Abstract
There is increasing evidence that genetic risk variants for non-syndromic cleft lip/palate (nsCL/P) are also associated with normal-range variation in facial morphology. However, previous analyses are mostly limited to candidate SNPs and findings have not been consistently replicated. Here, we used polygenic risk scores (PRS) to test for genetic overlap between nsCL/P and seven biologically relevant facial phenotypes. Where evidence was found of genetic overlap, we used bidirectional Mendelian randomization (MR) to test the hypothesis that genetic liability to nsCL/P is causally related to implicated facial phenotypes. Across 5,804 individuals of European ancestry from two studies, we found strong evidence, using PRS, of genetic overlap between nsCL/P and philtrum width; a 1 S.D. increase in nsCL/P PRS was associated with a 0.10 mm decrease in philtrum width (95% C.I. 0.054, 0.146; P = 2x10-5). Follow-up MR analyses supported a causal relationship; genetic variants for nsCL/P homogeneously cause decreased philtrum width. In addition to the primary analysis, we also identified two novel risk loci for philtrum width at 5q22.2 and 7p15.2 in our Genome-wide Association Study (GWAS) of 6,136 individuals. Our results support a liability threshold model of inheritance for nsCL/P, related to abnormalities in development of the philtrum.
Author summary
Non-syndromic cleft lip/palate (nsCL/P) is a birth defect, primarily affecting the upper lip and hard palate. Individuals with nsCL/P and their unaffected family members sometimes present with other minor craniofacial anomalies and may have differences in facial morphology compared to the general population. Here, we investigate the shared genetic relationship between nsCL/P and facial morphology in a sample of around 6,000 Europeans from two different studies. We demonstrate that genetic risk factors for nsCL/P are associated with decreased philtrum width in unaffected individuals from the general population and that the relationship is likely causal. This finding is important because it demonstrates that nsCL/P, which is often treated as a dichotomous trait, may have a continuous dimension and offers insight into potential biological mechanisms that result in nsCL/P.
Introduction
Orofacial clefts are malformations characterised by a failure of fusion between adjacent facial structures in the embryo [1]. Cleft lip with/without cleft palate (CL/P) is a sub-type of orofacial cleft, consisting of individuals presenting with a cleft of the upper lip, with or without a cleft of the palate. Approximately 70% of CL/P cases are non-syndromic, where the facial cleft is not accompanied by other apparent developmental or physical abnormalities [2]. The non-syndromic form of CL/P (nsCL/P) is a multifactorial trait with both genetic and environmental risk factors [1]. A possible polygenic threshold model of inheritance is supported by the identification of more than 20 common genetic risk variants for nsCL/P from genome-wide association studies (GWAS) [3–9] and single nucleotide polymorphism (SNP) heritability estimates of around 30% [6].
Facial morphology in the general population is also likely to be highly polygenic; genome-wide significant loci have been found for multiple facial phenotypes across diverse ethnic populations [10–14]. In some cases, the genes associated with normal-range variation in facial shape have also been implicated in nsCL/P (e.g. MAFB) [12]. Likewise, previous studies using candidate SNPs have found overlap between nsCL/P risk loci and facial phenotypes in the general population [11, 15, 16]. For example, the strongest nsCL/P GWAS signal, intergenic variant rs987525 on chromosome 8q24, was found to be associated with more than half of the 48 facial phenotypes studied in a European population [11] while in a Han Chinese population, rs642961 in IRF6 (a major nsCL/P-associated gene) strongly predicted lip-shape variation in females [16]. However, associations between nsCL/P genetic variants and facial morphology were not consistently replicated, possibly because of methodological differences in measuring facial phenotypes or population differences between cohorts [10].
The use of individual markers to demonstrate genetic overlap between two phenotypes has notable limitations; a large number of statistical tests are introduced, and interpretation is difficult when some SNPs show an association and others do not. Polygenic risk scores (PRS) involve incorporating multiple markers, including those not reaching genome-wide significance, into a genetic score that serves as a proxy for a trait [17]. PRS have been previously shown to be suitable predictors for nsCL/P [6] suggesting they can be used to estimate genetic overlap between nsCL/P and normal-range facial morphology.
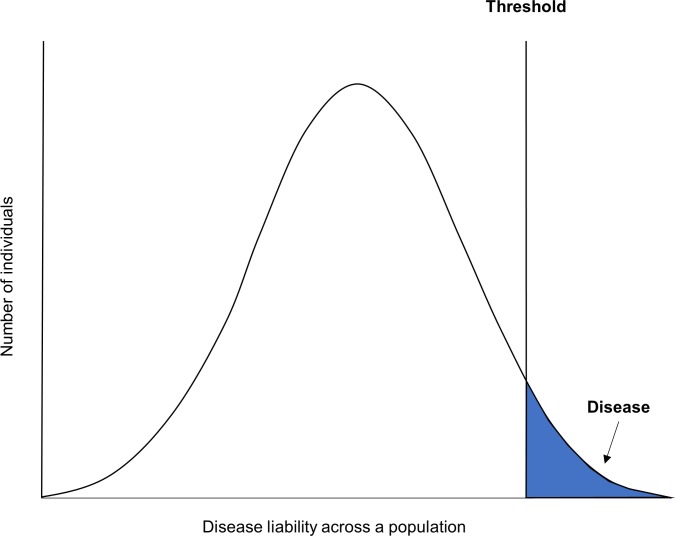
Interpreting genetic overlap between nsCL/P and a facial phenotype is difficult because the development of the face and development of an orofacial cleft are largely synchronous. One possibility is that differences in the facial phenotype are a sub-phenotypic manifestation of genetic liability to nsCL/P (see Fig 1). The inheritance of dichotomous traits can be modelled on the liability scale, where every individual has an underlying normally distributed liability to the trait determined by genes, environment and chance. Individuals above a liability threshold develop the trait, while increased liability may cause related phenotypic differences in individuals without the trait [18–20]. For example, increased liability to developing a cleft palate (CP) has been hypothesised to be associated with delayed movement of the palatal shelf, which may in turn result in a CP, dependant on other factors such as shelf and head width [20].
Fig 1. Liability threshold model for nsCL/P.
Shown is an illustration of a liability threshold model for nsCL/P. Every individual has a normally distributed liability to nsCL/P, determined by genes, environment and chance. Individuals over the liability threshold develop nsCL/P, with the area under the curve past the threshold equal to the trait incidence. We are hypothesising that liability to nsCL/P, specifically genetic liability to nsCL/P, may be associated with differences in facial morphology across the general population.
In order to evaluate the coherence of the liability-related sub-phenotype model, we apply the principles of Mendelian randomization (MR). MR is an instrumental variable approach, testing causality of an “exposure” and an outcome by using genetic instruments to mimic a randomised controlled trial [21]. MR relies on several strict assumptions; firstly, genetic variants must be robustly associated with the exposure (in this instance, genetic liability to nsCL/P); secondly, the variants cannot influence the outcome through a pathway independent of the exposure; and thirdly, the variants should not be associated with confounders of the relationship between the exposure and the outcome [22]. If these assumptions are met, bidirectional MR can be used to test the hypothesis that genetic liability to nsCL/P is causally related to facial morphology [22].
In the absence of large-scale publicly available GWAS summary data for nsCL/P, we used individual level genotype data from the International Cleft Consortium to Identify Genes and Interactions Controlling Oral Clefts (ICC) and GWAS summary statistics from the Bonn-II study [8] to replicate the meta-analysis GWAS summary statistics from the previously published Ludwig et al 2012 GWAS [3]. Next, we investigated genetic overlap between nsCL/P and normal-range facial morphology in the general population, using PRS derived from the GWAS summary statistics. Finally, in the instance of genetic overlap, we used bidirectional MR to explore the relationship between nsCL/P and implicated facial phenotypes. A flowchart detailing the primary analyses is contained in Fig 2.
Fig 2. Flowchart detailing primary analyses.
This figure outlines the primary analyses and data-sets in the study: 1) the meta-analysis GWAS for nsCL/P, 2) testing the association of nsCL/P PRS with facial phenotypes in ALSPAC, 3) attempted replication of PRS analyses in 3DFN for implicated facial phenotypes, 4) the meta-analysis GWAS for implicated facial phenotypes and 5) bidirectional MR analyses for nsCL/P and implicated facial phenotypes.
Results
Genome-wide association study and genetic proxy for nsCL/P
We performed a GWAS of nsCL/P using the TDT on 638 parent-offspring trios and 178 offspring duos of European descent, and then meta-analysed our results with GWAS summary results previously published on 399 cases and 1,318 controls in the Bonn-II study [8]. This yielded comparable results to a previously published GWAS [3], which used a very similar data-set with slightly different quality control and analysis methods (S1 Table).
We also evaluated the predictive accuracy of nsCL/P that could be achieved using different PRS constructed from these summary data by comparing the strength of association at different inclusion thresholds of the PTDT. We determined that including independent SNPs that surpass a P-value threshold of 10−5 was the most predictive of nsCL/P liability in both European and Asian trios (S2 Table). Therefore, this threshold was used for generating polygenic risk scores from the meta-analysis summary statistics. SNPs included in the selected score are listed in S3 Table.
The prediction of facial morphology using PRS for nsCL/P
Prior to testing the performance of our nsCL/P PRS on predicting facial morphology, we calculated the minimum genetic correlation required to detect an association between the PRS and the facial phenotypes. We found that the minimum genetic correlation required ranged from 0.17 to 0.28 with differences attributable to different heritability estimates across the facial phenotypes (S4 Table).
We evaluated the performance of our nsCL/P PRS for prediction of seven facial morphological traits. Facial distances used in the analysis are shown in Fig 3. We found evidence of an association between the nsCL/P PRS and philtrum width in the ALSPAC children, where a 1 S.D. increase in nsCL/P PRS was associated with a 0.07 mm decrease in philtrum width (95% C.I. 0.02, 0.13; P = 0.014) (Table 1).
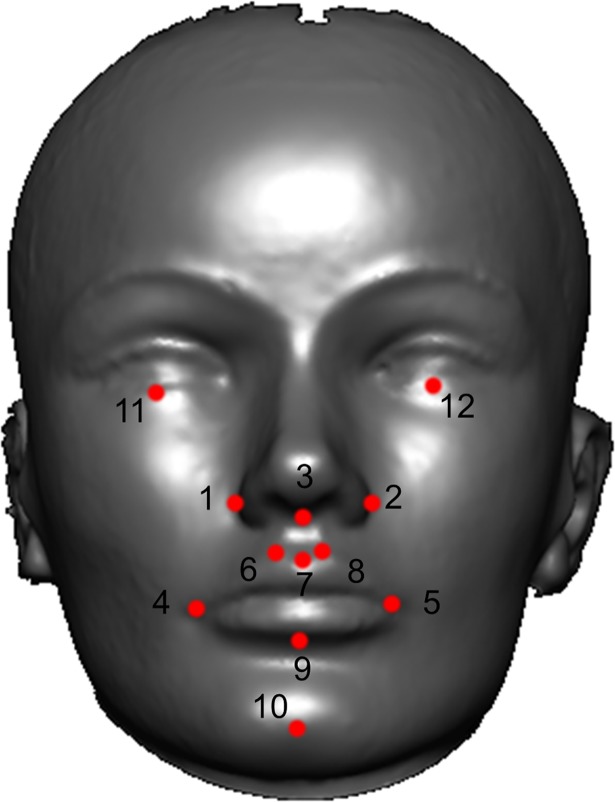
Fig 3. Facial morphological distances of interest.
This figure shows the 12 facial landmarks that were used to generate the facial phenotypes tested for association with the nsCL/P PRS. Facial phenotypes were defined as the 3D Euclidean distance between the following landmarks (Nasal width: 1–2, Nasal-lip distance: 3–7, Lip width: 4–5, Philtrum width: 6–8, Lip height: 7–9, Lip-chin distance: 9–10 and inter-palpebrale width: 11–12).
Table 1. Association of nsCL/P PRS with facial phenotypes in ALSPAC children.
| 3D facial Euclidean distances in ALSPAC | ALSPAC children (N = 3707) | |
|---|---|---|
| Beta (95% C.I.) (mm) | P-value | |
| Distance between subnasale and labiale superius (Nasal-lip) |
-0.25 (-2.16, 1.65) | 0.79 |
| Distance between labiale inferius and pogonion (Lip-chin) |
-0.02 (-0.10, 0.06) | 0.64 |
| Distance between left and right palpebrale inferius (Mid-point of eyes) | -0.08 (-0.17, 0.01) | 0.09 |
| Distance between left and right alare (Nasal width) | -0.01 (-0.08, 0.06) | 0.75 |
| Distance between labiales inferius and superius (lip height) |
0.02 (-0.05, 0.10) | 0.53 |
| Distance between left and right crista philtri (philtrum width) |
-0.07 (-0.13, -0.02) | 0.014 |
| Distance between left and right cheilion (lip width) |
-0.02 (-0.15, 0.10) | 0.70 |
We attempted to replicate this finding in the 3DFN study and found a consistent effect of 1 S.D. increase in nsCL/P PRS being associated with a 0.14 mm decrease in philtrum width (95% C.I. 0.07, 0.21; P = 1.7x10-4). Meta-analysing these results; indicated that a 1 S.D. increase in nsCL/P PRS is associated with a 0.10 mm decrease in philtrum width (95% C.I. 0.054, 0.146; P = 2x10-5).
GWAS of philtrum width
To generate SNP-philtrum width association information for MR analyses, we performed GWAS of philtrum width in both ALSPAC and 3DFN separately, before meta-analysing. The combined sample included 6,136 individuals of recent European descent. We identified two novel chromosomal regions associated with philtrum width with genome-wide significance at 5q22.2 (lowest P value for rs255877, P = 3.8x10-10), within the non-coding RNA intronic region of an uncategorised gene ENSG00000232633, and 7p15.2 (rs2522825, P = 1.4x10-8), an intergenic SNP near HOXA1 (S5 Table). We found some evidence that the two lead SNPs may be eQTLs for nearby genes (S6 Table). The two lead SNPs of the genome-wide significant loci, rs255877 and rs2522825, were used as genetic variants associated with philtrum width in subsequent MR analyses. The GWAS summary statistics are available at the University of Bristol data repository, data.bris, at https://doi.org/10.5523/bris.1kz9y0moa8sgj2lxlk53mdmlbj [23].
Bidirectional mendelian randomization
We used MR to investigate the possible causal mechanism that would give rise to the genetic overlap between nsCL/P and philtrum width.
Firstly, we determined whether genetic variants contributing to liability of nsCL/P cause changes in philtrum width, by testing SNPs strongly associated with nsCL/P for association with philtrum width. A 1-unit log odd increase in liability to nsCL/P was associated with a 0.11mm (95% C.I. 0.04, 0.19; P = 0.0036) decrease in philtrum width. Sensitivity analyses suggested there was no strong evidence for pleiotropy or heterogeneity and validated the consistency of the instrument. Leave-one-SNP-out analysis showed consistent effect estimates after exclusion of each SNP (Table 2).
Table 2. Causal estimates of genetic liability for nsCL/P on philtrum width using mendelian randomization.
| Test | Interpretation | Estimate (95% C.I.) | P-value |
|---|---|---|---|
| Inverse variance weighted | Primary causal estimate1 | -0.11 (-0.19, -0.04) | 0.0036 |
| Heterogeneity of Inverse variance weighted | Balanced pleiotropy | N/A | 0.36 |
| MR-Egger | Intercept test for directional pleiotropy2 | -0.01 (-0.11, 0.10) | 0.93 |
| Regression estimate1 | -0.10 (-0.33, 0.13) | 0.43 | |
| Weighted median | Consistency1 | -0.12 (-0.21, -0.04) | 0.0043 |
| Weighted mode | Consistency1 | -0.12 (-0.21, -0.03) | 0.049 |
| Leave-one out rs1873147 | Additive model1 | -0.11 (-0.20, -0.02) | 0.017 |
| Leave-one out rs227731 | Additive model1 | -0.10 (-0.16, -0.03) | 0.007 |
| Leave-one out rs7078160 | Additive model1 | -0.13 (-0.20, -0.06) | 0.0001 |
| Leave-one out rs7590268 | Additive model1 | -0.11 (-0.20, -0.02) | 0.013 |
| Leave-one out rs8001641 | Additive model1 | -0.13 (-0.21, -0.04) | 0.0030 |
| Leave-one out rs987525 | Additive model1 | -0.10 (-0.22, 0.01) | 0.084 |
1 Units: mm change in philtrum width per 1-unit log odd increase in liability to nsCL/P.
2 Units: Average pleiotropic effect of a nsCL/P genetic variant on philtrum width.
Secondly, we determined whether genetic variants associated with philtrum width also affect liability to nsCL/P, by testing two independent SNPs associated with philtrum width at genome-wide significance (derived in the ALSPAC and 3DFN cohorts) for association with nsCL/P. Utilising strong LD proxies (S7 Table), weak evidence was found of an association between philtrum width-associated variants and liability to nsCL/P (LogOR = 0.30; 95% C.I. -0.26, 0.86; P = 0.30). Sensitivity analyses for pleiotropy were limited, with only 2 SNPs.
Thirdly, we used the MR-Steiger test of directionality to test the direction of effect between philtrum width and liability to nsCL/P. The results suggested that the true direction of effect is that genetic variants contributing to liability to nsCL/P cause changes in philtrum width (P <10−10).
Interpretation of bidirectional mendelian randomization
The rationale for interpretation of the bidirectional MR analysis is contained in Fig 4. Strong evidence was found for genetic liability to nsCL/P causing decreased philtrum width, weak evidence was found for heterogeneity or assumption violations in the forward-MR, and weak evidence was found for the reverse-MR of philtrum width-associated variants on liability to nsCL/P. Therefore, we conclude that the most likely explanation for the genetic overlap between nsCL/P and philtrum width is that genetic liability to nsCL/P is causally related to decreased philtrum width.
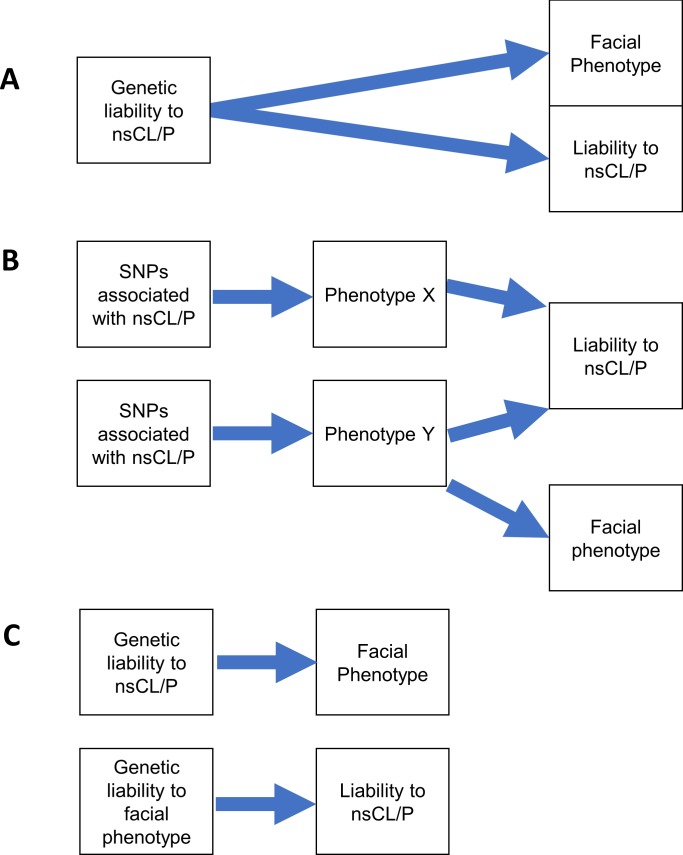
Fig 4. Interpretation of bidirectional MR.
(A) SNPs associated with nsCL/P have a homogeneous effect on the facial phenotype with weak evidence for the reverse direction MR. We would conclude that genetic liability to nsCL/P causes both increased liability to nsCL/P (in conjunction with the environment and chance) and changes in the facial phenotype. (B) SNPs associated with nsCL/P have a heterogeneous effect on the facial phenotype. In this instance, there is weak evidence for genetic liability to nsCL/P causing changes in the facial phenotype because liability assumes a consistent effect. We would conclude that an unknown confounder Y affects the facial phenotype and liability to nsCL/P independently. (C) SNPs associated with nsCL/P have a homogeneous effect on the facial phenotype AND SNPs associated with the facial phenotype cause increased liability to nsCL/P. In this instance, there are two possibilities. The first possibility is that the genetic instruments for the facial phenotype are weak (e.g. only one SNP) and so the causal effect estimate of the facial phenotype on liability to nsCL/P is imprecise. The second possibility is that nsCL/P and the facial phenotype have a substantial genetic correlation, which would require further investigation. Here, the results of the Steiger test are useful, as they can infer the most likely direction of effect between nsCL/P and implicated facial phenotypes.
Discussion
In this manuscript, we have shown that there is genetic overlap between nsCL/P and normal-range variation in philtrum width, and furthermore, that genetic risk SNPs for nsCL/P consistently cause decreased philtrum width in the general population. Notably there was weak evidence for genetic overlap between nsCL/P and upper lip width despite the observational correlation between the widths of the upper lip and philtrum.
There are two main implications of these results. First, our findings demonstrate the aetiological relevance of the formation of the philtrum to nsCL/P. The medial nasal and maxillary processes are responsible for development of the upper lip and philtrum [24]. Developmental anomalies within these processes may result in a cleft lip [25] and our findings show that even when there is successful fusion, as in our study populations, the genetic variants which give rise to a CL/P cause decreased philtrum width. Secondly, the non-heterogeneous additive effect of common nsCL/P risk variants, on a related phenotype in the general population, supports a polygenic threshold model of inheritance for nsCL/P.
Although previous studies have looked at nsCL/P related sub-phenotypes, this study uses causal inference methods to more formally investigate the relationship. Our identification of phenotypic differences related to nsCL/P liability are consistent with previous studies [26–31] observing sub-clinical facial phenotypes in individuals with nsCL/P and their unaffected family members, particularly a previous study which observed reduced philtrum width in unaffected parents of individuals with nsCL/P [31]. A polygenic threshold model of inheritance related to development of the philtrum is consistent with a previously proposed mechanism for the inheritance of cleft palate [20], the identification of numerous common nsCL/P genetic risk variants [3–7] and estimation of a substantial SNP heritability for nsCL/P [6]. We do not replicate associations between nsCL/P and other facial morphological dimensions found in previous studies [11, 15, 31] using candidate SNPs but note that polygenic risk score methods are methodologically distinct and are used to investigate a different research question to single SNP analyses.
We extend the investigation of the association between nsCL/P and facial morphology in two important ways. We demonstrate that the association is present not only in unaffected family members but also in the general population, and use MR to demonstrate that this relationship is present on the liability scale. Conventionally MR is used to test possible causal effects of a modifiable continuous exposure such as cholesterol or alcohol on disease outcomes [32, 33]. Here we exploit the principles of MR to test the threshold hypothesis, by inferring a causal relationship between genetic variants contributing to liability of nsCL/P and philtrum width in a non-clinical population. We interpret this causal relationship as evidence that smaller philtrum width is a sub-phenotypic manifestation attributable to the same genetic variants that cause nsCL/P.
In addition to investigating the relationship between facial morphology and nsCL/P, we also performed the first GWAS of philtrum width, and identified two novel genome-wide significant loci. Notably one of the loci, rs2522825 at 7p15.2, was associated with gene expression at several nearby genes in the homeobox gene family, which are known to play important roles in embryonic development [34, 35].
The causal inference made in this study was achieved through the use of two independent cohorts as discovery and replication samples which greatly reduces the risk of false positives and demonstrates that results can be generalised to different populations. Detailed facial phenotyping data on a large number of individuals in our cohorts along with other detailed phenotype and genotype data enabled us to identify philtrum width as being the most relevant facial morphological feature from amongst seven biologically likely candidates. Statistical power does limit the detection of other features that may have mechanistic relationships with smaller effect sizes (S4 Table).
In this study, we combined CL/P and cleft lip only (CLO), however there is evidence suggesting that there are distinct aetiological differences between these traits, [5, 36, 37] which could reduce our statistical power, and complicates interpretation. For example, the philtrum may be more related to CLO, but we did not have sufficient data to compare nsCL/P subtype differences. An additional limitation is that there are few well-characterised genetic risk loci for philtrum width, so our MR analysis testing if genetic variants associated with a narrow philtrum width also affect liability of nsCL/P, may be underpowered.
We conclude that genetic liability to nsCL/P is causally related to variation in philtrum width and that this finding supports a polygenic threshold model of inheritance for nsCL/P, related to abnormalities in development of the philtrum. Further research looking at the relationship between genetic liability for nsCL/P and severity of cleft would provide further evidence for the polygenic threshold model.
Materials and methods
Study participants
International cleft consortium (ICC)
Data were used on parent-child cleft trios from the ICC (dbGaP Study Accession phs000094.v1.p1) [38, 39] which includes genotype data from a wide array of geographical locations across North America, Europe and Asia. The data-set included 2,029 parent-offspring trios, 401 parent-offspring pairs, 88 single cleft cases and 25 assorted extended families. Of the 2,543 children with an orofacial cleft; 1,988 presented with nsCL/P while 582 presented with an isolated cleft palate (CPO) and 21 presented with an “unknown cleft”.
Analysis was restricted to trios with a proband diagnosed with nsCL/P. 654 of the parent-offspring trios and 164 of the parent-offspring pairs were of European descent and were used in the primary analyses. 759 parent-offspring trios and 159 parent-offspring pairs of Asian descent were included in secondary analyses.
GWAS genotypes and phenotypes are available at dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000094.v1.p1)
ALSPAC
We used data on children from the Avon Longitudinal Study of Parents and Children (ALSPAC), a longitudinal study that recruited pregnant women living in the former county of Avon (UK) with expected delivery dates between 1 April 1991 and 31 December 1992. The initial number of enrolled pregnancies was 14,541, which resulted in 14,062 live births and 13,988 children alive at the age of 1. When the oldest children were approximately 7 years of age, the initial sample was boosted with eligible cases who had failed to join the study originally. For analyses of children after the age of 7, the total possible sample size is 15,247 pregnancies, resulting in 14,775 live births. Full details of the enrolment have been documented elsewhere [40, 41]. Data have been gathered from the mother and her partner (during pregnancy and post birth) and the children (post birth) from self-report questionnaires and clinical sessions. Ethics approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committee. The study website contains details of all available data through a searchable data dictionary (http://www.bristol.ac.uk/alspac/researchers/access/).
3D facial norms database
The 3D Facial Norms Database (3DFN) has been described in detail previously [42]. In brief, we used data from the 3DFN, a database of controls for craniofacial research. 2,454 unrelated individuals of recent European descent aged between 3 and 40 years were recruited from 4 sites across the USA and screened for a history of craniofacial conditions. 3D-derived anthropometric measurements, 3D facial surface images and genotype data were derived from each study participant.
GWAS genotypes and phenotypes are available at dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000949.v1.p1)).
Facial phenotyping
ALSPAC
ALSPAC children were invited to a clinic at the age of 15 years and 5,253 attended, where two high-resolution facial images were taken by Konica Minolta Vivid 900 laser scanners. 4,747 individuals had usable images (506 individuals did not complete the assessment, or the scans were of poor quality and consequently excluded). The coordinates of 22 facial landmarks were derived from the scans. Further methodological details are contained in a previous publication [10].
Distances between facial landmarks were computed by calculating the Euclidean distance between the 3D coordinates. To alleviate multiple testing issues, this study chose to test 7 distances that were either tested previously or have biological relevance to nsCL/P (S8 Table).
3D facial norms database
A methodological description of the facial phenotyping has been previously described in detail [42]. In brief, 3DFN study participants had their facial surfaces captured via 3D stereo-photogrammetry using either a two-pod 3dMDface or a multi-pod 3dMDcranial system. Captures were inspected to ensure 3D surface quality and additional captures were obtained if necessary. Similar to ALSPAC, a set of standard facial landmarks were collected from each 3D facial image and linear distances were calculated between the landmark coordinates.
Genotyping
ICC dbGaP
Of 7,347 DNA samples from study subjects genotyped using the Illumina Human610_Quadv1_B array SNP genotyping platform, scans from 7,089 subjects passed QC for unexpected relatedness, gender errors and missingness (>5%) and were released on dbGAP (phs000094.v1.p1). Pre-dbGaP release, SNPs in sample-chromosome combinations with a chromosomal anomaly (e.g. aneuploidy) were excluded. Post dbGaP release, SNPs were excluded for missingness (>5%), MAF (<5%) and HWE (P < 0.05) leaving 490,493 SNPs using PLINK [43].
ALSPAC
A total of 9,912 ALSPAC children were genotyped using the Illumina HumanHap550 quad genome-wide SNP genotyping platform. Individuals were excluded from further analysis based on having incorrect gender assignments; minimal or excessive heterozygosity (0.345 for the Sanger data and 0.330 for the LabCorp data); disproportionate levels of individual missingness (>3%); evidence of cryptic relatedness (>10% IBD) and being of non-European ancestry (as detected by a multidimensional scaling analysis seeded with HapMap 2 individuals). The resulting post-quality control dataset contained 8,237 individuals. The post-quality control ALSPAC children were combined with the ALSPAC mothers cohort (described in detail previously [44]) and imputed together using a subset of markers common to both the mothers and the children. The combined sample was pre-phased using ShapeIT (v2.r644) [45] and imputed to the 1000 Genomes reference panel (Phase 1, Version3) [46] using IMPUTE3 V2.2.2 [47]. After removing SNPs with MAF (<0.1) and INFO (<0.8), genotype data were available for 8,099,747 SNPs.
3D facial norms database
In collaboration with the Center for Inherited Disease Research (CIDR), 2,454 subjects in the 3DFN database were genotyped using a genome-wide association array including 964,193 SNPs from the Illumina OmniExpress+exome v1.2 array and an additional 4,322 SNPs from previous craniofacial genetic studies. Imputation was performed using the 1000 Genomes reference panel (phase 3) [42].
Statistical analysis
nsCL/P meta-analysis genome-wide association study
The transmission disequilibrium test (TDT) [48] evaluates the frequency with which parental alleles are transmitted to affected offspring to test genetic linkage in the presence of genetic association. The TDT was run on 638 parent-offspring trios and 178 parent-offspring duos of European descent from the ICC to determine genome-wide genetic variation associated with nsCL/P using PLINK [43].
The Bonn-II study [8] summary statistics from a case-control GWAS of 399 nsCL/P cases and 1,318 controls were meta-analysed, in terms of effect size and standard error, with the TDT GWAS summary statistics using METAL [49], based on a previously described protocol for combining TDT and case-control studies [50]. The final sample consisted of 1215 cases and 2772 parental and unrelated controls.
Polygenic risk score analysis
P-value inclusion threshold determination and PRS construction
We started by estimating the most appropriate P-value inclusion threshold for the nsCL/P PRS. The Bonn-II study summary statistics were used to construct PRS with different P-value inclusion thresholds in nsCL/P trios from the ICC. Analysis was performed separately in the Asian and European trios. The Polygenic-Transmission Disequilibrium Test (PTDT) [51] was then used to measure over-transmission of polygenic risk scores from unaffected parents to affected offspring and thereby select the most predictive P-value inclusion threshold. The P-value inclusion threshold was selected based on the most predictive threshold in the European trios, with results from the Asian trios treated as a sensitivity analysis. Parents with any form of orofacial cleft were removed from this analysis.
Next, using ALSPAC as a reference panel for linkage disequilibrium, PLINK was used to prune and clump the nsCL/P meta-analysis summary statistics (r2<0.1 and 250 kb) using the most predictive P-value threshold. The PRS were then constructed in the ALSPAC sample.
Power calculations for PRS analysis were performed using AVENGEME [17, 52]. Assuming 80% power and an alpha level of 0.05, we estimated the minimum genetic covariance required between nsCL/P and the 3D face-shape distances, for an association between the PRS and the face-shape distances to be detectable. Parameters used in power calculations are contained in S9 Table. The genetic covariance estimates were then converted to genetic correlation estimates using Genome-wide complex trait analysis (GCTA) [53] heritability estimates of the facial morphology variables derived in ALSPAC.
Association of nsCL/P PRS with facial phenotypes in ALSPAC
Of the 4,747 ALSPAC children with face-shape scans, 3,941 individuals had genotype data. GCTA [53] was used to prune these individuals for relatedness (IBS < 0.05) and the final sample with complete covariates included 3,707 individuals. The association between facial phenotypes and the nsCL/P PRS was measured in the final sample using a linear regression adjusted for sex, age at clinic visit, height at clinic visit and the first four principal components. Effect sizes were reported per standard deviation increase in PRS.
Replication in 3D facial norms database
Distances with some evidence of an association (P < 0.05) in the ALSPAC children were followed up for replication in an independent cohort (3DFN). 2,429 3DFN individuals had genotype and face-shape data. 332 individuals were removed due to missing SNPs in the PRS. The final sample consisted of 2,097 individuals. The association between implicated facial measurements and the nsCL/P PRS was measured using a linear regression adjusted for sex, age, height and the first 4 principal components. Effect sizes were reported per standard deviation increase in PRS.
Bidirectional mendelian randomization analysis
A bidirectional two-sample Mendelian randomization analysis was performed using the TwoSampleMR R package [54], testing both the forward direction (the effect of genetic risk variants for nsCL/P on implicated facial measurements) and the reverse direction (the effect of genetic risk variants for implicated facial measurements on liability to nsCL/P). The Inverse-Variance Weighted method was used as the primary analysis. Several sensitivity analyses were performed to test the assumptions of MR; the heterogeneity test was used to measure balanced pleiotropy, MR-Egger [55] was used to test for directional pleiotropy, the weighted median method [56] was used to test if the result is consistent assuming that at least half of the variants are valid and the weighted mode method [57] was used to test if the result is consistent assuming that the most common effect is valid. The Steiger test [58] was used to determine the likely direction of effect.
GWAS summary statistics for nsCL/P and implicated facial phenotypes
MR analysis required relevant SNP association information with respect to both nsCL/P and implicated facial measurements. SNP information relevant to nsCL/P was extracted from the nsCL/P meta-analysis summary statistics, previously described.
For implicated facial phenotypes, GWAS were performed using PLINK [43] in both ALSPAC (3,707 individuals) and the 3DFN study (2,429 individuals with genotype and face-shape data), using the same covariates as previously described in the polygenic risk score analysis. Summary statistics were then meta-analysed using METAL [49] with the combined sample including 6,136 individuals. SNP information relevant to implicated facial phenotypes was then extracted from the ALSPAC/3DFN meta-analysis summary statistics.
The ALSPAC/3DFN meta-analysis GWAS summary statistics of implicated facial phenotypes were subsequently analysed and functionally annotated [59] with the potential overlap between philtrum-width associated SNPs and expression quantitative trait loci (eQTLs) investigated using the Genotype-Tissue Expression (GTEx) catalogue [60].
Genetic risk variants for nsCL/P and implicated facial phenotypes
For the forward direction, genetic instruments for nsCL/P are SNPs that are strongly associated with nsCL/P. 6 well-characterised genome-wide significant nsCL/P SNPs in Europeans were taken from a previous study [3].
Information on the 6 nsCL/P SNPs is contained in S10 Table.
For the reverse direction, genetic instruments for implicated facial phenotypes are SNPs that are strongly associated with the implicated facial phenotypes. We LD clumped (r2<0.001 within 500KB) the ALSPAC/3DFN meta-analysis summary statistics to generate independent instruments for the MR analysis. LD proxies (r2>0.9) were used for SNPs unavailable in the nsCL/P summary statistics and were generated using LDlink and LDproxy [61] using the 1000 Genomes CEU/GBR populations as the reference panel [46].
Interpreting bidirectional mendelian randomization analysis
The results of the bidirectional MR and relevant sensitivity analyses were used to infer the likelihood of the liability-related sub-phenotype model. Three distinct possibilities were considered to explain the association between nsCL/P PRS and implicated facial phenotypes (which were detailed previously in Fig 4).
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We are extremely grateful to all the families who took part in the ICC, ALSPAC, Bonn-II and 3DFN studies as well as the many individuals involved in the running of the studies and recruitment, which include midwives, interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses.
Data Availability
The nsCL/P summary statistics used in this study were constructed using data downloaded from dbGAP. Therefore, the authors are unable to make the complete summary statistics publicly available without appropriate permission. However, replication of the analyses in the manuscript is possible because the nsCL/P-related SNPs used analyses are listed along with betas, standard errors and effect alleles in the supplementary material of the manuscript. The ICC nsCL/P genotype dataset was obtained from dbGaP at [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000094.v1.p1] through dbGaP accession number [phs000094.v1.p1]. ALSPAC genotype and phenotype data are available to researchers meeting the access criteria at http://www.bristol.ac.uk/alspac/researchers/access/. The 3D Facial Norms Database GWAS genotypes and phenotypes are available at dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000949.v1.p1). The ALSPAC/3DFN philtrum width GWAS summary statistics are available at the University of Bristol data repository, data.bris, at https://doi.org/10.5523/bris.1kz9y0moa8sgj2lxlk53mdmlbj.
Funding Statement
LJH, GCS, GDS, BSP, ES, GH and SJL are funded by the UK Medical Research Council (https://www.mrc.ac.uk/; Grant number MC_UU_12013). GCS, JRS, ES and SJL and the Cleft Collective are funded by the Scar Free Foundation (http://scarfree.org.uk/; REC approval 13/SW/0064). The UK Medical Research Council and the Wellcome Trust (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. MKL, JRS, MLM, EF and SMW are funded by the National Institute for Dental and Craniofacial Research (http://www.nidcr.nih.gov/), which provided funding for 3DFN related analyses, through grants U01- DE020078, R01-DE027023, and R01-DE016148. Funding support for the ICC study was provided by several previous grants from the National Institute of Dental and Craniofacial Research (NIDCR). Funding for individual investigators include: R21-DE-013707 and R01-DE-014581 (Beaty); R37-DE-08559 and P50-DE-016215 (Murray, Marazita) and the Iowa Comprehensive Program to Investigate Craniofacial and Dental Anomalies (Murray); R01-DE-09886 (Marazita), R01-DE-012472 (Marazita), R01-DE-014677 (Marazita), R01-DE-016148 (Marazita), R21-DE016930 (Marazita); R01-DE-013939 (Scott). Parts of this research were supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Wilcox, Lie) and additional recruitment was supported by the Smile Train Foundation for recruitment in China (Jabs, Beaty, Shi) and a grant from the Korean government (Jee). The genome-wide association study, also known the Cleft Consortium, is part of the Gene Environment Association Studies (GENEVA) program of the trans-NIH Genes, Environment and Health Initiative [GEI] supported by U01-DE-018993. Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is fully funded through a federal contract from the National Institutes of Health (NIH) to The Johns Hopkins University, contract number HHSN268200782096C. Funds for genotyping were provided by the NIDCR through CIDR’s NIH contract. Assistance with genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01-HG-004446) and by the National Center for Biotechnology Information (NCBI). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nature Reviews Genetics. 2011;12(3):167 10.1038/nrg2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murray J. Gene/environment causes of cleft lip and/or palate. Clinical Genetics. 2002;61(4):248–56. [DOI] [PubMed] [Google Scholar]
- 3.Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nature Genetics. 2012;44(9):968–71. 10.1038/ng.2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu Y, Zuo X, He M, Gao J, Fu Y, Qin C, et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nature Communications. 2017;8:14364 10.1038/ncomms14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ludwig KU, Ahmed ST, Böhmer AC, Sangani NB, Varghese S, Klamt J, et al. Meta-analysis reveals genome-wide significance at 15q13 for nonsyndromic clefting of both the lip and the palate, and functional analyses implicate GREM1 as a plausible causative gene. PLoS Genetics. 2016;12(3):e1005914 10.1371/journal.pgen.1005914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ludwig KU, Böhmer AC, Bowes J, Nikolić M, Ishorst N, Wyatt N, et al. Imputation of orofacial clefting data identifies novel risk loci and sheds light on the genetic background of cleft lip±cleft palate and cleft palate only. Human Molecular Genetics. 2017;26(4):829–42. 10.1093/hmg/ddx012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leslie EJ, Carlson JC, Shaffer JR, Feingold E, Wehby G, Laurie CA, et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24. 2, 17q23 and 19q13. Human Molecular Genetics. 2016;25(13):2862–72. 10.1093/hmg/ddw104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nature Genetics. 2010;42(1):24 10.1038/ng.506 [DOI] [PubMed] [Google Scholar]
- 9.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature Genetics. 2010;42(6):525 10.1038/ng.580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paternoster L, Zhurov AI, Toma AM, Kemp JP, Pourcain BS, Timpson NJ, et al. Genome-wide association study of three-dimensional facial morphology identifies a variant in PAX3 associated with nasion position. The American Journal of Human Genetics. 2012;90(3):478–85. 10.1016/j.ajhg.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Van Der Lijn F, Schurmann C, Zhu G, Chakravarty MM, Hysi PG, et al. A genome-wide association study identifies five loci influencing facial morphology in Europeans. PLoS Genetics. 2012;8(9):e1002932 10.1371/journal.pgen.1002932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaffer JR, Orlova E, Lee MK, Leslie EJ, Raffensperger ZD, Heike CL, et al. Genome-wide association study reveals multiple loci influencing normal human facial morphology. PLoS Genetics. 2016;12(8):e1006149 10.1371/journal.pgen.1006149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JB, Manyama M, Kimwaga E, Mathayo J, Larson JR, Liberton DK, et al. Genomewide association study of African children identifies association of SCHIP1 and PDE8A with facial size and shape. PLoS Genetics. 2016;12(8):e1006174 10.1371/journal.pgen.1006174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adhikari K, Fuentes-Guajardo M, Quinto-Sánchez M, Mendoza-Revilla J, Chacón-Duque JC, Acuña-Alonzo V, et al. A genome-wide association scan implicates DCHS2, RUNX2, GLI3, PAX1 and EDAR in human facial variation. Nature Communications. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehringer S, Van Der Lijn F, Liu F, Günther M, Sinigerova S, Nowak S, et al. Genetic determination of human facial morphology: links between cleft-lips and normal variation. European Journal of Human Genetics. 2011;19(11):1192 10.1038/ejhg.2011.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng S, Tan J, Hu S, Zhou H, Guo J, Jin L, et al. Detecting genetic association of common human facial morphological variation using high density 3D image registration. PLoS Computational Biology. 2013;9(12):e1003375 10.1371/journal.pcbi.1003375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genetics. 2013;9(3):e1003348 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dempster ER, Lerner IM. Heritability of threshold characters. Genetics. 1950;35(2):212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Annals of Human Genetics. 1965;29(1):51–76. [Google Scholar]
- 20.Fraser F. The multifactorial/threshold concept—uses and misuses. Teratology. 1976;14(3):267–80. 10.1002/tera.1420140302 [DOI] [PubMed] [Google Scholar]
- 21.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? International Journal of Epidemiology. 2003;32(1):1–22. [DOI] [PubMed] [Google Scholar]
- 22.Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Smith GD. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. The American Journal of Clinical Nutrition. 2016;103(4):965–78. 10.3945/ajcn.115.118216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GWAS meta analysis of philtrium width. [Internet]. 2018. Available from: 10.5523/bris.1kz9y0moa8sgj2lxlk53mdmlbj. [DOI]
- 24.Som P, Naidich T. Illustrated review of the embryology and development of the facial region, part 1: early face and lateral nasal cavities. American Journal of Neuroradiology. 2013;34(12):2233–40. 10.3174/ajnr.A3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang R, Bush JO, Lidral AC. Development of the upper lip: morphogenetic and molecular mechanisms. Developmental Dynamics. 2006;235(5):1152–66. 10.1002/dvdy.20646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marazita M. Subclinical features in non‐syndromic cleft lip with or without cleft palate (CL/P): review of the evidence that subepithelial orbicularis oris muscle defects are part of an expanded phenotype for CL/P*. Orthodontics & Craniofacial Research. 2007;10(2):82–7. [DOI] [PubMed] [Google Scholar]
- 27.Neiswanger K, Weinberg SM, Rogers CR, Brandon CA, Cooper ME, Bardi KM, et al. Orbicularis oris muscle defects as an expanded phenotypic feature in nonsyndromic cleft lip with or without cleft palate. American Journal of Medical Genetics Part A. 2007;143(11):1143–9. [DOI] [PubMed] [Google Scholar]
- 28.Aspinall A, Raj S, Jugessur A, Marazita M, Savarirayan R, Kilpatrick N. Expanding the cleft phenotype: the dental characteristics of unaffected parents of Australian children with non‐syndromic cleft lip and palate. International Journal of Paediatric Dentistry. 2014;24(4):286–92. 10.1111/ipd.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menezes R, Vieira AR. Dental anomalies as part of the cleft spectrum. The Cleft Palate-Craniofacial Journal. 2008;45(4):414–9. 10.1597/07-064.1 [DOI] [PubMed] [Google Scholar]
- 30.Stanier P, Moore GE. Genetics of cleft lip and palate: syndromic genes contribute to the incidence of non-syndromic clefts. Human Molecular Genetics. 2004;13(suppl 1):R73–R81. [DOI] [PubMed] [Google Scholar]
- 31.Weinberg S, Naidoo S, Bardi K, Brandon C, Neiswanger K, Resick J, et al. Face shape of unaffected parents with cleft affected offspring: combining three‐dimensional surface imaging and geometric morphometrics. Orthodontics & Craniofacial Research. 2009;12(4):271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ference BA, Yoo W, Alesh I, Mahajan N, Mirowska KK, Mewada A, et al. Effect of long-term exposure to lower low-density lipoprotein cholesterol beginning early in life on the risk of coronary heart disease: a Mendelian randomization analysis. Journal of the American College of Cardiology. 2012;60(25):2631–9. 10.1016/j.jacc.2012.09.017 [DOI] [PubMed] [Google Scholar]
- 33.Holmes MV, Dale CE, Zuccolo L, Silverwood RJ, Guo Y, Ye Z, et al. Association between alcohol and cardiovascular disease: Mendelian randomisation analysis based on individual participant data. BMJ. 2014;349:g4164 10.1136/bmj.g4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boncinelli E. Homeobox genes and disease. Current Opinion in Genetics & Development. 1997;7(3):331–7. [DOI] [PubMed] [Google Scholar]
- 35.Holland PW, Booth HAF, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biology. 2007;5(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharp GC, Ho K, Davies A, Stergiakouli E, Humphries K, McArdle W, et al. Distinct DNA methylation profiles in subtypes of orofacial cleft. Clinical Epigenetics. 2017;9(1):63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie EJ, Carlson JC, Shaffer JR, Butali A, Buxó CJ, Castilla EE, et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Human Genetics. 2017;136(3):275–86. 10.1007/s00439-016-1754-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International Consortium to Identify Genes and Interactions Controlling Oral Clefts 2010. Available from: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000094.v1.p1.
- 39.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nature Genetics. 2007;39(10):1181–6. 10.1038/ng1007-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golding P, Jones and the ALSPAC Study Team. ALSPAC–the avon longitudinal study of parents and children. Paediatric and Perinatal Epidemiology. 2001;15(1):74–87. [DOI] [PubMed] [Google Scholar]
- 41.Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, et al. Cohort profile: the ‘children of the 90s’—the index offspring of the Avon Longitudinal Study of Parents and Children. International Journal of Epidemiology. 2012:111–27. 10.1093/ije/dys064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinberg SM, Raffensperger ZD, Kesterke MJ, Heike CL, Cunningham ML, Hecht JT, et al. The 3D Facial Norms Database: Part 1. A web-based craniofacial anthropometric and image repository for the clinical and research community. The Cleft Palate-Craniofacial Journal. 2016;53(6):e185–e97. 10.1597/15-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, et al. Cohort profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. International journal of epidemiology. 2012;42(1):97–110. 10.1093/ije/dys066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nature Methods. 2012;9(2):179–81. [DOI] [PubMed] [Google Scholar]
- 46.Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics. 2009;5(6):e1000529 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM). The American Journal of Human Genetics. 1993;52(3):506 [PMC free article] [PubMed] [Google Scholar]
- 49.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kazeem G, Farrall M. Integrating case‐control and TDT studies. Annals of Human Genetics. 2005;69(3):329–35. [DOI] [PubMed] [Google Scholar]
- 51.Weiner DJ, Wigdor EM, Ripke S, Walters RK, Kosmicki JA, Grove J, et al. Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nature Genetics. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palla L, Dudbridge F. A fast method that uses polygenic scores to estimate the variance explained by genome-wide marker panels and the proportion of variants affecting a trait. The American Journal of Human Genetics. 2015;97(2):250–9. 10.1016/j.ajhg.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. The American Journal of Human Genetics. 2011;88(1):76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemani G, Zheng J, Wade KH, Laurin C, Elsworth B, Burgess S, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016:078972. [Google Scholar]
- 55.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International Journal of Epidemiology. 2015;44(2):512–25. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genetic Epidemiology. 2016;40(4):304–14. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hartwig FP, Smith GD, Bowden J. Robust inference in two-sample Mendelian randomisation via the zero modal pleiotropy assumption. bioRxiv. 2017:126102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemani G, Tilling K, Smith GD. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS genetics. 2017;13(11):e1007081 10.1371/journal.pgen.1007081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe K, Taskesen E, Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nature Communications. 2017;8(1):1826 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lonsdale J, Thomas J, Salvatore M, Phillips R, Lo E, Shad S, et al. The genotype-tissue expression (GTEx) project. Nature Genetics. 2013;45(6):580–5. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31(21):3555–7. 10.1093/bioinformatics/btv402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The nsCL/P summary statistics used in this study were constructed using data downloaded from dbGAP. Therefore, the authors are unable to make the complete summary statistics publicly available without appropriate permission. However, replication of the analyses in the manuscript is possible because the nsCL/P-related SNPs used analyses are listed along with betas, standard errors and effect alleles in the supplementary material of the manuscript. The ICC nsCL/P genotype dataset was obtained from dbGaP at [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000094.v1.p1] through dbGaP accession number [phs000094.v1.p1]. ALSPAC genotype and phenotype data are available to researchers meeting the access criteria at http://www.bristol.ac.uk/alspac/researchers/access/. The 3D Facial Norms Database GWAS genotypes and phenotypes are available at dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000949.v1.p1). The ALSPAC/3DFN philtrum width GWAS summary statistics are available at the University of Bristol data repository, data.bris, at https://doi.org/10.5523/bris.1kz9y0moa8sgj2lxlk53mdmlbj.