Abstract
Poly(ADP-ribosyl)ation is a common post-translational modification that mediates a wide variety of cellular processes including DNA damage repair, chromatin regulation, transcription, and apoptosis. The difficulty associated with accessing poly(ADP-ribose) (PAR) in a homogeneous form has been an impediment to understanding the interactions of PAR with poly(ADP-ribose) glycohydrolase (PARG) and other binding proteins. Here we describe the chemical synthesis of the ADP-ribose dimer, and we use this compound to obtain the first human PARG substrate-enzyme co-crystal structure. Chemical synthesis of PAR is an attractive alternative to traditional enzymatic synthesis and fractionation, allowing access to products such as dimeric ADP-ribose, which has been detected but never isolated from natural sources. Additionally, we describe the synthesis of an alkynylated dimer and demonstrate that this compound can be used to synthesize PAR probes including biotin and fluorophore-labeled compounds. The fluorescently labeled ADP-ribose dimer was then utilized in a general fluorescence polarization-based PAR-protein binding assay. Finally, we use intermediates of our synthesis to access various PAR fragments and evaluation of these compounds as substrates for PARG reveals the minimal features for substrate recognition and enzymatic cleavage. Homogeneous PAR oligomers and unnatural variants produced from chemical synthesis will allow for further detailed structural and biochemical studies on the interaction of PAR with its many protein binding partners.
Introduction
Poly(ADP-ribosyl)ation is a critically important post-translational modification affecting a wide variety of cellular processes1,2. In cells, poly(ADP-ribose) (PAR) is produced by PAR polymerases (PARPs) from β-NAD+ in response to DNA damage. PAR recruits a number of proteins responsible for DNA repair, and is ultimately processed to ADP-ribose by PAR glycohydrolase (PARG) (Figure 1a). The list of proteins with PAR-binding activity is ever-growing; PAR has been experimentally shown to bind proteins through at least four distinct protein binding domains, and is believed to interact with >500 proteins3–7. The details of PAR-protein interactions on a molecular level, however, are poorly understood, primarily due to the difficulty associated with obtaining PAR in homogeneous and pure form. For use in vitro, PAR can be enzymatically produced by PARP from β-NAD+ to afford polymers ranging from 2-200 units in length (Figure 1a) and subsequently fractionated down to narrow distributions by anion exchange chromatography8,9. While important advances in this process have been made, enzymatic synthesis and fractionation remains challenging, yielding small quantities after repeated ion-exchange purification10,11. Additionally, while in vitro enzymatic syntheses of PAR commonly produces long chain polymers (>50 units)10,11, this distribution is not representative of PAR produced in whole cells and in vivo where shorter oligomers (2-20 units) make up a greater percentage10,12–14. The ADP-ribose dimer (1, Figure 1b) represents the simplest form of PAR that still retains the structural features of the polymer (multiple pyrophosphates and an α-ribosyl linkage between ADP-ribose units), and is produced in only small quantities in vitro15. Though it has been described as an “ideal ligand for x-ray co-crystal structure determination”16 and detected by a variety of analytical methods including MS analysis17 there are no reports where this compound has been isolated from natural sources or enzymatically or chemically synthesized.
Figure 1.
(a) Synthesis and degradation of poly(ADP-Ribose), an important cellular signaling molecule and (b) ADP-ribose dimer (1) and propargyl ADP-ribose dimer (2) and retrosynthetic disconnections
If short PAR oligomers such as the ADP-ribose dimer (1) could be accessed through a modular chemical route, this would afford these valuable compounds in purities and quantities that are not attainable by enzymatic production. Additionally, chemical synthesis could allow for facile modification of PAR oligomers, providing access to unnatural compounds such as the propargyl ADP-ribose dimer (2). Compounds such as 2 could be used in combination with click chemistry to append various biochemical probes enabling further interrogation of PAR biochemistry.
As a target for chemical synthesis, compounds such as 1 and 2 specifically and PAR more generally present several challenges. Construction of the α-glycosidic linkage between the ribofuranose and the adenosine 2’-hydroxyl involves creation of a 1,2-cis-furanoside, a major challenge of synthetic carbohydrate chemistry18. Additionally, nucleosides are traditionally challenging substrates for chemical glycosylation due to the reactivity of the nitrogenous base19,20. While two important chemical syntheses of 2’-O- α -ribosylated adenosine exist20,21, drawbacks of both approaches related to their orthogonality and efficiency inspired us to devise our own alternative route. A second challenge is the presence of the repeating pyrophosphate unit. Methods to construct pyrophosphates are often low-yielding and involve long reaction times22. To overcome these challenges, we envisioned accessing the ADP-ribose dimer (1) and propargyl ADP-ribose dimer (2) from pyrophosphate-forming couplings of protected ribose phosphates 3 and 4, respectively, phosphorylated disaccharide 5, and protected adenosine monophosphate 6 (Figure 1b). Disaccharide 5 would be formed from an α-selective glycosylation of a protected adenosine by an orthogonally protected ribose.
Results
Chemical Synthesis and Enzymatic Evaluation of an ADP-Ribose Dimer
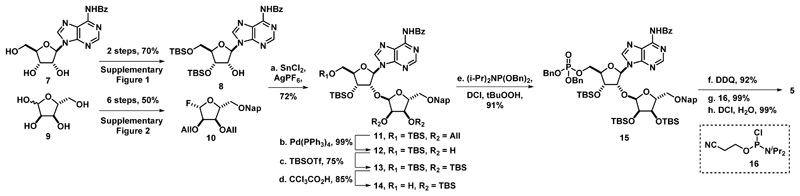
Attention was first turned toward selective glycosylation of adenosine (Scheme 1). To control selectivity, N-benzoyladenosine (7) was silylated in a two step sequence to afford 5’, 3’-silyl isomer 8 in ~15:1 selectivity over the 5’,2’ silylated isomer (Supplementary Figure 1). Next, D-ribose (9) was efficiently converted to β-glycosyl fluoride 10 in a six step sequence involving introduction of the small and non-participating allyl ether protecting groups for the C-2 and C-3 alcohols of ribose, the 2-naphthylmethyl ether group for protection of the C-5 alcohol, and conversion of the C-1 alcohol to the glycosyl fluoride23 to allow for activation under mild conditions (Supplementary Figure 2). Disaccharide 11 was synthesized by activation of glycosyl donor 10 with tin(II) chloride and silver(I) salts in the presence of glycosyl acceptor chlorophosphate which can then condense with phosphate 6 and afford pyrophosphate diester 17 upon deprotection of cyanoethyl group of the intermediate pyrophosphate triester with DBU. Use of this coupling method afforded the product in good yield (72%) and allowed for the reaction to be completed quickly (<1 h). Use of this H-phosphonate coupling allowed for easier and more scalable synthesis of compound 17 than the more commonly used phosphomorphilodate and phosphorimidazolide methods to form pyrophosphate diesters22. Hydrogenolysis of the benzyl groups of pyrophosphate 17 afforded deprotected phosphate 18. Attempts to form the second pyrophosphate with the H-phosphonate method described above were unsuccessful, leading us to synthesize (Supplementary Figure 4) and activate compound 3 as the phosphorimidazolide. Condensation of the phosphorimidazolide of 3 with phosphate 18 allowed for formation of the second pyrophosphate to afford fully protected dimer 19. Global deprotection of compound 19 gave the ADP-ribose dimer (1) in good yield after HPLC purification. To confirm the structure of dimer 1, full 2D NMR spectroscopy data was obtained for compound 1 as well as the preceding intermediates (see supplementary information).
Scheme 1. Synthesis of phosphorylated disaccharide.
(a) SnCl2 (2.2 eq), AgPF6 (2.2 eq), 4 Å MS, CH2Cl2, -78°C to 4 °C, 12 h, 72%, (b) Pd(PPh3)4 (20 mol%), N,N-dimethylbarbituric acid (10 eq), MeOH, 16 h, 99%, (c) TBSOTf (3 eq), DMAP (0.2 eq), EtNiPr2 (6 eq), CH2Cl2, 0°C to rt, 2 h, 75%, (d) CCl3CO2H (46 eq), THF/H2O (1/1), 0°C, 5 h, 85%, (e) (i-Pr)2NP(OBn)2 (1.2 eq), 4,5-dicyanoimidazole (1.2 eq), CH2Cl2/CH3CN (4/1), rt, 2 h, then: tBuOOH (5 eq), 0°C to rt, 3 h, 91% (f) DDQ (1.5 eq), CH2Cl2/H2O (4/1), 0°C, 6 h, 92% (g) Cl(i-Pr)2NPO(CH2)2CN (16) (1.2 eq), EtNiPr2 (1.2 eq), THF, -78°C to rt, 3 h, 99% (h) H2O (3 eq), 4,5-dicyanoimidazole (1.2 eq), CH3CN, rt, 2 h, 99%.
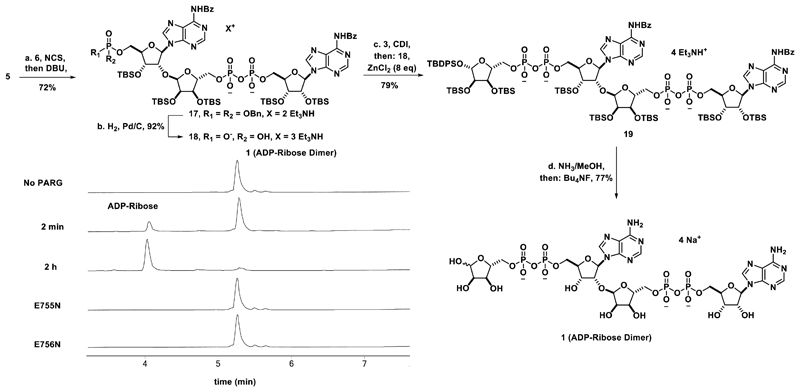
The processing of the ADP-ribose dimer (1) by PARG was next examined (Scheme 2, inset). Treatment of dimer 1 with human PARG at a concentration of 7.5 nM allowed for complete conversion of 1 to ADP-ribose in approximately two hours (Scheme 2 and Supplementary Figure 5) as detected by LC/MS. No processing of 1 was observed by the catalytically inactive E755N and E756N human PARG mutants29, even at 10-fold higher concentrations of enzyme and longer reaction times (4 h) (Scheme 2). Additionally, dimer 1 was shown to be a competent substrate for PARG from other organisms (T. thermophilia, B. taurus, and T. curvata30) as well as for ADP-ribosylhydrolase 3 (ARH3), another enzyme with PAR processing activity31 (Supplementary Figure 6).
Scheme 2. Synthesis of the ADP-ribose dimer (1) and processing by PARG.
(a) N-chlorosuccinimide (4 eq), EtNiPr2 (4 eq), CH3CN, rt, 20 min, then DBU (10 eq), CH3CN, rt, 20 min, then C-18 chromatography and cation exchange, 72%, (b) H2, Pd/C, Et3N (20 eq), tBuOH/H2O, rt, 16 h, 92% (c) 3, N,N-carbonyldiimidazole (10 eq), Et3N (4 eq), Pyridine, rt, 2 h, then: 18, ZnCl2 (8 eq), DMF, rt, 96 h, then: EDTA (Et3NH+ form) and C-18 chromatography, 79%, (d) NH3/MeOH (7M), rt, 20 h, then: Bu4NF (33 eq), THF, rt, 3 h, then: precipitation, C-18 ion pairing chromatography, and cation exchange, 77%. Inset: Processing of 100 μM 1 by 7.5 nM Human PARG shown by HPLC. Compound 1 is converted to ADP-ribose in 2 h by WT PARG while the catalytically inactive mutants (E755N and E756N) do not process 1 at 10-fold higher enzyme concentration at 4 h.
Human PARG Co-Crystal Structure
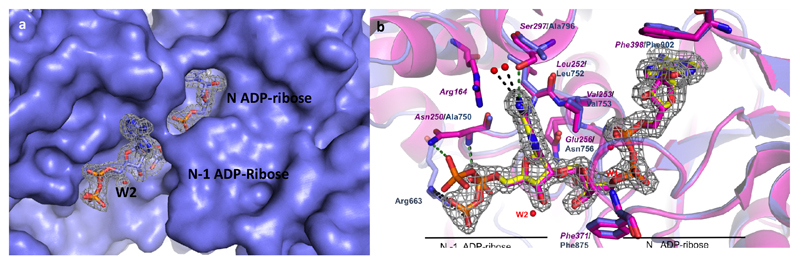
The inability of mutant PARG to process 1 afforded the opportunity to determine the 3D structure of the PARG mutant with this novel substrate. While X-ray structures of human PARG exist29, there are no co-crystal structures of human PARG with a PAR oligomer. Dimer 1 was co-crystallized with a catalytically inactive mutant (E756N) human PARG. The E756N mutant is devoid of PARG enzymatic activity without alteration in the shape of the substrate binding pocket29, making it an excellent candidate for crystallization studies. The complex reported here (Figure 2a) was solved to 1.9 Å resolution and represents the first crystal structure of human PARG in complex with a PAR oligomer of any length.
Figure 2.
(a) The surface representation of human PARG with bound 1 in the active site. The 2Fo-Fc electron density corresponding to the ordered region of dimer 1 is shown in a grey mesh at contour level of 1σ. (b) Overlay of protozoan (4L2H, pink) and human PARG (this work, blue) structures bound to a PAR fragment and dimer 1, respectively. Please note that due to the flexibility observed for the human N-1 β-phosphate group, two different β-phosphate conformations can be assigned, only one has been retained for clarity of the figure. Also, the N-1 ribose moiety is not shown as this portion of the structure is disordered.
In comparison with the published structure of mutant T. thermophila PARG in complex with PAR oligomers (PDB ID: 4L2H, oligomers obtained through enzymatic synthesis and isolation)17, there are similarities but also important differences. In particular, the interactions with the N ADP-ribose unit (Figure 2b) are retained in both structures. However, the enzyme—substrate complex reported herein allows the first view of the ribose-ribose O-glycosidic linkage and the N-1 adenine in the human system, and it is here where differences with the T. thermophila enzyme are observed. Specifically, due to the absence of a residue homologous to Arg164, present in the protozoan enzyme, and due to the protozoan Ser297 being replaced by Ala796 in the human enzyme, the N-1 adenine group in human PARG appears to be significantly more solvent exposed than in its protozoan orthologue and thus makes fewer contacts with the enzyme (Figure 2b). The N-1 adenine group interacts with the amide nitrogen of the conserved Leu752 in human PARG (Leu252 in protozoan PARG), while the β-phosphate forms H-bonds with the conserved Ala750 (Asn250 in protozoan PARG) (Figure 2b). In the human PARG structure the N-1 β-phosphate is positioned differently to its counterpart in protozoan PARG and it forms H-bonds with the side chain of the Arg663 (located in the human PARG accessory domain17), which is conserved in a majority of metazoan PARGs. Both the human and T. thermophila PARG-PAR structures suggest that binding of branched PAR fragments appears to be discouraged due to the steric hindrance around the 2”-OH position of the N-1 ribose.
Synthesis of Derivatives of the ADP-Ribose Dimer and Development of a PAR Binding Assay
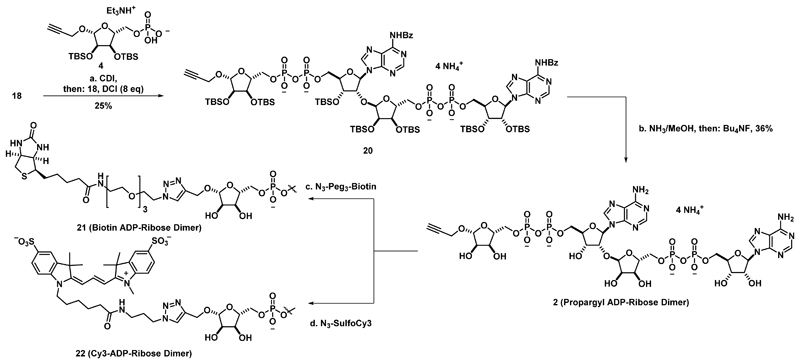
Using modifications of our synthetic route, variants of 1 were constructed to provide tools to interrogate PAR biochemistry. Specifically, analogues amenable to functionalization via bioorthogonal click chemistry were constructed such that these compounds could be functionalized with various probes. Towards this end, alkyne ribose phosphate 4 was synthesized (Supplementary Figure 7), activated as the corresponding phosphorimidazolide, and coupled to phosphate 18 (Scheme 3) to furnish protected propargyl ADP-ribose dimer 20. Global deprotection provided propargyl ADP-ribose dimer 2. This derivative contains a non-reducing sugar and the non-natural β-glycosidic linkage at the N-1 ribose. To demonstrate its utility as a generally taggable PAR derivative, compound 2 was treated with a biotin azide under azide-alkyne Husigen cycloaddition conditions to afford biotinylated ADP-ribose dimer 21. Similar chemistry was performed with compound 2 and sulfo-Cy3 azide to afford Cy3 ADP-ribose dimer 22, a fluorescent PAR derivative.
Scheme 3. Synthesis of ADP-Ribose Dimer Derivatives.
(a) 4, N,N-carbonyldiimidazole (5 eq), Et3N (2 eq), CH3CN, rt, 2 h, then: 18, 4,5-dicyanoimidazole (8 eq), rt, 16 h, 25%. (b) NH3/MeOH (7M), rt, 14 h, then: Bu4NF, THF, rt, 5 h, then: precipitation, C-18 ion pairing chromatography, and cation exchange, 36%. (c) Biotin-Peg3-N3 (2 eq), CuSO4.5H2O (2 eq), Cu wire, H2O, rt, 16 h (d) SulfoCy3-N3, CuSO4.5H2O (0.6 eq), Cu Wire, H2O, rt, 16 h.
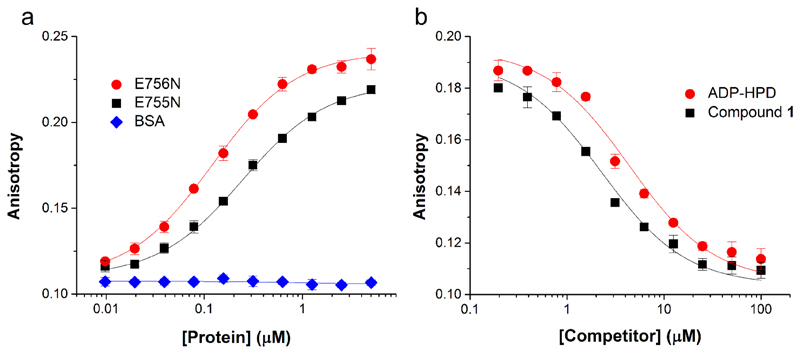
Using fluorescent dimer 22, we sought to develop a general fluorescence polarization-based PAR-protein binding assay. Incubation of compound 22 with E756N and E755N human PARG provided binding curves (Figure 3a) that were used to calculate dissociation constants. Compound 22 binds to E756N and E755N human PARG (Figure 3a) with KD values of 83±7 nM and 208±14 nM, respectively (see supporting information for calculation). Fluorescent dimer 22 does not bind proteins devoid of PAR binding activity such as bovine serum albumin (Figure 3a) even at high concentrations of protein (Supplementary Figure 8a). Binding of compound 22 to E756N PARG can be disrupted by pre-incubation of the protein with compound 1 or the PARG inhibitor ADP-HPD32–35 (Figure 3b). Interestingly, it was found that ADP-HPD has substantially lower affinity for the E755N PARG mutant both relative to compound 1 and relative to its affinity for the E756N mutant (Figure 3b and Supplementary Figure 8b). This data could suggest that the E756N mutant more accurately mimics the wild type enzyme and is a better candidate for crystallography than E755N human PARG.
Figure 3.
Analysis of PAR-Protein Binding by Fluorescence Polarization (a) Binding of E755N (black squares) and E756N (red circles) PARGs but not bovine serum albumin (blue diamonds) to compound 22 (7.5 nM). (b) Compound 22 binding to E756N mutant PARG (250 nM) can be blocked by pretreatment with unlabeled ligand (compound 1) or the PARG inhibitor ADP-HPD.
Synthesis and Evaluation of Fragments of the ADP-Ribose Dimer
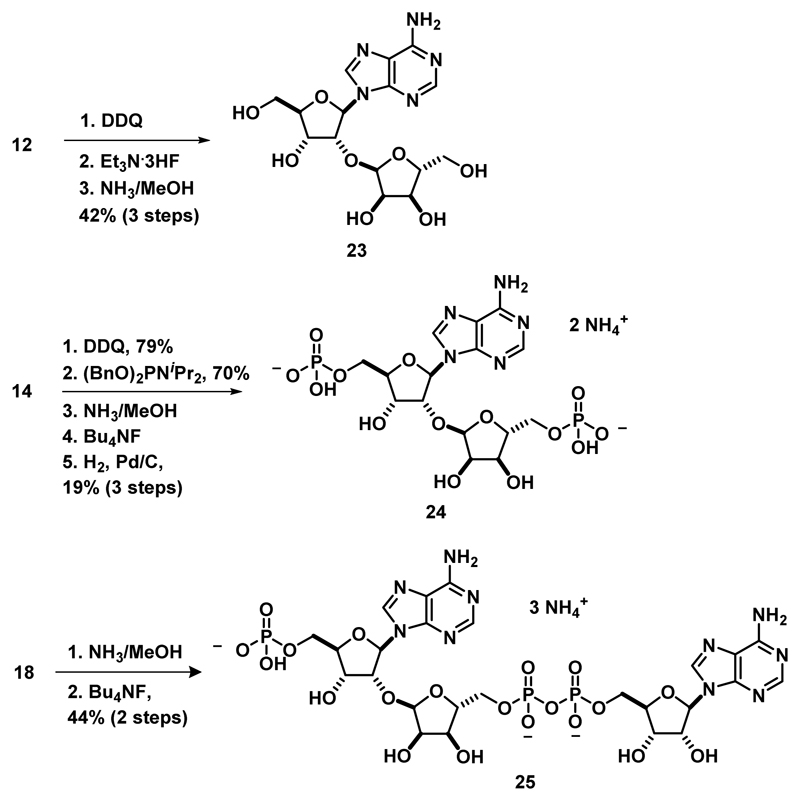
Another benefit of a modular synthesis of ADP-ribose oligomers is the ability to divert key intermediates to create simple fragments of PAR oligomers and use these fragments to investigate PARG substrate specificity. Towards this end, compound 12 was globally deprotected to reveal the fully deprotected disaccharide 2320,21 (Scheme 4). Additionally, the 2-naphthyl methyl group of compound 14 was cleaved selectively and phosphorylated at both primary alcohols to give bisphosphate 24 (Scheme 4, known as iso-ADP-ribose36) upon global deprotection (for a detailed synthetic scheme see Supplementary Figure 9). This represents the first chemical synthesis of this widely used molecule36–39, and we provide the first detailed characterization for this compound including full 2D NMR spectroscopic analysis (see supporting information). Finally, global deprotection of 18 furnished compound 25 (Scheme 4), a fragment retaining the N ADP-ribose unit and N-1 AMP unit, while lacking the N-1 ribose moiety.
Scheme 4. Synthesis of simplified PAR fragments.
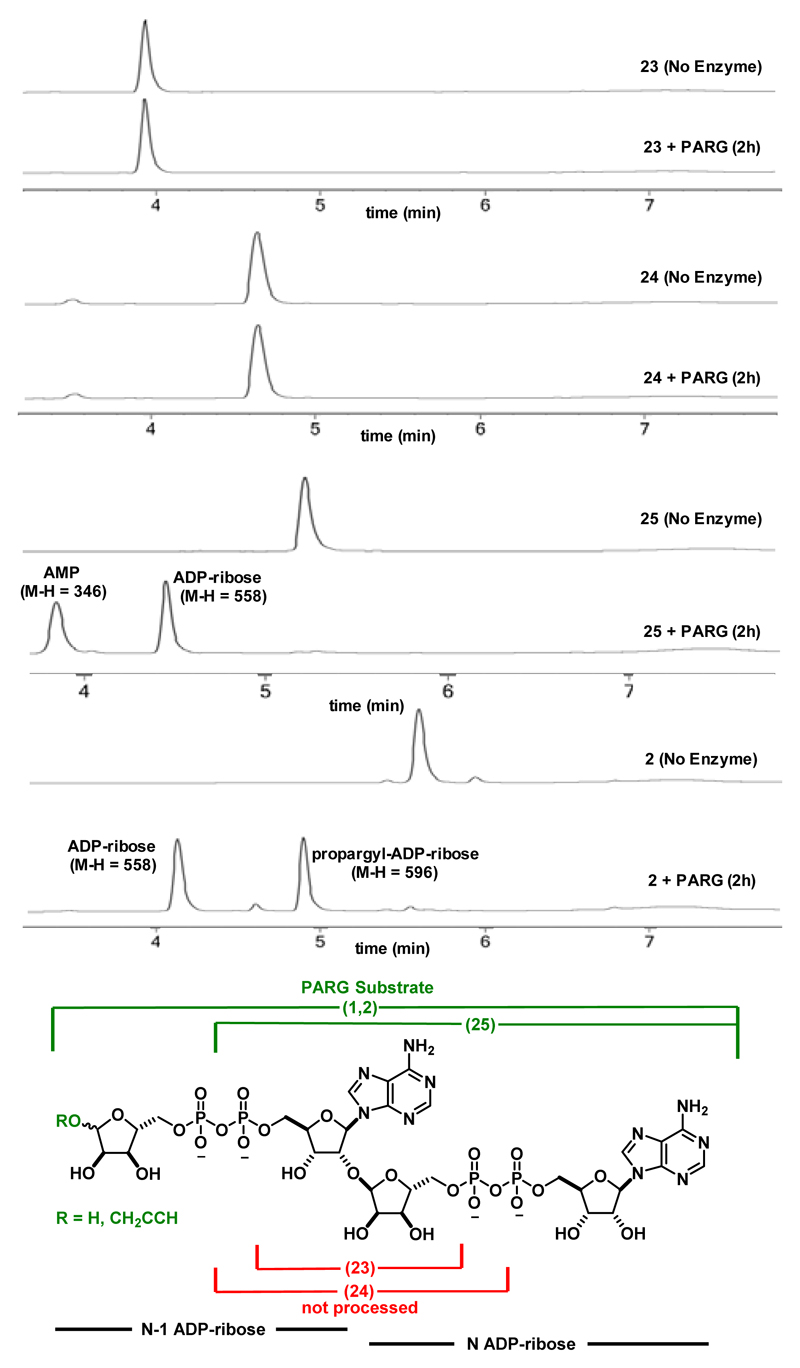
The ability of PARG to process these dimer fragments was next examined (Figure 4). We found that PARG was unable to process the truncated substrates 23 and 24 at 25 nM (as shown in Figure 4), and even at enzyme concentrations of 100 nM no processing was observed (data not shown). Building out from the glycosidic linkage, PARG was able to process compound 25 to adenosine monophosphate and ADP-ribose, indicating that PARG is tolerant to truncated substrates in the N-1 region of the polymer. Finally, in addition to truncations in the N-1 portion of the polymer, unnatural modifications of the natural substrate are tolerated as PARG was able to process the propargyl ADP-ribose dimer (2) to ADP-ribose and propargyl ADP-ribose (Figure 4). Additionally, the α-glycosidic linkage of the propargyl group of compound 2 was not hydrolyzed. That PARG was able to process compounds 1, 2 and 25 with similar efficiency but not compounds 23 and 24 suggests that the N ADP-ribose motif is essential for PAR recognition, binding, and processing but PARG appears more tolerant to substitutions in the N-1 region.
Figure 4.
LC/MS assay of enzymatic cleavage of PAR derivatives by T. thermophilia PARG
Discussion
We present here the first total chemical synthesis of the ADP-ribose dimer 1. This represents the first time that this compound has been obtained and characterized and represents an alternative to accessing PAR oligomers via enzymatic synthesis and fractionation. Key to the synthesis of 1 has been the development of a scalable α-selective glycosylation of adenosine and the development of an efficient H-phosphonate coupling method for pyrophosphate formation. In addition to the chemistry presented here, the H-phosphonate coupling method has found wide use in our laboratory as a fast, high-yielding, and convenient method to form pyrophosphate diesters for a wide variety of substrates. We have found that reactions using this coupling method scale easier than traditional phosphoimidazolide and phosphomorphilodate chemistry as this method does not require the addition and subsequent removal of metal additives such as ZnCl2.
The ability to access compound 1 enabled the first crystal structure of human PARG bound to a cognate substrate. Consistent with the previously reported17 T. thermophilia structure, the N ADP-ribose unit appears to make the most contacts with the protein. We found the N-1 adenine ring in the human structure to be more solvent exposed than in the T. thermophilia structure and found changes to the orientation of the N-1 pyrophosphate moiety due to hydrogen bonds with Arg663 not present in the T. thermophilia structure. Chemical synthesis of PAR oligomers should facilitate crystallography experiments with proteins other than PARG and should enable understanding of PAR-protein interactions on a molecular level.
The modular nature of our synthetic route allows intermediates such as 12, 14, and 18 to be easily diverted to the synthesis of PAR fragments. Experiments with PARG show that the N ADP-ribose unit of PAR is essential for binding and recognition by PARG, but that truncations or unnatural modifications are tolerated in the N-1 unit (summarized in Figure 4). This finding is consistent with the PARG-PAR crystal structures where the N unit is bound most tightly (Figure 2). The somewhat promiscuous nature of PARG demonstrated here could enable the generation of alternative PARG substrates and inhibitors inspired by ADP-ribose.
The synthetic route to the ADP-ribose dimer (1) served as a blueprint for the synthesis of other short PAR oligomers, providing access to unnatural PAR variants including the propargyl ADP-ribose dimer (2). The ability of compound 2 to be used as a generally taggable PAR derivative will enable the synthesis of a variety of biochemical probes through click chemistry, as demonstrated by the synthesis of the biotinylated ADP-ribose dimer (21) and the fluorescent ADP-ribose dimer (22).
Fluorescent ADP-ribose dimer 22 should be a valuable tool for PAR binding assays as well as for high-throughput screening campaigns. The development of PARG inhibitors has been hindered by a lack of convenient methods for high-throughput screening, as traditionally PARG activity is monitored by a radiometric TLC assay40 or immunochemical dot blot assay41,42. The approach described herein provides quantitative binding data and can be performed rapidly in a high-throughput format. Additionally, as fluorescence polarization does not require enzymatic activity as a readout and merely measures binding of compound 22 to various proteins, it can in principle be applied to find inhibitors of PAR-protein interactions for which there is not an immediate enzymatic readout of the interaction.
Supplementary Material
Supporting Information. Supplementary Figures detailing synthesis of compounds 3, 4, 6, 8, 10, and 24 as well as additional PARG processing and fluorescence polarization data. Crystallographic data. Experimental procedures and characterization of chemical products. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank the University of Illinois for supporting this work. M.J.L. is a member of the National Institutes of Health Chemistry–Biology Interface Training Grant (NRSA 1-T-32-GM070421). We are grateful to B. Yestrepsky for synthesis of ADP-HPD, B. Drown and D. Leys for advice and J. Tucker for materials. Work in the I.A. laboratory is supported by the Wellcome Trust and the European Research Council.
References
- (1).Malanga M, Althaus FR. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- (2).Schreiber V, Dantzer F, Amé J-C, de Murcia G. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- (3).Krietsch J, Rouleau M, Pic É, Ethier C, Dawson TM, Dawson VL, Masson J-Y, Poirier GG, Gagné J-P. Mol Aspects Med. 2013;34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- (6).Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- (7).Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, et al. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Andrabi SAS, Kim NSN, Yu S-WS, Wang HH, Koh DWD, Sasaki MM, Klaus JAJ, Otsuka TT, Zhang ZZ, Koehler RCR, Hurn PDP, et al. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Nucleic Acids Res. 2007;35:e143–e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Kiehlbauch CCC, Aboul-Ela NN, Jacobson ELE, Ringer DPD, Jacobson MKM. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- (11).Tan ES, Krukenberg KA, Mitchison TJ. Anal Biochem. 2012;428:126–136. doi: 10.1016/j.ab.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Malanga M, Bachmann S, Panzeter PL, Zweifel B, Althaus FR. Anal Biochem. 1995;228:245–251. doi: 10.1006/abio.1995.1346. [DOI] [PubMed] [Google Scholar]
- (13).Kleczkowska HE, Malanga M, Szumiel I, Althaus FR. Int J Radiat Biol. 2002;78:527–534. doi: 10.1080/095530002317577349. [DOI] [PubMed] [Google Scholar]
- (14).Malanga M, Althaus FR. J Biol Chem. 1994;269:17691–17696. [PubMed] [Google Scholar]
- (15).Popp O, Veith S, Fahrer J, Bohr VA, Bürkle A, Mangerich A. ACS Chem Biol. 2013;8:179–188. doi: 10.1021/cb300363g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Oberoi J, Richards MW, Crumpler S, Brown N, Blagg J, Bayliss R. J Biol Chem. 2010;285:39348–39358. doi: 10.1074/jbc.M110.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Barkauskaite E, Brassington A, Tan ES, Warwicker J, Dunstan MS, Banos B, Lafite P, Ahel M, Mitchison TJ, Ahel I, Leys D. Nat Commun. 2013;4:1–8. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Demchenko AV. Handbook of Chemical Glycosylation. John Wiley & Sons; 2008. [Google Scholar]
- (19).Knapp S, Gore VK. J Org Chem. 1996;61:6744–6747. doi: 10.1021/jo960727z. [DOI] [PubMed] [Google Scholar]
- (20).Mikhailov SN, Kulikova IV, Nauwelaerts K, Herdewijn P. Tetrahedron. 2008;64:2871–2876. [Google Scholar]
- (21).van der Heden van Noort GJ, Overkleeft HS, van der Marel GA, Filippov DV. Org Lett. 2011;13:2920–2923. doi: 10.1021/ol200971z. [DOI] [PubMed] [Google Scholar]
- (22).Wagner GK, Pesnot T, Field RA. Nat Prod Rep. 2009;26:1172–1194. doi: 10.1039/b909621n. [DOI] [PubMed] [Google Scholar]
- (23).Mukaiyama T, Murai Y, Shoda S. Chemistry Letters. 1981;10:431–432. [Google Scholar]
- (24).Lear MJ, Yoshimura F, Hirama M. Angew Chem Int Ed. 2001;40:946–949. [PubMed] [Google Scholar]
- (25).Larsen CH, Ridgway BH, Shaw JT, Smith DM, Woerpel KA. J Am Chem Soc. 2005;127:10879–10884. doi: 10.1021/ja0524043. [DOI] [PubMed] [Google Scholar]
- (26).van Rijssel ER, van Delft P, Lodder G, Overkleeft HS, van der Marel GA, Filippov DV, Codée JDC. Angew Chem Int Ed. 2014;53:10381–10385. doi: 10.1002/anie.201405477. [DOI] [PubMed] [Google Scholar]
- (27).Atherton FR, Openshaw HT, Todd AR. J Chem Soc. 1945:660. [Google Scholar]
- (28).Le Corre SS, Berchel M, Couthon-Gourvès H, Haelters J-P, Jaffrès P-A. Beilstein J Org Chem. 2014;10:1166–1196. doi: 10.3762/bjoc.10.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Tucker JA, Bennett N, Brassington C, Durant ST, Hassall G, Holdgate G, McAlister M, Nissink JWM, Truman C, Watson M. PLoS ONE. 2012;7:e50889. doi: 10.1371/journal.pone.0050889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Slade D, Dunstan MS, Barkauskaite E, Weston R, Lafite P, Dixon N, Ahel M, Leys D, Ahel I. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Niere M, Mashimo M, Agledal L, Dölle C, Kasamatsu A, Kato J, Moss J, Ziegler M. J Biol Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Slama JT, Aboul-Ela N, Jacobson MK. J Med Chem. 1995;38:4332–4336. doi: 10.1021/jm00021a023. [DOI] [PubMed] [Google Scholar]
- (33).Slama JT, Aboul-Ela N, Goli DM, Cheesman BV, Simmons AM, Jacobson MK. J Med Chem. 1995;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- (34).Koh DW, Coyle DL, Mehta N, Ramsinghani S, Kim H, Slama JT, Jacobson MK. J Med Chem. 2003;46:4322–4332. doi: 10.1021/jm020541u. [DOI] [PubMed] [Google Scholar]
- (35).Patel C, Koh DW, Jacobson MK, Oliveira MA. Biochem J. 2005;388:493–500. doi: 10.1042/BJ20040942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wang Z, Michaud GA, Cheng Z, Zhang Y, Hinds TR, Fan E, Cong F, Xu W. Genes Dev. 2012;26:235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wang Z, Gagné J-P, Poirier GG, Xu W. PLoS ONE. 2014;9:e86010. doi: 10.1371/journal.pone.0086010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Zhang F, Chen Y, Li M, Yu X. Proc Natl Acad Sci USA. 2014;111:7278–7283. doi: 10.1073/pnas.1318367111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).DaRosa PA, Wang Z, Jiang X, Pruneda JN, Cong F, Klevit RE, Xu W. Nature. 2015;517:223–226. doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Ménard L, Poirier GG. Biochem Cell Biol. 1987;65:668–673. doi: 10.1139/o87-088. [DOI] [PubMed] [Google Scholar]
- (41).Bachir Affar El, Duriez PJ, Shah RG, Sallmann FR, Bourassa S, Kupper J-H, Bürkle A, Poirier GG. Anal Biochem. 1998;259:280–283. doi: 10.1006/abio.1998.2664. [DOI] [PubMed] [Google Scholar]
- (42).Okita N, Ashizawa D, Ohta R, Abe H, Tanuma S-I. Biochem Biophys Res Commun. 2010;392:485–489. doi: 10.1016/j.bbrc.2010.01.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.