Abstract
Genome-wide functional genomic screens utilizing the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 system have proven to be a powerful tool for systematic genomic perturbation in mammalian cells and provide an alternative to previous screens utilizing RNA interference technology. The wide availability of these libraries through public plasmid repositories as well as the decreasing cost and speed in quantifying these screens using high-throughput next-generation sequencing (NGS) allows for the adoption of the technology in a variety of laboratories interested in diverse biologic questions. Here, we describe the protocol to generate next-generation sequencing libraries from genome-wide CRISPR genomic screens.
Keywords: CRISPR, Cas9, Genome-wide screen, Genome engineering, GeCKO
1 Introduction
Initially discovered as a prokaryotic adaptive immune system, the bacterial type II clustered regularly interspaced short palindrome repeats (CRISPR) and their associated proteins (Cas9) system have been adapted into mammalian cells to generate a simple and efficient means of generating targeted loss-of-function mutations [1–7]. Cas9 proteins generate precise double-strand breaks at target loci determined by short guide RNAs (sgRNAs). In the absence of a homologous template, these breaks are repaired by the error-prone non-homologous end-joining (NHEJ) mechanism leading to the production of indels. By targeting coding regions, knockout libraries can be generated due to the introduction of indels that lead to frame-shifts. The combination of the simplicity of the CRISPR-Cas9 system, in which the short 20 base pair sgRNA sequence confers the targeting specificity of the Cas9 nuclease, economic oligonucleotide synthesis using pooled microarray synthesis, and the ability to use NGS sequencing for quantifying readout has enabled the use of pooled genome-scale loss-of-function CRISPR screens to systematically interrogate gene function in different biologic processes. Pooled CRISPR screening has been used to identify genes essential in different genetic contexts [8–10] as well as to identify genes involved in drug or toxin resistance [11–14]. CRISPR screens appear to be more robust in comparison to previous screens utilizing RNA interference technology with short-hairpin RNAs (shRNAs) which were hampered by off-target effects and incomplete genetic knockdown [10].
The most widely adopted system thus far has been the Genome-Scale CRISPR Knock Out (GeCKO) library generated by the Feng Zhang lab and distributed on Addgene and the current protocol is adapted from the methods outlined by this lab which can be found online at http://genome-engineering.org/gecko/. The GeCKO library is available targeting either mouse or human coding genes and is available as a 1 plasmid system where sgRNAs and Cas9 protein are encoded on the same plasmid as well as a 2 plasmid system where sgRNAs and Cas9 protein are on separate lentiviral expression plasmids. Each library contains two sublibraries (A and B) with each sublibrary containing 3 sgRNAs targeting each coding gene. A similar library was also generated by the Sabatini and Lander labs targeting human protein coding genes (Addgene #1000000067) as well as targeted sublibraries (Addgene #51043–51048). Second generation libraries built by improving the specificity of sgRNA guides include the Toronto knockout (TKO) library generated by the Moffat lab (Addgene #1000000069) and the Brunello (Addgene #73178 or #73179)/Brie (Addgene #73632 or #73633) libraries from the Broad Institute Genetic Pertubation Platform (GPP) lab. The TKO and Brunello libraries target human coding genes, while the Brie library targets mouse coding genes. The Brunello and Brie libraries are also available in a smaller format targeting the kinome (Addgene #75312–75317). Screens done with these libraries could be sequenced in similar manner by changing primer sequences, but the current protocol is for generating NGS libraries from a GeCKO v2 screen for sequencing on Illumina NextSeq 500.
2 Materials
2.1 Genome-Wide CRISPR-Cas9 Lentiviral Pooled Library
2.1.1 Plasmid Library Amplification
GeCKOv2 library plasmid (Addgene pooled library #1000000048 or #1000000049).
Endura electrocompetent cells with Recovery Medium (Lucigen Middleton, WI, USA).
LB agar bacterial plates.
Ampicillin 100 mg/mL: made in sterile water and sterilized by filtration (0.2 μm filter).
Incubator 32 °C.
Electroporator (i.e., Gene Pulsar II; Bio-Rad, Hercules, CA, USA).
Cell scraper (Corning, Corning, NY, USA).
DNA Maxi prep kit.
2.1.2 Lentiviral Library Packaging in 293FT HEK Cells
Lipofectamine Transfection Reagent (ThermoFisher Scientific, Waltham, MA, USA).
PLUS Reagent (ThermoFisher Scientific).
pMD2.G plasmid (Addgene #12259).
psPAX2 plasmid (Addgene #12260).
Opti-MEM (ThermoFisher Scientific).
Amicon Ultra-15 Centrifugal Filter Unit with Utracel-100 membrane (EMD Millipore, Temecula, CA, USA).
DMEM/F-12, HEPES (ThermoFisher Scientific).
Fetal Bovine Serum.
0.25% Trypsin-EDTA (ThermoFisher Scientific).
293FT human embryonic kidney cells.
Humidified incubator, with cells maintained at 37 °C and 5% CO2.
Millex-HP Syringe Filter, 0.45 μm, PES (EMD Millipore).
Table top centrifuge.
2.1.3 Functional Titration of Pooled Lentiviral CRISPR Library
CellTiter-Glo Luminescent Cell Viability Assay (Promega, Fitchburg, WI, USA).
Puromycin Dihydrochloride (ThermoFisher Scientific).
Luminescence plate reader (i.e., Synergy 2; BioTek, Winooski, VT, USA).
Polybrene (EMD Millipore).
2.2 Genomic DNA Extraction
Nanodrop 2000 (ThermoFisher Scientific).
2.2.1 Option A
gDNA Lysis Buffer: 50 mM Tris, 50 mM EDTA, 1% SDS, pH 8. To 75 mL of water add 5 mL of 1 M Tris pH 8.0, 10 mL of 0.5 M EDTA pH 8.0, and 5 mL of 10% SDS.
RNase A (100 mg): add 10 mL of gDNA Lysis Buffer to 100 mg of RNase A for 10 mg/mL stock solution. Store at 4 °C.
Proteinase K (Qiagen, Hilden, Germany).
Ammonium Acetate 7.5 M (Sigma, St. Louis, MO, USA): Dissolve 57.81 g of ammonium acetate in water to a final volume of 100 mL. Sterilize by filtration (0.2 μm filter). Store at 4 °C.
Isopropanol.
Ethanol.
Incubator 55 °C.
Table top centrifuge.
2.2.2 Option B
QIAamp Blood Midi/Maxi kit (Qiagen) or Quick-gDNA MidiPrep (Zymo Research, Irvine, CA, USA).
Microcentrifuge or vacuum manifold.
2.3 PCR NGS Library Prep
NEBNext Q5 Hot Start HiFI PCR Master Mix (New England Biolabs, Ipswich, MA, USA) or Herculase II Fusion DNA polymerase (Agilent Technologies, Santa Clara, CA, USA) (see Note 1).
PCR grade water.
Thermocycler.
PCR1 F primer: AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG.
PCR1 R primer: TCTACTATTCTTTCCCCTGCACTGTTGTGGGCGATGTGCGCTCTG.
QIAquick PCR purification kit (Qiagen) or Wizard SV PCR cleanup kit (Promega).
-
PCR2 F:
Primer no. lllumina P5 and lllumina Seq Stagger Inline barcode Priming site F01 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT t AAGTAGAG tcttgtggaaaggacgaaacaccg F02 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT at ACACGATC tcttgtggaaaggacgaaacaccg F03 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT gat CGCGCGGT tcttgtggaaaggacgaaacaccg F04 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT cgat CATGATCG tcttgtggaaaggacgaaacaccg F05 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT tcgat CGTTACCA tcttgtggaaaggacgaaacaccg F06 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT atcgat TCCTTGGT tcttgtggaaaggacgaaacaccg F07 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT gatcgat AACGCATT tcttgtggaaaggacgaaacaccg F08 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT cgatcgat ACAGGTAT tcttgtggaaaggacgaaacaccg F09 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT acgatcgat AGGTAAGG tcttgtggaaaggacgaaacaccg F10 AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT t AACAATGG tcttgtggaaaggacgaaacaccg -
PCR2 R primers:
Primer no. lllumina P7 Index barcode lllumina seq R Stagger Priming site R01 CAAGCAGAAGACGGCATACGAGAT AAGTAGAG GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT t TCTACTATTCTTTCCCCTGCACTGT R02 CAAGCAGAAGACGGCATACGAGAT ACACGATC GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT at TCTACTATTCTTTCCCCTGCACTGT R03 CAAGCAGAAGACGGCATACGAGAT CGCGCGGT GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT gat TCTACTATTCTTTCCCCTGCACTGT R04 CAAGCAGAAGACGGCATACGAGAT CATGATCG GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT cgat TCTACTATTCTTTCCCCTGCACTGT R05 CAAGCAGAAGACGGCATACGAGAT CGTTACCA GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT tcgat TCTACTATTCTTTCCCCTGCACTGT R06 CAAGCAGAAGACGGCATACGAGAT TCCTTGGT GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT atcgat TCTACTATTCTTTCCCCTGCACTGT R07 CAAGCAGAAGACGGCATACGAGAT AACGCATT GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT gatcgat TCTACTATTCTTTCCCCTGCACTGT R08 CAAGCAGAAGACGGCATACGAGAT ACAGGTAT GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT cgatcgat TCTACTATTCTTTCCCCTGCACTGT R09 CAAGCAGAAGACGGCATACGAGAT AGGTAAGG GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT acgatcgat TCTACTATTCTTTCCCCTGCACTGT RIO CAAGCAGAAGACGGCATACGAGAT AACAATGG GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT t TCTACTATTCTTTCCCCTGCACTGT
3 Methods
The following method is the protocol for generating sequencing libraries for quantification of sgRNA abundance from a genome-wide CRISPR screen using the lentiviral GeCKOv2 library in mammalian cells or tissue on an Illumina NextSeq 500. The general protocol can be adapted for sequencing of custom sgRNA pooled libraries or other ready-made libraries from Addgene.
3.1 Screen Design
Forward (or functional) genomic screens can fundamentally be carried out in two ways. One is dropout (or negative selection) screening in which a gene knockout results in a selective disadvantage in that cell such as decreased proliferation in cancer cells. The other is enrichment (or positive selection) screening in which gene knockout results in selective advantage for cells such as drug or toxin resistance.
After the appropriate phenotype has been selected and a dropout or resistance screen has been designed, two other critical factors to take into account in the design of genome-wide screens is (1) the depth of coverage of each sgRNA guide and (2) the number of sgRNA guides per gene. Experience from previous genome-wide shRNA screens suggested that dropout screens require much higher coverage than resistance screens with the recommendation for at least 500–1000× coverage of each individual element for dropout screens to obtain enough signal over noise [15]. Because of the different mechanisms of CRISPR-Cas9 resulting in gene knockouts rather than knockdown, it seems that signal-to-noise ratio is higher than in shRNA screens and the initial CRISPR-Cas9 genomic dropout screens have been carried out at a minimum of 200–300× coverage [9, 11]. For shRNA screens, because of the significant off-target effects of shRNAs, one typically needed to include many shRNAs per gene (typically 5–25 shRNAs per gene) to overcome the noise generated by off-target effects. CRISPR screening appears to have less off-target effects, and the reason to include multiple sgRNAs per gene for CRISPR screening appears to be the variability in the efficiency of each individual sgRNA rather than the off-target noise such that increasing the amount of sgRNAs appears to increase the sensitivity of the screen rather than the specificity [10, 16]. One of the largest hurdles in performing genome-wide screening is providing for adequate coverage of the library by using appropriate amounts of starting cells and maintaining that coverage throughout the experiment. For example, if using the entire human GeCKOv2 library containing 122,411 sgRNA guides, one would need to transduce 2 × 108 cells at 30% efficiency and maintain 6 × 107 cells at each passage and harvesting time point for each biological replicate. This amount of cells could be prohibitive in certain situations. An alternative emerging strategy is to decrease the amount of sgRNAs performed at the genome-wide level (to 3–4 sgRNAs) in the primary screen and to use relaxed cutoffs for hit selection and perform smaller targeted secondary screens using additional sgRNAs per gene [16].
3.2 Genome-Wide CRISPR-Cas9 Lentiviral Pooled Library
3.2.1 Plasmid Library Amplification
Obtain GeCKOv2 library from Addgene delivered as half-libraries (A and B each with three sgRNAs per gene) in 20 μL at a concentration of 50 ng/μL.
Follow provided protocol from Addgene and Zhang lab for amplification of library using pooled electroporation. (https://www.addgene.org/static/cms/filer_public/b5/fd/b5fde702-d02c-4973-806f-24ac28b2a15a/geckov20_library_amplification_protocol_1.pdf).
3.2.2 Lentiviral Library Packaging in 293FT HEK Cells
Culture 293FT cells in DMEM with 10% FBS (D10 media) without antibiotics, prepare a 10 cm plate about 90% confluent day of transfection.
Wash the cells with 3 mL room temperature PBS.
Aspirate PBS, add 3 mL Trypsin/EDTA and incubate at 37 °C for 2 min.
Add 3 mL D10 to inactivate Trypsin/EDTA and collect the cells by centrifugation (1000 rpm for 10 min).
Resuspend the cells in 5 mL pre-warmed DMEM media without serum.
Plate the cells in a 10 cm tissue culture plate and place in an incubator.
Prepare plasmid/PLUS mixture by adding 900 μL room temperature Opti-MEM in a sterile 1.5 mL microcentrifuge tube. Add 12 μg of amplified GeCKO library plasmid, 9 μg psPAX2 plasmid, 6 μg pMD2.G plasmid, and 48 μL of PLUS reagent.
Prepare lipofectamine mixture by adding 60 μL of lipofectamine reagent to 900 μL of room temperature Opti-MEM in a sterile 1.5 mL microcentrifuge tube.
Vortex mixtures briefly and incubate at room temperature for 5 min.
Combine plasmid/PLUS mixture with lipofectamine mixture. Vortex briefly and incubate at room temperature for 15 min.
Add plasmid/lipofectamine mixture dropwise to cell suspension from step 6 and place in an incubator for 4–6 h.
Aspirate media and replace with 10 mL fresh D10 media.
Harvest the supernatant after 60 h into a 50 mL conical tube.
Remove cellular debris by centrifugation at 1600 × g at 4 °C for 10 min.
Filter the supernatant through a 0.45 μm syringe filter.
Concentrate lentivirus using Amicon Ultra-15 centrifugal filter by adding filtered viral supernatant into reservoir and centrifuging at 2850 × g for 40 min at 4 °C.
Aliquot virus and store frozen at −80 °C.
3.2.3 Functional Titration of Pooled Lentiviral CRISPR Library
Harvest target cells and count cells and resuspend cells at a concentration of 2–3 × 106 cells/mL in appropriate media for the cell type.
Add 1 mL cells into each well of a 12-well tissue culture plate and add polybrene to a final concentration of 8 μg/mL per well.
Add different amounts of concentrated virus (usually in the range of 0.5–20 μL for virus generated from GeCKOv2 1 plasmid system, titers are generally higher if using the 2 plasmid system) to each well.
Spinfect by centrifugation at 700 × g for 1–2 h at 32 °C.
Remove media by aspiration and replace with fresh media without polybrene.
Incubate for 6 h or overnight then collect cells by washing gently with 500 μL PBS per well.
Aspirate PBS and add 500 μL Trypsin/EDTA per well and incubate until cells detach. Inactivate by adding 1 mL appropriate media for cell type with serum per well.
Centrifuge the cells at 1000 rpm for 5 min and aspirate media.
Resuspend in cells collected from each well in 10 mL of appropriate media and add 1 mL of cell suspension to duplicate wells in a 6-well tissue culture plate. Add another 3 mL of appropriate media to each well.
In each duplicate, add puromycin to one replicate well at a concentration that results in no surviving cells after 3 days.
After 3 days of puromycin selection count viable cells using CellTiterGlo by aspirating media and adding CellTiterGlo (diluted 1:1 in PBS) to each well.
Cover the plates with foil and shake for 2 min followed by incubation for 10 min at room temperature.
Read luminescence on plate reader and compare with control untreated plate.
Choose the concentration of virus resulting in 20–40% of cell survival compared to untreated replicate well (this corresponds to a MOI of around 0.3–0.4).
Scale up to necessary amount of cells to conduct primary screen by increasing the number of wells spinfected with 2–3 × 106 cells/well.
3.3 Harvesting Genomic DNA
For each time point and replicate in the screen, collect and freeze the amount of cells required to maintain predetermined coverage (6 × 107 cells for 500× coverage of full GeCKOv2 human library) at −20 °C in 15 mL conical tubes with up to 200 mg tissue or 3 × 107 cells per tube.
Thaw cells until the pellet can be dislodged easily by flicking tube, proceed to genomic DNA extraction using salt precipitation (option A) or commercial silica-membrane kits (option B).
3.3.1 Option A
The salt precipitation method used in the screen conducted by the Zhang lab [17] provides high yield genomic DNA with consistent results in sequencing. This option is more cost-effective and simpler when extracting DNA from large amounts of cells or tissues compared to using the commercial silica membrane kits.
For 100–200 mg of tissue or 3 × 107–5 × 107 cells in a 15 mL conical tube, add 6 mL gDNA lysis buffer and 30 μL of proteinase K to tissue/cells.
Incubate at 55 °C overnight.
After complete lysing of cells/tissue, add 30 μL of 10 mg/mL RNase A.
Incubate at 37 °C for 30 min, then cool on ice.
Add 2 mL of cold 7.5 M ammonium acetate and vortex for 20 s.
Centrifuge >4000 × g for 10 min.
Transfer the supernatant to a new tube and add 6 mL 100% isopropanol. Mix by inverting tube 50 times.
Centrifuge >4000 × g for 10 min. Discard the supernatant, add 6 mL of 70% ethanol, and mix by inverting tube ten times.
Centrifuge >4000 × g for 5 min, discard the supernatant by decanting and aspirating with P200 tip.
Air dry 10–30 min until the pellet becomes slightly translucent.
Resuspend in 500 μL of TE.
Incubate at 65 °C for 1 h, then at room temperature overnight.
Measure gDNA concentration on nanodrop 2000 next day.
3.3.2 Option B
Extract gDNA from frozen cell pellets using QIAamp Blood midi kit according to the manufacturer’s protocol.
Elute gDNA in EB or TE buffer and incubate at room temperature overnight.
Measure gDNA concentration on nanodrop 2000 next day.
3.4 PCR NGS Library Prep
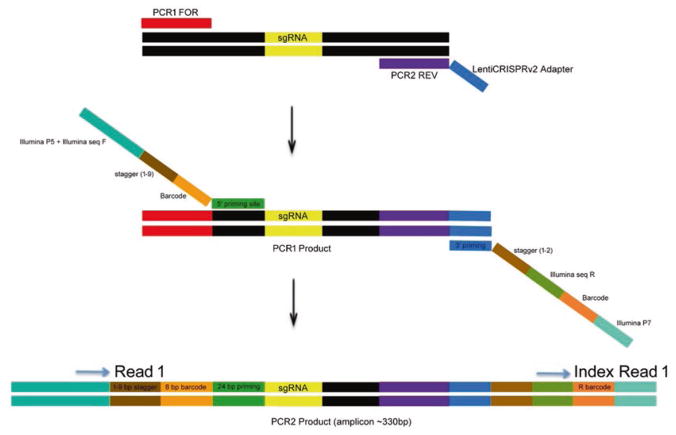
NGS libraries are generated by a two-step PCR where the first PCR amplifies the sgRNA region utilizing primers recognizing constant lentiviral integration sequence and a second PCR adds illumina i5 and i7 sequences as well as barcodes for multiplexing directly in front of the variable 20 bp sgRNA sequences (Fig. 1). sgRNA libraries generated with these primers can then be sequenced on Illumina NextSeq (1 × 75) single indexed read.
Fig. 1.
Schematic representation of two step PCR for the generation of NGS libraries (Subheading 3.4)
3.4.1 PCR1
It is important to maintain library coverage during the library preparation. For the generation of libraries for the amplified plasmid, 10 ng of starting material is more than adequate. For libraries generated from harvested genomic DNA, using the estimation of 7 μg of gDNA per 106 cells, an adequate number of PCR1 reactions will need to be performed. Using Herculase II Fusion DNA Polymerase, we can generally use up to 10 μg of gDNA per 100 μL PCR reaction. Using NEBNext Q5 Hot start HiFi Polymerase, we can generally use up to 5 μg of gDNA per 100 μL PCR reaction. So for maintaining 500× coverage of a full GeCKOv2 library, one would need to perform 40 separate 100 μL PCR1 reactions with Herculase II Fusion DNA Polymerase and 85 separate 100 μL PCR1 reactions with NEBNext Q5 Hot Start HiFi Polymerase.
PCR1 primers can be ordered from IDT at the smallest scale (25 nM, standard desalted) and resuspended in TE for a final concentration of 10 μM.
Prepare 100 μL PCR reactions as follows: (a) For Herculase II Fusion DNA Polymerase: prepare reactions on ice: 20 μL Herculase 5× Buffer; 1 μL dNTP (100 mM, 25 mM each dNTP); 2.5 μL PCR1 F primer (10 μM); 2.5 μL PCR1 R primer (10 μM); 1 μL Herculase II Fusion Enzyme; 10 ng plasmid or up to 10 μg genomic DNA; PCR grade water to 100 μL total volume. (b) For NEBNext Q5 HotStart HiFi PCR Mastermix: 50 μL NEBNext Q5 HotStart HiFi PCR mastermix; 1.5 μL PCR1 F primer (10 μM); 1.5 μL PCR1 R primer (10 μM); 10 ng plasmid or up to 5 μg genomic DNA; PCR grade water to 100 μL total volume.
-
Place the tubes in a thermocycler and run the following programs:
Purify PCR products using QIAquick PCR purification or Wizard SV PCR cleanup. Resuspend each PCR reaction in 100 μL of TE. Pool PCR1 reactions together for each experimental condition and use 10 μL of PCR1 product as a template for PCR2.
3.4.2 PCR2
To maintain library complexity, we will need to perform one PCR2 reaction per 104 constructs in library (so 13 reactions for GeCKOv2 A and B library for each experimental condition) with technical duplicates.
PCR2 primers are longer and more costly than PCR1 primers and can be synthesized by IDT as Ultramer DNA Oligos at 4nmole scale and resuspended in TE to a final concentration of 10 μM.
-
Assemble 50 μL PCR2 reactions using 10 μL of purified pooled PCR1 product as template. Each experimental condition can be barcoded with unique F and R primers for multiplexing on the same NextSeq flowcell.
For Herculase II Fusion DNA Polymerase (prepare reactions on ice): 10 μL Herculase 5× Buffer; 0.5 μL dNTP (100 mM, 25 mM each dNTP); 1.25 μL PCR2 F primer (10 μM) (F01–F10); 1.25 μL PCR2 R primer (10 μM) (R01–R10); 0.5 μL Herculase II Fusion Enzyme; 10 μL PCR1 product; PCR grade water to 50 μL total volume.
For NEBNext Q5 HotStart HiFi PCR Mastermix: 25 μL NEBNext Q5 HotStart HiFi PCR mastermix; 1.5 μL PCR2 F primer (10 μM) (F01–F10); 1.5 μL PCR2 R primer (10 μM) (R01–R10); 10 μL PCR1 product; PCR grade water to 50 μL total volume.
-
Place the tubes in a thermocycler and run the following programs:
Purify PCR products using QIAquick PCR purification or Wizard SV PCR cleanup. Resuspend each PCR reaction in 50 μL of TE and pool products with the same barcode but keeping technical replicates separate.
Run PCR product on 2% E-gel and gel purify approx. 370 bp band.
Quantify by Qubit or bioanalyzer and pool barcoded samples for sequencing on Illumina NextSeq 500.
Acknowledgments
We thank the Rana lab members, John Shimishata, and Steven Head at The Scripps Research Institute Genomic Core. This work was supported in part by grants from the National Institutes of Health to T.M.R., E.Y, is supported by the National Cancer Institute of the National Institutes of Health under award number T32CA121938. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
It is important to use a high-fidelity, GC unbiased polymerase to obtain accurate NGS libraries for quantification. In addition to Herculase II Fusion and Q5, many labs have had success with Kapa HiFi enzyme. Due to its GC bias, we would avoid using Phusion polymerases. We have found Herculase II Fusion is able to tolerate the largest amount of genomic DNA. In order to help with the large amounts of genomic DNA, one can also supplement additional MgCl2 up to a final concentration of 4 mM.
Some optimization of cycle numbers is required for PCR1 and PCR2. Given the quantitative goal of NGS library generation, keeping the PCR reactions in the linear range is desired. For Herculase II Fusion, we have found that around 18–20 cycles of PCR1 and 18–20 cycles of PCR2 is a good place to start. Excessive cycle numbers of PCR2 results in a large MW smear seen on agarose gels. Using Q5 enzyme, even lower cycle numbers can be used with around 15–18 cycles for PCR1 and 12–15 cycles for PCR2.
References
- 1.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome-engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 5.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;4:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sternberg SH, Doudna JA. Expanding the biologist’s toolkit with CRISPR-Cas0. Mol Cell. 2015;58:568–574. doi: 10.1016/j.molcel.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, Mero P, Dirks P, Sidhu S, Roth FP, Rissland OS, Durocher D, Angers S, Moffat J. High-resolution CRISPR screen reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Morgens DW, Deans RM, Li A, Bassik MC. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat Biotechnol. 2016;34:634–636. doi: 10.1038/nbt.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Gulmaraes C, Panning B, Pioegh HL, Bassik MC, Qi LS, Kampmann M, Weissman JS. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virreira Winter S, Zychlinsky A, Bardoel BW. Genome-wide CRISPR screen reveals novel host factors required for Staphylococcus aureus α-hemolysin-mediated toxicity. Sci Rep. 2016;6:24242. doi: 10.1038/srep24242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kampmann M, Bassik MC, Weissman JS. Functional genomics platform for pooled screening and mammalian genetic interaction maps. Nat Protoc. 2014;9:1825–1847. doi: 10.1038/nprot.2014.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, Lee H, Zhang F, Sharp PA. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]