a sheet into a sphere might still be latent: starfish zygotes in which the fertilization envelope has been removed undergo multiple rounds of vertical cleavage to give rise to a flat epithelial sheet that subsequently folds into a spherical blastula and thereafter develops into a normal starfish larva (Kadokawa et al. 1986). In most other animals (eumetazoans and non-calcareous sponges) with non-flagellated zygotes, this constraint is thought to have been overcome, as cleavage can occur along all cellular axes and thereby directly produce a spherical blastula (Arendt and Nübler-Jung 1997).
The ancestry of animal cell differentiation
Alternative hypotheses for the origin of animal cell types: temporal-to-spatial transition and division of labor
Choanoflagellate rosettes and the multicellular forms of ichthyosporeans and filastereans are not known to undergo spatial cell differentiation. This contrasts with the “complex multicellularity” (Knoll 2011) of animals, which are mosaics of different cell types with controlled spatial distribution. The origin of cell types thus represents one of the key steps in the evolution of animals from their microeukaryote ancestors. Two main hypotheses have been put forward to explain the evolution of animal cell differentiation: the temporal-to-spatial transition (TST) hypothesis (Zakhvatkin 1949; Mikhailov et al. 2009; Sebé-Pedrós et al. 2017) and the division of labor (DOL) hypothesis (Mackie 1970; Arendt 2008). The TST hypothesis proposes that cell differentiation predated multicellularity, building upon the observation that many modern microbial eukaryotes can switch between different cell phenotypes over their life history (Figure 5). In this scenario, some of these temporally alternating phenotypes were converted into spatially segregated cell types in animal ancestors. The DOL hypothesis starts from the observation that individual microbial eukaryotes execute in a single cell multiple functions that are accomplished by different cell types in animals (Figure 6), including perception, movement, feeding, and division. In the DOL hypothesis, the bulk of cell differentiation evolved after multicellularity, by differential loss of function from multifunctional ancestral cell types. The TST and the DOL hypotheses are complementary rather than mutually exclusive, and it is plausible that both processes contributed to the early evolution of animal cell types.
Figure 5. Temporally alternating cell types in protozoans.

(A) The heterolobosean excavate Naegleria gruberi can switch between a flagellated swimmer phenotype and a deformable crawler (“amoeboid”) phenotype. Redrawn from (Fritz-Laylin et al. 2010). (B) Cell types and life history transitions in the choanoflagellate S. rosetta, from (Dayel et al. 2011; Levin and King 2013). Main panel depicts the dynamic asexual life history of S. rosetta whereas the inset indicates its sexual cycle. Dotted lines indicated inferred transitions that have not been directly observed.
Figure 6. The division of labor hypothesis.

(A) Cellular modules present in the choanoflagellate S. rosetta. On the right: choanoflagellate orthologs of the selector transcription factors that control these modules in animals, on top of a list of choanoflagellate orthologs of their animal targets (Supplementary Table 2, Supplementary Figure 3). No terminal selector is indicated for the secretion apparatus, as there seems to be no known neural terminal selector with a choanoflagellate ortholog. Dotted lines indicate that it is unknown whether the choanoflagellate transcription factors control the same genes as their animal orthologs, except for the Myc::Max complex for which regulation is indicated by computational predictions (Brown et al. 2008) and electrophoretic mobility shift assays (Young et al. 2011). (B) Cellular modules shown in panel A are segregated into distinct cell types in animals (here, the putative cell type complement of stem-eumetazoans is illustrated based on the cell types shared by cnidarians and bilaterians (Fautin and Mariscal 1991; Schmidt-Rhaesa 2007; Arendt et al. 2015)) and terminal selector transcription factors specify distinct cell types.
Testing the TST hypothesis
Many single-celled eukaryotes have temporally alternating cell phenotypes (Figure 5). The distinct morphologies, transcriptomes (Fairclough et al. 2013; Sebé-Pedrós et al. 2013b; de Mendoza et al. 2015), proteomes (Sebé-Pedrós et al. 2016b), and chromatin states (Sebé-Pedrós et al. 2016a) of these phenotypes in single-celled holozoans suggest that they represent stable cell types, like those of animals, rather than instances of short-term phenotypic plasticity. Are some of these cell types homologous to those of animals, as the TST hypothesis postulates? A clear example of temporal-to-spatial transition is meiosis, which clearly predated animals and dates back to the last common eukaryotic ancestor (Ramesh et al. 2005). In choanoflagellates, gametes competent to undergo cell-cell fusion directly transdifferentiate from haploid solitary cells (Figure 5B) (Levin and King 2013; Woznica et al. 2017), while in all extant animals meiosis and gametogenesis start from diploid cells integrated within the adult organism (Nielsen 2012).
Another temporal switch frequently found in protozoa that has been proposed to have been co-opted in animal cell differentiation is the alternation between a flagellated phenotype (allowing locomotion in an aqueous environment by flagellar beating) and a deformable “amoeboid” crawling phenotype that navigates solid environments by actin-mediated deformations of the cell body (Figure 5A). These two phenotypes and associated genes are broadly distributed in the eukaryotic tree of life (Supplementary Figure 2), suggesting that the last common eukaryotic ancestor might have been able to switch between both (Fritz-Laylin et al. 2010, 2017). If so, this switch could have existed in the last single-celled ancestors of animals.
Suggestively, virtually all animals combine static epithelial cells (often bearing a flagellum and lining the body surface) with deformable crawling cells that patrol tissues by actin-mediated locomotion. These migratory interstitial cells usually function as phagocytes and quickly accumulate upon infection, wound repair or allograft rejection. They are called archaeocytes in sponges (Cheng et al. 1968; Alié et al. 2015), amoebocytes in cnidarians (Patterson and Landolt 1979; Olano and Bigger 2000; Couch et al. 2013) and hemocytes or macrophages in bilaterians (Hartenstein 2006; Schmidt-Rhaesa 2007). If the interstitial phagocytes of these large animal clades are homologous (which remains to be tested), they might have evolved from the crawling phase of the Urchoanozoan or one of its descendants (Mendoza et al. 2002; Arendt et al. 2015).
How could one test the homology between animal and single-celled holozoan cell types? In the past few years, the comparison of transcriptomes has emerged as a promising approach for investigating cell type homology (Arendt 2005, 2008; Lauri et al. 2014). Central to these comparisons are the “terminal selector” transcription factors that directly implement cell phenotypes by sitting directly above large batteries of differentiation genes (Arendt et al. 2016; Hobert 2016). This approach can potentially be extended to single-celled holozoans: do terminal selector transcription factors play a role in establishing and maintaining their cell types? If so, how do they compare to their animal counterparts? These questions are still open, but intriguing potential case studies can already be identified. For deformable crawlers, a candidate is the Runx family of transcription factors (Coffman 2003), which were likely present as a single copy in urmetazoan and urholozoan progenitors (Rennert et al. 2003; Sullivan et al. 2008; de Mendoza et al. 2013). Runx transcription factors specify circulating cells in animals that move by actin-mediated crawling (Pancer et al. 1999; Otto et al. 2003; Waltzer et al. 2003; Burns et al. 2005) and directly promote cell motility (Leong et al. 2010; Zusso et al. 2012; Lie-A-Ling et al. 2014; VanOudenhove et al. 2016). High levels of Runx transcription have been detected by RNA-seq in the archaeocytes of the sponge Ephydatia fluviatilis (Alié et al. 2015), suggesting that the link between this transcription factor family and the crawling cell phenotype might be ancient in animals. Runx is also present in the genomes of ichthyosporeans and filastereans (de Mendoza et al. 2013), which both have a crawling (“amoeboid”) phase. The runx gene is significantly upregulated during the crawling phase of the ichthyosporean Creolimax (de Mendoza et al. 2015) and, in Capsaspora, predicted Runx targets show a significant enrichment for genes encoding actin cytoskeleton and other proteins involved in crawling (Sebé-Pedrós et al. 2016a).
Another intriguing transcription factor of stem-holozoan ancestry is Brachyury, which is upregulated in Capsaspora amoebae and is predicted to regulate homologs of genes controlled by Brachyury in mouse, including those required for cell motility (Sebé-Pedrós et al. 2016a). In animals, Brachyury is often involved in the motility of embryonic (but not adult) cells (Yanagisawa et al. 1981; Gross and McClay 2001). Direct mechanistic studies of these and other transcription factors in both single-celled holozoans and non-bilaterian animals, as well as more extensive molecular characterization of the relevant cell types in a broad sampling of phylogenetically relevant species, will help in testing these hypotheses and in revealing other potential instances of TST.
Testing the DOL hypothesis
The phenotypic comparison of animal cells with free-living microeukaryotes offers some direct observations in support of the DOL hypothesis. Several cellular features that are constitutively present in choanoflagellates are restricted to a subset of animal cell types. These include a motile flagellum, filopodia, contractile stress fibers, polarized secretion (Burkhardt et al. 2011), and mitotic machinery (Figure 6). In support of the antiquity of these cellular modules, many of the effector genes that implement these phenotypes in animals are conserved in choanoflagellates and other holozoans (Figure 6A). Intriguingly, the same is true of the selector transcription factors that control expression of these modules in animals (Figure 6A) – for example FoxJl and RFX for flagella (see Figure 6 and Supplementary Table 2 for other examples). It is currently unknown whether the targets of most of these transcription factors are the same in choanoflagellates as in animals. In the case of Myc, bioinformatic data suggest that the choanoflagellate ortholog triggers heightened synthesis of ribosomal genes (Brown et al. 2008), as it does in animals (in line with translation being the rate-limiting step during cell proliferation (Klumpp et al. 2013)), and that RFX controls at least a few flagellar genes in the choanoflagellate M. brevicollis (Piasecki et al. 2010).
Detailed comparisons of these cellular modules and of their upstream transcription factors with the cell type-specific modules and circuits of animals will be crucial for further testing of the DOL hypothesis. Transcription factor targets (Figure 6) could be determined experimentally by ChIP-seq and/or loss-of-function experiments in choanoflagellates and other single-celled holozoans. If they are at the top of the same regulatory networks as in animals, this would further support the DOL hypothesis, and would suggest a mechanistic basis for division of labor. Indeed, it would imply that the transcriptional networks of single-celled holozoan ancestors were modular, with different transcription factors sitting upstream of functionally distinct gene modules. Such an architecture would have made animal ancestors ‘pre-adapted’ to evolving division of labor once multicellularity evolved, as a module could be selectively ‘shut down’ in a cell by inhibiting its upstream transcription factor with minimal impact on the expression of other modules. Intriguingly, and consistent with this hypothesis, a modular architecture of gene regulatory networks, with functionally related genes or operons being controlled by the same transcription factors, has often been found in single-celled organisms including Escherichia coli, Bacillus subtilis (Shen-Orr et al. 2002; Madan Babu and Teichmann 2003; Fadda et al. 2009) and yeast (Tavazoie et al. 1999; Tanay et al. 2004). The advantage of such a modular architecture in single-celled organisms might include modulating the total abundancy of a module (depending for example on environmental signals), ensuring stoichiometry between its components, or allowing regeneration of a given part of the cell. For example, choanoflagellates regenerate their flagellum after each division or when recovering from microtubule-depolymerizing treatment (Froesler and Leadbeater 2009). In the unicellular alga Chlamydomonas, flagellum regeneration involves a coordinated rise in flagellar gene transcription (Keller et al. 1984) and investigation of promoter motifs suggests shared regulation of these flagellar genes by yet unidentified, specific transcription factors (Stolc et al. 2005).
Currently, the evidence for the TST and DOL scenarios remains primarily descriptive and restricted to a few candidate genes. It will be crucial to ground future comparative studies in mechanistic insights to more fully test these two hypotheses. This endeavor should benefit from ongoing efforts to obtain unbiased transcriptomes and proteomes for animal cell types, notably by single-cell approaches (Achim et al. 2015; Wagner et al. 2016; Villani et al. 2017; Regev et al. 2017). Analyzed with refined methods for phylogenetic reconstruction (Liang et al. 2015; Kin et al. 2015; Musser and Wagner 2015), such datasets should help illuminate our understanding of the origin and evolution of animal cell differentiation.
Conclusion and outlooks
The first animals likely developed through serial division of flagellated, bacterivorous cells that sported a microvillar collar. Cell types then evolved through a combination of innovation, division of labor, and spatial juxtaposition of temporally segregated cell types. Determining the relative contributions of these different mechanisms, and the interrelationships among different cell types in diverse animals will not only help us understand our evolutionary origins, but help to deepen our understanding of the structure and function of animal cells themselves. Indeed, insights into evolutionary relationships have already led to unexpected discoveries in cell biology – e.g., comparative characterizations of ciliary proteomes have, by identifying an evolutionarily conserved core, revealed central players of clinical relevance (Li et al. 2004).
In the future, it will be indispensable to develop functional molecular and cellular techniques in a broader range of early-branching animals and single-celled holozoans, and to expand observational research to clarify uncertainties concerning, for example, the life cycles of additional choanoflagellates or the embryology of sponges, ctenophores and placozoans. Transcriptome sequencing of diverse cell types from choanoflagellates and early branching animals may also help to reveal the relative importance of division of labor as opposed to temporal-to-spatial transitions in the evolution of animal cell differentiation. By integrating these lines of research, we hope to gain a clearer picture of how our protozoan ancestors broke free of the microbial world and founded the animal kingdom.
Supplementary Material
Acknowledgements
We thank all members of the King lab for discussion of the ideas presented, David Booth, Ben Larson, Scott Nichols, Iñaki Ruiz-Trillo, Monika Sigg and three anonymous reviewers for critical reading of the manuscript, Patrick Keeling for sharing the figure panel on eukaryote phylogeny, Daniel J. Richter and Cédric Berney for discussions on defining Choanozoa. T.B. has been supported by the EMBO long-term fellowship ALTF 1474–2016 and by the long-term Human Science Frontier Program fellowship LT000053/2017.
Glossary
- Clade
(n.) a group of organisms composed of a common ancestor and all its past and present descendants. For example, animals are a clade.
- Coenocyte
(n.) a cell with multiple nuclei enclosed within a single plasma membrane, produced by serial nuclear division (karyokinesis) without cytokinesis.
- Monophyletic
(adj.) a monophyletic group comprises a common ancestor and all its descendants. “Monophyletic group” is a synonym of “clade”.
- Paraphyletic
(adj.) a paraphyletic group comprises a common ancestor and only some its descendants. For example, protozoans are a paraphyletic group (as the last common ancestor of all protozoans was also the ancestor of animals, plants, fungi and other multicellular groups).
- Synapomorphy
(n.) an evolutionarily derived feature unique to a clade. For example, the collar complex is a synapomorphy of choanozoans.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abedin M, King N (2008) The premetazoan ancestry of cadherins. Science 319:946–948. doi: 10.1126/science.1151084 [DOI] [PubMed] [Google Scholar]
- Abedin M, King N (2010) Diverse evolutionary paths to cell adhesion. Trends Cell Biol 20:734–742. doi: 10.1016/j.tcb.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achim K, Pettit J-B, Saraiva LR, et al. (2015) High-throughput spatial mapping of single-cell RNA-seq data to tissue of origin. Nat Biotechnol 33:503–509. doi: 10.1038/nbt.3209 [DOI] [PubMed] [Google Scholar]
- Adamska M (2016) Sponges as the rosetta stone of colonial-to-multicellular transition In: Niklas KJ, Newman SA (eds) Multicellularity. Origins and Evolution. MIT Press, pp 185–200 [Google Scholar]
- Alegado RA, Brown LW, Cao S, et al. (2012) A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. eLife 1:e00013. doi: 10.7554/eLife.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alié A, Hayashi T, Sugimura I, et al. (2015) The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci U S A 112:E7093–7100. doi: 10.1073/pnas.1514789112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D (2004) Comparative aspects of gastrulation In: Stern C (ed) Gastrulation. From cells to embryos. Cold Spring Harbor Laboratory Press, pp 679–693 [Google Scholar]
- Arendt D (2008) The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9:868–882. doi: 10.1038/nrg2416 [DOI] [PubMed] [Google Scholar]
- Arendt D (2005) Genes and homology in nervous system evolution: comparing gene functions, expression patterns, and cell type molecular fingerprints. Theory Biosci 124:185–197. doi: 10.1016/j.thbio.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Arendt D, Benito-Gutierrez E, Brunet T, Marlow H (2015) Gastric pouches and the mucociliary sole: setting the stage for nervous system evolution. Philos Trans R Soc Lond B Biol Sci. doi: 10.1098/rstb.2015.0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Musser JM, Baker CVH, et al. (2016) The origin and evolution of cell types. Nat Rev Genet 17:744–757. doi: 10.1038/nrg.2016.127 [DOI] [PubMed] [Google Scholar]
- Arendt D, Nübler-Jung K (1997) Dorsal or ventral: similarities in fate maps and gastrulation patterns in annelids, arthropods and chrodates. Mech Dev 61:7–21. [DOI] [PubMed] [Google Scholar]
- Benabentos R, Hirose S, Sucgang R, et al. (2009) Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr Biol 19:567–572. doi: 10.1016/j.cub.2009.02.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benayahu Y, Berner T, Achituv Y (1989) Development of planulae within a mesogleal coat in the soft coral Heteroxenia fuscescens. Mar Biol 100:203–210. [Google Scholar]
- Bergman K, Goodenough UW, Goodenough DA, et al. (1975) Gametic differentiation in Chlamydomonas reinhardtii. II. Flagellar membranes and the agglutination reaction. J Cell Biol 67:606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JT (1998) The origins of multicellularity. Integr Biol 1:27–36. [Google Scholar]
- Boraas ME, Seale DB, Boxhorn JE (1998) Phagotrophy by a flagellate selects for colonial prey: a possible origin of multicellularity. Evol Ecol 12:153–164. [Google Scholar]
- Borner J, Rehm P, Schill RO, et al. (2014) A transcriptome approach to ecdysozoan phylogeny. Mol Phylogenet Evol 80:79–87. doi: 10.1016/j.ympev.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ (2012) Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol 22:1123–1127. doi: 10.1016/j.cub.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Brown SJ, Cole MD, Erives AJ (2008) Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 9:442. doi: 10.1186/1471-2164-9-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd GE, Jensen S (2017) The origin of the animals and a “Savannah” hypothesis for early bilaterian evolution. Biol Rev Camb Philos Soc 92:446–473. doi: 10.1111/brv.12239 [DOI] [PubMed] [Google Scholar]
- Burkhardt P, Stegmann CM, Cooper B, et al. (2011) Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci U S A 108:15264–15269. doi: 10.1073/pnas.1106189108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, et al. (2005) Hematopoietic stem cell fate is established by the Notch–Runx pathway. Genes Dev 19:2331–2342. doi: 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss LW (1988) The Evolution of Individuality. Princeton University Press [Google Scholar]
- Cannon JT, Vellutini BC, Smith J, et al. (2016) Xenacoelomorpha is the sister group to Nephrozoa. Nature 530:89–93. doi: 10.1038/nature16520 [DOI] [PubMed] [Google Scholar]
- Carr M, Leadbeater BSC, Hassan R, et al. (2008) Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A 105:16641–16646. doi: 10.1073/pnas.0801667105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr M, Richter DJ, Fozouni P, et al. (2017) A six-gene phylogeny provides new insights into choanoflagellate evolution. Mol Phylogenet Evol 107:166–178. doi: 10.1016/j.ympev.2016.10.011 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (2017) Origin of animal multicellularity: precursors, causes, consequences-the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Philos Trans R Soc Lond B Biol Sci. doi: 10.1098/rstb.2015.0476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE-Y (2006) Phylogeny and megasystematics of phagotrophic heterokonts (kingdom Chromista). J Mol Evol 62:388–420. doi: 10.1007/s00239-004-0353-8 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE-Y, Oates B (2004) Molecular phylogeny of Amoebozoa and the evolutionary significance of the unikont Phalansterium. Eur J Protistol 40:21–48. [Google Scholar]
- Cavalier-Smith T, Patterson DJ, Larsen J (1991) Cell diversification in heterotrophic flagellates In: The biology of free-living heterotrophic flagellates. Clarendon Press, pp 113–131 [Google Scholar]
- Chang ES, Neuhof M, Rubinstein ND, et al. (2015) Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc Natl Acad Sci U S A 112:14912–14917. doi: 10.1073/pnas.1511468112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng TC, Yee HW, Rifkin E, Kramer MD (1968) Studies on the internal defense mechanisms of sponges: III. Cellular reactions in Terpios zeteki to implanted heterologous biological materials. J Invertebr Pathol 12:29–35. [Google Scholar]
- Coffman JA (2003) Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int 27:315–324. doi: 10.1016/S1065-6995(03)00018-0 [DOI] [PubMed] [Google Scholar]
- Couch CS, Weil E, Harvell CD (2013) Temporal dynamics and plasticity in the cellular immune response of the sea fan coral, Gorgonia ventalina. Mar Biol 160:2449–2460. [Google Scholar]
- Crawford BJ, Campbell SS (1993) The microvilli and hyaline layer of embryonic asteroid epithelial collar cells: a sensory structure to determine the position of locomotory cilia? Anat Rec 236:697–709. [DOI] [PubMed] [Google Scholar]
- Curtis ASG (1962) Pattern and mechanism in the reaggregation of sponges. Nature 196:245–248. [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, et al. (2011) Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol 357:73–82. doi: 10.1016/j.ydbio.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Sebé-Pedrós A, Šestak MS, et al. (2013) Transcription factor evolution in eukaryotes and the assembly of the regulatory toolkit in multicellular lineages. Proc Natl Acad Sci U S A 110:E4858–4866. doi: 10.1073/pnas.1311818110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mendoza A, Suga H, Permanyer J, et al. (2015) Complex transcriptional regulation and independent evolution of fungal-like traits in a relative of animals. eLife 4:e08904. doi: 10.7554/eLife.08904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas LM, Li L, Yang Y, Dranginis AM (2007) Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot Cell 6:2214–2221. doi: 10.1128/EC.00284-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Kawabe Y, Schilde C, et al. (2015) The Evolution of Aggregative Multicellularity and Cell-Cell Communication in the Dictyostelia. J Mol Biol 427:3722–3733. doi: 10.1016/j.jmb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Schilde C, Birgersson E, et al. (2014) The cyclic AMP phosphodiesterase RegA critically regulates encystation in social and pathogenic amoebas. Cell Signal 26:453–459. doi: 10.1016/j.cellsig.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboscq O, Tuzet O (1939) Les diverses formes des choanocytes des éponges calcaires hétérocoeles et leur signification. Arch Zool Exp Gen 80:353–388. [Google Scholar]
- Ereskovsky AV (2010) The comparative embryology of sponges. Springer Science & Business Media [Google Scholar]
- Fadda A, Fierro AC, Lemmens K, et al. (2009) Inferring the transcriptional network of Bacillus subtilis. Mol Biosyst 5:1840–1852. doi: 10.1039/B907310H [DOI] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, et al. (2013) Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol 14:R15. doi: 10.1186/gb-2013-14-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N (2010) Multicellular development in a choanoflagellate. Curr Biol 20:R875–876. doi: 10.1016/j.cub.2010.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fautin DG, Mariscal RN (1991) Cnidaria: Anthozoa In: Microscopic Anatomy of Invertebrates Volume 2: Placozoa, Porifera, Cnidaria, and Ctenophora, Harrison Frederick W. and Ruppert Edward E. (ed.). Wiley-Liss, [Google Scholar]
- Fidalgo M, Barrales RR, Ibeas JI, Jimenez J (2006) Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A 103:11228–11233. doi: 10.1073/pnas.0601713103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierro-González JC, White MD, Silva JC, Plachta N (2013) Cadherin-dependent filopodia control preimplantation embryo compaction. Nat Cell Biol 15:1424–1433. doi: 10.1038/ncb2875 [DOI] [PubMed] [Google Scholar]
- Franzen W (1988) Oogenesis and larval development of Scypha ciliata (Porifera, Calcarea). Zoomorphology 107:349–357. [Google Scholar]
- Fraser BR, Zalik SE (1977) Lectin-mediated agglutination of amphibian embryonic cells. J Cell Sci 27:227–243. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Lord SJ, Mullins RD (2017) WASP and SCAR are evolutionarily conserved in actin-filled pseudopod-based motility. J Cell Biol. doi: 10.1083/jcb.201701074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, et al. (2010) The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140:631–642. doi: 10.1016/j.cell.2010.01.032 [DOI] [PubMed] [Google Scholar]
- Froesler J, Leadbeater BS (2009) Role of the cytoskeleton in choanoflagellate lorica assembly. J Eukaryot Microbiol 56:167–173. [DOI] [PubMed] [Google Scholar]
- Garrone R (1969) Une formation paracristalline d’ARN intranucléaire dans les choanocytes de l’eponge Haliclona rosea O.S. (Démosponge, Haploscléride). Comptes-Rendus Academie Sci Paris 269:2219–2221. [Google Scholar]
- Gilbert OM, Foster KR, Mehdiabadi NJ, et al. (2007) High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc Natl Acad Sci U S A 104:8913–8917. doi: 10.1073/pnas.0702723104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G, Lawal HM, Felder M, et al. (2016) The multicellularity genes of dictyostelid social amoebas. Nat Commun 7:12085. doi: 10.1038/ncomms12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg WM, Taylor GT (1989) Cellular structure and ultrastructure of the black coral Antipathes aperta: 2. The gastrodermis and its collar cells. J Morphol 202:255–269. [DOI] [PubMed] [Google Scholar]
- Gonobobleva E, Maldonado M (2009) Choanocyte ultrastructure in Halisarca dujardini (Demospongiae, Halisarcida). J Morphol 270:615–627. doi: 10.1002/jmor.10709 [DOI] [PubMed] [Google Scholar]
- Grassé P-P (1973) Traité de zoologie: Anatomie, Systématique, Biologie Spongiaires: anatomie, physiologie, systématique, écologie. Masson [Google Scholar]
- Grau-Bové X, Torruella G, Donachie S, et al. (2017) Dynamics of genomic innovation in the unicellular ancestry of animals. eLife. doi: 10.7554/eLife.26036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JM, McClay DR (2001) The Role of Brachyury (T) during Gastrulation Movements in the Sea Urchin Lytechinus variegatus. Dev Biol 239:132–147. doi: 10.1006/dbio.2001.0426 [DOI] [PubMed] [Google Scholar]
- Haeckel E (1874) Memoirs: the Gastraea-Theory, the phylogenetic classification of the animal kingdom and the homology of the germ-lamellæ. J Cell Sci 2:142–165. [Google Scholar]
- Haeckel E (1892) The history of creation, 2 vols. Kegan Paul, Trench, Trubner & Co. Ltd [Google Scholar]
- Hanschen ER, Marriage TN, Ferris PJ, et al. (2016) The Gonium pectorale genome demonstrates co-option of cell cycle regulation during the evolution of multicellularity. Nat Commun 7:11370. doi: 10.1038/ncomms11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H, Zalik SE (1985) Studies on the endogenous galactose-binding lectin during early development of the embryo of Xenopus laevis. J Cell Sci 79:105–117. [DOI] [PubMed] [Google Scholar]
- Hartenstein V (2006) Blood Cells and Blood Cell Development in the Animal Kingdom. Annu Rev Cell Dev Biol 22:677–712. doi: 10.1146/annurev.cellbio.22.010605.093317 [DOI] [PubMed] [Google Scholar]
- Hehenberger E, Tikhonenkov DV, Kolisko M, et al. (2017) Novel Predators Reshape Holozoan Phylogeny and Reveal the Presence of a Two-Component Signaling System in the Ancestor of Animals. Curr Biol. doi: 10.1016/j.cub.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Hernandez-Nicaise M-L (1991) Ctenophora In: Harrison FW, Ruppert EE (eds) Microscopic anatomy of invertebrates. Wiley-Liss, pp 359–418 [Google Scholar]
- Hibberd DJ (1983) Ultrastructure of the colonial colourless zooflagellates Phalansterium digitatum Stein (Phalansteriida ord. nov.) and Spongomonas uvella Stein (Spongomonadida ord. nov.). Protistologica 19:523–535. [Google Scholar]
- Hibberd Dj (1975) Observations on the ultrastructure of the choanoflagellate Codosiga botrytis (Ehr.) Saville-Kent with special reference to the flagellar apparatus. J Cell Sci 17:191–219. [DOI] [PubMed] [Google Scholar]
- Hirakow R, Kajita N (1994) Electron microscopic study of the development of amphioxus, Branchiostoma belcheri tsingtauense: the neurula and larva. Kaibogaku Zasshi 69:1–13. [PubMed] [Google Scholar]
- Hirose S, Benabentos R, Ho H-I, et al. (2011) Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science 333:467–470. doi: 10.1126/science.1203903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O (2016) Terminal Selectors of Neuronal Identity. Curr Top Dev Biol 116:455–475. doi: 10.1016/bs.ctdb.2015.12.007 [DOI] [PubMed] [Google Scholar]
- Höhn S, Hallmann A (2016) Distinct shape-shifting regimes of bowl-shaped cell sheets - embryonic inversion in the multicellular green alga Pleodorina. BMC Dev Biol 16:35. doi: 10.1186/s12861-016-0134-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James-Clark H (1867) IV.—Conclusive proofs of the animality of the ciliate sponges, and of their affinities with the Infusoria flagellata. J Nat Hist 19:13–18. [Google Scholar]
- Karpov SA, Coupe SJ (1998) A revision of choanoflagellate genera Kentrosiga Schiller, 1953 and Desmarella Kent, 1880. Acta Protozool 37:23–28. [Google Scholar]
- Karpov SA, Leadbeater BS (1998) Cytoskeleton structure and composition in choanoflagellates. J Eukaryot Microbiol 45:361–367. [Google Scholar]
- Kawabe Y, Schilde C, Du Q, Schaap P (2015) A conserved signalling pathway for amoebozoan encystation that was co-opted for multicellular development. Sci Rep 5:9644. doi: 10.1038/srep09644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, et al. (2014) The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLOS Biol 12:e1001889. doi: 10.1371/journal.pbio.1001889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller LR, Schloss JA, Silflow CD, Rosenbaum JL (1984) Transcription of alpha- and beta-tubulin genes in vitro in isolated Chlamydomonas reinhardi nuclei. J Cell Biol 98:1138–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin K, Nnamani MC, Lynch VJ, et al. (2015) Cell-type phylogenetics and the origin of endometrial stromal cells. Cell Rep 10:1398–1409. doi: 10.1016/j.celrep.2015.01.062 [DOI] [PubMed] [Google Scholar]
- King N (2004) The unicellular ancestry of animal development. Dev Cell 7:313–325. doi: 10.1016/j.devcel.2004.08.010 [DOI] [PubMed] [Google Scholar]
- King N, Carroll SB (2001) A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A 98:15032–15037. doi: 10.1073/pnas.261477698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Rokas A (2017) Embracing uncertainty in reconstructing early animal evolution. Current Biology (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, et al. (2008) The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451:783–788. doi: 10.1038/nature06617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, Scott M, Pedersen S, Hwa T (2013) Molecular crowding limits translation and cell growth. Proc Natl Acad Sci 110:16754–16759. doi: 10.1073/pnas.1310377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll AH (2011) The multiple origins of complex multicellularity. Annu Rev Earth Planet Sci 39:217–239. [Google Scholar]
- Koschwanez JH, Foster KR, Murray AW (2011) Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol 9:e1001122. doi: 10.1371/journal.pbio.1001122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda MJ, Robbins PW (1991) Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J Biol Chem 266:19758–19767. [PubMed] [Google Scholar]
- Kuzdzal-Fick JJ, Fox SA, Strassmann JE, Queller DC (2011) High relatedness is necessary and sufficient to maintain multicellularity in Dictyostelium. Science 334:1548–1551. doi: 10.1126/science.1213272 [DOI] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, et al. (2008) Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol 25:980–996. doi: 10.1093/molbev/msn047 [DOI] [PubMed] [Google Scholar]
- Larsen J (1985) Ultrastructure and taxonomy of Actinomonas pusilla, a heterotrophic member of the Pedinellales (Chrysophyceae). Br Phycol J 20:341–355. [Google Scholar]
- Laumer CE, Bekkouche N, Kerbl A, et al. (2015) Spiralian phylogeny informs the evolution of microscopic lineages. Curr Biol 25:2000–2006. doi: 10.1016/j.cub.2015.06.068 [DOI] [PubMed] [Google Scholar]
- Lauri A, Brunet T, Handberg-Thorsager M, et al. (2014) Development of the annelid axochord: insights into notochord evolution. Science 345:1365–1368. doi: 10.1126/science.1253396 [DOI] [PubMed] [Google Scholar]
- Leadbeater BS (2006) The “mystery” of the flagellar vane in choanoflagellates. Nova Hedwig Beih 130:213. [Google Scholar]
- Leadbeater BS (1983) Life-history and ultrastructure of a new marine species of Proterospongia (Choanoflagellida). J Mar Biol Assoc U K 63:135–160. [Google Scholar]
- Leadbeater BSC (2014) The Choanoflagellates: Evolution, Biology and Ecology. Cambridge University Press [Google Scholar]
- Lee JK, Buckhaults P, Wilkes C, et al. (1997) Cloning and expression of a Xenopus laevis oocyte lectin and characterization of its mRNA levels during early development. Glycobiology 7:367–372. doi: 10.1093/glycob/7.3.367 [DOI] [PubMed] [Google Scholar]
- Leong DT, Lim J, Goh X, et al. (2010) Cancer-related ectopic expression of the bone-related transcription factor RUNX2 in non-osseous metastatic tumor cells is linked to cell proliferation and motility. Breast Cancer Res 12:R89. doi: 10.1186/bcr2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin TC, Greaney AJ, Wetzel L, King N (2014) The Rosetteless gene controls development in the choanoflagellate S. rosetta. eLife. doi: 10.7554/eLife.04070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin TC, King N (2013) Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Curr Biol 23:2176–2180. doi: 10.1016/j.cub.2013.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leys SP, Ereskovsky AV (2006) Embryogenesis and larval differentiation in sponges. Can J Zool 84:262–287. [Google Scholar]
- Leys SP, Hill A (2012) 1 The Physiology and Molecular Biology of Sponge Tissues. Adv Mar Biol 62:1. [DOI] [PubMed] [Google Scholar]
- Leys SP, Nichols SA, Adams EDM (2009) Epithelia and integration in sponges. Integr Comp Biol 49:167–177. doi: 10.1093/icb/icp038 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, et al. (2004) Comparative Genomics Identifies a Flagellar and Basal Body Proteome that Includes the BBS5 Human Disease Gene. Cell 117:541–552. doi: 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Liang C, FANTOM Consortium, Forrest ARR, Wagner GP (2015) The statistical geometry of transcriptome divergence in cell-type evolution and cancer. Nat Commun 6:6066. doi: 10.1038/ncomms7066 [DOI] [PubMed] [Google Scholar]
- Lie-A-Ling M, Marinopoulou E, Li Y, et al. (2014) RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 124:e11–e20. doi: 10.1182/blood-2014-04-572958 [DOI] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM (1996) FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J Bacteriol 178:7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackie GO (1970) Neuroid conduction and the evolution of conducting tissues. Q Rev Biol 45:319–332. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA (2003) Evolution of transcription factors and the gene regulatory network in Escherichia coli. Nucleic Acids Res 31:1234–1244. doi: 10.1093/nar/gkg210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah JL, Christensen-Dalsgaard KK, Leys SP (2014) Choanoflagellate and choanocyte collar-flagellar systems and the assumption of homology. Evol Dev 16:25–37. doi: 10.1111/ede.12060 [DOI] [PubMed] [Google Scholar]
- Maldonado M (2004) Choanoflagellates, choanocytes, and animal multicellularity. Invertebr Biol 123:1–22. [Google Scholar]
- Margulis L (1981) Symbiosis in Cell Evolution. W. H. Freeman, San Francisco [Google Scholar]
- Mehl D, Reiswig HM (1991) The presence of flagellar vanes in choanomeres of Porifera and their possible phylogenetic implications. J Zool Syst Evol Res 29:312–319. [Google Scholar]
- Mendoza L, Taylor JW, Ajello L (2002) The class mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu Rev Microbiol 56:315–344. doi: 10.1146/annurev.micro.56.012302.160950 [DOI] [PubMed] [Google Scholar]
- Metzger MJ, Reinisch C, Sherry J, Goff SP (2015) Horizontal transmission of clonal cancer cells causes leukemia in soft-shell clams. Cell 161:255–263. doi: 10.1016/j.cell.2015.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod RE (2007) Evolution of individuality during the transition from unicellular to multicellular life. Proc Natl Acad Sci U S A 104 Suppl 1:8613–8618. doi: 10.1073/pnas.0701489104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhailov KV, Konstantinova AV, Nikitin MA, et al. (2009) The origin of Metazoa: a transition from temporal to spatial cell differentiation. BioEssays 31:758–768. doi: 10.1002/bies.200800214 [DOI] [PubMed] [Google Scholar]
- Musser JM, Wagner GP (2015) Character trees from transcriptome data: Origin and individuation of morphological characters and the so-called “species signal.” J Exp Zoolog B Mol Dev Evol 324:588–604. doi: 10.1002/jez.b.22636 [DOI] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, et al. (2012) Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/β-catenin complex. Proc Natl Acad Sci 109:13046–13051. doi: 10.1073/pnas.1120685109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C (2012) Animal Evolution: Interrelationships of the Living Phyla, 3rd edition Oxford University Press, Oxford ; New York [Google Scholar]
- Nielsen C (2008) Six major steps in animal evolution: are we derived sponge larvae? Evol Dev 10:241–257. doi: 10.1111/j.1525-142X.2008.00231.x [DOI] [PubMed] [Google Scholar]
- Nørrevang A, Wingstrand KG (1970) On the occurrence and structure of choanocyte-like cells in some echinoderms. Acta Zool 51:249–270. [Google Scholar]
- Olano CT, Bigger CH (2000) Phagocytic activities of the gorgonian coral Swiftia exserta. J Invertebr Pathol 76:176–184. doi: 10.1006/jipa.2000.4974 [DOI] [PubMed] [Google Scholar]
- Ott DW, Oldham-Ott CK, Rybalka N, Friedl T (2015) Xanthophyte, Eustigmatophyte, and Raphidophyte Algae. In: Freshwater algae of North America Ecology and classification, JD Wehr, RG Sheath, JP Kociolek (ed.). Academic Press, pp 485–536 [Google Scholar]
- Otto F, Lübbert M, Stock M (2003) Upstream and downstream targets of RUNX proteins. J Cell Biochem 89:9–18. doi: 10.1002/jcb.10491 [DOI] [PubMed] [Google Scholar]
- Oud B, Guadalupe-Medina V, Nijkamp JF, et al. (2013) Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 110:E4223–4231. doi: 10.1073/pnas.1305949110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Outenreath RL, Roberson MM, Barondes SH (1988) Endogenous lectin secretion into the extracellular matrix of early embryos of Xenopus laevis. Dev Biol 125:187–194. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Rast JP, Davidson EH (1999) Origins of immunity: transcription factors and homologues of effector genes of the vertebrate immune system expressed in sea urchin coelomocytes. Immunogenetics 49:773–786. doi: 10.1007/s002510050551 [DOI] [PubMed] [Google Scholar]
- Patterson DJ, Fenchel T (1985) Insights into the evolution of heliozoa (Protozoa, Sarcodina) as provided by ultrastructural studies on a new species of flagellate from the genus Pteridomonas. Biol J Linn Soc 24:381–403. [Google Scholar]
- Patterson MJ, Landolt ML (1979) Cellular reaction to injury in the anthozoan Anthopleura elegantissima. J Invertebr Pathol 33:189–196. [Google Scholar]
- Peña JF, Alié A, Richter DJ, et al. (2016) Conserved expression of vertebrate microvillar gene homologs in choanocytes of freshwater sponges. EvoDevo 7:13. doi: 10.1186/s13227-016-0050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen JB (1929) Beitrage zur Kenntnis der Flagellatengeisseln. Bot Tidsskr 40:373–389. [Google Scholar]
- Piasecki BP, Burghoorn J, Swoboda P (2010) Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci U S A 107:12969–12974. doi: 10.1073/pnas.0914241107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh MA, Malik S-B, Logsdon JM Jr. (2005) A Phylogenomic Inventory of Meiotic Genes: Evidence for Sex in Giardia and an Early Eukaryotic Origin of Meiosis. Curr Biol 15:185–191. doi: 10.1016/j.cub.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Ratcliff WC, Denison RF, Borrello M, Travisano M (2012) Experimental evolution of multicellularity. Proc Natl Acad Sci U S A 109:1595–1600. doi: 10.1073/pnas.1115323109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Fankhauser JD, Rogers DW, et al. (2015) Origins of multicellular evolvability in snowflake yeast. Nat Commun 6:6102. doi: 10.1038/ncomms7102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Herron MD, Howell K, et al. (2013) Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat Commun 4:2742. doi: 10.1038/ncomms3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA (2005) Evolution of Plasmodesmata In: Oparka KJ (ed) Plasmodesmata. Blackwell Publishing Ltd, pp 33–52 [Google Scholar]
- Ray J Shinnick T, Lerner R (1979) A mutation altering the function of a carbohydrate binding protein blocks cell-cell cohesion in developing Dictyostelium discoideum. Nature 279:215–221. [DOI] [PubMed] [Google Scholar]
- Regev A, Teichmann S, Lander ES, et al. (2017) The Human Cell Atlas. bioRxiv 121202. doi: 10.1101/121202 [DOI] [Google Scholar]
- Rennert J, Coffman JA, Mushegian AR, Robertson AJ (2003) The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol 3:4. doi: 10.1186/1471-2148-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter DJ, King N (2013) The genomic and cellular foundations of animal origins. Annu Rev Genet 47:509–537. doi: 10.1146/annurev-genet-111212-133456 [DOI] [PubMed] [Google Scholar]
- Rieger RM (1976) Monociliated epidermal cells in Gastrotricha: significance for concepts of early metazoan evolution. J Zool Syst Evol Res 14:198–226. [Google Scholar]
- Ritchie AV, van Es S, Fouquet C, Schaap P (2008) From drought sensing to developmental control: evolution of cyclic AMP signaling in social amoebas. Mol Biol Evol 25:2109–2118. doi: 10.1093/molbev/msn156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, Hammel JU, Haen KM, et al. (2011) RNA interference in marine and freshwater sponges: actin knockdown in Tethya wilhelma and Ephydatia muelleri by ingested dsRNA expressing bacteria. BMC Biotechnol 11:67. doi: 10.1186/1472-6750-11-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson MM, Barondes SH (1983) Xenopus laevis lectin is localized at several sites in Xenopus oocytes, eggs, and embryos. J Cell Biol 97:1875–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A (2008) The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet 42:235–251. doi: 10.1146/annurev.genet.42.110807.091513 [DOI] [PubMed] [Google Scholar]
- Roper M, Dayel MJ, Pepper RE, Koehl M a. R (2013) Cooperatively generated stresslet flows supply fresh fluid to multicellular choanoflagellate colonies. Phys Rev Lett 110:228104. doi: 10.1103/PhysRevLett.110.228104 [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, et al. (2008) A phylogenomic investigation into the origin of metazoa. Mol Biol Evol 25:664–672. doi: 10.1093/molbev/msn006 [DOI] [PubMed] [Google Scholar]
- Salas-Vidaı E, Lomeli H (2004) Imaging filopodia dynamics in the mouse blastocyst. Dev Biol 265:75–89. [DOI] [PubMed] [Google Scholar]
- Salvini-Plawen LV (1978) On the origin and evolution of the lower Metazoa. J Zool Syst Evol Res 16:40–87. [Google Scholar]
- Santorelli LA, Thompson CRL, Villegas E, et al. (2008) Facultative cheater mutants reveal the genetic complexity of cooperation in social amoebae. Nature 451:1107–1110. doi: 10.1038/nature06558 [DOI] [PubMed] [Google Scholar]
- Schmidt-Rhaesa A (2007) The Evolution of Organ Systems. Oxford University Press [Google Scholar]
- Sebé-Pedrós A, Ballaré C, Parra-Acero H, et al. (2016a) The Dynamic Regulatory Genome of Capsaspora and the Origin of Animal Multicellularity. Cell 165:1224–1237. doi: 10.1016/j.cell.2016.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Burkhardt P, Sánchez-Pons N, et al. (2013a) Insights into the origin of metazoan filopodia and microvilli. Mol Biol Evol 30:2013–2023. doi: 10.1093/molbev/mst110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I (2017) The origin of Metazoa: a unicellular perspective. Nat Rev Genet. doi: 10.1038/nrg.2017.21 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Irimia M, Del Campo J, et al. (2013b) Regulated aggregative multicellularity in a close unicellular relative of metazoa. eLife 2:e01287. doi: 10.7554/eLife.01287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Peña MI, Capella-Gutiérrez S, et al. (2016b) High-Throughput Proteomics Reveals the Unicellular Roots of Animal Phosphosignaling and Cell Differentiation. Dev Cell 39:186–197. doi: 10.1016/j.devcel.2016.09.019 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, Roger AJ, Lang FB, et al. (2010) Ancient origin of the integrin-mediated adhesion and signaling machinery. Proc Natl Acad Sci 107:10142–10147. doi: 10.1073/pnas.1002257107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, et al. (2008) Multigene phylogeny of choanozoa and the origin of animals. PloS One 3:e2098. doi: 10.1371/journal.pone.0002098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31:64–68. doi: 10.1038/ng881 [DOI] [PubMed] [Google Scholar]
- Shinnick TM, Lerner RA (1980) The cbpA gene: role of the 26,000-dalton carbohydrate-binding protein in intercellular cohesion of developing Dictyostelium iscoideum cells. Proc Natl Acad Sci U S A 77:4788–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares EV (2011) Flocculation in Saccharomyces cerevisiae: a review. J Appl Microbiol 110:1–18. doi: 10.1111/j.1365-2672.2010.04897.x [DOI] [PubMed] [Google Scholar]
- Souza GM, da Silva AM, Kuspa A (1999) Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development 126:3263–3274. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, et al. (2010) The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466:720–726. doi: 10.1038/nature09201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc V, Samanta MP, Tongprasit W, Marshall WF (2005) Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci U S A 102:3703–3707. doi: 10.1073/pnas.0408358102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassmann JE, Zhu Y, Queller DC (2000) Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature 408:965–967. doi: 10.1038/35050087 [DOI] [PubMed] [Google Scholar]
- Stratford M (1992) Lectin-mediated aggregation of yeasts--yeast flocculation. Biotechnol Genet Eng Rev 10:283–341. [DOI] [PubMed] [Google Scholar]
- Struck TH, Wey-Fabrizius AR, Golombek A, et al. (2014) Platyzoan paraphyly based on phylogenomic data supports a noncoelomate ancestry of spiralia. Mol Biol Evol 31:1833–1849. doi: 10.1093/molbev/msu143 [DOI] [PubMed] [Google Scholar]
- Sucgang R, Kuo A, Tian X, et al. (2011) Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol 12:R20. doi: 10.1186/gb-2011-12-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Chen Z, de Mendoza A, et al. (2013) The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun 4:2325. doi: 10.1038/ncomms3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Ruiz-Trillo I (2013) Development of ichthyosporeans sheds light on the origin of metazoan multicellularity. Dev Biol 377:284–292. doi: 10.1016/j.ydbio.2013.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JC, Sher D, Eisenstein M, et al. (2008) The evolutionary origin of the Runx/CBFbeta transcription factors – Studies of the most basal metazoans. BMC Evol Biol 8:228. doi: 10.1186/1471-2148-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Gold D, Hartenstein V (2013) Stem cells and lineages of the intestine: a developmental and evolutionary perspective. Dev Genes Evol 223:85–102. doi: 10.1007/s00427-012-0422-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanay A, Sharan R, Kupiec M, Shamir R (2004) Revealing modularity and organization in the yeast molecular network by integrated analysis of highly heterogeneous genomewide data. Proc Natl Acad Sci U S A 101:2981–2986. doi: 10.1073/pnas.0308661100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie S, Hughes JD, Campbell MJ, et al. (1999) Systematic determination of genetic network architecture. Nat Genet 22:281–285. doi: 10.1038/10343 [DOI] [PubMed] [Google Scholar]
- Telford MJ, Budd GE, Philippe H (2015) Phylogenomic Insights into Animal Evolution. Curr Biol 25:R876–887. doi: 10.1016/j.cub.2015.07.060 [DOI] [PubMed] [Google Scholar]
- Torruella G, de Mendoza A, Grau-Bové X, et al. (2015) Phylogenomics Reveals Convergent Evolution of Lifestyles in Close Relatives of Animals and Fungi. Curr Biol 25:2404–2410. doi: 10.1016/j.cub.2015.07.053 [DOI] [PubMed] [Google Scholar]
- Vacquier VD (1968) The connection of blastomeres of sea urchin embryos by filopodia. Exp Cell Res 52:571–581. [DOI] [PubMed] [Google Scholar]
- Valentine JW (2006) On the Origin of Phyla. University of Chicago Press [Google Scholar]
- VanOudenhove JJ, Medina R, Ghule PN, et al. (2016) Transient RUNX1 Expression during Early Mesendodermal Differentiation of hESCs Promotes Epithelial to Mesenchymal Transition through TGFB2 Signaling. Stem Cell Rep 7:884–896. doi: 10.1016/j.stemcr.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villani A-C, Satija R, Reynolds G, et al. (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. doi: 10.1126/science.aah4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlk W (1938) Über den bau der Geissel. Arch Für Protistenkd 90:448–488. [Google Scholar]
- Wagner A, Regev A, Yosef N (2016) Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol 34:1145–1160. doi: 10.1038/nbt.3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK (1993) Monophyletic origins of the metazoa: an evolutionary link with fungi. Science 260:340–342. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Ferjoux G, Bataille L, Haenlin M (2003) Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J 22:6516–6525. doi: 10.1093/emboj/cdg622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenfels N (1982) Bau und Funktion des Süßwasserschwamms Ephydatia fluviatilis L.(Porifera). Zoomorphology 100:75–87. [Google Scholar]
- Wickerham LJ (1958) Sexual agglutination of heterothallic yeasts in diverse taxonomic areas. Science 128:1504–1505. [DOI] [PubMed] [Google Scholar]
- Williams F, Tew HA, Paul CE, Adams JC (2014) The predicted secretomes of Monosiga brevicollis and Capsaspora owczarzaki, close unicellular relatives of metazoans, reveal new insights into the evolution of the metazoan extracellular matrix. Matrix Biol 37:60–68. doi: 10.1016/j.matbio.2014.02.002 [DOI] [PubMed] [Google Scholar]
- Woollacott RM, Pinto RL (1995) Flagellar basal apparatus and its utility in phylogenetic analyses of the Porifera. J Morphol 226:247–265. [DOI] [PubMed] [Google Scholar]
- Woznica A, Cantley AM, Beemelmanns C, et al. (2016) Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc Natl Acad Sci U S A 113:7894–7899. doi: 10.1073/pnas.1605015113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woznica A, Gerdt JP, Hulett R, et al. (2017) Mating in the closest living relatives of animals is induced by a bacterial chondroitinase. Cell (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa KO, Fujimoto H, Urushihara H (1981) Effects of the Brachyury (T) mutation on morphogenetic movement in the mouse embryo. Dev Biol 87:242–248. doi: 10.1016/0012-1606(81)90147-0 [DOI] [PubMed] [Google Scholar]
- Young SL, Diolaiti D, Conacci-Sorrell M, et al. (2011) Premetazoan ancestry of the Myc-Max network. Mol Biol Evol 28:2961–2971. doi: 10.1093/molbev/msr132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhvatkin AA (1949) The comparative embryology of the low invertebrates. In: Sources and method of the origin of metazoan development. Soviet Science [Google Scholar]
- Zusso M, Methot L, Lo R, et al. (2012) Regulation of Postnatal Forebrain Amoeboid Microglial Cell Proliferation and Development by the Transcription Factor Runx1. J Neurosci 32:11285–11298. doi: 10.1523/JNEUROSCI.6182-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


