Abstract
While influenza virus infection remains a concerning disease for public health, the roles of individual cytokines during the immune response to influenza infection are not fully understood. We have identified IL-36γ as a key mediator of immune protection during both high- and low-pathogenesis influenza infection. Il36g mRNA is upregulated in the lung following influenza infection and mice lacking IL-36γ have greatly increased morbidity and mortality upon infection with either H1N1 or H3N2 influenza. The increased severity of influenza infection in IL-36γ-deficient mice is associated with increased viral titers, higher levels of proinflammatory cytokines early in infection, and more diffuse pathology late in the disease course. Interestingly, the increased severity of disease in IL-36γ-deficient mice correlates with a rapid loss of alveolar macrophages following infection. We find that the alveolar macrophages from naïve IL-36γ-KO mice have higher expression of M2-like surface markers compared to WT mice, and show increased apoptosis within 24 hours of infection. Finally, transfer of WT alveolar macrophages to IL-36γ-KO mice restores protection against lethal influenza challenge to levels observed in WT mice. Together, these data identify a critical role for IL-36γ in immunity against influenza virus and demonstrate the importance of IL-36γ signaling for alveolar macrophage survival during infection.
INTRODUCTION
Despite the broad availability of licensed vaccines, influenza viral illness remains a major threat to public health, particularly among children and the elderly (1, 2). In severe cases, influenza infection can cause primary viral pneumonia or sensitize the host to secondary bacterial infection, and can also exacerbate underlying health problems including congestive heart failure, chronic obstructive pulmonary disease, and asthma. One reason for the continued public health burden of influenza is the imperfect antigenic match between the vaccine and circulating influenza strains that results in only 50–80 % protection in the best of years (2). In the absence of effective cellular or humoral memory responses, the innate defenses of the respiratory tract serve as the only mechanisms to control the spread of virus in the early phases of infection.
Influenza infection of the respiratory epithelium leads to the activation of antiviral responses via the extracellular and endosomal TLR receptors or the intracellular receptors RIG-I and NLRP3, causing cells to release interferons as well as pro-inflammatory and chemotactic factors (3–6). Interferons serve to limit protein translation within infected cells and decrease the permissibility of uninfected cells to the virus during the mobilization and recruitment of immune cells from the bone marrow and circulation, which can take 4–5 days (7–9). Several innate cell types have been shown to be key to controlling influenza infection. NK cells can recognize infected cells via NKp46 binding to surface hemagglutinin, leading to lysis of cells that are displaying viral proteins (8). Neutrophils can also serve to promote viral clearance in severe cases of influenza, but neutrophils, along with monocytes, and have been shown to contribute to immune pathology during some models of influenza infection (10, 11). In contrast, among the earliest responders to influenza infection in the lung are tissue-resident alveolar macrophages. In mice, these cells have been shown to decrease the infection of type I alveolar epithelial cells during the initial 48 hours of infection, before other innate cell types arrive in the lung (12). Depletion of alveolar macrophages by either genetic knockout or liposomal clondronate treatment increases morbidity, mortality, and viral load during influenza infection (12, 13). Therefore, lung alveolar macrophages play a critical role as one of the few lung-resident innate immune cell types capable of responding to influenza infection during the initial stages of infection, prior to immune recognition that leads to the initial inflammatory burst alerting the systemic immune system to a lung infection.
Severe influenza infections are characterized by viral infection throughout the upper and lower respiratory tract (14). Consequences of increased viral titers throughout the respiratory tract include hyperactive cytokine responses, and patients hospitalized with severe influenza showed significant increases in pro-inflammatory cytokines (15). Elevated expression of type I interferons, TNF-α, IL-1, IL-6, IL-18, and IL-33 have been observed in pathogenic influenza infections, suggesting that inhibition of these cytokines may ameliorate influenza pathology and improve host survival (16). However, deletion or blockade of pro-inflammatory cytokine pathways including IFNα, IFNγ, TNF, IL-1, and IL-6 has shown mixed results in mice and humans (9, 17–24). General immunosuppression with corticosteroids has also shown little to no effect on patient outcome in clinical trials (24–26). These failures have renewed efforts to identify novel pro-inflammatory host mediators that contribute to influenza pathogenesis. One such family of pro-inflammatory mediators are the IL-36 family of cytokines, IL-36α, -β, and -γ, which are recently-discovered members of the IL-1 family. These proteins have been shown to be important in several different inflammatory conditions including asthma, colitis, and psoriasis (27–29). In addition, IL-36γ administration has been shown to increase mucus production and neutrophil migration into the lungs of mice (28). A recent study showed that IL-36γ-deficient mice were more susceptible to pulmonary S. pneumonia infection, demonstrating an important role for IL-36γ in lung immunity (30). In addition, deletion of IL-36 receptor has been shown to improve survival during lethal influenza challenge in mice (31). However, the role of individual IL-36 cytokines in immunity against respiratory virus infections has not been investigated.
In the present study, we investigated the regulation of IL-36 cytokines during mild and severe influenza infections in mice. Il36g mRNA was significantly increased in the lung during the initial stages of influenza infection, suggesting that inhibition of IL-36γ production may prevent excessive inflammation during severe influenza infection and improve host survival. Surprisingly, infection of IL-36γ-deficient mice with influenza virus showed the opposite effect, as IL-36γ-deficient mice had significantly increased weight loss, mortality, and viral titers during both mild and severe influenza infection. Analysis of innate immune cell subsets in the lung revealed a rapid and dramatic loss of lung alveolar macrophages in IL-36γ-deficient mice compared to their wild-type counterparts. Finally, the decreased survival in IL-36γ-deficient mice could be rescued through the transfer of wild-type alveolar macrophages into the lung prior to influenza infection. Overall, these data demonstrate an unexpected protective role for IL-36γ during influenza infection by promoting the survival of lung alveolar macrophages.
MATERIALS AND METHODS
Mice
C57BL/6J (WT) and B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) were purchased from The Jackson Laboratory and bred in house under specific pathogen free conditions at Emory University. For some experiments WT and CD45.1 mice were crossed to produce CD45.1+CD45.2+ F1 offspring. Cyropreserved sperm of the line B6;129S5-Il1f9tm1Lex/Mmucd (IL-36γ KO) was purchased from the Knockout Mouse Project, injected into a C57Bl6/J oocyte and backcrossed in house (32). SNP analysis was performed by Jackson Laboratories to confirm backcrossing to the C57BL/6J strain (Supplementary Table 1). Intranasal infection with influenza A/HKx31 (H3N2) at 30,000 or 300,000 50% egg infectious dose (EID50) and PR8 (H1N1) at 450 PFU was performed under isoflurane anesthesia. For all mortality experiments, mice were monitored with daily weighing and euthanized when they reached 25% weight loss. All experiments were completed in accordance with the Institutional Animal Care and Use Committee guidelines of Emory University.
Whole lung mRNA isolation for viral titers and il36 qPCR
Infected mice were euthanized by 2,2,2-tribromoethanol overdose and brachial exsanguination. Lungs were harvested into RNA later (Sigma) and stored at 4 degrees C overnight and then at −80 degrees C until processing. Lungs were homogenized in Trizol (Ambion) by either Tissue Tearor or Bead Bug according to the manufacturer’s directions. RNA was purified from the homogenized lung using Ambion spin columns by a modified manufacturer’s protocol. cDNA was synthesized from RNA using reverse transcriptase (Applied Biosystems) and viral titers were determined by digital droplet PCR and Quantasoft software (Biorad) using primers and probes directed at the Influenza NP gene, which is conserved between x31 and PR8. The sequences were X-31 NP Forward: 5’-GCA TGC CAT TCT GCC GCA TT-3’, X-31 NP Reverse: 5’-GCT GAT TTG GCC CGC AGA TG-3’, Probe: 5’- /5HEX/TA+G T+CT +CCA TAT TTT CAT T+G+G AA+G C/3BHQ_1/ −3’. Expression levels of IL-36 family members were determined by RT-PCR on a Biorad CFX96 Real time system and CFX manager v3.1 using primers against Il36a, Il36b, Il36g, and Hprt. Sequences were Il36a forward: 5’-TAG TGG GTG TAG TTC TGT AGT GTG-3’, Il36a reverse: 5’-GTT CGT CTC AAG AGT GTC CAG ATA T-3’, Il36b forward: 5’-ACA AAA AGC CTT TCT GTT CTA TCA T-3’, Il36b reverse: 5’-CCA TGT ATT TAC TTC TCA GAC T-3’, Il36g forward: 5’-AGA GTA ACC CCA GTC AGC GTG-3’, Il36g reverse: 5’-AGG GTG GTG GTA CAA ATC CAA-3’, Hprt forward: 5’-CAC AGG ACT AGA ACA CCT GC-3’, Hprt reverse: 5’-GCT GGT GAA AAG GAC CTC T-3’. For Il36g expression in sorted immune cells, cDNA was synthesized with the iScript cDNA synthesis kit before RT-PCR (Biorad). RT-PCR Data were analyzed using CFX Manager before export to Microsoft Excel.
Lung harvest for pathological examination
Mice were euthanized as above and the ribcage carefully removed. A small incision was made in the trachea and the lungs were inflated with 0.75 mL formalin. The lungs, heart, and thymus were removed en bloc and placed into a cassette and then into formalin jars. Lungs were embedded in paraffin, sectioned and stained with hematoxylin and eosin by the Winship Research Pathology Core Lab at Emory University. Lungs were scored by a pathologist blinded to treatment conditions.
Isolation of immune cells from lungs, flow cytometry, and cytokine analysis
For isolation and quantification of immune cells from lungs, mice were euthanized as above. Bronchoalveolar lavage was performed using 1 mL of Compete RPMI delivered via an 18-gauge IV catheter into a small incision into the trachea. The airways were washed five times before the lungs were harvested into HBSS. Lungs were finely minced with scissors before digestion (30 minutes, 37 degrees C) with warm HBSS containing 4E5 units/mL DNase (Sigma) and 500 mg/L Collagenase D (Roche). Lungs were further homogenized via swishing with a 3-mL syringe before and after digestion and every 10 minutes during digestion. Following digestion, immune cells were purified via 40 %/80 % isotonic Percoll gradient centrifugation. Live/dead staining was performed with Zombie Yellow, Zombie Ultraviolet or Zombie Near Infrared (Biolegend) and Fc blocking was done with anti-CD16/32 clone 2.4G2 in FACS buffer consisting of 2% w/v BSA and 0.05 % w/v sodium azide in PBS. The hybridoma expressing 2.4G2 was purchased from ATCC and cultured in our lab. The antibody was purified from the supernatant by standard techniques. Cells were stained with fluorescently labeled antibodies purchased from Abcam, Biolegend, BD Biosciences or eBioscience in FACS buffer. Antibodies were anti-CD45.2-A488 [104, Biolegend], anti-CD45.2-A647 [104, Biolegend], anti-CD86-PE [GL1, Biolegend] anti-CD11c-PE/CF594 [HL3, BD Biosciences], anti-CD103-PerCP/Cy5.5 [2E7, Biolegend], anti-I-A/I-E-PE/Cy7 [M5/114.15.2, Biolegend], anti-CD45R-APC [RA3–6B3, Biolegend], anti-Ly6C-APC/Cy7 [AL-21, BDBiosciences], anti-SiglecF-BV421 [E50–2440, BD Biosciences], anti-Ly6G-BV510, [1A8, Biolegend], anti-NK1.1-BV650 [PK136, Biolegend], anti-CD11b-BV711 [M1/70, Biolegend], anti-CD90.2-BV785 [30-H12, Biolegend], anti-CD206-A700 [C068C2, Biolegend], anti-CD200R-PE [OX-110, Biolegend] anti-TREM2-FITC [78.18, Abcam], anti-CCR5-PerCP/e710 [7A4, eBioscience], and anti-CD45.1-BUV395 [A20, BD Biosciences]. Cells were sorted on a BD Aria II or were fixed with 1 % paraformaldehyde before analysis on a BD LSRII or BD Fortessa X20. Annexin V (Biolegend) staining was performed according to manufacturer’s instructions immediately before flow cytometric analysis. Cell numbers were determined by counting live cells on a hemocytometer in the presence of trypan blue (Sigma). Flow cytometry data was initially analyzed using FlowJo v10.0.8r1 before exporting data to Microsoft Excel for further calculations. For cytokine analysis, the first BAL pull was kept separate and spun at 700 x g for 5 minutes to remove cells. The supernatant was stored at −80 degrees C while the cells were combined with the remaining BAL pulled. Cytokine levels in the BAL supernatant were determined by a Biolegend LEGENDPlex Mouse Inflammation Panel by manufacturer’s instructions.
Cell isolation and adoptive transfer
For transfer of alveolar macrophages, lungs were harvested from naïve B6 or IL-36γ-KO mice and digested (30 minutes, 37 degrees C) with Collagenase type II (625 units/mL, Worthington) and DNase (4E5 units/mL, Sigma) and swished as above. Percoll purification was performed as above. Alveolar macrophages were isolated via positive selection on CD11c and SiglecF with MACS columns per manufacturer’s instructions (Miltenyi). The bound fraction was collected and an aliquot was stained to determine purity. 500,000 alveolar macrophages in 50 µL PBS were administered intranasally to recipient mice under isoflurane anesthesia. WT controls received PBS intranasally. For survival experiments, mice were infected with high dose x31 the following day and monitored daily for 14 days. For next day analysis of transfer efficiency or phenotypic analysis of KO alveolar macrophages transferred to WT mice, the mice were not infected.
Statistics
Statistical analysis was performed in GraphPad Prism and the tests used to determine significance are listed in the appropriate figure legends. Asterisks in the figures denote significance (* p≤0.05, ** p<0.01, *** p<0.001, **** p<0.0001).
RESULTS
IL-36γ is induced during influenza infection and protects against morbidity and mortality
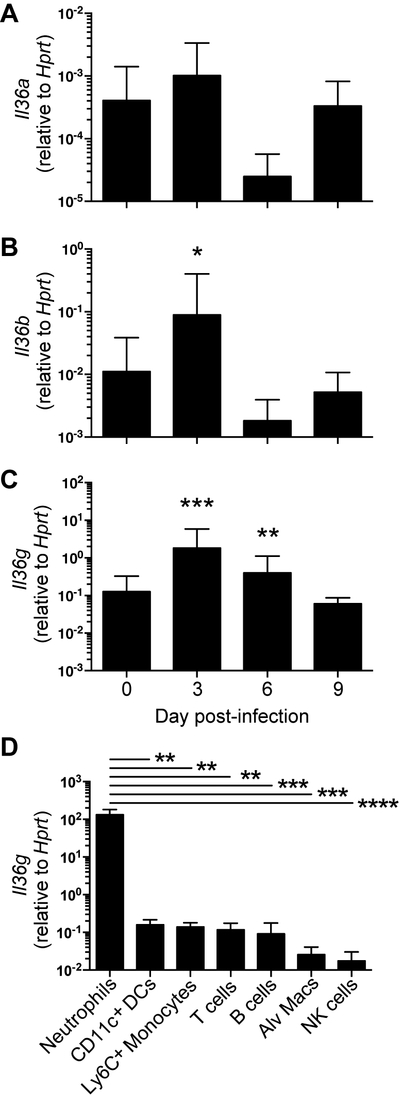
To determine the expression level of IL-36 family members following influenza infection, B6 mice were infected with influenza A/HK-x31 (H3N2) and expression of the IL-36 cytokine family genes Il36a, Il36b, and Il36g in whole lungs were measured by RT-PCR (Figure 1A-C). Il36a expression was not altered following infection, and Il36b was significantly increased only on day 3 following influenza infection. We observed Il36g expression in the lung was significantly increased by day 3 after infection, and increased expression of Il36g was maintained through day 6. In addition, expression levels of Il36g in naïve (d0) whole lungs were higher than either Il36a or Il36b (Supplemental Figure 1). Il36g expression has previously been reported in bronchial epithelial cells following infection with respiratory viruses or bacteria, but whether immune cells produce the cytokine during influenza infection is not known. (30, 33). As Il36g expression has been observed in multiple immune cell types in different models of inflammation, we examined several types of leukocytes for Il36g RNA 6 days after influenza infection. Surprisingly, among immune cells examined, neutrophils were the highest producers of Il36g mRNA at day 6 post-infection (Figure 1D). These data show that IL-36 family members, most notably IL-36γ, are induced in the lung following influenza infection and suggest a potential role of IL-36γ in mediating influenza immunity.
Figure 1: RT-PCR of whole lung homogenate or sorted cells for expression of Il36 cytokines.
The expression levels of (A) Il36a, (B) Il36b, and (C) Il36g were determined by real-time PCR relative to the housekeeping gene Hprt in naïve C57Bl/6J mice and at days 3, 6, and 9 post-infection with 30,000 EID50 of influenza A/HK-x31. Data are combined from three independent replicates of at least 5 mice per group. Males and females were used in equal proportions. Expression levels of Il36g (D) from several immune cell types as indicated below the x axis. Asterisks indicate significantly different expression from uninfected lungs by Student’s t test on logarithmically transformed data (A-C) or ANOVA followed by Holm-Sidak mulitiple comparisons test on log-transformed data (D).
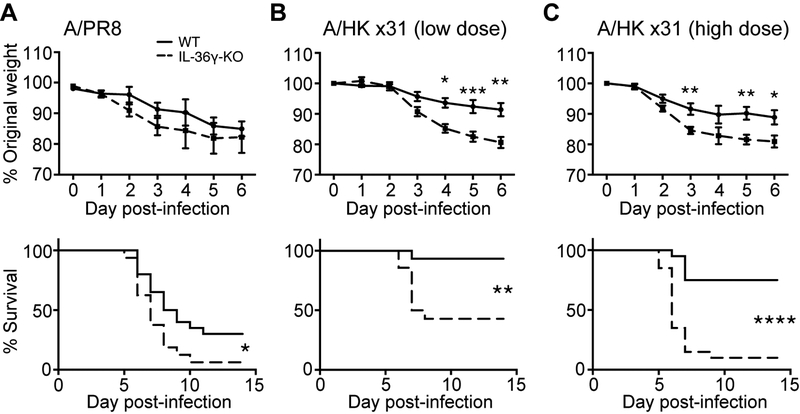
Highly pathogenic influenza infections are associated with high levels of pro-inflammatory cytokines such as TNFα and IL-1, and some studies have shown that blocking these cytokines limited immunopathology and improved survival in murine models (20, 21). Given the pro-inflammatory role for IL-36γ in other diseases, we examined whether deletion of Il36g could decrease morbidity and mortality following influenza A/PR8 (H1N1) infection. As shown in Figure 2A, IL-36γ-KO mice had significantly greater mortality following infection, indicating that IL-36γ had a protective rather than pathogenic role in influenza immunity. To confirm a protective role for IL-36γ during influenza infection, we measured survival following infection with the less pathogenic influenza x31 (H3N2) virus. Compared to WT mice, IL-36γ-KO mice showed significantly increased weight loss beginning on day 3 post-infection, as well as significantly greater mortality, with two different doses of influenza x31 (Figures 2B and 2C). Together, these data demonstrate a surprising protective role for IL-36γ in improving survival following influenza infection with different strains and doses.
Figure 2: Survival and weight loss of WT and IL-36γ-KO mice following influenza infection.
Age- and sex-matched C57Bl/6J (solid lines) and Il36g−/− mice (dashed lines) were infected with (A) influenza PR8, (B) 30,000 EID50 ×31, or (C) 300,000 EID50 ×31 and monitored daily for weight loss. Data are combined from 2–3 independent replicate experiments of at least 10 mice per group per replicate. Male and female mice were used in equal proportions. Asterisks indicate significant differences in survival by the log-rank test or differences in weight loss by Student’s t test.
The absence of IL-36γ results in increased viral replication and immunopathology
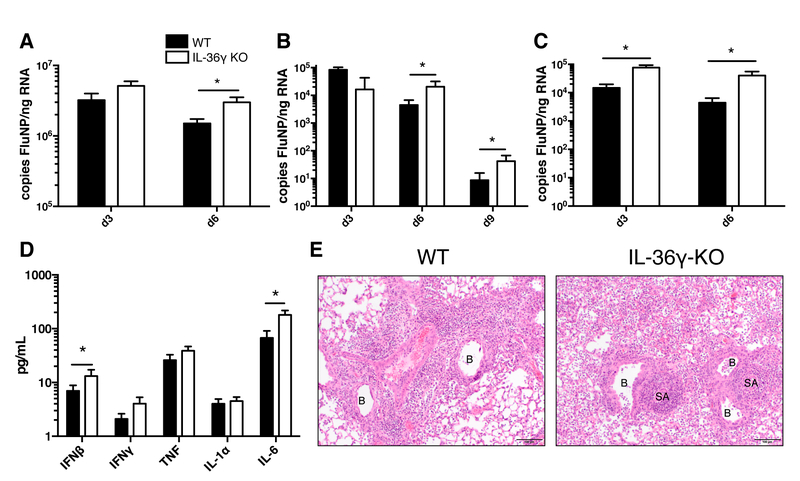
We next examined whether the decreased survival in mice lacking IL-36γ was associated decreased control of viral replication during influenza infection. As shown in Figure 3A, we found that IL-36γ-KO mice have increased viral load by digital droplet PCR 6 days after infection with PR8. Likewise, following infection with a low dose of x31 influenza, IL-36γ-KO mice have increased viral load at days 6 and 9 post-infection (Figure 3B). Mice which were infected with the high dose of x31 had increased viral titers as soon as 3 days post-infection and continued to have increased titers until 6 days post-infection, when the majority of the mice reach humane endpoints (Figure 3C). We also analyzed the cytokines present in the bronchoalveolar lavage (BAL) fluid of WT and IL-36γ-KO mice infected with high dose x31, since these mice have a large difference in viral titers early in infection. We found that compared to WT mice, the BAL of IL-36γ-KO mice had increased interferon β and IL-6, but not interferon γ, tumor necrosis factor, or IL-1α (Figure 3D). To further understand the role of IL-36γ during influenza infection, formalin-fixed lungs from WT and IL-36γ-KO mice infected with high-dose x31 were evaluated for inflammation and damage by an independent pathologist blinded to treatment conditions. On day 6 post-infection, IL-36γ-KO mice had more diffuse immune infiltrate around the bronchioles compared to WT mice (Figure 3E). Together, these data point to a role for IL-36γ in controlling the early viral replication and limiting the resulting immunopathology following influenza infection.
Figure 3: Viral titers, cytokine levels, and pathology of WT and IL-36γ-KO mice following influenza infection.
Age- and sex-matched C57Bl/6J (filled bars) and Il36g−/− mice (open bars) were infected with (A) influenza PR8, (B) 30,000 EID50 ×31, or (C) 300,000 EID50 ×31 and lungs were analyzed for influenza NP copy number. (D) Bronchoalveolar lavage fluid was analyzed for expression of proinflammatory cytokines on day 3 post-infection in C57Bl/6J (filled bars) and Il36g−/− mice (open bars). (E) Example images of formalin-fixed, paraffin-embedded, H&E-stained lungs from C57Bl/6J and Il36g−/− mice. Inflamed bronchioles (B) and small arterioles (SA) are noted on the images. For A-D, data are combined from 2–3 independent replicate experiments of at least 5 mice per group per replicate. Males and females were used in equal proportions. Asterisks indicate significant differences by Student’s t test on log-transformed data.
Alveolar macrophages are rapidly depleted following influenza infection in IL-36γ-deficient mice
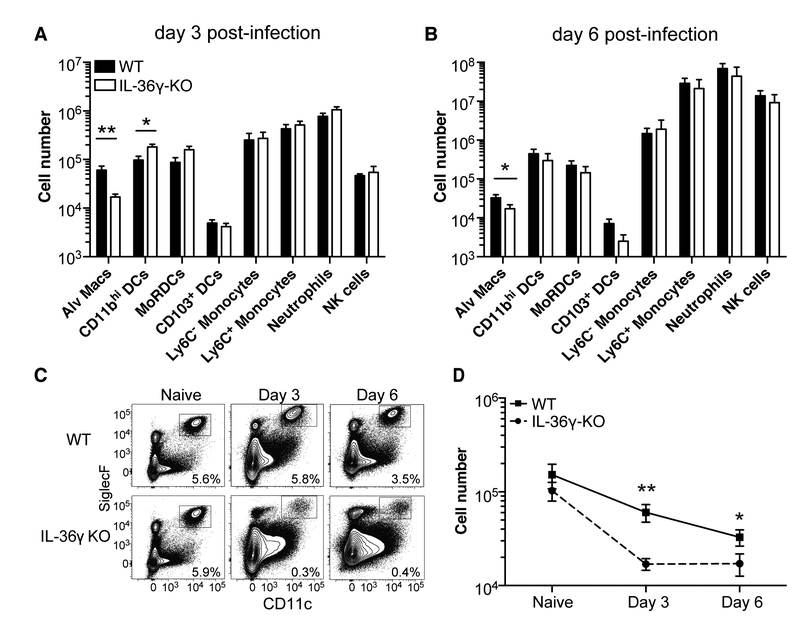
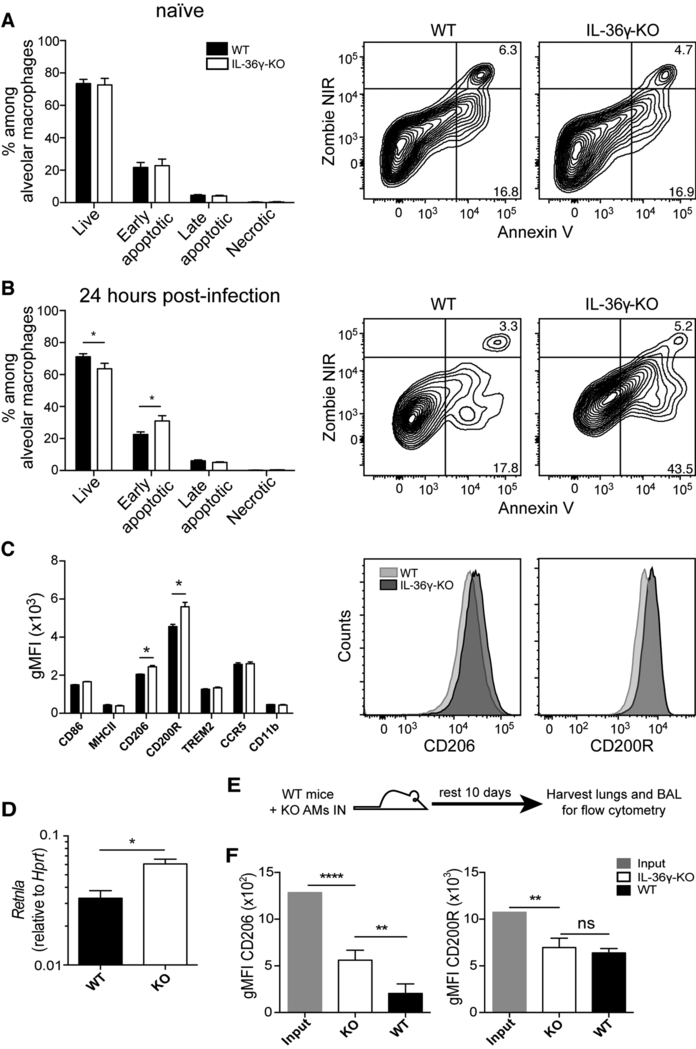
The increased morbidity and viral titers observed in IL-36γ-KO mice were evident within three days post-infection, suggesting a defect in early innate immunity. As many different immune cell types contribute to the early response during influenza infection (6), we examined multiple innate and adaptive immune cell subsets in WT and IL-36γ-KO mice following influenza x31 infection using a 13-color flow cytometry panel (Supplemental Figure 2). As shown in Figure 4A and 4B, the number of alveolar macrophages was significantly decreased in IL-36γ-KO mice on days 3 and 6 post-infection, whereas no other immune cell subsets examined showed any differences across both time points. Although there were increased numbers of CD11bhi DCs in IL-36γ-KO mice on day 3 post-infection, this difference was not evident at day 6, and seemed unlikely to account for the increased severity of influenza infection in IL-36γ-KO mice. To determine whether the decrease in alveolar macrophages was the result of a homeostatic defect in IL-36γ-KO mice, we next compared the frequency and number of lung alveolar macrophages in WT and IL-36γ-KO mice prior to, or following, influenza infection. As shown in Figures 4C and 4D, alveolar macrophage frequency and numbers are similar in naïve WT and IL-36γ-KO mice, but rapidly decline in IL-36γ-KO mice following infection. Thus, in the absence of IL-36γ, lung alveolar macrophages are rapidly lost following influenza infection.
Figure 4: Analysis of innate immune subsets in the lung by flow cytometry.
C57Bl/6J (filled bars) and Il36g−/− mice (open bars) were infected with 300,000 EID50 ×31 influenza and the numbers of different immune cell subsets in the lung were assessed at (A) day 3 and (B) day 6 post-infection. (C) Representative flow plots of live CD45.2+ cells are shown with gates denoting alveolar macrophages. (D) The number of alveolar macrophages in C57Bl/6J (solid line) and Il36g−/− mice (dashed line) over time following influenza infection. Data are combined from two-three independent replicates of at least 5 mice per group per replicate. Males and females were used in equal proportions. Asterisks indicate significant differences in cell number by Student’s t test on log-transformed data.
Alveolar macrophages from IL-36γ-KO mice have an M2-like phenotype
We next investigated the potential mechanism governing alveolar macrophage loss in IL-36γ-KO mice following influenza infection. In naïve mice, there was no difference in the frequency of live or early apoptotic alveolar macrophages by Annexin V and live/dead staining (Figure 5A). However, within one day of influenza infection, we observed a significant decrease in live alveolar macrophages in IL-36γ-KO mice compared to WT mice, with a corresponding increase in early apoptotic cells (Figure 5B). To better understand this difference in apoptosis we examined if there were differences in M1/M2 macrophage skewing between WT and IL-36γ-KO mice, as M1-like macrophages are more resistant to apoptosis (34). We examined the surface expression of classical M1 proteins CD86 and MHCII, M2 proteins CD206 and CD200R, the pro-survival proteins TREM2 and CCR5, as well as CD11b on alveolar macrophages form WT and IL-36γ-KO mice (35–37). Alveolar macrophages isolated from naïve IL-36γ-KO mice had a significant increase in the expression of CD206 and CD200R, markers associated with an M2-like macrophage (Figure 5C). To confirm this result, we sorted out alveolar macrophages from naïve WT and IL-36γ-KO mice and performed RT-PCR for the hallmark M2 gene Retnla. We found that IL-36γ-KO alveolar macrophages had greater expression of Retnla compared to their WT counterparts, supporting our flow cytometric results (Figure 5D). We considered the possibility that IL-36γ may have a role in the conditioning of alveolar macrophages and impact macrophage polarization. Thus, we isolated and transferred alveolar macrophages from IL-36γ-KO into the lungs of WT mice and allowed the cells to rest for 10 days (Figure 5E). When the phenotype of the transferred cells was compared to the pre-transfer phenotype, levels of surface CD200R and CD206 decreased over the 10 days in the presence of IL-36γ (Figure 5F). In fact, CD200R expression was equivalent to the endogenous WT alveolar macrophages while CD206 expression remained slightly elevated. Together, these data show the loss of alveolar macrophages in IL-36γ KO mice following influenza infection is associated with a rapid increase in apoptosis, that alveolar macrophages in IL-36γ-KO mice have a phenotype consistent with a more M2-like macrophage polarization, and that IL-36γ plays a role in alveolar macrophage polarization that may contribute to their decline following influenza infection.
Figure 5: Apoptosis and surface marker expression of WT and IL-36γ-KO mice.
The apoptosis of alveolar macrophages from C57Bl/6J (filled bars) and Il36g−/− mice (open bars) was measured using live/dead and Annexin V staining in (A) naïve mice or (B) 24 hours post-infection. Representative staining is shown gated on alveolar macrophages. (C) Phenotype of alveolar macrophages in naïve C57Bl/6J and Il36g−/− mice for markers associated with M1 and M2 macrophages subsets. (D) Expression of Retnla in alveolar macrophages isolated from naïve WT (black bars) and Il36g−/− (open bars) alveolar macrophages. (E and F) Geometric mean fluorescence intensity of CD206 and CD200R on input Il36g−/− alveolar macrophages (grey bars), Il36g−/− alveolar macrophages after 10 days in a WT mouse (open bars, or the host alveolar macrophages (black bars). Data are combined from 2–3 independent replicates of at least 5 mice per group per replicate. Males and females were used in equal proportions. Asterisks indicates significant differences in cell number by ANOVA followed by Tukey’s multiple comparisons test (A and B), Student’s t test (C, D WT vs KO), or the one-sample T test (D, input vs KO).
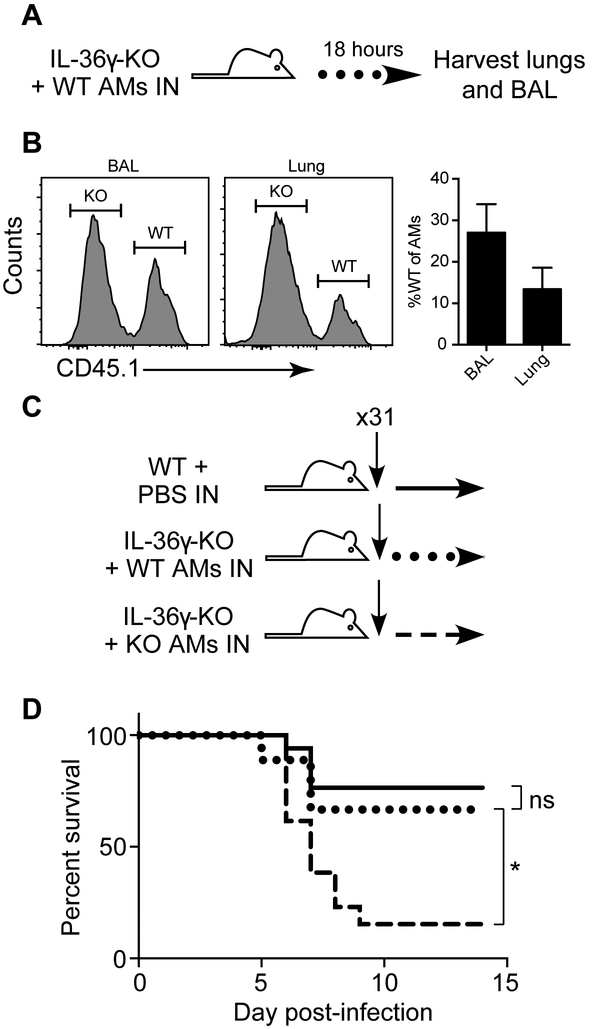
Transfer of wild-type alveolar macrophages restores protection against influenza infection in IL-36γ-deficient mice
Alveolar macrophages are known to be critical for the early control of viral replication and protection against severe influenza infection (12, 13). Therefore, we sought to determine if complementing IL-36γ-KO mice with WT alveolar macrophages prior to influenza infection would be sufficient to protect against severe influenza disease. To confirm our method transferred alveolar macrophages in physiologically relevant numbers, WT alveolar macrophages were transferred to IL-36γ-KO mice and the BAL and lung were harvested the next day (Figure 6A). We found that 30% of alveolar macrophages in the BAL and 15% of those in the lung were transferred WT cells, confirming that physiological numbers of cells were transferred (Figure 6B). Next, we isolated alveolar macrophages from the lungs of naïve WT or IL-36γ-KO mice, and transferred equal numbers intranasally to separate cohorts of IL-36γ-KO mice one day prior to influenza infection (Figure 6C). As shown in Figure 6D, IL-36γ-KO mice that received WT alveolar macrophages showed a significant increase in survival compared to IL-36γ-KO mice that received IL-36γ-KO alveolar macrophages. The survival of IL-36γ-KO mice that received WT alveolar macrophages was equivalent to WT mice. Thus, the defect in influenza immunity observed in IL-36γ-KO mice resulting in severe disease can be rescued by the presence of WT alveolar macrophages.
Figure 6: Transfer of WT alveolar macrophages rescues mortality in IL-36γ-KO mice.
(A) Il36g−/− mice were transferred with 5×105 purified WT alveolar macrophages intranasally and (B) lungs were harvested the next day for identification of the transferred cells on the congenic marker CD45.1. Representative staining gated on live alveolar macrophages and percentage WT (transferred cells) are shown. (C) Age- and sex-matched Il36g−/− mice were transferred with 5×105 purified Il36g−/− alveolar macrophages intranasally (dashed line), and a matched cohort of Il36g−/− mice (dotted line) received 5×105 purified WT alveolar macrophages intranasally. C57Bl/6J (solid line) mice were mock transferred with PBS. Data are combined from 2 independent replicate experiments of at least 4 mice per group. All recipients were male mice to prevent rejection on Y antigens. Asterisk indicate a significant difference in survival by the log-rank test.
DISCUSSION
We have shown that IL-36γ is uniquely upregulated among IL-36 cytokines during the first 6 days of influenza infection. Mice lacking IL-36γ have increased morbidity and mortality following influenza infection and this increase is associated with increased viral titers, higher levels of inflammatory cytokines in the airways, and more diffuse pathology in IL-36γ-KO mice. Analysis of innate cell subsets revealed a significant decrease in alveolar macrophages in IL-36γ-KO mice as soon as 3 days post-infection that continues through day 6 post infection. Compared to their WT counterparts, alveolar macrophages in IL-36γ-KO mice were skewed towards an M2-like phenotype by surface marker and gene expression prior to influenza infection and showed increased apoptosis within 24 hours of infection. Transfer of WT alveolar macrophages into the lungs of IL-36γ-KO mice increased survival compared to IL-36γ-KO mice that received IL-36γ-KO alveolar macrophages, and the survival of WT-transferred IL-36γ-KO mice was equivalent to mock-transferred WT mice.
Early control of viral replication in the lung is key to preventing dissemination of the virus throughout the lung. Several cell types have been shown to have a role in the innate immune response to influenza, including neutrophils, natural killer cells, and alveolar macrophages. However, of these, only alveolar macrophages are situated within the respiratory tract prior to infection and can respond immediately without the need for recruitment from the vasculature or bone marrow (7–9). Our data and previously published studies indicate a role for alveolar macrophages in controlling viral replication during the first three days of infection, before neutrophils and natural killer cells arrive to the lungs en masse. IL-36γ-KO mice, which rapidly lose alveolar macrophages during influenza infection, have increased viral titers, presumably leading to increased inflammation and immunopathology. Indeed, we detected more IL-6 and IFNβ in the airways of IL-36γ-KO mice at day 3 post-infection. A more robust cytokine response early during infection correlated with more diffuse immune infiltrate late in the infection, presumably leading to an acute respiratory distress syndrome (ARDS)-like state in IL-36γ-KO mice and increased mortality.
Cytokines function as key factors for communication between immune cells during influenza infection. The outcome of influenza infection is not only dependent on the presence or absence of particular cytokines, but also their levels within the tissue. For example, production of IFNα is correlated with symptom onset in humans and can boost the production of other inflammatory mediators, but in mice, deletion of its receptor in mice leads to increased IL-8 production, increased neutrophil recruitment, and greater mortality upon lethal influenza challenge (17, 38). Another example of the importance of maintaining a balanced cytokine response for proper immune function comes from studies of IL-1. Deletion of the IL-1 receptor leads to decreased pathology and neutrophil recruitment, but also delays viral clearance and increases mortality (39). IL-1 is thus key to survival of influenza infection, but also contributes to the development of ARDS in humans (40). We find that lack of IL-36γ has a similar effect to deletion of IL-1R on the outcome of influenza infection, albeit by a different mechanism. In both cases, the mice without the cytokine pathway of interest have an extended viremia and suffer greater morbidity and mortality. However, in IL-36γ-KO mice, alveolar macrophages are rapidly lost from the lung due to increased apoptosis. This removes a key blockade to early viral replication and induces greater proinflammatory cytokines (similar to observations in IFNαR-KO mice) leading to increased pathology as the infection progresses. We find that in naïve mice, Il36g mRNA is produced at much higher levels than either Il36a or Il36b and other reports have found that IL-36γ protein is detectable in the BAL of naïve mice (30, 41). This baseline production of IL-36γ may act on alveolar macrophages to maintain a more M1-like phenotype in the absence of infection. Following influenza infection, neutrophils have the highest expression of Il36g among immune cells, which differs from the dogmatic production of the cytokine by epithelium, dendritic cells, or CD4+ T cells (42). Il36g production by neutrophils has only been observed in experimental autoimmune encephalitis, a mouse model of multiple sclerosis, and it is notable that full length IL-36γ is cleaved to its more active form by neutrophil-derived elastase and proteinase-3 (43, 44). As neutrophils produce both the cytokine and activating protease, it is possible that neutrophils release cleaved IL-36γ, but confirming that hypothesis requires further work. While our data suggest that baseline production of IL-36γ is important for macrophage polarization in naïve mice, it is also possible the IL-36γ produced by neutrophils contributes to the survival of alveolar macrophages during the early stages of influenza infection.
IL-36 family cytokines have been shown to have a key role in pulmonary immune responses to bacterial and viral infection as well as the development of asthma. In mice, a SNP regulating expression of IL-36γ determines the asthma sensitivity or resistance of two inbred mouse lines, and house dust mite administration causes rapid expression of IL-36γ in experimental asthma models (28, 45). Respiratory infection of mice with either Gram-positive or Gram-negative bacteria induce production of IL-36γ, and antibody blockade or genetic deletion of Il36g causes greater morbidity and mortality after infection with S. pneumoniae (30). In this model, mice lacking IL-36γ had greater bacterial load in the lung as well as systemic dissemination. In agreement with our observed increase in surface expression of M2-like markers in naïve mice, that study also found that interstitial and alveolar macrophages from the lungs of IL-36γ-KO mice had decreased expression of classical M1-like genes. Based on our PCR and transfer experiments, IL-36γ from a macrophage-extrinsic source is acting on the alveolar macrophages to maintain a more M1-like profile, as transfer of IL-36γ-KO alveolar macrophages to WT hosts reduces their expression of M2-like markers. While the skewing of IL-36γ-KO alveolar macrophages towards an M2-like cell was not complete, it would explain the decreased survival and increased apoptosis of the cells following influenza infection. Collectively, these data, point towards a key role for IL-36γ in macrophage polarization in the lung.
Deletion of IL-36R led to increased survival and decreased pathology in a lethal influenza infection with PR8(31). Mice lacking IL-36R also had an extended viremia, but decreased pathology and pro-inflammatory cytokines compared to WT mice. Analysis of immune cell subsets in the lungs of IL-36R-KO mice showed decreased neutrophils and monocytes following infection, in contrast to our findings in IL-36γ-KO mice. The differences in survival and cell recruitment between IL-36R-KO and IL-36γ-KO mice following influenza infection may indicate differential roles for the various IL-36 family members in the immune response. Despite similar EC50 values in vitro, IL-36γ has weaker affinity than IL-36α (KD= 1800 vs 480 nM, respectively) for IL-36 receptor, possibly affecting how the ligands signal through their receptor (46, 47). Higgens et al. showed differential effects of IL-36α and IL-36β in conditioning MDDCs for T cell stimulation (48). Aoyagi et al. showed that IL-36R-KO and IL-36γ-KO, but not IL-36α-KO mice are protected during lethal pulmonary infection with P. aeruginosa (41). These data together with our findings suggest differential roles for IL-36 family members during infection. It is possible that IL-36γ acts to oppose signaling from another IL-36 family member, or has an effect in the lungs of naïve mice which is only seen with genetic deletion of the cytokine and not of the receptor. Future studies are necessary to dissect the roles of individual IL-36 family members in regulating lung immune function during homeostasis and following infection.
In conclusion, we have observed that the rapid apoptosis of alveolar macrophages in IL-36γ-KO mice following influenza infection results in increased viral load, increased proinflammatory cytokines, and enhanced immunopathology leading to decreased survival. Importantly, this defect can be corrected by the transfer of wild-type alveolar macrophages into the lungs of IL-36γ-KO mice. Overall, we demonstrate a critical role for IL-36γ in the immune response to influenza infection.
Supplementary Material
Acknowledgements
We thank Dr. Tamas Nagy, DVM, PhD for pathological examination of mouse lungs. This paper utilized shared resources at Emory University including the Emory+Children’s Flow Cytometry Core and the Winship Research Pathology Core Lab. We would also like to thank Sarah Hayward and Emily Cartwright for helpful discussion and Laurel Lawrence for invaluable help in managing our animal colony.
Financial Support
Funding for this project was provided by National Institutes of Health grant R01HL122559 (to JEK) and Emory University. PRD was supported by National Institutes of Health grant F31AI124611, and SRM was supported by National Institutes of Health grant F30HL118954.
REFERENCES
- 1.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, and Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289: 179–186. [DOI] [PubMed] [Google Scholar]
- 2.Dolin R 2012. Chapter 187. Influenza In Harrison’s Principles of Internal Medicine, 18 ed. Longo DL, Fauci AS, Kasper DL, Hauser SL, Jameson J, and Loscalzo J, eds. McGraw-Hill, New York. [Google Scholar]
- 3.Loo Y-M, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, and Gale M. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol 82: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichinohe T 2010. Respective roles of TLR, RIG-I and NLRP3 in influenza virus infection and immunity: impact on vaccine design. Expert Rev Vaccines 9: 1315–1324. [DOI] [PubMed] [Google Scholar]
- 5.Iwasaki A, and Pillai PS. 2014. Innate immunity to influenza virus infection. Nat. Rev. Immunol 14: 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulendran B, and Maddur MS. 2014. Innate Immune Sensing and Response to Influenza In Current Topics in Microbiology and Immunology. Current Topics in Microbiology and Immunology Springer Berlin; Heidelberg, Berlin, Heidelberg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim K, Hyun Y-M, Lambert-Emo K, Capece T, Bae S, Miller R, Topham DJ, and Kim M. 2015. Neutrophil trails guide influenza-specific CD8⁺ T cells in the airways. Science 349: aaa4352–aaa4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz-Cherry S 2015. Role of NK cells in influenza infection. Curr. Top. Microbiol. Immunol 386: 109–120. [DOI] [PubMed] [Google Scholar]
- 9.Weiss ID, Wald O, Wald H, Beider K, Abraham M, Galun E, Nagler A, and Peled A. 2010. IFN-gamma treatment at early stages of influenza virus infection protects mice from death in a NK cell-dependent manner. J. Interferon Cytokine Res 30: 439–449. [DOI] [PubMed] [Google Scholar]
- 10.Lin KL, Suzuki Y, Nakano H, Ramsburg E, and Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J. Immunol 180: 2562–2572. [DOI] [PubMed] [Google Scholar]
- 11.Camp JV, and Jonsson CB. 2017. A Role for Neutrophils in Viral Respiratory Disease. Front Immunol 8: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardani A, Boulton A, Kim TS, and Braciale TJ. 2017. Alveolar Macrophages Prevent Lethal Influenza Pneumonia By Inhibiting Infection Of Type-1 Alveolar Epithelial Cells. PLoS Pathog. 13: e1006140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider C, Nobs SP, Heer AK, Kurrer M, Klinke G, van Rooijen N, Vogel J, and Kopf M. 2014. Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 10: e1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taubenberger JK, and Morens DM. 2008. The pathology of influenza virus infections. Annu Rev Pathol 3: 499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TKT, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen K-Y, Writing Committee of the World Health Organization (WHO) Consultation on Human Influenza A/H5. 2005. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med 353: 1374–1385. [DOI] [PubMed] [Google Scholar]
- 16.Guo X-ZJ, and Thomas PG. 2017. New fronts emerge in the influenza cytokine storm. Semin Immunopathol 39: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seo S-U, Kwon H-J, Ko H-J, Byun Y-H, Seong BL, Uematsu S, Akira S, and Kweon M-N. 2011. Type I interferon signaling regulates Ly6C(hi) monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 7: e1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, and Muster T. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252: 324–330. [DOI] [PubMed] [Google Scholar]
- 19.Bot A, Bot S, and Bona CA. 1998. Protective role of gamma interferon during the recall response to influenza virus. J. Virol 72: 6637–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrone LA, Szretter KJ, Katz JM, Mizgerd JP, and Tumpey TM. 2010. Mice lacking both TNF and IL-1 receptors exhibit reduced lung inflammation and delay in onset of death following infection with a highly virulent H5N1 virus. J. Infect. Dis 202: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hussell T, Pennycook A, and Openshaw PJ. 2001. Inhibition of tumor necrosis factor reduces the severity of virus-specific lung immunopathology. Eur. J. Immunol 31: 2566–2573. [DOI] [PubMed] [Google Scholar]
- 22.Salomon R, Hoffmann E, and Webster RG. 2007. Inhibition of the cytokine response does not protect against lethal H5N1 influenza infection. Proc Natl Acad Sci USA 104: 12479–12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dienz O, Rud JG, Eaton SM, Lanthier PA, Burg E, Drew A, Bunn J, Suratt BT, Haynes L, and Rincon M. 2012. Essential role of IL-6 in protection against H1N1 influenza virus by promoting neutrophil survival in the lung. Mucosal Immunology 5: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teijaro JR 2014. The Role of Cytokine Responses During Influenza Virus Pathogenesis and Potential Therapeutic Options In. Current Topics in Microbiology and Immunology Springer Berlin Heidelberg, Berlin, Heidelberg: 1–20. [DOI] [PubMed] [Google Scholar]
- 25.Brun-Buisson C, Richard J-CM, Mercat A, Thiébaut ACM, Brochard L, REVA-SRLF A/H1N1v 2009 Registry Group. 2011. Early corticosteroids in severe influenza A/H1N1 pneumonia and acute respiratory distress syndrome. Am J Respir Crit Care Med 183: 1200–1206. [DOI] [PubMed] [Google Scholar]
- 26.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, and Katze MG. 2012. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev 76: 16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medina-Contreras O, Harusato A, Nishio H, Flannigan KL, Ngo V, Leoni G, Neumann P-A, Geem D, Lili LN, Ramadas RA, Chassaing B, Gewirtz AT, Kohlmeier JE, Parkos CA, Towne JE, Nusrat A, and Denning TL. 2015. Cutting Edge: IL-36 Receptor Promotes Resolution of Intestinal Damage. The Journal of Immunology 196: 1501312–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramadas RA, Ewart SL, Medoff BD, and LeVine AM. 2011. Interleukin-1 family member 9 stimulates chemokine production and neutrophil influx in mouse lungs. Am J Respir Cell Mol Biol 44: 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tortola L, Rosenwald E, Abel B, Blumberg H, Schäfer M, Coyle AJ, Renauld J-C, Werner S, Kisielow J, and Kopf M. 2012. Psoriasiform dermatitis is driven by IL-36-mediated DC-keratinocyte crosstalk. J. Clin. Invest 122: 3965–3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kovach MA, Singer B, Martinez-Colon G, Newstead MW, Zeng X, Mancuso P, Moore TA, Kunkel SL, Peters-Golden M, Moore BB, and Standiford TJ. 2017. IL-36γ is a crucial proximal component of protective type-1-mediated lung mucosal immunity in Gram-positive and -negative bacterial pneumonia. Mucosal Immunology 119: 1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoyagi T, Newstead MW, Zeng X, Kunkel SL, Kaku M, and Standiford TJ. 2017. IL-36 receptor deletion attenuates lung injury and decreases mortality in murine influenza pneumonia. Mucosal Immunology 10: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harusato A, Abo H, Ngo VL, Yi SW, Mitsutake K, Osuka S, Kohlmeier JE, Li JD, Gewirtz AT, Nusrat A, and Denning TL. 2017. IL-36γ signaling controls the induced regulatory T cell-Th9 cell balance via NFκB activation and STAT transcription factors. Mucosal Immunology 10: 1455–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bochkov YA, Hanson KM, Keles S, Brockman-Schneider RA, Jarjour NN, and Gern JE. 2010. Rhinovirus-induced modulation of gene expression in bronchial epithelial cells from subjects with asthma. Mucosal Immunology 3: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pachulec E, Abdelwahed Bagga RB, Chevallier L, O’Donnell H, Guillas C, Jaubert J, Montagutelli X, Carniel E, and Demeure CE. 2017. Enhanced Macrophage M1 Polarization and Resistance to Apoptosis Enable Resistance to Plague. J. Infect. Dis. 216: 761–770. [DOI] [PubMed] [Google Scholar]
- 35.Rőszer T 2015. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediators Inflamm. 2015: 816460–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu K, Byers DE, Jin X, Agapov E, Alexander-Brett J, Patel AC, Cella M, Gilfilan S, Colonna M, Kober DL, Brett TJ, and Holtzman MJ. 2015. TREM-2 promotes macrophage survival and lung disease after respiratory viral infection. Journal of Experimental Medicine 212: 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyner JW, Uchida O, Kajiwara N, Kim EY, Patel AC, O’Sullivan MP, Walter MJ, Schwendener RA, Cook DN, Danoff TM, and Holtzman MJ. 2005. CCL5-CCR5 interaction provides antiapoptotic signals for macrophage survival during viral infection. Nature Medicine 11: 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, and Straus SE. 1998. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J. Clin. Invest 101: 643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz N, Kurrer M, Bachmann MF, and Kopf M. 2005. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J. Virol 79: 6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pugin J, Ricou B, Steinberg KP, Suter PM, and Martin TR. 1996. Proinflammatory activity in bronchoalveolar lavage fluids from patients with ARDS, a prominent role for interleukin-1. Am J Respir Crit Care Med 153: 1850–1856. [DOI] [PubMed] [Google Scholar]
- 41.Aoyagi T, Newstead MW, Zeng X, Nanjo Y, Peters-Golden M, Kaku M, and Standiford TJ. 2017. Interleukin-36γ and IL-36 receptor signaling mediate impaired host immunity and lung injury in cytotoxic Pseudomonas aeruginosa pulmonary infection: Role of prostaglandin E2. PLoS Pathog. 13: e1006737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gabay C, and Towne JE. 2015. Regulation and function of interleukin-36 cytokines in homeostasis and pathological conditions. Journal of Leukocyte Biology 1–8. [DOI] [PubMed] [Google Scholar]
- 43.Bozoyan L, Dumas A, Patenaude A, and Vallières L. 2015. Interleukin-36γ is expressed by neutrophils and can activate microglia, but has no role in experimental autoimmune encephalomyelitis. J Neuroinflammation 12: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henry CM, Sullivan GP, Clancy DM, Afonina IS, Kulms D, and Martin SJ. 2016. Neutrophil-Derived Proteases Escalate Inflammation through Activation of IL-36 Family Cytokines. CellReports 14: 1–16. [DOI] [PubMed] [Google Scholar]
- 45.Ramadas RA, Li X, Shubitowski DM, Samineni S, Wills-Karp M, and Ewart SL. 2006. IL-1 Receptor antagonist as a positional candidate gene in a murine model of allergic asthma. Immunogenetics 58: 851–855. [DOI] [PubMed] [Google Scholar]
- 46.Towne JE, Renshaw BR, Douangpanya J, Lipsky BP, Shen M, Gabel CA, and Sims JE. 2011. Interleukin-36 (IL-36) ligands require processing for full agonist (IL-36α, IL-36β, and IL-36γ) or antagonist (IL-36Ra) activity. J. Biol. Chem 286: 42594–42602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou L, Todorovic V, Kakavas S, Sielaff B, Medina L, Wang L, Sadhukhan R, Stockmann H, Richardson PL, DiGiammarino E, Sun C, and Scott V. 2017. Quantitative ligand and receptor binding studies reveal the mechanism of interleukin-36 (IL-36) pathway activation. J. Biol. Chem jbc.M117.805739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Higgins J, Mutamba S, Mahida Y, Barrow P, and Foster N. 2015. IL-36α induces maturation of Th1-inducing human MDDC and synergises with IFN-γ to induce high surface expression of CD14 and CD11c. Hum. Immunol 76: 1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.