Abstract
Malaria parasites replicate within a parasitophorous vacuole (PV) in red blood cells (RBC). Progeny merozoites egress upon rupture of first the PV membrane (PVM) then poration and rupture of the RBC membrane (RBCM). Egress is protease-dependent1, but none of the effector molecules that mediate membrane rupture has been identified and it is unknown how sequential rupture of the two membranes is controlled. Minutes before egress, the parasite serine protease SUB1 is discharged into the PV2–6 where it cleaves multiple substrates2,5,7–9 including SERA6, a putative cysteine protease10–12. Here we show that Plasmodium falciparum parasites lacking SUB1 undergo none of the morphological transformations that precede egress and fail to rupture the PVM. In contrast, PVM rupture and RBCM poration occur normally in SERA6-null parasites but RBCM rupture does not occur. Complementation studies show that SERA6 is an enzyme that requires processing by SUB1 for its function. RBCM rupture is associated with SERA6-dependent proteolytic cleavage within the actin-binding domain of the major RBC cytoskeletal protein β-spectrin. We conclude that SUB1 and SERA6 play distinct, essential roles in a coordinated proteolytic cascade that enables sequential rupture of the two bounding membranes and culminates in RBCM disruption through rapid, precise, SERA6-mediated disassembly of the RBC cytoskeleton.
Malaria, caused by parasitic protozoa of the genus Plasmodium, causes over 400,000 deaths per annum. With widespread resistance to most antimalarial drugs, there is a need to better understand the biology of the parasite, especially the species responsible for most fatalities, Plasmodium falciparum. Asexual blood stage parasites proliferate within RBC. Following each cycle of intracellular development (lasting ˜48 h in P. falciparum) the PVM and RBCM rupture to allow egress of merozoites which invade fresh RBC.
Egress comprises several rapidly successive steps. Following biogenesis of intracellular merozoites by segmentation of the mature schizont, the PVM becomes permeable, allowing mixing of the contents of the PV and residual RBC cytoplasm6. Within the ensuing minutes, the parasite cGMP-dependent protein kinase PKG is activated to trigger discharge of SUB1, a subtilisin-like protease, from merozoite secretory organelles called exonemes3,4. In the PV lumen, SUB1 proteolytically modifies several merozoite surface and PV proteins2,5,7,8, including SERA6, which is cleaved to release a central domain with homology to papain-like (clan CA, family C1) cysteine peptidases10. Within ˜10 minutes of SUB1 discharge, the PV abruptly swells whilst the entire infected RBC transforms from an irregular to a roughly spherical ‘flower’ or rounded-up structure13,14. The PVM then fragments into multilamellar vesicles, closely followed by collapse and poration (permeabilisation) of the RBCM6,14,15. Within seconds the RBCM ruptures, allowing merozoite release16. Inhibitors of PKG block SUB1 discharge and all stages of egress subsequent to the initial PVM permeabilisation step3,5,6,17. In contrast, the broad-spectrum cysteine protease inhibitor epoxysuccinyl-L-leucylamido(4-guanidino)butane (E64), which does not inhibit SUB1 activity3, prevents neither PVM rupture nor RBCM collapse and poration, but blocks the final step of RBCM rupture6,14,15,18.
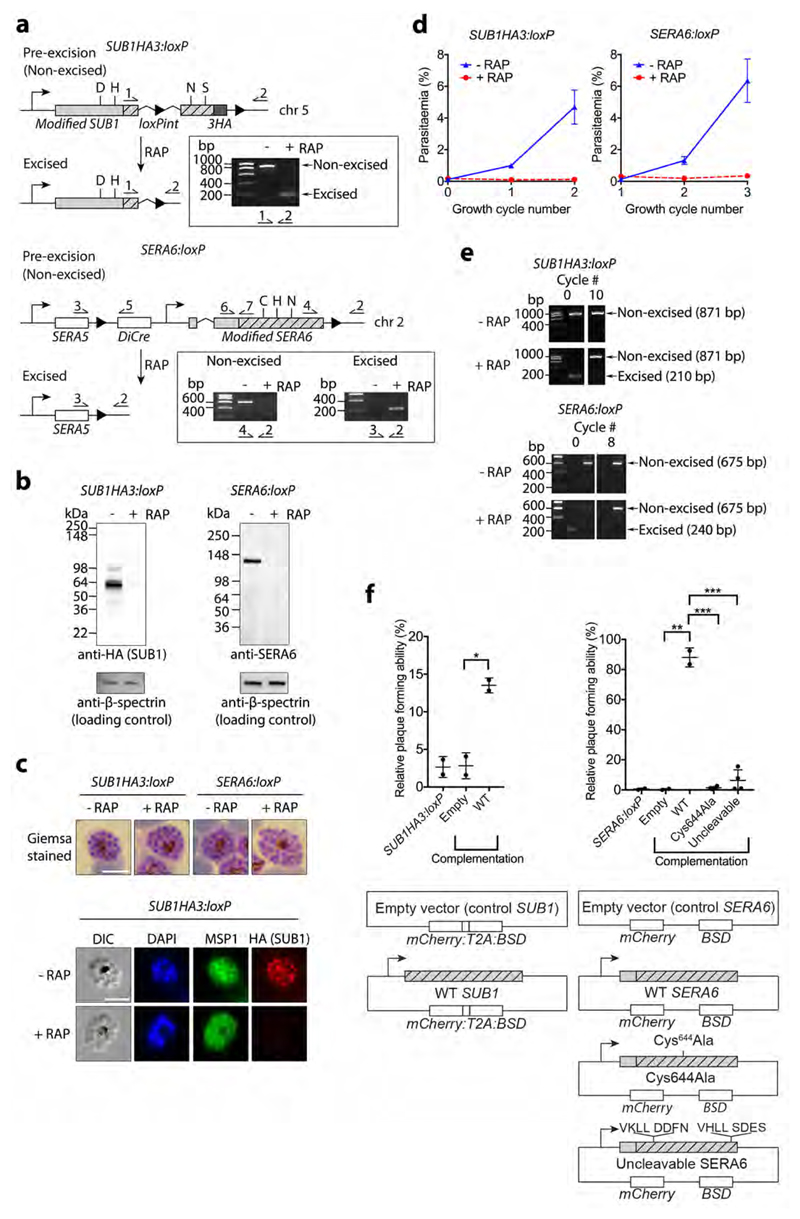
We used the rapamycin (RAP)-inducible dimerizable Cre recombinase (DiCre) system19,20 to conditionally excise either a segment of the SUB1 gene encoding crucial catalytic residues, or the entire SERA6 coding sequence (Fig. 1a). In each case, PCR (Fig. 1a) and Western blot (Fig. 1b and Supplementary Fig. 1) demonstrated rapid and efficient RAP-induced excision of the floxed DNA sequences and ablation of SUB1 or SERA6 expression. Immunofluorescence analysis (IFA) confirmed loss of SUB1 in 99.8% of schizonts (of 5,056 examined) by the end of the erythrocytic cycle (cycle 0) in which the parasites were RAP-treated (Fig. 1c). Both SUB1-null (ΔSUB1) and ΔSERA6 parasites formed morphologically normal schizonts at the end of cycle 0, showing that neither gene is required for intracellular development (Fig. 1c). However, over the ensuing erythrocytic cycles there was a dramatic reduction in replication rates of the RAP-treated cultures (Fig. 1d). Monitoring over 8-10 erythrocytic cycles showed that the initially minor population of non-excised parasites gradually overgrew these cultures whilst the ΔSUB1 or ΔSERA6 parasites disappeared (Fig. 1e), indicating a severe defect. To further assess the impact of gene disruption we used a plaque assay12 which captures successive rounds of replication by individual parasite clones. Substantial reductions in plaque numbers were observed in RAP-treated cultures (Fig. 1f and reference 12), and the few plaques generated were found to arise from the small population of non-excised parasites (Supplementary Fig. 2 and reference 12). These results suggested that both the SUB1 and SERA6 genes are required for in vitro parasite growth.
Figure 1. SUB1 and SERA6 are essential for asexual blood stage P. falciparum growth.
a, Architecture of floxed loci in SUB1HA3:loxP and SERA6:loxP parasites. Introduced loxP sites (arrowheads), recodonised sequence (hatched), HA3 epitope and known (SUB1) or predicted (SERA6) catalytic residues are indicated. Outcomes of rapamycin (RAP)-induced DiCre-mediated excision and positions of primers (half arrows) used for diagnostic PCR are indicated (see Supplementary Table 1 for primer sequences). Insets, PCR (representative of 4 independent experiments) confirming efficient gene excision by the end of cycle 0, ˜44 h following mock-treatment (-RAP) or RAP-treatment (+RAP) of ‘ring’-stage parasites. b, Western blots (representative of 2 independent experiments) showing ablation of SUB1 and SERA6 expression in cycle 0 schizonts. c, Light microscopic and IFA images of mature cycle 0 schizonts, showing normal parasite development and RAP-induced loss of SUB1HA3 expression (representative of 6 independent experiments). Loss of SERA6 expression could not be similarly confirmed by IFA due to C-terminal tagging of SERA6 being unsuccessful and the lack of suitable SERA6-specific antibodies. Scale bar, 5 μm. DAPI, 4,6-diamidino-2-phenylindole. d, Replication of mock- and RAP-treated SUB1HA3:loxP and SERA6:loxP parasites over 2 erythrocytic cycles. Parasitaemia values (quantified by FACS) are averages from 2 biological replicates in different blood sources. Error bars, ±SD. e, PCR showing loss of ΔSUB1 (1 experiment) and ΔSERA6 (representative of 2 independent experiments) parasites and outgrowth of non-excised parasites upon extended passage of RAP-treated cultures. f, Dot plots showing relative plaque forming ability (ratio of plaque numbers produced by RAP-treated cultures to those produced by mock-treated cultures, x100) of SUB1HA3:loxP and SERA6:loxP parasites without or following transfection with the indicated episomal expression plasmids. Statistical significance was determined by two-tailed t-test: SUB1HA3:loxPint: Empty vs WT complementation (t=7.702, d.f.=2, p=0.0164, 95% CI 4.636 to 16.73) n=2. SERA6:loxP: Empty vs WT complementation (t=19.65, d.f.=2, p=0.0026, 95% CI 68.39 to 107) n=2; WT vs Cys644Ala complementation (t=30.96, d.f.=4, p<0.0001, 95% CI -94.44 to -78.81) n=3; WT vs Uncleavable complementation (t=13.8, d.f.=4, p=0.0002, 95% CI -98.14 to -65.25) n=3. In all plots, central horizontal bar, mean. Error bars, ±SD. Significance levels: p≤0.001, ***; p≤0.01, **; p≤0.05, *.
To confirm that loss of viability was a consequence of gene disruption, plasmids for episomal expression of wild-type (WT) SUB1 or SERA6 transgenes were introduced into the (non-RAP-treated) SUB1HA3:loxP or SERA6:loxP parasites respectively. The resulting lines were RAP-treated to disrupt the chromosomal genes, then immediately analysed by plaque assay in comparison with RAP-treated control lines harbouring ‘empty’ plasmid. As shown in Fig. 1f, lines carrying episomal WT SUB1 or SERA6 transgenes produced significantly more plaques following disruption of the chromosomal genes than similarly-treated parasites harbouring empty plasmid. Parasites expanded from plaques produced by RAP-treated parasites carrying the episomal SUB1 or SERA6 transgenes had lost the respective chromosomal gene as expected and so were likely relying solely on the episomal gene copies (Supplementary Fig. 3). Crucially, the ΔSERA6 growth defect could not be rescued by a mutant SERA6 transgene possessing an Ala substitution of the predicted catalytic Cys644 codon (expected to ablate enzyme activity10) (Fig. 1f). Similarly, the ΔSERA6 defect was not complemented by a SERA6 mutant in which the SUB1 processing sites flanking the papain-like domain were modified by Leu substitutions of the P1 and P2 residues upstream of each scissile bond; these mutations prevent SUB1-mediated cleavage10 (Fig. 1f). Collectively, these findings confirmed that SUB1 and SERA6 are indispensable and indicated that SERA6 is an enzyme that requires proteolytic processing by SUB1 to perform its function.
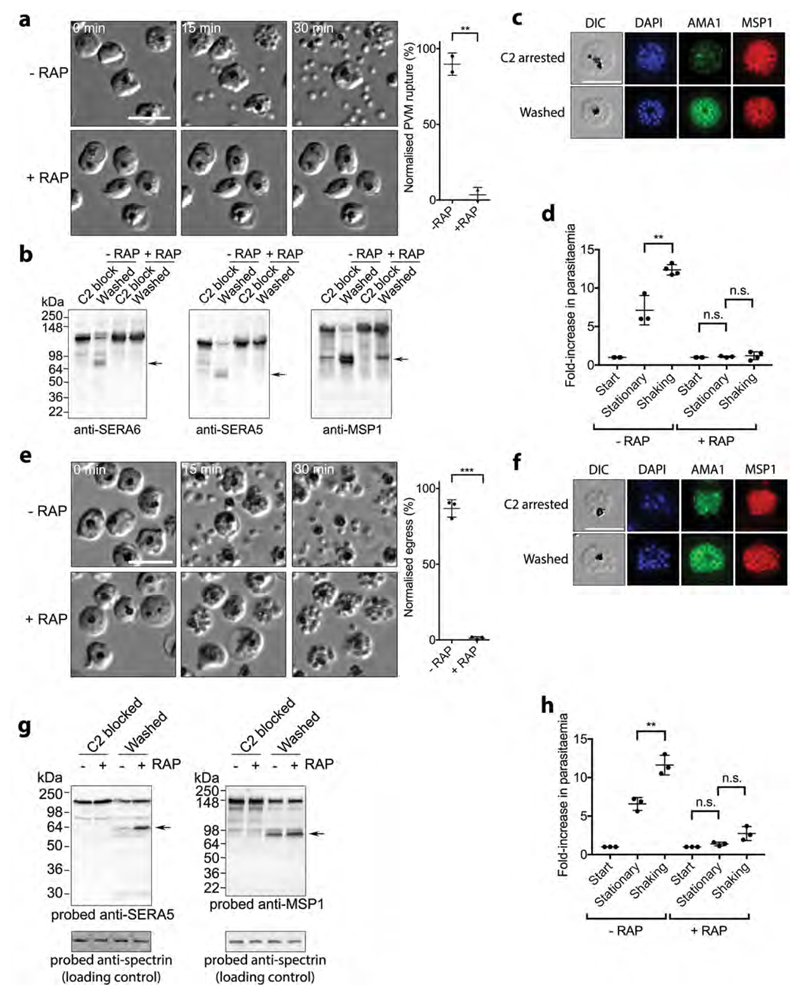
To examine egress of ΔSUB1 and ΔSERA6 parasites, mature schizonts were incubated with the PKG inhibitor (4-[7-[(dimethylamino)methyl]-2-(4-fluorphenyl)imidazo[1,2-α]pyridine-3-yl]pyrimidin-2-amine (compound 2; C2), which reversibly blocks development just prior to egress, resulting in accumulation of ‘stalled’ segmented schizonts. Wash-out of the inhibitor allows rapid progress to rupture, enabling live microscopic examination of multiple egress events over the ensuing ˜10-30 minutes3,5,9. As shown in Fig. 2a and Supplementary Movie 1, ΔSUB1 parasites underwent none of the morphological changes associated with egress, with no signs of PVM swelling, rounding up, apparent PVM rupture (as indicated by loss of differential interference contrast and increased mobility of the intracellular merozoites), or RBCM rupture. Indeed, the ΔSUB1 phenotype was indistinguishable from that produced by continued incubation with C2, suggesting that SUB1 is required for all the egress-related transformations that follow PKG activation. This egress defect was completely rescued by the WT SUB1 transgene (Supplementary Movie 2). As expected, proteolytic processing of the SUB1 substrates SERA52, SERA6 and merozoite surface protein-1 (MSP1)5,7 was ablated in the ΔSUB1 parasites (Fig. 2b). However, IFA using antibodies to AMA1, a protein released onto the merozoite surface from micronemes (which are distinct from the exonemes in which SUB1 is stored2,3) showed that microneme discharge occurred in the trapped ΔSUB1 merozoites (Fig. 2c). Like SUB1 discharge, AMA1 discharge is blocked by PKG inhibitors3, so this confirmed reversal of the C2-mediated PKG inhibition in these experiments.
Figure 2. SUB1 and SERA6 play distinct, sequential roles at egress.
a, Left, stills from time-lapse DIC microscopic examination of control (-RAP) and RAP-treated (ΔSUB1) SUB1HA3:loxP schizonts following removal of C2; elapsed time indicated. Scale bar, 20 μm. Right, quantitation of PVM rupture in control and RAP-treated SUB1HA3:loxP schizont populations, collated from 5 videos of each from 2 independent experiments (total number of observed PVM rupture events in control parasites, 226). PVM rupture is normalised to that in the controls (100% egress). Statistical significance determined by two-tailed t-test: -RAP vs +RAP (t=13.84, d.f.=2, p=0.0052, 95% CI -113.2 to -59.5); p≤0.01, **. b, Processing of SUB1 substrates is ablated in ΔSUB1 parasites. Western blot of C2-blocked SUB1HA3:loxP schizonts, or 30 min after washing away C2. Processed forms of SUB1 substrates are arrowed. c, Microneme discharge in ΔSUB1 parasites. IFA of C2-arrested parasites compared to 30 min after washing away C2. Translocation of AMA1 to the intracellular merozoite surface is evident in the washed parasites. Scale bar, 10 μm. d, Invasion by control and RAP-treated SUB1HA3:loxP parasites under static and shaking conditions. Statistical significance by two-tailed t-test: -RAP stationary vs -RAP shaking (t=5.233, d.f.=5, p=0.0034, 95% CI 2.666 to 7.813) n=4; +RAP start vs +RAP stationary (t=1.722, d.f.=5, p=0.1456, 95% CI -0.04104 to 0.2077) n=4; +RAP stationary vs +RAP shaking (t=0.4585, d.f.=5, p=0.6658, 95% CI -0.641 to 0.9193) n=4. Results shown are from 4 biological replicate experiments (some dots are overlaid). e, Left, time-lapse DIC microscopic stills of control and RAP-treated SERA6:loxP schizonts following C2 removal. Scale bar, 20 μm. Right, quantitation of RBCM rupture. Data collated from 8 videos each of control and RAP-treated parasites, from 3 independent experiments (total number of observed rupture events in control parasites, 568). RBCM rupture is normalised to that in the controls (100% egress). Statistical significance by two-tailed t-test: -RAP vs +RAP (t=25.39, d.f.=4, p<0.0001, 95% CI -95.07 to -76.33) n = 3; p≤0.001, ***. f, Microneme discharge in arrested ΔSERA6 parasites. IFA of C2-arrested parasites compared with 30 min after washing away C2. g, Disruption of the SERA6 gene has no effect on processing of SUB1 substrates. h, Invasion efficiencies of the ΔSERA6 parasites under static and shaking conditions. Statistical significance by two-tailed t-test: -RAP stationary vs -RAP shaking (t=5.674, d.f.=4, p=0.0048, 95% CI 2.57 to 7.496) n=3; +RAP start vs +RAP stationary (t=2.741, d.f.=4, p=0.0518, 95% CI -0.004807 to 0.7581) n=3; +RAP stationary vs +RAP shaking (t=2.526, d.f.=4, p=0.0649, 95% CI -0.1348 to 2.855) n=3; p≤0.01, **. Results shown are from 3 biological replicate experiments. In all plots, central bar, mean. Error bars, ±SD. Experiments in panels b, c, f and g were repeated twice, with reproducible results.
P. falciparum is generally maintained in static culture. To address whether invasion-competent merozoites could be liberated by fluid shear stress, SUB1HA3:loxP schizonts were cultured overnight with fresh RBC under vigorously shaking conditions. As expected21, shaking enhanced increases in parasitaemia in mock-treated SUB1HA3:loxP cultures (Fig. 2d), likely due to more efficient merozoite dissemination and RBC invasion. However, shaking had no impact on the low proliferation rate of RAP-treated (ΔSUB1) parasites, indicating that SUB1 is essential for release of invasive merozoites.
In contrast to the ΔSUB1 phenotype, ΔSERA6 schizonts displayed normal rounding up as well as the increases in merozoite visibility and mobility thought to indicate PVM rupture; however, RBCM rupture did not occur (Fig. 2e and Supplementary Movie 3). Introduction of the complementing WT SERA6 expression plasmid restored egress (Supplementary Movie 4). IFA confirmed microneme discharge in theΔSERA6 schizonts (Fig. 2f) whilst Western blot revealed normal SUB1 activity (Fig. 2g). Similar to the ΔSUB1 parasites, shaking did not enhance the replicative capacity of ΔSERA6 parasites (Fig. 2h).
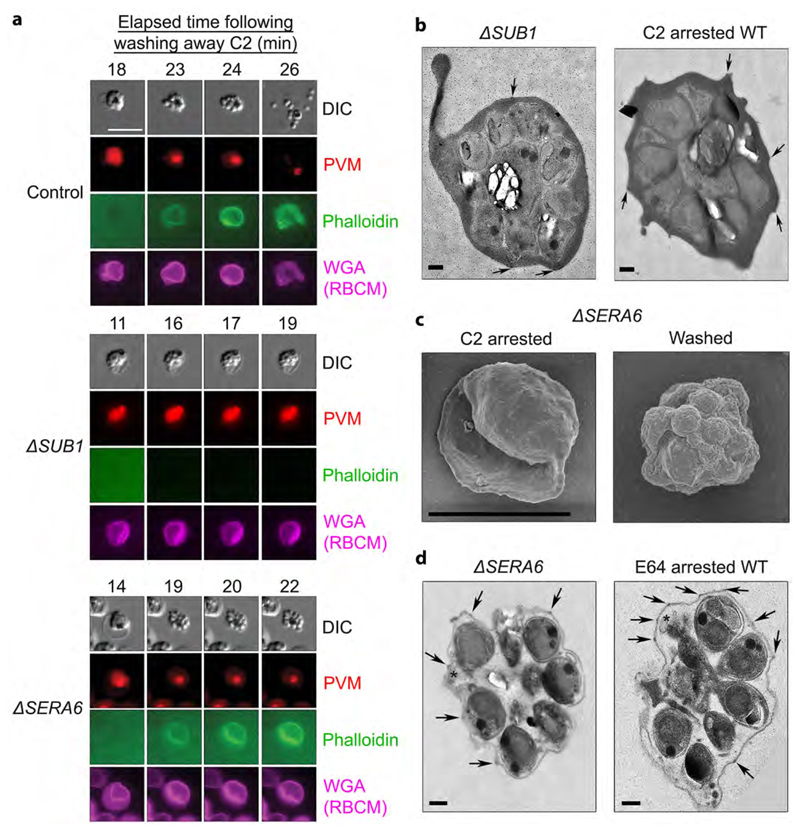
Upon PVM lysis, but just prior to RBCM rupture, the RBCM suddenly becomes permeable to the F-actin binding peptide phalloidin9,14,22. To definitively establish the fate of the PVM in the mutants and to examine the timing and efficiency of RBCM poration, SUB1HA3:loxP and SERA6:loxP parasites were transfected prior to RAP-treatment with a plasmid for constitutive expression of the PVM protein EXP123 fused to mCherry, fluorescently labelling the PVM (Supplementary Fig. 4). Simultaneous DIC and fluorescence video-microscopy in the presence of fluorescent wheat germ agglutinin (which labels the RBCM) and fluorescent phalloidin, confirmed that neither PVM rupture nor RBCM poration took place in ΔSUB1 parasites (Fig. 3a and Supplementary Movie 5). In contrast, RBCM poration occurred normally in the ΔSERA6 parasites upon PVM rupture. Examination of the arrested ΔSUB1 parasites by transmission electron microscopy (TEM) confirmed an intact PVM and RBCM indistinguishable from C2-arrested schizonts (Fig. 3b and Supplementary Fig. 5). In contrast, scanning EM (Fig. 3c) and TEM (Fig. 3d and Supplementary Fig. 5) of arrested ΔSERA6 parasites revealed merozoites and PVM fragments within an intact but collapsed and evacuated RBCM, as previously observed in WT parasites arrested by E646.
Figure 3. SUB1 is required for PVM disruption and RBCM poration, whereas the ΔSERA6 phenotype mimics egress arrest with the cysteine protease inhibitor E64.
a, Stills from simultaneous time-lapse DIC and fluorescence microscopic examination of typical control WT, ΔSUB1 and ΔSERA6 schizonts at the indicated intervals following removal of the egress inhibitor C2. PVM rupture and RBCM poration (indicated by access of phalloidin to the host RBC cytoskeleton) occurs in the ΔSERA6 parasites but not in the ΔSUB1 parasites, whilst RBCM rupture occurs in neither mutant. Scale bar, 10 μm. b, TEM micrographs of an arrested ΔSUB1 schizont and a C2-arrested control cell, showing that the trapped merozoites are surrounded in both cases by an intact PVM and RBCM. Knob structures characteristic of the parasite-infected RBCM6 are indicated on its outer surface (arrow heads). The black dots are gold fiducials added for tomography. Scale bar, 500 nm. c, SEM images of ΔSERA6 schizonts before and 30 min following C2 removal, showing collapse of the RBCM around the intracellular merozoites in the washed sample. Scale bar, 5 μm. d, TEM micrographs of an arrested ΔSERA6 schizont and an E64-arrested control cell, showing in both remnants of ruptured PVM (asterisks) adjacent to the trapped merozoites. Knobs are highlighted as above (arrow heads). Scale bar, 500 nm. All experiments were repeated twice, with reproducible results.
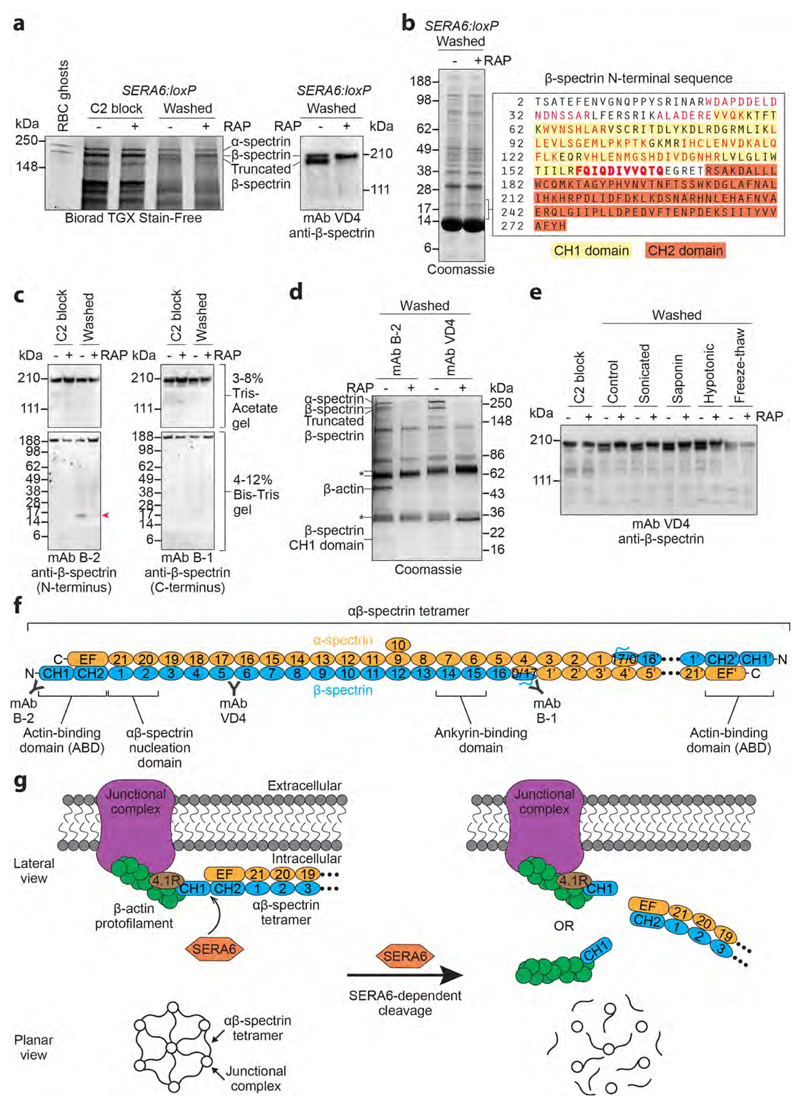
Our observation that egress proceeded normally in the ΔSERA6 parasites up to the point of RBCM rupture suggested that SERA6 mediates RBCM breakdown. To test this model and further dissect the ΔSERA6 defect, we performed a proteomic comparison of mock- and RAP-treated SERA6:loxP schizonts. SDS PAGE (Fig. 4a) detected a high molecular mass species that appeared within 20 minutes in soluble fractions of mock-treated parasites allowed to undergo egress, identified as a truncated form of the major RBC cytoskeleton protein β-spectrin. Further quantitative tandem mass spectrometry (LC-MS/MS) analysis revealed the concomitant appearance of one or more lower molecular mass polypeptides comprising the N-terminal calponin homology (CH) domain of the β-spectrin actin-binding domain (ABD) (Fig. 4b and Supplementary Fig. 6). This suggested that RBCM rupture is associated with proteolytic cleavage of β-spectrin near its N-terminus and release of the cleavage products from the cytoskeleton (which is generally insoluble in aqueous buffers). Western blot (Fig. 4c) and pull-downs using spectrin-specific monoclonal antibodies (mAbs) combined with LC-MS/MS (Fig. 4d and Supplementary Fig. 7) confirmed and extended this, showing that RBCM rupture is accompanied by extensive SERA6-dependent cleavage of β-spectrin at two closely-spaced sites (Gln167-Glu168 and Gln165-Thr166) between the CH1 and CH2 domains of the ABD, releasing the CH1 domain as a ˜17 kDa fragment and resulting in solubilisation of the truncated β-spectrin along with some α-spectrin. The released CH1 domain co-purified with human β-actin, likely also derived from the RBC cytoskeleton (Fig. 4d and Supplementary Fig. 8). No proteolytic cleavage of the other major RBC cytoskeletal components α-spectrin and protein 4.1R was detectable, although limited SERA6-dependent cleavage of ankyrin was evident (Supplementary Fig. 9). Strikingly, β-spectrin cleavage was never observed in ΔSERA6 parasites, even upon mechanical, hypotonic, freeze-thaw or detergent-mediated rupture of the schizonts (Fig. 4e), showing that cleavage was not due to lysis per se and implicating SERA6 as the enzyme responsible. The β-spectrin CH1 domain mediates key interactions between each end of the α2β2-spectrin tetramers that constitute the bulk of the cytoskeleton, and short β-actin filaments (together with protein 4.1R) at the junctional complexes that link the RBC cytoskeleton to its plasma membrane, providing the latter with structural integrity (Fig. 4f)24–26. Cleavage is therefore predicted to unravel the cytoskeleton with resulting RBCM destabilisation (Fig. 4g). Spectrin tetramers also bind the plasma membrane through interactions with the ankyrin complex24, so the limited cleavage of ankyrin might facilitate release of the cleavage products and associated proteins from the cytoskeletal complex.
Figure 4. RBCM rupture is associated with rapid, SERA6-dependent cleavage of host RBC cytoskeleton β-spectrin within its actin-binding domain.
a, SDS PAGE showing appearance upon egress of mock-treated (-RAP) SERA6:loxP schizonts of a high molecular mass species identified by Western blot and LC-MS/MS as truncated β-spectrin (reproducible in 15 independent experiments). b, Peptides (red) identified by LC-MS/MS (3 technical replicate runs from a single biological experiment) of tryptic digests of polypeptide(s) enriched in the mock-treated SERA6:loxP schizont extract in the indicated region of the gel (˜15-20 kDa), indicating egress-associated, SERA6-dependent cleavage of β-spectrin. A semi-tryptic peptide likely representing the C-terminus of the polypeptide(s) is in bold (Supplementary Fig. 6 shows fragmentation spectra). Calculated mass of the β-spectrin sequence (UniProtKB P11277) from Thr2-Gln167 is 19,251 Da. CH1, CH2, calponin homology domains. c, Appearance of a ˜17 kDa N-terminal fragment of β-spectrin (arrowed) upon egress of mock-treated SERA6:loxP schizonts (reproducible in 4 independent experiments). d, Pull-down of cytoskeletal components from soluble extracts of egressing SERA6:loxP schizonts. Annotated species, including co-precipitating β-actin, were identified by LC-MS/MS or Western blot (reproducible in 3 independent experiments). Peptide fingerprinting of the ˜17 kDa β-spectrin CH1 domain was as in b. The presence of α-spectrin and full-length and truncated β-spectrin in pull-downs from the -RAP extracts indicates their SERA6-dependent dissociation from the normally insoluble cytoskeleton. Antibody heavy and light chains, asterisked. e, Fate of β-spectrin in SERA6:loxP schizonts following washing away a C2 block (control) or with additional treatment by the indicated disruption methods. Cleavage never occurred in the absence of SERA6 (Western blot representative of 2 independent experiments). f, Architecture of RBC cytoskeleton spectrin heterotetramer, comprising 2 antiparallel αβ-spectrin heterodimers linked head-to-head (the right-hand dimer is abbreviated for clarity) which cross-link β-actin-containing junctional complexes24. Spectrin repeat domains are numbered. Other structural features and positions of epitopes recognised by mAbs B-1, B-2 and VD4 are indicated. g, Top, SERA6-dependent cleavage of β-spectrin should release each end of the αβ-spectrin tetramer from its cognate junctional complex. The cleaved CH1 domain may be released with actin still bound. Bottom, predicted global effect of SERA6-dependent cleavage on the cytoskeleton.
Our findings ascribe the physico-mechanical processes underlying malarial egress to two parasite proteases that act rapidly and sequentially within the same, PKG-regulated pathway. SUB1 is required for all the structural changes following PKG activation, including rounding up, PVM lysis, RBCM poration and RBCM rupture. SERA6 is not required for PVM rupture or RBCM poration, but accomplishes the final step of RBCM rupture primarily through targeted cleavage of β-spectrin at a site that is essential for cytoskeletal stability. PVM rupture is unlikely to be directly mediated by protease activity, so SUB1 may regulate this by activating one or more membrane lytic effectors that mediate PVM rupture, as well as perhaps RBCM poration. These could include pore-forming proteins or phospholipases, both implicated in egress of other parasite developmental stages22,27,28. That SERA6 function requires SUB1-mediated processing is consistent with processing representing activation of SERA6, as previously suggested10. The striking similarity between the ΔSERA6 phenotype and that produced by treatment with E646,14,18 supports this and suggests that SERA6 is the major target of E64 in schizonts. Importantly, our study proves that host RBC calpain-1, previously implicated by others in egress29, is not sufficient for RBCM rupture since its expression should be unmodified in ΔSERA6 parasites.
All Plasmodium species, including the other major pathogens P. vivax and P. knowlesi, express orthologues of SUB1 and SERA6. Drugs that inhibit these proteases, particularly if combined with inhibitors of PKG30, would target consecutive, interdependent steps in the egress pathway and so could form a new class of antimalarial designed to prevent parasite proliferation and disease.
Methods
Reagents and antibodies
Anonymised human blood was obtained from the UK National Blood Transfusion Service. The antifolate WR99210 was from Jacobus Pharmaceuticals (New Jersey, USA). Blasticidin, rapamycin and E64 (Sigma) were used as described previously9,19. Compound 2 was kindly provided by Dr Simon Osborne (LifeArc, SBC Open Innovation Campus, Stevenage UK); stocks (10 mM) were stored in DMSO at -20°C and used throughout at a final concentration of 1 μM. Alexa Fluor 488 phalloidin and Alexa Fluor 647-conjugated WGA was from Thermofisher. The β-spectrin-specific mAbs B-1, B-2 and VD4, the α-spectrin-specific mAb 17C7, and mAbs B-11 and 8C3 specific for protein 4.1R and ankyrin respectively were all from Santa Cruz Biotechnology. Monoclonal antibody 7.7 (anti-EXP2) was from the European Malaria Reagent Repository (http://www.malariaresearch.eu/), contributed by Jana McBride. The polyclonal anti-mCherry antibody (ab167453) was from Abcam. The P. falciparum MSP1-specific mAb 89.1 has been described previously31, as have rabbit antisera to P. falciparum SERA59, SERA610, SUB132 and AMA133. Phusion high-fidelity DNA polymerase and restriction enzymes were from New England BioLabs, and a Rapid DNA Ligation Kit (Roche) was used for DNA ligation.
Parasite maintenance, synchronization and transfection
The DiCre-expressing P. falciparum clone 1G5DC19 was maintained at 37°C in human RBC in RPMI 1640 medium containing Albumax (Invitrogen) supplemented with 2 mM L-glutamine. Cultures were routinely monitored by microscopic examination of Giemsa-stained thin blood films and synchronised by standard procedures34. As required, mature schizonts were isolated by centrifugation over cushions of 70% (v/v) isotonic Percoll (GE Healthcare Life Sciences) as described34. Invasion assays were performed as previously described3,5, either in static culture or in a shaking incubator revolving at 225 rpm. For transfection of plasmid constructs, ˜108 Percoll-enriched schizonts were suspended in 100 μl of P3 primary cell solution containing 10 μg of DNA and electroporated with an Amaxa™ P3 primary cell 4D Nucleofector™ X Kit L (Lonza), using program FP158 as previously described5. Growth medium was supplemented ˜20 h post transfection with WR99210 (2.5 nM) or blasticidin (2 μg/ml). Once sustained growth of drug-resistant parasites was observed, drug cycling was used to enrich for genomic integration of plasmid constructs as previously described2. Transgenic parasite clones SUB1HA3:loxP and SERA6:loxP were obtained by limiting dilution cloning in microplates at a 0.1-0.3 parasite per well. Parasite genomic DNA (gDNA) for genotype analysis was extracted using a Qiagen DNeasy Bood and Tissue kit and analyzed by PCR using Kappa 2G Fast HotStart ReadyMix (Kappa Biosciences).
Immunofluorescence and Western blot
For IFA, air-dried thin films of parasite cultures were fixed in paraformaldehyde, permeabilized, then probed with relevant primary antibodies as described previously10. Secondary Alexa Fluor 488- or 594-conjugated antibodies specific for human, rabbit or mouse IgG (Invitrogen), or Alexa Fluor 594-conjugated streptavidin (Invitrogen) were used at a dilution of 1:1000. Samples were mounted in Vectashield antifade mounting medium (Vector Laboratories) containing DAPI. Images were acquired using a Nikon Eclipse and NIS Elements software (Nikon, Japan), using identical exposure conditions for all samples being compared. Western blots were performed as described previously33. For detection of HA3-tagged SUB1, the rat anti-HA mAb 3F10 (Sigma) was used at a 1:1000 dilution, followed by biotin-conjugated anti-rat antibody (Roche) (1:8,000 dilution), then horseradish peroxidase (HRP)-conjugated streptavidin (Sigma) (1:10,000 dilution). Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used according to the manufacturer’s instructions, and blots were visualised and documented using a ChemiDoc Imager (Bio-Rad) with Image Lab software (BioRad).
Generation of integration, complementation and expression plasmids
SUB1HA3:loxP and SERA6:loxP parasite clones were generated by single crossover homologous recombination into the 1G5DC genome using integration plasmids pHH1_SUB1HA3_loxP and pHH1_S6chimera_loxP respectively. In both cases, correct transcriptional regulation of the modified gene was assured by placement of the 3’ UTR of the P. berghei dihydrofolate reductase thymidylate synthase (PbDT) gene downstream of the floxed coding sequence19. To target the SUB1 gene, a chimeric gene fragment with a native 5’ portion and a recodonised 3’ segment was constructed by first amplifying the 5’ sequence from P. falciparum 3D7 gDNA using primers JT-S1endo-F and JT-S1CO-R, whilst the recodonised region was amplified from plasmid pFastBac-sPfSUB1wt32 using primers JT-S1CO-F and JT-S1synth-R. The amplicons were then fused in frame by inclusion of both in a fresh PCR reaction including primers JT-S1endo-F and JT-S1synth-R, and the product cloned into pGEM-T (Promega). A segment of this fragment was then excised using HindIII and KpnI and replaced with a similarly-digested loxP-containing synthetic intron (loxPint)20 (Geneart). This intermediate vector was digested with HpaI and Xho I to liberate the SUB1 sequence which was ligated into pHH1_sera5_LoxP119 digested with the same enzymes to generate pHH1_SUB1HA3_loxP. Integration of this construct into the 1G5DC SUB1 locus by homologous recombination was detected by diagnostic PCR with primers JT111-1p and JT111-2p, and the floxed or excised SUB1 locus was detected by PCR using primers JT111-1p and JT111-3p.
To generate pHH1_S6chimera_loxP, a chimeric SERA6 gene fragment was excised from plasmid MWS36 by digestion with HpaI and NcoI. Full details of plasmid MWS36 will be provided in a separate manuscript (M. Shea and M. Blackman, in preparation). The chimeric SERA6 sequence comprised a 920 bp 5’ portion of endogenous coding sequence starting from within the first intron followed by synthetic recodonised SERA6 cDNA sequence extending to the stop codon. Plasmid pHH1_sera5_LoxP1 was digested with HindIII, ‘blunted’ with T4 DNA polymerase, then further digested with NcoI before ligation to the chimeric SERA6 fragment excised from MWS36, generating pHH1_S6chimera_loxP. Integration of this construct by homologous recombination into the 1G5DC genome (which already contains a single genomic loxP site upstream of the SERA6 locus19) was designed to introduce a second loxP site downstream of the SERA6 stop codon. Correct integration was detected by diagnostic PCR using primers SERA6-5’UTRb and SERA6-37, whilst the floxed or excised SERA6 locus was detected with primers S65’UTRb-2 and S6EndoEx2Rev.
For generation of plasmid constructs designed for transgenic expression of SERA6, a chimeric coding sequence and native SERA6 promoter was excised from plasmid MW28 and ligated into SalI and NcoI digested plasmid pDC-mCherry-MCS (a modification of pDC2-mCherry9), giving rise to pDC2-wtSERA6 (WT SERA6). The chimeric SERA6 sequence comprised 979 bp of putative promoter sequence upstream of the native SERA6 ATG start codon, as well as 477 bp of the 5’ segment of the coding sequence (including the first intron) and a synthetic recodonised SERA6 cDNA10 encoding the remainder of the ORF. Full details of plasmid MWS28 will be provided in a separate manuscript (M. Shea and M. Blackman, in preparation). Site-directed mutagenesis and sub-cloning steps were then used as previously described10 to generate identical constructs containing di-Leu substitutions of the P1 and P2 positions at the SUB1 processing sites 1 and 2 in SERA6, as well as a Cys644Ala substitution of the active site nucleophile, giving rise to plasmids pDC2-SERA6-uncleavable (Uncleavable SERA6) and pDC2-SERA6_Alamut (Cys644Ala) respectively.
For transgenic expression of WT SUB1 the pDC-mCherry-MCS plasmid was modified such that the blasticidin deaminase (BSD) drug selection cassette and mCherry reporter gene were expressed from a single promoter by the use of the ribosomal T2A skip peptide. To do this, the BSD cassette was excised from pDC2-mCherry_MCS with ApaI and SacI and the backbone re-ligated. The mCherry ORF was then excised by digestion with AvrII and XhoI and replaced with a synthetic gBlock® (IDT) comprising the mCherry and BSD ORFs separated by the T2A sequence. This resulted in construct pDC2-mCherryT2ABSD_MCS in which these ORFs remained under control of the constitutive P. falciparum calmodulin (CAM) promoter (which remained from the original mCherry expression cassette). This vector was then linearized with SnaBI. The P. falciparum SUB1 promoter sequence was amplified from P. falciparum 3D7 gDNA using primers PfSUB1_prom_for_infu and PfSUB1_prom_rev_infu. Primers PfSUB1_synth_for_infu and PfSUB1_synth_rev_infu were used to amplify the recodonised synthetic SUB1 ORF from pFastBac-sPfSUB1wt and primers PbDT3UTR-for_infu and PbDT3UTR-for_infu were used to amplify the PbDT 3’ UTR from pDC2-mCherry_MCS. Primers included complementary overhangs such that all 3 fragments could then be finally assembled into the linearized pDC2-mCherryT2ABSD_MCS backbone in a single step using an InFusion® HD Cloning Kit (Clontech), generating pDC2-mCherryT2ABSD_wtSUB1 (WT SUB1).
For episomal transgenic expression of an EXP1mCherry fusion protein (to fluorescently label the PVM), a synthetic intronless DNA fragment encoding mCherry fused to the C-terminus of the P. falciparum EXP1 (PlasmoDB ID PF3D7_1121600) via a polyglycine-alanine linker (GAGGGGGGGGA) was obtained from Geneart. This was sub-cloned into vector pCR-Blunt using the ZeroBlunt PCR cloning kit (Invitrogen). The resulting plasmid was digested with AvrII and XhoI before ligating the EXP1mCherry fragment into pDC-mCherry-MCS in the place of the mCherry ORF, generating pDC2-EXP1-mCherry.
Parasitaemia quantitation by flow cytometry
Parasites were fixed in 4% paraformaldehyde, 0.02% glutaraldehyde for 30 min at 37°C, diluted five-fold in phosphate-buffered saline, then stored at 4°C until required. Cells were stained with Hoechst 33342 (diluted 1:10,000) for 30 min at 37°C, then parasitaemia determined using a Fortress or FACS Aria (BD) flow cytometer as previously described9. Briefly, samples were initially screened using forward and side scatter parameters and gated for RBC (Supplementary Fig. 10). Ultraviolet light with a violet filter (450/50 nm) was then used to determine the proportion of infected cells in 100,000 RBC.
Time-lapse DIC and fluorescence video microscopy
Viewing chambers for live microscopy were constructed as described previously3,5 by adhering 22 x 64 mm borosilicate glass coverslips to microscope slides. Mature Percoll-enriched schizonts were incubated for 3-4 h at 37°C in complete medium supplemented with C2 (1 μM), then ˜5 x 107 schizonts were rapidly washed twice in gassed warm complete medium lacking C2, pelleting at 1,800 x g for 1 min. The cells were suspended in 50 μl of the same medium and introduced into the pre-warmed viewing chamber on a temperature-controlled microscope stage held at 37°C on a Nikon Eclipse Ni-E wide-field microscope fitted with a Hamamatsu C11440 digital camera and Nikon N Plan Apo λ 100x/1.45NA oil immersion objective. Images (DIC alone or simultaneous DIC and fluorescence) were taken at 5-10 s intervals over a total of 20-60 min, then annotated and exported as TIFFs, AVI or QuickTime movies using Nikon NIS-Elements software.
Parasite plaque assays
Plaque assays were performed by dispensing parasite cultures (200 μl at a haematocrit of 0.75%) into flat-bottomed 96-well microplates, as described12. Plates were imaged 14-16 days later to detect plaque formation, using an Epson Perfection V750 Pro high resolution flat-bed scanner in top-down transmission light mode. When required, parasites from wells containing a single plaque were expanded by transferring initially to round-bottomed microplate wells to aid medium changes, before further expansion into culture flasks.
Scanning electron microscopy
Mature arrested ΔSERA6 schizonts, or WT control schizonts allowed to reach the point of egress in the presence of E64 (50 μM) were fixed in 2.5% glutaraldehyde, washed, osmicated (1% OsO4 for 16 h), dehydrated, critical point dried and sputter coated with 5 nm gold for scanning EM. Images were collected on a JEOL JSM 7610F with 2.6 kV accelerating voltage.
Transmission electron microscopy
Mature schizonts were fixed for 5 min at 37°C in 2% formaldehyde (ΔSERA6 and E64-arrested schizonts) or 2% formaldehyde 1% glutaraldehyde (ΔSUB1 and C2-arrested WT schizonts), pelleted by centrifugation, mixed with 20% (w/v) dextran in complete medium containing bakers’ yeast, then frozen using a HPM100 high-pressure freezer (Leica). Vitrified cells were freeze-substituted using a EM AFS2 (Leica) into Lowicryl HM20 resin (EMS) with 0.2% (w/v) uranyl acetate and cut into 250 nm or 120 nm sections using a UC7 microtome (Leica). Sections were placed on glow-discharged carbon-coated copper London Finder grids (EMS) with 10 nm Protein A-Au fiducials (EMS) and post-stained with 0.2% (w/v) uranyl acetate and 4% (w/v) lead citrate. Images and tomograms were recorded using a Model 2040 dual-axis tomography holder (Fischione Instruments) on a Tecnai F20 200 kV field emission gun electron microscope (FEI) equipped with a DE20 camera (Direct Electron), or a Tecnai T12 120 kV field emission gun electron microscope (FEI) equipped with a 4kx4k Ultrascan 4000 CCD camera (Gatan). Dual-axis tilt series were acquired from -60° to +60° with an increment of 2° using SerialEM35 and processed using IMOD36 with nonlinear anisotropic diffusion filtering.
Proteomic analysis and pull-downs
Mature mock- or RAP-treated mature SERA6:loxP schizonts were incubated for ˜4 h at 37°C in complete medium supplemented with C2, then washed once in gassed, warm protein-free medium containing C2 before rapidly washing twice in similar medium lacking C2, pelleting at 1,800 x g for 1 min. The cells were suspended at high density (˜1 x 109/ml) in warm protein-free medium lacking C2 and incubated for just 20 min at 37°C to allow progress to egress. The entire samples were then snap-frozen in liquid N2.
For global proteomic analysis, samples were thawed by the addition of an equal volume of ice-cold water containing protease inhibitors (cOmplete™ Protease Inhibitor Cocktail, Sigma-Aldrich), followed by centrifugation at 16,000 × g for 10 min at 4°C to separate soluble and insoluble fractions. These were immediately fractionated by SDS PAGE on NuPAGE 4-12% Bis-Tris gels (Invitrogen). Gels were stained with Quick Blue Coomassie (Triple Red) then the entire gel cut into 24 equally-sized slices and proteins in excised slices subjected to tryptic digestion37. LC-MS/MS analysis was as described below.
For pull-down analysis, frozen schizont preparations were thawed in the presence of 1% (v/v) Nonidet® P40 (CAS 68412-54-4, Santa Cruz Biotechnology), 10 mM EDTA and protease inhibitors (cOmplete™ Protease Inhibitor Cocktail, Sigma-Aldrich) and extracted at 4°C for 1 h with intermittent vortexing. The extracts were clarified by centrifugation at 16,000 × g for 10 min at 4°C, filtered through 0.22 μm PVDF centrifugal filter units (MilliporeSigma), then incubated with ˜2 μg of the relevant anti-spectrin mAb for 1.5 h at 4°C with gentle mixing. Precipitation of immune complexes with Protein G Sepharose™ 4 Fast Flow (GE Healthcare) followed the manufacturer’s protocol. Bound complexes were subjected to SDS PAGE and stained bands of interest excised and analysed by tryptic digestion and LC-MS/MS.
Mass spectrometry (LC-MS/MS)
Tryptic digests were chromatographically resolved using an Ultimate 3000 RSLCnano (Dionex) with an EASY-Spray column (2 μm particle size, PepMap C18, 100 Å pore size, 50 cm x 75 μm ID; Thermo Scientific). Spectra were acquired using an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific) acquiring the top 10 most intense ions in data dependent acquisition mode with CID fragmentation at 35% normalised collision energy. For targeted work, data were acquired using an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) using a mixture of data dependent fragmentation and three targeted fragmentations (545.3006 Da, 659.8537 Da and 830.9363 Da for peptides FQIQDIVVQ, FQIQDIVVQTQ and FQIQDIVVQTQEGR, respectively) over a 3 sec cycle time. Dynamic exclusion was employed throughout to prevent repeat sampling of data dependent fragmentation.
Data were searched using Mascot (Matrix Science) against the UniProt database, using trypsin or semi-trypsin as the cleavage enzyme, with a fixed carbamidomethylation modification (+57.021 Da) and variable methionine oxidation (+ 15.994 Da). A 10 ppm mass tolerance filter was applied for peptides with charge states +2 or above. Mascot search results were imported into Skyline38 to generate a spectral library for further quantitative processing of β-spectrin peptides.
Statistical analysis
Prism 7 (GraphPad) was used for all statistical analysis. All experiments were performed at least twice, and statistical analysis was carried out by unpaired t-test of biological replicate or triplicate experiments. A p value of <0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by funding to M.J.B. from the Francis Crick Institute (https://www.crick.ac.uk/) which receives its core funding from Cancer Research UK (FC001043; https://www.cancerresearchuk.org), the UK Medical Research Council (FC001043; https://www.mrc.ac.uk/), and the Wellcome Trust (FC001043; https://wellcome.ac.uk/). J.A.T. and M.S.Y.T. were in receipt of Crick PhD studentships, and V.L.H. was supported by Gatan BBSRC CASE PhD studentship BB/F016948/1. The work was also supported by MRC project grants G1100013 and MR/P010288/1 (H.R.S., M.J.B., R.A.F.), Wellcome equipment grants 101488, 079605 and 086018 (H.R.S., M.J.B., R.A.F.), and Wellcome ISSF2 funding to the London School of Hygiene & Tropical Medicine.
Footnotes
Data availability
The data supporting the findings of this study are available within the paper and its Supplementary Information and are also available from the corresponding authors upon request.
Author contributions
J.A.T. performed all P. falciparum genetic manipulations and phenotype analysis. M.S.Y.T. performed phenotype analysis and parasite manipulation. F.H. performed parasite manipulation. G.V.B. and R.A.F. performed SEM. C.B., T.R.U. and V.L.H. performed and interpreted TEM. A.B., M.S.Y.T. and B.S. performed and interpreted proteomic analysis. J.A.T., M.S.Y.T, B.S., H.R.S. and M.J.B. conceived the study, designed experiments, interpreted results and wrote the manuscript.
References
- 1.Blackman MJ. Malarial proteases and host cell egress: an 'emerging' cascade. Cell Microbiol. 2008;10:1925–1934. doi: 10.1111/j.1462-5822.2008.01176.x. CMI1176 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeoh S, et al. Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell. 2007;131:1072–1083. doi: 10.1016/j.cell.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 3.Collins CR, et al. Malaria parasite cGMP-dependent protein kinase regulates blood stage merozoite secretory organelle discharge and egress. PLoS Pathog. 2013;9:e1003344. doi: 10.1371/journal.ppat.1003344. PPATHOGENS-D-12-03129 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Withers-Martinez C, et al. The malaria parasite egress protease SUB1 is a calcium-dependent redox switch subtilisin. Nat Commun. 2014;5:3726. doi: 10.1038/ncomms4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, et al. Processing of Plasmodium falciparum Merozoite Surface Protein MSP1 Activates a Spectrin-Binding Function Enabling Parasite Egress from RBCs. Cell Host Microbe. 2015;18:433–444. doi: 10.1016/j.chom.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale VL, et al. Parasitophorous vacuole poration precedes its rupture and rapid host erythrocyte cytoskeleton collapse in Plasmodium falciparum egress. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1619441114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koussis K, et al. A multifunctional serine protease primes the malaria parasite for red blood cell invasion. EMBO J. 2009;28:725–735. doi: 10.1038/emboj.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silmon de Monerri NC, et al. Global identification of multiple substrates for Plasmodium falciparum SUB1, an essential malarial processing protease. Infect Immun. 2011;79:1086–1097. doi: 10.1128/IAI.00902-10. IAI.00902-10 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins CR, Hackett F, Atid J, Tan MSY, Blackman MJ. The Plasmodium falciparum pseudoprotease SERA5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes. PLoS Pathog. 2017;13:e1006453. doi: 10.1371/journal.ppat.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruecker A, et al. Proteolytic activation of the essential parasitophorous vacuole cysteine protease SERA6 accompanies malaria parasite egress from its host erythrocyte. J Biol Chem. 2012;287:37949–37963. doi: 10.1074/jbc.M112.400820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller SK, et al. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J Biol Chem. 2002;277:47524–47532. doi: 10.1074/jbc.M206974200. [DOI] [PubMed] [Google Scholar]
- 12.Thomas JA, et al. Development and Application of a Simple Plaque Assay for the Human Malaria Parasite Plasmodium falciparum. PloS one. 2016;11:e0157873. doi: 10.1371/journal.pone.0157873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glushakova S, Yin D, Li T, Zimmerberg J. Membrane transformation during malaria parasite release from human red blood cells. Current biology : CB. 2005;15:1645–1650. doi: 10.1016/j.cub.2005.07.067. [DOI] [PubMed] [Google Scholar]
- 14.Glushakova S, et al. New stages in the program of malaria parasite egress imaged in normal and sickle erythrocytes. Current Biology : CB. 2010;20:1117–1121. doi: 10.1016/j.cub.2010.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickham ME, Culvenor JG, Cowman AF. Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J Biol Chem. 2003;278:37658–37663. doi: 10.1074/jbc.M305252200. [DOI] [PubMed] [Google Scholar]
- 16.Abkarian M, Massiera G, Berry L, Roques M, Braun-Breton C. A novel mechanism for egress of malarial parasites from red blood cells. Blood. 2011;117:4118–4124. doi: 10.1182/blood-2010-08-299883. [DOI] [PubMed] [Google Scholar]
- 17.Taylor HM, et al. The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryot Cell. 2010;9:37–45. doi: 10.1128/EC.00186-09. EC.00186-09 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glushakova S, Mazar J, Hohmann-Marriott MF, Hama E, Zimmerberg J. Irreversible effect of cysteine protease inhibitors on the release of malaria parasites from infected erythrocytes. Cell Microbiol. 2009;11:95–105. doi: 10.1111/j.1462-5822.2008.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins CR, et al. Robust inducible Cre recombinase activity in the human malaria parasite Plasmodium falciparum enables efficient gene deletion within a single asexual erythrocytic growth cycle. Mol Microbiol. 2013;88:687–701. doi: 10.1111/mmi.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones ML, et al. A versatile strategy for rapid conditional genome engineering using loxP sites in a small synthetic intron in Plasmodium falciparum. Sci Rep. 2016;6 doi: 10.1038/srep21800. 21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ribacke U, et al. Improved in vitro culture of Plasmodium falciparum permits establishment of clinical isolates with preserved multiplication, invasion and rosetting phenotypes. PloS one. 2013;8:e69781. doi: 10.1371/journal.pone.0069781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wirth CC, et al. Perforin-like protein PPLP2 permeabilizes the red blood cell membrane during egress of Plasmodium falciparum gametocytes. Cell Microbiol. 2014;16:709–733. doi: 10.1111/cmi.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmons D, Woollett G, Bergin-Cartwright M, Kay D, Scaife J. A malaria protein exported into a new compartment within the host erythrocyte. EMBO J. 1987;6:485–491. doi: 10.1002/j.1460-2075.1987.tb04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lux SEt. Anatomy of the red cell membrane skeleton: unanswered questions. Blood. 2016;127:187–199. doi: 10.1182/blood-2014-12-512772. [DOI] [PubMed] [Google Scholar]
- 25.An X, et al. Identification and functional characterization of protein 4.1R and actin-binding sites in erythrocyte beta spectrin: regulation of the interactions by phosphatidylinositol-4,5-bisphosphate. Biochemistry. 2005;44:10681–10688. doi: 10.1021/bi047331z. [DOI] [PubMed] [Google Scholar]
- 26.Karinch AM, Zimmer WE, Goodman SR. The identification and sequence of the actin-binding domain of human red blood cell beta-spectrin. J Biol Chem. 1990;265:11833–11840. [PubMed] [Google Scholar]
- 27.Deligianni E, et al. A perforin-like protein mediates disruption of the erythrocyte membrane during egress of Plasmodium berghei male gametocytes. Cell Microbiol. 2013;15:1438–1455. doi: 10.1111/cmi.12131. [DOI] [PubMed] [Google Scholar]
- 28.Burda PC, et al. A Plasmodium phospholipase is involved in disruption of the liver stage parasitophorous vacuole membrane. PLoS Pathog. 2015;11:e1004760. doi: 10.1371/journal.ppat.1004760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandramohanadas R, et al. Apicomplexan parasites co-opt host calpains to facilitate their escape from infected cells. Science. 2009;324:794–797. doi: 10.1126/science.1171085. 1171085 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker DA, et al. A potent series targeting the malarial cGMP-dependent protein kinase clears infection and blocks transmission. Nat Commun. 2017;8:430. doi: 10.1038/s41467-017-00572-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holder AA, Freeman RR. Biosynthesis and processing of a Plasmodium falciparum schizont antigen recognized by immune serum and a monoclonal antibody. J Exp Med. 1982;156:1528–1538. doi: 10.1084/jem.156.5.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Withers-Martinez C, et al. Expression of recombinant Plasmodium falciparum subtilisin-like protease-1 in insect cells: Characterization, comparison with the parasite protease, and homology modelling. J Biol Chem. 2002;277:29698–29709. doi: 10.1074/jbc.M203088200. [DOI] [PubMed] [Google Scholar]
- 33.Collins CR, Withers-Martinez C, Hackett F, Blackman MJ. An inhibitory antibody blocks interactions between components of the malarial invasion machinery. PLoS Pathog. 2009;5:e1000273. doi: 10.1371/journal.ppat.1000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blackman MJ. Purification of Plasmodium falciparum merozoites for analysis of the processing of merozoite surface protein-1. Methods Cell Biol. 1994;45:213–220. doi: 10.1016/s0091-679x(08)61853-1. [DOI] [PubMed] [Google Scholar]
- 35.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 37.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 38.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.