Abstract
Objective
Brain-gut-microbiota interactions may play an important role in human health and behavior. However, while rodent models have demonstrated effects of the gut microbiota on emotional, nociceptive and social behaviors, there is little translational human evidence to date. In this study we identify brain and behavioral characteristics of healthy women clustered by gut microbiota profiles.
Methods
Forty women supplied fecal samples for 16s rRNA profiling. Microbial clusters were identified using Partitioning Around Medoids. Functional magnetic resonance imaging was acquired. Microbiota-based group differences were analyzed in response to affective images. Structural and diffusion tensor imaging provided gray matter metrics (volume, cortical thickness, mean curvature, surface area) as well as fiber density between regions. A sparse Partial Least Square-Discrimination Analysis was applied to discriminate microbiota-clusters using white and gray matter metrics.
Results
Two bacterial genus-based clusters were identified, one with greater Bacteroides abundance (n=33), one with greater Prevotella abundance (n=7). The Prevotella group showed less hippocampal activity viewing negative valences images. White and gray matter imaging discriminated the two clusters, with accuracy of 66.7% and 87.2% respectively. The Prevotella cluster was associated with differences in emotional, attentional, and sensory processing regions. For gray matter, the Bacteroides cluster showed greater prominence in the cerebellum, frontal regions, and the hippocampus.
Conclusions
These results support the concept of brain-gut-microbiota interactions in healthy humans. Further examination of the interaction between gut microbes, brain and affect in humans is needed to inform preclinical reports that microbial modulation may affect mood and behavior.
INTRODUCTION
Comprised of trillions of organisms and responsible for numerous biologically important processes, the human microbiota has a role in health and disease that is increasingly evident (1, 2). One area of particular interest is the role of the gut microbiota within the gut-brain axis and their relationship to emotional processing (3, 4). While spatially separated from the brain by both the intestinal epithelial barrier and the blood brain barrier, there is bidirectional communication between the gut microbiota and the central nervous system (CNS) via the vagus nerve, the immune system, and neuroactive metabolites released into systemic circulation (5).
Evidence for the influence of the microbiota on the CNS has been plentiful in the preclinical literature and suggests that beyond brain development, the microbiota can influence behavior and affect (5, 6). Animals raised in germ-free environments exhibit altered brain chemistry as well as changes in behavior, with increased risk taking, reduced anxiety, and decreased sociability (3, 5, 7–10). Some of the biochemical changes resulting from germ-free status are irreversible, even after colonization of the animals with normal gut microbiota later in life. Other abnormalities, such as anxiety behavior, can be ameliorated after reconstitution of the gut microbiota (7).
Along with the preclinical evidence, emerging evidence in humans suggests that while some aspects of the microbiota’s influence on the CNS are likely to be established early as traits, other aspects may be malleable and are vulnerable to environmental factors (11, 12). For example, in the only study to date showing interactions between brain structure and gut microbes, results suggested that obese patients had brain structural changes in the hypothalamus and caudate nucleus associated with specific microbiota profiles and changes in cognitive function. However, in this cross-sectional design, it was not clear whether these changes may have been a risk factor for, or a result of, obesity status (13). Small studies of probiotic interventions have shown modest effects on mood and variable effects on cognition, leading to speculations that gut bacteria may be evolutionarily programmed to improve our moods, thus making us more social and prone to activities that allow person-to-person transmission of the organisms (14–17). We have previously demonstrated in healthy women that 4-week ingestion of a fermented milk product with probiotics can shift functional brain responses to an emotional attention task towards lower reactivity in viscerosensory, somatosensory, and affective regions, providing more direct evidence in humans that brain function can be affected by modulation of the gut microbiota (18).
In this study, using fecal samples, magnetic resonance imaging (MRI), and an emotion induction task obtained in conjunction with our previous study (18), we aimed to identify whether the gastrointestinal microbiota composition in healthy women was associated with characteristics of brain structure, brain structural white-matter connectivity, and brain function as measured by affective response to emotionally valenced images.
METHODS
Subjects
Healthy, non-obese women aged 18–55 were recruited by advertisement. A medical history, physical exam, and standardized psychiatric screening exam (Mini International Neuropsychiatric Interview Plus 5.0)(19) were performed. Exclusion criteria included: use of antibiotics or probiotics in the month prior, active pain disorder, active medical or psychiatric illness, tobacco dependence, pregnancy or lactation, exercise >8 hours/week, metallic implants, claustrophobia, body mass index >30 or <18. Subjects taking central nervous system (antidepressants including serotonin reuptake inhibitors and serotonin/norepinephrine reuptake inhibitors, sedatives or anxiolytics) medications or use of opioid analgesics were excluded. Informed consent was obtained from all subjects. Data acquisition was performed between January 2009 and December 2012. All procedures complied with the principles of the Declaration of Helsinki and were approved by the Institutional Review Board at UCLA.
Behavioral and Clinical Measures
Questionnaires were completed before MRI scans. A 2-week electronic diary was used to assess gastrointestinal symptoms prior to MRI. Subjects were excluded if they reported abdominal pain or discomfort on 2 or more days, abnormal stool form (Bristol stool scale 1, 6, or 7) or stool frequency of less than 3 bowel movements per week or greater than 3 bowel movements per day (20). The Positive Affect Negative Affect Schedule (PANAS) was used to measure positive and negative affect (21, 22). The Hospital Anxiety and Depression Scale (23) was used to assess active mood symptoms and the Spielberger Trait Anxiety Inventory (24) was collected.
Stool collection and 16S rRNA pyrosequencing
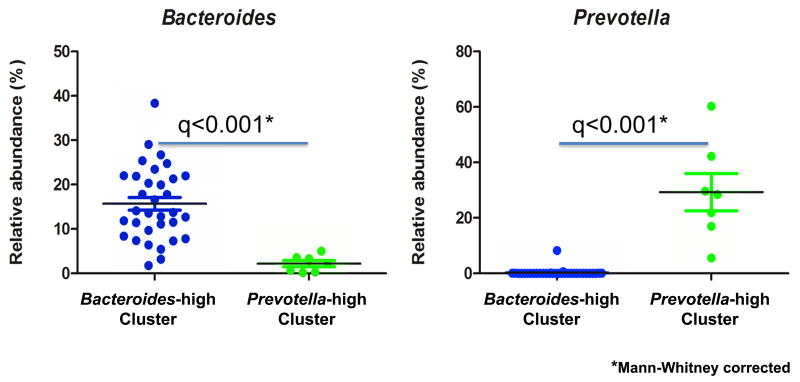
Subjects were provided stool collection kits with a sterile collection container, ice packs and a freezer bag for transport. They were instructed to collect the stool on the day prior to their MRI visit, freeze it in their home freezer and then transport it on ice packs to the Research Center where the stool was frozen at −80 degrees until analysis. Samples were shipped frozen to the laboratory facilities at Danone Research (Palaiseau, France) where it underwent 16S rRNA profiling using 454 pyrosequencing (V5–V6 variable regions). V5 and V6 hypervariable 16S ribosomal RNA (rRNA) regions were amplified using primers 784F and 1061R (25). Sequencing was performed by DNAVision SA (Charleroi, Belgium) on a 454 Life Sciences Genome Sequencer FLX instrument (Roche) using titanium chemistry and primer A. Analyses were performed using Quantitative Insights Into Microbial Ecology (QIIME) v1.7 (26). Reads were filtered according to the following quality criteria: size between 150 and 500nt, quality above 25 over a 50 base pairs window, no mismatch authorized in primers and barcode sequences, and absence of polymers larger than 6nt. Remaining sequences were denoised using default parameters and clustered into Operational Taxonomic Units (OTUs) defined at 97% identity using CD-HIT (27). Representative sequences for each OTU were aligned and taxonomically assigned using Greengenes (August 2011). ChimeraSlayer was used to discard potential chimeric sequences (28) leading to 5943 +/− 3100 (mean +/− SD) reads per sample. Normalization was performed using relative abundances. Beta diversity was performed on weighted Unifrac distances using rarefaction of 1770 sequences per sample. To cluster the samples based on weighted distance matrix, we used the Partitioning Around Medoids (PAM) algorithm in the R package ‘Cluster’. The optimal number of clusters was two, as it had the highest Rousseeuw's Silhouette internal cluster quality index (SI=.36), calculated using the R package ‘Cluster’ (29). The resulting microbial clusters showed one with greater Prevotella abundance (Prevotella-high, n=7) and one with greater Bacteroides abundance (Bacteroides-high, n=32) (Figure 1), SI=.36. The variance accounted for by the two components that defined the microbiota clusters totaled 57.2% (Component 1=39.5%, Component 2= 17.7%). These two clusters were utilized for the analyses described below. To identify demographic factors which may confound the cluster designations, t-tests were performed for age, body mass index, anxiety and depression (HAD score), and positive and negative affect using Statistical Package for the Social Sciences (SPSS) software (version 19).
Figure 1. Identification of bacterial clusters.
Plot of the relative abundance of Bacteroides and Prevotella are shown for each cluster, Bacteroides-high (N=32) and Prevotella-high (N=7).
MRI Data Acquisition
Subjects were scanned on a 3-Tesla Siemens Trio after a sagittal scout was used to position the head. Structural scans were acquired using a high-resolution 3-dimensional T1-weighted, sagittal magnetization-prepared rapid gradient echo (MP-RAGE) protocol: Repetition Time (TR) = 20ms, echo time (TE) = 3.00ms, flip angle (FA) = 25°, field of view (FOV) = 256 mm, acquisition matrix = 256×256, slice thickness 1mm, voxel size 1×1×1mm3. Functional scans were acquired in transverse orientation, interleaved, with TR = 2500ms, TE = 26ms, FA = 90°, FOV = 200mm, acquisition matrix of 64×64mm, slice thickness 3.0mm, and voxel size 3.1×3.1×3.0mm3. Diffusion weighted MRIs (DTI) were acquired in 64 non-collinear directions with b = 1000 s/mm2 images with the following protocol: TR = 7000ms, TE = 93ms, and FOV = 190mm with an acquisition matrix of 96×96, and a slice thickness of 2mm to produce 2×2×2mm3 isotropic voxels.
Functional MRI (fMRI) and emotion induction task
Functional MRI was performed in three runs. During each run the subjects viewed mood inducing negative, positive or neutral valence pictures presented in four blocks of the same valence. Negative and positive runs were counterbalanced with the neutral run in the second position. Each run contained 24 pictures of the same valence presented in 4 blocks. Pictures were presented for 5 seconds each, and a 30 second interblock interval (during which a crosshair was presented) separated each block. The images were selected from the International Affective Picture Set to have moderate arousal and valence (9 point scale) based on the mean scores for women.(30). Arousal and valence scores are presented in Table 1. They were converted to black and white and were luminosity adjusted.
Table 1.
Mean valence and arousal of IAPS pictures based on means from adult women.
| IAPS scores | Positive images | Negative images | Neutral images | |||
|---|---|---|---|---|---|---|
| valence | arousal | valence | arousal | valence | arousal | |
| Mean | 7.30 | 4.50 | 2.84 | 5.05 | 5.13 | 5.00 |
| Standard deviation | .53 | .84 | .57 | .89 | .55 | .81 |
The first 2 volumes were discarded to allow for stabilization of the magnetic field. The remaining functional images were slice-time and motion corrected, spatially normalized to the Montreal Neurologic Institute template, and spatially smoothed with an 8mm3 Gaussian kernel using SPM8 (Welcome Department of Cognitive Neurology, London, UK). The experiment was analyzed in a block design. A first-level fixed effects general linear model (GLM) was applied in SPM8 to determine blood oxygen level dependent (BOLD) activity by specifying as regressors, the 30 second baseline and the three valence conditions (negative, positive, neutral). At the subject-level, regressors for the conditions were convolved with a canonical hemodynamic response function to estimate brain activity. Individual brain responses were determined by subtracting estimated brain activity during the neutral valence image condition from estimates of brain activity during the positive and negative affect conditions. To determine microbiota-based cluster group differences, a GLM was implemented at the second-level using a region of interest (ROI) approach. ROIs defined by the Destrieux atlas (31) included the extended emotional arousal network (amygdala, anterior cingulate and hippocampus). An initial cluster defining threshold of p<.001 was applied and cluster based extent threshold significance was p<.05 after controlling for family wise error as implemented in SPM (32).
Structural MRI Analysis
Data pre-processing workflows for the MRI data were designed and created in collaboration with the University of Southern California, Laboratory of Neuroimaging (LONI) Pipeline (pipeline.loni.usc.edu). T1-image segmentation and regional parcellation were conducted using FreeSurfer (33, 34) following the nomenclature described in (31). Based on the Destrieux and Harvard-Oxford atlases, for each cerebral hemisphere, a set of 74 cortical structures were labeled in addition to 7 subcortical structures, the cerebellum and the brain stem, resulting in a complete set of 165 parcellations for the entire brain. Four representative morphological measures were computed for each cortical parcellation: gray matter volume (GMV), surface area (SA), mean cortical thickness (CT), and mean curvature (MC) (35, 36). For subcortical regions only volume was computed.
Diffusion tensors were computed and rotationally re-oriented at each voxel. Tensor-valued images were linearly realigned based on tri-linear interpolation of log-transformed tensors as described in (37) and resampled to an isotropic voxel resolution (2×2×2mm3). White matter connectivity for each subject was estimated between the 165 brain regions using DTI fiber tractography performed via the Fiber Assignment by Continuous Tracking algorithm (38) using TrackVis software. The final estimate of white matter connectivity between each of the brain regions was determined based on the number of fiber tracts intersecting each region, normalized by the total number of fiber tracts within the entire brain (39–42).
Sparse partial least squares Discriminant analysis (sPLS-DA)
A sPLS-DA was used to determine if the brain signatures (gray matter morphology and white-matter connections) could discriminate microbial cluster membership. sPLS-DA simultaneously performs feature selection and modelling and achieves sparsity using lasso penalization (43). sPLS-DA operates using a supervised framework to find orthogonal components, linear combinations of a limited set of variables (brain features) that predict class membership. We refer to each component as discriminatory “brain signature”. sPLS-DA was performed using the R package mixOmics version 5.1(http://www.R-project.org).
Features entered in the sPLS-DA models
The predictive power of brain morphometry and DTI white-matter connectivity were assessed separately. Covariates for all models included age and total GMV. To investigate morphometry, GMV, SA, CT, and MC estimates for the 165 brain regions were entered as predictors. For DTI white-matter connectivity data, subject-specific matrices indexing relative fiber density between the 165 regions were transformed to 1 dimensional matrices containing 13,530 unique connectivities (upper triangle from the initial matrix). These matrices were then concatenated across subjects and entered into the sPLS-DA. As an initial data reduction step, near zero variance predictors were dropped resulting in 2751 predictors for the anatomical based discrimination.
sPLS-DA model specifications and development
For each model, the number of components to identify was fixed at 2 (42, 44). To select the optimal number of features for each component we estimated the 5-fold classification error with respect to a range of number of features (10 to 200 by units of 10). This process was repeated 50 times and the results averaged. This “tuning” procedure indicated that two components comprised of 10 brain features each would be optimal for both the DTI and Morphometric models.
Model summary indices
The discriminatory brain signatures were summarized using variable loadings and VIP coefficients. Each variable has an associated “loading” indexing the relative importance of that variable in the brain signature for group discrimination (42). Variable importance in projection (VIP) scores is a standardized measure that represents contribution of each feature relative to the variance explained by all selected brain signatures (44). Usually, predictors with VIP coefficients greater than one are considered particularly important for discrimination (42). Graphical displays illustrated the discriminative abilities of the algorithms (44). The accuracy of the final models were assessed using leave-one-out cross-validation. This overall error rate or accuracy reflects the number of correct predictions from all the predictions made. During this cross-validation procedure, we calculate the stability of the selected variables comprising a component if a training set is altered (44). Stability is computed by calculating the frequency of selected variables across the cross validation runs. During this cross validation step, it was noted that the classification accuracy for of both models decreased with 2 components, therefore the second component for each model was dropped. PLS-DA is discovery-based method that enables generation of novel hypotheses. Post hoc exploratory evaluation of the specific role of Prevotella and Bacteriodes on specific brain regions and affect was performed by correlating bacterial abundances with brain metrics and PANAS scores (See Tables S1–S3, Supplemental Digital Content 1).
RESULTS
Cluster Characteristics
The total sample of 40 healthy females included 7 subjects in the Prevotella-high group and 33 subjects in the Bacteroides-high group (Figure 1). The mean age of the subjects was 28.89 years, SD 9.87; body mass index (BMI) was normal with a mean of 23.33, SD = 2.69; anxiety and depression symptoms were within normal ranges (mean 3.48, SD = 2.44 and 1.2, SD = 1.81, respectively), and normalized trait anxiety was in the mild to moderate range with a mean of 43.03, SD = 8.87. No differences between the microbiota clusters were identified for age (t = 1.58, p = .12), BMI (t = 1.11, p= .27), anxiety (t = 1.08, p = .29), depression (t = .09, p = .93), or trait anxiety (t = .61, p= .54). Mean PANAS scores for positive and negative affect at baseline and after viewing positive and negative images are shown in Table 2. No difference in baseline positive or negative affect on the PANAS was seen between groups (positive affect t=−.84, p=.41; negative affect t=.16, p=.87). The PANAS questionnaire data was missing for one subject in the Bacteroides-high group so that subject was excluded from the analysis of affect. One subject in the Bacteroides-high group had imaging data that failed quality control measures and was excluded, leaving a total of 39 subjects analyzed for neuroimaging.
Table 2.
Mean and standard deviation (SD) PANAS scores for microbiota based clusters are shown at baseline, after viewing positive IAPS images and after viewing negative IAPS images.
| Prevotella-high group | Bacteroides-high group | |||||||
|---|---|---|---|---|---|---|---|---|
| Positive Affect | Negative Affect | Positive Affect | Negative Affect | |||||
| mean | SD | mean | SD | mean | SD | mean | SD | |
| Baseline | 22.57 | 10.18 | 10.86 | 2.27 | 19.79 | 7.40 | 11.07 | 3.31 |
| Post positive images | 29.57 | 8.34 | 10.57 | 1.51 | 25.93 | 9.60 | 10.28 | .73 |
| Post negative images | 20.43 | 6.58 | 21.86 | 10.19 | 18.24 | 6.29 | 15.90 | 3.76 |
Microbiota cluster-related group differences in emotional response
Changes in positive and negative affect scores (PANAS) obtained at baseline and after each valence block confirmed appropriate affect response to the positive and negative valence blocks. The Prevotella-high cluster group had higher negative affect after viewing the negative valence picture block, p=.012 (21.86, SD=10.2 vs. 15.9, SD=3.8). There was no difference between groups after viewing positive pictures, p=.36 (29.57, SD=8.3 vs. 25.93, SD=9.6). A significantly lower BOLD activity in the right hippocampus during negative stimuli was observed in the Prevotella-high group compared to the Bacteroides-high group (p =.041, familywise error corrected; Z=4.4, cluster size =16). No differences were seen in the other regions during negative emotion condition and no regions of interest were different in the positive emotion condition.
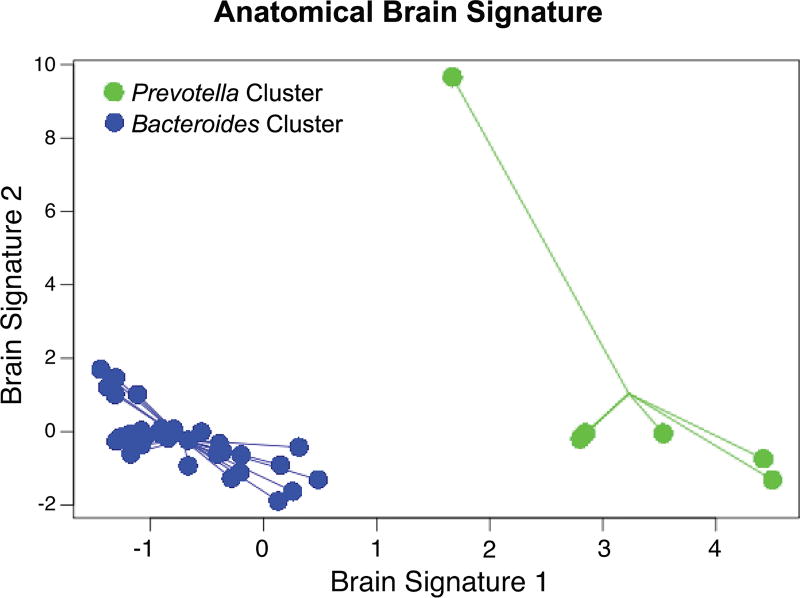
Anatomical white-matter signatures associated with microbiota group clusters
One brain signature comprised of 10 white matter connectivities was able to discriminate between the Bacteroides-high and Prevotella-high clusters. Figure 2 demonstrates this discrimination by plotting individuals with respect to their scores on the signature. This anatomical signature accounted for 85% of the variance in group differences. Overall classification accuracy was 66.7%. This anatomical signature is comprised of 10 brain white-matter connectivities with all connections showing less fiber density in the Bacteroides-high cluster (Table 3). Based on the VIP coefficients, the connectivities having the most explanatory power were between emotional regulation (amygdala, anterior cingulate cortex) and basal ganglia regions as well as attentional (right middle frontal gyrus) and sensory (the right central sulcus) regions. Other sensory connectivities included thalamic connections with the pericallosal sulci and the temporal pole, and connectivity between the left central sulcus and the left posterior midcingulate cortex. Several connections with the temporal gyrus were also major contributors in the prediction. The stability analysis indicated that 7 of 10 selected brain features were quite reliable with >90% stability (See Table 3).
Figure 2. Classification of the white matter connections based on the microbiota group clusters.
Depicts the discriminative abilities of the white-matter classifier based on the bacterial clusters with an overall accuracy of 66.7%. The samples from each group are connected based on the centroid.
Table 3. Brain signature regions: white matter.
This table shows the white-matter connections differentiating the microbiota-based clusters.
Brain region connections with positive loading scores are stronger in the Prevotella-high group.
| Brain Region 1 | ↔ | Brain Region 2 | VIP | Loadings | Stability Indices |
|
|---|---|---|---|---|---|---|
| Middle Frontal Gyrus | Right | Central Sulcus (Rolando's Fissure) | Right | 33.089 | 0.631 | 1.000 |
| Amygdala | Left | Caudate | Right | 22.492 | 0.429 | 0.949 |
| Pallidum | Right | Anterior Cingulate Gyrus/Sulcus | Right | 18.132 | 0.346 | 0.949 |
| Lateral Occipito-Temporal Gyrus (Fusiform Gyrus) | Right | Inferior Temporal Gyrus | Right | 18.017 | 0.344 | 0.949 |
| Anterior Transverse Collateral Sulcus | Right | Inferior Temporal Sulcus | Right | 17.006 | 0.324 | 0.923 |
| Inferior Temporal Sulcus | Left | Superior Temporal Sulcus (Parallel Sulcus) | Right | 12.598 | 0.240 | 0.897 |
| Thalamus | Right | Pericallosal Sulcus (S of Corpus Callosum) | Left | 5.866 | 0.112 | 0.897 |
| Posterior ramus of the lateral sulcus/fissure | Right | Temporal Pole | Right | 3.365 | 0.064 | 0.795 |
| Thalamus | Left | Temporal Pole | Right | 1.464 | 0.028 | 0.641 |
| Posterior Mid Cingulate Gyrus/Sulcus | Left | Central Sulcus (Rolando's fissure) | Left | 1.026 | 0.020 | 0.462 |
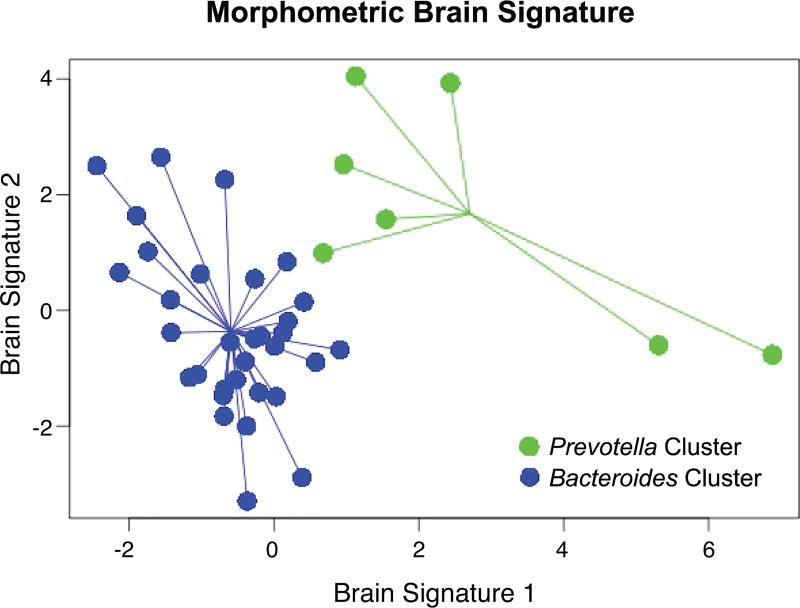
Morphological gray matter signatures associated with microbiota group clusters
Three of the four gray matter characteristics (GMV, CT, MC) discriminated the two clusters. One signature comprised of 10 gray matter metrics demonstrated good discriminative ability. Figure 3 plots the individual scores on the brain signature. Overall classification accuracy was 87.2%. Table 4 contains the list of selected gray matter metrics for each brain signature along with their loadings, VIP coefficients and stability indices. The first brain signature explained 52.8% and the second explained 21.8% of the variability in microbiota cluster discrimination. The variables with the most explanatory power included volume of the cerebellum and the hippocampus and the cortical thickness of the frontomarginal gyrus and the anterior insula, which were larger in the Bacteroides-high cluster. In the Prevotella-high cluster the nucleus accumbens had a greater volume, and two regions, the subparietal sulcus and the superior occipital gyrus, showed greater mean curvature. High reliability was observed for 8 of 10 brain morphometric features contributing to the model.
Figure 3. Classification of the gray matter connections based on the microbiota group clusters.
Depicts the discriminative abilities of the gray-matter classifier based on the bacterial clusters with an overall accuracy of 87.2%. The samples from each group are connected based on the centroid.
Table 4. Brain signature regions: gray matter.
This table shows the gray-matter regions differentiating the microbiota-based clusters.
Brain regions with positive loading scores are stronger in the Prevotella-high group.
| Brain Region | Hemisphere | Gray Matter Metric |
VIP | Loading | Stability Indices |
|---|---|---|---|---|---|
| Cerebellum | Left | Volume | 16.870 | −0.682 | 1.000 |
| Fronto-Marginal Gyrus/Sulcus | Right | Cortical Thickness | 11.284 | −0.456 | 1.000 |
| Cerebellum | Right | Volume | 7.978 | −0.322 | 0.974 |
| Anterior Occipital Sulcus/Preoccipital Notch | Right | Volume | 6.160 | −0.249 | 0.974 |
| Nucleus Accumbens | Left | Volume | 6.130 | 0.248 | 0.949 |
| Anterior Insula | Left | Cortical Thickness | 4.549 | −0.184 | 0.923 |
| Inferior Temporal Sulcus | Right | Cortical Thickness | 4.469 | −0.181 | 0.923 |
| Hippocampus | Right | Volume | 3.428 | −0.139 | 0.974 |
| Subparietal Sulcus | Left | Mean Curvature | 2.370 | 0.096 | 0.821 |
| Superior Occipital Gyrus | Left | Mean Curvature | 1.686 | 0.068 | 0.667 |
DISCUSSION
To date, most of our understanding of the interaction between the microbiota and the brain has come from rodent models, in which the gut microbiota is linked to brain signaling mechanisms and affective behavioral phenotypes, such as anxiety or depression-like behavior. The relationship between gut microbial community structure, brain structure and emotional processing observed in this study is consistent with some of these preclinical findings.
When healthy women were clustered by their stool microbiota composition into two groups, the groups showed differential response to negatively valenced images, with heightened increases associated with negative affect in the cluster with greater abundance of Prevotella. This tendency towards greater behavioral responses to negative valenced stimuli was associated with both functional and structural differences in the hippocampus. The hippocampus, a brain region involved in emotion regulation, had lower volume and showed less BOLD response during negative image viewing. Reduced hippocampal engagement to negative imagery may be associated with increased emotional arousal. Such changes have been suggested to result in less specificity of encoding the contextual details of incoming stimuli, a deficit seen in the setting of several psychiatric disorders, including depression, post traumatic stress disorder, and borderline personality disorder (45–47). While the subjects in this study were healthy, it is possible that the patterns which emerge from the microbial clustering represent vulnerability factors.
The interaction between behavior, functional response, and microbiota was supported by the structural brain signatures, which describe the microbial clusters. The observed lower hippocampal volume and higher volume of the nucleus accumbens in the Prevotella-high group are consistent with greater reactivity to an affective stimulus, with similar volumetric changes described in some studies of mood disorder (48, 49). The Prevotella-high group was associated with greater white matter connectivity in limbic-cortical-striatal-pallidal-thalamic circuitry (e.g., anterior cingulate cortex to pallidum, amygdala to caudate), and a smaller hippocampal volume compared to the Bacteriodes-high group, both findings also seen in depression (50). This group also showed differences in regions associated with attention and sensory processing. Specifically, they showed more connectivity between the central sulcus, which borders the primary somatosensory and motor cortices, and the middle frontal gyrus, a region at the convergence of the dorsal and ventral attentional networks (51); more dense white matter tracts were seen between the splenium and thalamus, which along with less cortical thickness in the anterior insula, suggests altered sensory processing. Higher connectivity in the Prevotella group was also noted between multiple regions of the temporal lobe associated with visual conceptual/semantic processing (e.g., fusiform gyrus, inferior temporal gyrus, collateral sulcus, temporal pole) (52, 53).
To our knowledge, this is the first report of behavioral and neurobiological differences related to microbial composition in healthy humans. Although these groups were identified using an unsupervised approach based on microbial composition, the identified two clusters of subjects defined by the genera Bacteriodes and Prevotella are similar to clusters previously identified across diverse populations (54–56). These clusters have been described variously as enterotypes or enterogradients, though this nomenclature has been controversial both because it implies a fixed characteristic of a individual or group and because not all of the clusters are reproducible across studies and analytic techniques (57). Of the reported “enterotypes” the most robust groupings appear to be those based on Bacteroides and Prevotella with frequency in the Western population being dominated by the Bacteroides group, as is seen in the current sample (54–56). In previous studies these groupings have been independent of age, sex, body mass index or nationality and are shared between monozygotic twins at a high rate (58). Based on a diet questionnaire recalling food intake over the past year (Food Frequency Questionnaire) (59) but not a short term recall, a diet more plentiful in saturated fat and animal protein was associated with the Bacteroides group and a diet rich in plant based fiber with the Prevotella group (56) suggesting an important role for long term diet. Further, in a short term study using a controlled dietary intervention with either high fiber/low fat or high fat/low fiber, individuals failed to change from one “enterotype” to another, despite rapid changes in individual bacterial species (56). Beyond these descriptions, the relative abundance of Bacteroides and Prevotella has not been firmly associated with any specific health or physiologic phenotype and the functional pathways associated with the microbiota clusters are not clearly defined. While “enterotypes” have been described to be fairly stable, there has been a published report of a single healthy individual moving between “enterotypes” over time, without a specific intervention to induce such change (60). Such changes could certainly be mirrored or even induced by changes in brain function and by affective responses, but changes in structure would not be expected on this temporal scale. Therefore, if an interaction between diet and the brain signatures described in the current study exists, it could be based on long term diet, or potentially on dietary factors in early life, while the brain is developing the cortico-limbic and sensory circuitry featured in the brain signatures.
While the findings of this study are consistent with reports from rodent models of brain-gut-microbiota interactions, caution should be used in the interpretation of this proof of concept study, as it is based on a small sample of physically and psychologically healthy women, and the results may or may not be generalizable to other populations. Due to the preponderance of individuals in the Western population with lower Prevotella abundance, supported in the distribution seen in this sample, the Prevotella group analyzed is very small. Conclusions drawn from this sample will require further validation in larger samples. Further, it is unclear whether the propensity towards developing a more negative affect during this paradigm is a marker of personality traits, a risk factor for developing clinically relevant negative mood states, or just a healthy variant. Although sPLS-DA was chosen specifically because of its ability to deal with small samples, biases due to the unbalanced nature of the samples may exist. For the fMRI task analysis, non-parametric testing for cluster-based inference may have resulted in alternative p-values and better control over family-wise error rate. Importantly, relatively few brain biomarkers were able to predict microbial subgroup membership better than chance and the variables selected for the solutions make biological sense in terms of previous preclinical data on neurobiological-microbiota associations. Ultimately, future studies are needed to test the generalizability of the solution and the estimates of overall model accuracy to other samples.
Much work remains in defining the relationship between the microbiota and the brain, and several key questions are raised by the work presented here. Of primary interest is the directionality of the interaction between the brain and gut microbes in healthy humans. The concept that the gut microbiota can drive affective response is supported by preclinical work showing affect changes in rodents after manipulation of gut microbes using antibiotics, probiotics, or fecal transplants, and is also suggested by the changes exhibited in human brain networks after chronic probiotic ingestion (18, 61–63). However, the human gut microbiota is frequently affected by the intake of antibiotics, dietary changes, and other interventions without noting changes in mood or affective response. The clinical studies which have shown that such interventions affect mood symptoms in humans have shown very modest changes (14, 64–66). The alternate, more plausible hypothesis is that the brain influences gut microbiota via the autonomic nervous system, stress hormones, and centrally mediated immune modulation. Therefore, the same central drivers that create negative affect may also modulate the gastrointestinal milieu which secondarily shapes microbial composition. Whether the microbiota-brain connections we see in this study reflect the modulation of the gut microbiota structure by the brain, or the influence of the microbiota on the brain and its affective responses, cannot be answered in this cross-sectional view. Careful longitudinal studies, including assessment of gut microbial community structure and microbial metabolomics, in conjunction with neuroimaging and behavioral testing is required to establish directionality and causality.
Supplementary Material
Supplemental Table 1. Baseline affect was correlated with individual abundance levels of Prevotella and Bacteroides. No significant correlations were noted.
Supplemental Table 2: Brain signature regions: white matter
This table shows the white-matter connections differentiating the microbiota-based clusters.
Brain region connections with positive loading scores are stronger in the Prevotella-high group.
Supplemental Table 3: Brain signature regions: gray matter
This table shows the gray-matter regions differentiating the microbiota-based clusters.
Brain regions with positive loading scores are stronger in the Prevotella-high group.
Acknowledgments
KirstenTillisch received grant funding for this project from Danone Research. Emeran A Mayer is member of Advisory Boards for Dannon and Danone, Muriel Derrien, Boris Le Nevé, and Denis Guyonnet, Rémi Brazeilles are or were employed by Danone Research at the time of the study.
Funding: This research was supported by grants from Danone Research and from the National Institutes of Health including R01 DK048351 (Mayer), P50 DK64539 (Mayer), P30 DK041301, R01 HD076756 (Labus), and pilot funds were provided for brain scanning by the Ahmanson-Lovelace Brain Mapping Center.
Abbreviations
- BMI
Body Mass Index
- BOLD
Blood Oxygen Level Dependent
- CD-HIT
Cluster Database at High Identity with Tolerance
- CNS
Central Nervous System
- CT
Cortical Thickness
- DTI
Diffusion Tensor Imaging
- FA
Flip Angle
- fMRI
Functional MRI
- FOV
Field of View
- GLM
General Linear Model
- GMV
Gray Matter Volume
- HAD
Hospital Anxiety And Depression Scale
- LONI
Laboratory of Neuroimaging
- MC
Mean Curvature
- MP-RAGE
Magnetization Prepared Rapid Gradient-Echo
- MRI
Magnetic Resonance Imaging
- OTU
Operational Taxonomic Units
- PAM
Partitioning Around Medoids
- PANAS
Positive Affect Negative Affect Schedule
- QIIME
Quantitative Insights Into Microbial Ecology
- ROI
Region of Interest
- rRNA
Ribosomal Ribonucleic Acid
- SA
Surface Area
- SD
Standard Deviation
- sPLS-DA
Sparse Partial Least Squares Discriminant Analysis
- SPM
Statistical Parametric Mapping
- SPSS
Statistical Package for the Social Sciences
- TE
Echo Time
- TR
Repetition Time
- UCLA
University of California, Los Angeles
- VIP
Variable Importance In Projection
Footnotes
Disclosures: The remaining authors disclose no conflicts.
References
- 1.Biedermann L, Rogler G. The intestinal microbiota: its role in health and disease. Eur J Pediatr. 2015;174:151–67. doi: 10.1007/s00431-014-2476-2. [DOI] [PubMed] [Google Scholar]
- 2.Moloney RD, Desbonnet L, Clarke G, Dinan TG, Cryan JF. The microbiome: stress, health and disease. Mamm Genome. 2014;25:49–74. doi: 10.1007/s00335-013-9488-5. [DOI] [PubMed] [Google Scholar]
- 3.Sherwin E, Rea K, Dinan TG, Cryan JF. A gut (microbiome) feeling about the brain. Curr Opin Gastroenterol. 2016;32:96–102. doi: 10.1097/MOG.0000000000000244. [DOI] [PubMed] [Google Scholar]
- 4.Dinan TG, Cryan JF. Melancholic microbes: a link between gut microbiota and depression? Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2013;25:713–9. doi: 10.1111/nmo.12198. [DOI] [PubMed] [Google Scholar]
- 5.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–38. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Aidy S, Stilling R, Dinan TG, Cryan JF. Microbiome to Brain: Unravelling the Multidirectional Axes of Communication. Adv Exp Med Biol. 2016;874:301–36. doi: 10.1007/978-3-319-20215-0_15. [DOI] [PubMed] [Google Scholar]
- 7.Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, Dinan TG, Cryan JF. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Molecular psychiatry. doi: 10.1038/mp.2012.77. [DOI] [PubMed] [Google Scholar]
- 8.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A. 2011;108:3047–52. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neufeld KM, Kang N, Bienenstock J, Foster JA. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroent Motil. 2011;23 doi: 10.1111/j.1365-2982.2010.01620.x. [DOI] [PubMed] [Google Scholar]
- 10.Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Molecular psychiatry. 2014;19:146–8. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.SM OM, Stilling RM, Dinan TG, Cryan JF. The microbiome and childhood diseases: Focus on brain-gut axis. Birth Defects Res C Embryo Today. 2015;105:296–313. doi: 10.1002/bdrc.21118. [DOI] [PubMed] [Google Scholar]
- 12.O'Mahony SM, Clarke G, Dinan TG, Cryan JF. Early-life adversity and brain development: Is the microbiome a missing piece of the puzzle? Neuroscience. 2015 doi: 10.1016/j.neuroscience.2015.09.068. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez-Real JM, Serino M, Blasco G, Puig J, Daunis-i-Estadella J, Ricart W, Burcelin R, Fernandez-Aranda F, Portero-Otin M. Gut Microbiota Interacts With Brain Microstructure and Function. The Journal of clinical endocrinology and metabolism. 2015;100:4505–13. doi: 10.1210/jc.2015-3076. [DOI] [PubMed] [Google Scholar]
- 14.Steenbergen L, Sellaro R, van Hemert S, Bosch JA, Colzato LS. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain, behavior, and immunity. 2015;48:258–64. doi: 10.1016/j.bbi.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Parashar A, Udayabanu M. Gut microbiota regulates key modulators of social behavior. Eur Neuropsychopharmacol. 2016;26:78–91. doi: 10.1016/j.euroneuro.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Dinan TG, Stilling RM, Stanton C, Cryan JF. Collective unconscious: how gut microbes shape human behavior. J Psychiatr Res. 2015;63:1–9. doi: 10.1016/j.jpsychires.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Montiel-Castro AJ, Gonzalez-Cervantes RM, Bravo-Ruiseco G, Pacheco-Lopez G. The microbiota-gut-brain axis: neurobehavioral correlates, health and sociality. Front Integr Neurosci. 2013;7:70. doi: 10.3389/fnint.2013.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, Mayer EA. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–401. 401, e1–4. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sheehan DV, Lecrubier Y, Sheehan KH, Amorim P, Janavs J, Weiller E, Hergueta T, Baker R, Dunbar GC. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. The Journal of clinical psychiatry. 1998;59(Suppl 20):22–33. quiz 4–57. [PubMed] [Google Scholar]
- 20.Riegler G, Esposito I. Bristol scale stool form. A still valid help in medical practice and clinical research. Tech Coloproctol. 2001;5:163–4. doi: 10.1007/s101510100019. [DOI] [PubMed] [Google Scholar]
- 21.Crawford JR, Henry JD. The positive and negative affect schedule (PANAS): construct validity, measurement properties and normative data in a large non-clinical sample. Br J Clin Psychol. 2004;43:245–65. doi: 10.1348/0144665031752934. [DOI] [PubMed] [Google Scholar]
- 22.Watson D, Clark LA, Tellegen A. Development and validation of brief measures of positive and negative affect: the PANAS scales. J Pers Soc Psychol. 1988;54:1063–70. doi: 10.1037//0022-3514.54.6.1063. [DOI] [PubMed] [Google Scholar]
- 23.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand. 1983;67:361–70. doi: 10.1111/j.1600-0447.1983.tb09716.x. [DOI] [PubMed] [Google Scholar]
- 24.Spielberger CD, Gorsuch RL, Lushene R, Vagg PR, Jacobs GA. Manual for the State-Trait Anxiety Inventory. Palo Alto: Consulting Psychologists Press; 1983. [Google Scholar]
- 25.Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. Plos One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–9. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 28.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methe B, DeSantis TZ, Human Microbiome C. Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R, Huttenhower C, Ley RE. A Guide to Enterotypes across the Human Body: Meta-Analysis of Microbial Community Structures in Human Microbiome Datasets. PLoS Comput Biol. 2013;9:e1002863. doi: 10.1371/journal.pcbi.1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang PJ, Bradley MM, Cuthbert BN. International affective picture system (IAPS): Affective ratings of pictures and instruction manual. Gainesville, FL: University of Florida; 2005. [Google Scholar]
- 31.Destrieux C, Fischl B, Dale A, Halgren E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage. 2010;53:1–15. doi: 10.1016/j.neuroimage.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo CW, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: pitfalls and recommendations. Neuroimage. 2014;91:412–9. doi: 10.1016/j.neuroimage.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 34.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis - I. Segmentation and surface reconstruction. NeuroImage. 1999;9:179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 35.Labus JS, Van Horn JD, Gupta A, Alaverdyan M, Torgerson C, Ashe-McNalley C, Irimia A, Hong JY, Naliboff B, Tillisch K, Mayer EA. Multivariate morphological brain signatures predict patients with chronic abdominal pain from healthy control subjects. Pain. 2015;156:1545–54. doi: 10.1097/j.pain.0000000000000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta A, Mayer EA, Sanmiguel CP, Van Horn JD, Woodworth D, Ellingson BM, Fling C, Love A, Tillisch K, Labus JS. Patterns of brain structural connectivity differentiate normal weight from overweight subjects. NeuroImage Clinical. 2015;7:506–17. doi: 10.1016/j.nicl.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang MC, Barysheva M, Toga AW, Medland SE, Hansell NK, James MR, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. BDNF gene effects on brain circuitry replicated in 455 twins. NeuroImage. 2011;55:448–54. doi: 10.1016/j.neuroimage.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–9. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 39.Irimia A, Chambers M, Wang B, Prastawa M, Vespa P, Hovda DA, Alger J, Gerig G, Toga A, Kikinis R, Van Horn J. Systematic Connectomic Analysis of White Matter Atrophy Associated with Severe Traumatic Brain Injury. J Neurotraum. 2012;29:A8–A9. [Google Scholar]
- 40.Irimia A, Chambers MC, Torgerson CM, Van Horn JD. Circular representation of human cortical networks for subject and population-level connectomic visualization. NeuroImage. 2012;60:1340–51. doi: 10.1016/j.neuroimage.2012.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irimia A, Van Horn JD. The structural, connectomic and network covariance of the human brain. NeuroImage. 2012;66C:489–99. doi: 10.1016/j.neuroimage.2012.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Cao KARD, Robert-Granie C, Besse P. A sparse PLS for variable selection when integrating omics data. Stat Appl Genet Mol Biol. 2008;7:35. doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 43.Le Cao KA, Martin PG, Robert-Granie C, Besse P. Sparse canonical methods for biological data integration: application to a cross-platform study. Bmc Bioinformatics. 2009;10:17. doi: 10.1186/1471-2105-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. Bmc Bioinformatics. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soloff PH, White R, Omari A, Ramaseshan K, Diwadkar VA. Affective context interferes with brain responses during cognitive processing in borderline personality disorder: fMRI evidence. Psychiatry research. 2015;233:23–35. doi: 10.1016/j.pscychresns.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee BT, Seok JH, Lee BC, Cho SW, Yoon BJ, Lee KU, Chae JH, Choi IG, Ham BJ. Neural correlates of affective processing in response to sad and angry facial stimuli in patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2008;32:778–85. doi: 10.1016/j.pnpbp.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Hayes JP, LaBar KS, McCarthy G, Selgrade E, Nasser J, Dolcos F, workgroup VM-AM. Morey RA. Reduced hippocampal and amygdala activity predicts memory distortions for trauma reminders in combat-related PTSD. J Psychiatr Res. 2011;45:660–9. doi: 10.1016/j.jpsychires.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irle E, Ruhleder M, Lange C, Seidler-Brandler U, Salzer S, Dechent P, Weniger G, Leibing E, Leichsenring F. Reduced amygdalar and hippocampal size in adults with generalized social phobia. J Psychiatry Neurosci. 2010;35:126–31. doi: 10.1503/jpn.090041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuhn S, Schubert F, Gallinat J. Structural correlates of trait anxiety: reduced thickness in medial orbitofrontal cortex accompanied by volume increase in nucleus accumbens. J Affect Disord. 2011;134:315–9. doi: 10.1016/j.jad.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Fang P, Zeng LL, Shen H, Wang L, Li B, Liu L, Hu D. Increased cortical-limbic anatomical network connectivity in major depression revealed by diffusion tensor imaging. Plos One. 2012;7:e45972. doi: 10.1371/journal.pone.0045972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Japee S, Holiday K, Satyshur MD, Mukai I, Ungerleider LG. A role of right middle frontal gyrus in reorienting of attention: a case study. Frontiers in systems neuroscience. 2015;9:23. doi: 10.3389/fnsys.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonner MF, Price AR. Where is the anterior temporal lobe and what does it do? The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:4213–5. doi: 10.1523/JNEUROSCI.0041-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lener MS, Kundu P, Wong E, Dewilde KE, Tang CY, Balchandani P, Murrough JW. Cortical abnormalities and association with symptom dimensions across the depressive spectrum. J Affect Disord. 2016;190:529–36. doi: 10.1016/j.jad.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M'Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roager HM, Licht TR, Poulsen SK, Larsen TM, Bahl MI. Microbial enterotypes, inferred by the prevotella-to-bacteroides ratio, remained stable during a 6-month randomized controlled diet intervention with the new nordic diet. Appl Environ Microbiol. 2014;80:1142–9. doi: 10.1128/AEM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knights D, Ward TL, McKinlay CE, Miller H, Gonzalez A, McDonald D, Knight R. Rethinking "enterotypes". Cell Host Microbe. 2014;16:433–7. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim MY, Rho M, Song YM, Lee K, Sung J, Ko G. Stability of gut enterotypes in Korean monozygotic twins and their association with biomarkers and diet. Sci Rep. 2014;4:7348. doi: 10.1038/srep07348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dreon DM, John EM, DiCiccio Y, Whittemore AS. Use of NHANES data to assign nutrient densities to food groups in a multiethnic diet history questionnaire. Nutr Cancer. 1993;20:223–30. doi: 10.1080/01635589309514290. [DOI] [PubMed] [Google Scholar]
- 60.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–7. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Desbonnet L, Clarke G, Traplin A, O'Sullivan O, Crispie F, Moloney RD, Cotter PD, Dinan TG, Cryan JF. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain, behavior, and immunity. 2015;48:165–73. doi: 10.1016/j.bbi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 62.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu XN, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol. 2004;558:263–75. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci U S A. 2011;108:16050–5. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takada M, Nishida K, Kataoka-Kato A, Gondo Y, Ishikawa H, Suda K, Kawai M, Hoshi R, Watanabe O, Igarashi T, Kuwano Y, Miyazaki K, Rokutan K. Probiotic Lactobacillus casei strain Shirota relieves stress-associated symptoms by modulating the gut-brain interaction in human and animal models. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society. 2016 doi: 10.1111/nmo.12804. [DOI] [PubMed] [Google Scholar]
- 65.Messaoudi M, Violle N, Bisson JF, Desor D, Javelot H, Rougeot C. Beneficial psychological effects of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in healthy human volunteers. Gut microbes. [Clinical Trial Research Support, Non-U.S. Gov't] 2011;2:256–61. doi: 10.4161/gmic.2.4.16108. [DOI] [PubMed] [Google Scholar]
- 66.Rao AV, Bested AC, Beaulne TM, Katzman MA, Iorio C, Berardi JM, Logan AC. A randomized, double-blind, placebo-controlled pilot study of a probiotic in emotional symptoms of chronic fatigue syndrome. Gut Pathog. 2009;1:6. doi: 10.1186/1757-4749-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Baseline affect was correlated with individual abundance levels of Prevotella and Bacteroides. No significant correlations were noted.
Supplemental Table 2: Brain signature regions: white matter
This table shows the white-matter connections differentiating the microbiota-based clusters.
Brain region connections with positive loading scores are stronger in the Prevotella-high group.
Supplemental Table 3: Brain signature regions: gray matter
This table shows the gray-matter regions differentiating the microbiota-based clusters.
Brain regions with positive loading scores are stronger in the Prevotella-high group.