Sialic acids (Nonulosonic acids)
Sialic acids (neuraminic acids) are a diverse family of 9 carbon (nonulosonic) α-keto acidic carbohydrates. The canonical sialic acid, 2-keto-3-deoxy-5-acetamido-D-glycero-D-galacto-nonulosonic acid, also known as N-acetylneuraminic acid (Neu5Ac) is the backbone on which a large number of known modifications are made (6). The Neu5Ac structure is typified by a 6 carbon carboxylic acid ring structure with a glycerol tail, an acetamido at the C-5 position and hydroxyl groups present on C-4, C-7, C-8, and C-9. Modifications occur primarily on the hydroxyl groups, with O-acetylation being the most common alteration, and substitutions have been shown to occur after the completion of the core structure (18). Other modifications such as O-methylation, O-lactylation, and O-sulfation add to the diversity of this molecule in vivo. Two structurally similar sialic acids, N-glycolylneuraminic acid (Neu5Gc), which differs from Neu5Ac by the presence of a hydroxyl group on the N-5 acetyl moiety, and 2-keto-3-deoxy -D-glycero-D-galacto-nonulosonic acid (KDN), a deaminated form of Neu5Ac also occur in nature and similar modifications are made to their core structure (6). These three main structures (Neu5Ac, Neu5Gc, and KDN) encompass the family of sialic acids due to their retention of the same stereochemical configuration of the 9-carbon backbone.
Significance of Sialic Acids in Vertebrates
Among metazoans, sialic acid is primarily limited to members of the deuterostome lineage of the phyla Chordata and Echinodermata. Neu5Ac is the most widely synthesized of the family, and the most studied form. Neu5Gc is common among mammals but conspicuously absent in humans, due to loss of the hydroxylase gene required for its formation (122). KDN was once thought to be exclusive to lower order vertebrates such as fish and amphibians, but recent studies have found it to be present in humans in an unbound form, and its presence in the gametes of fish as well as human ovarian cells and fetal serum may indicate that KDN plays a role in development (48). Sialic acids in both eukaryotes and prokaryotes are typically positioned at the terminal end of glycoconjugates allowing them to interact with the external environment and play a role in cell to cell communication as well as self-recognition. In particular, in eukaryotes the self-recognition function of sialic acid is shown in its role as a modulator of immune function. An example of this would include Factor H, which preferentially binds to C3b on cell surfaces containing sialic acid glycoconjugates, preventing the binding of Factor B and halting the alternative complement cascade (56). Sialic acids also bind to a family of cell surface proteins known as sialic acid-binding immunoglobulin-like lectins (Siglecs). This interaction between sialic acid glycoconjuates and Siglecs has been reported to dampen immune function in macrophages, natural killer cells, neutrophils, and B-cells (8, 21, 64, 88). The highest concentration of sialic acid in humans, however, is found in the brain, and is heavily utilized throughout the central nervous system (CNS), yielding the name neuraminic acid (neurons). Within the brain and CNS polysialic acid chains are associated with the neural cell adhesion molecules of neurons, gila cells and ganglions (99, 130). Lastly, a major reservoir of sialic acids is on mucosa surfaces. Sialic acid’s common name is derived from the Greek word sialon, meaning saliva, as it was first isolated in bovine submaxillary mucin (10). In addition to salivary mucins, sialic acid has also been shown to be present in intestinal, lung and vaginal mucin glycans (29, 102, 116). In intestinal mucin, over 65% of glycans contain sialic acid residues (97). The addition of sialic acid on mucin glycans is thought to play a role in protecting the underlying peptides from proteolysis, and it has also been implicated in playing a role in mucin mediated bacterial aggregation and hydroxyl radical scavenging (37, 85, 108)
Sialic Acid Biosynthesis
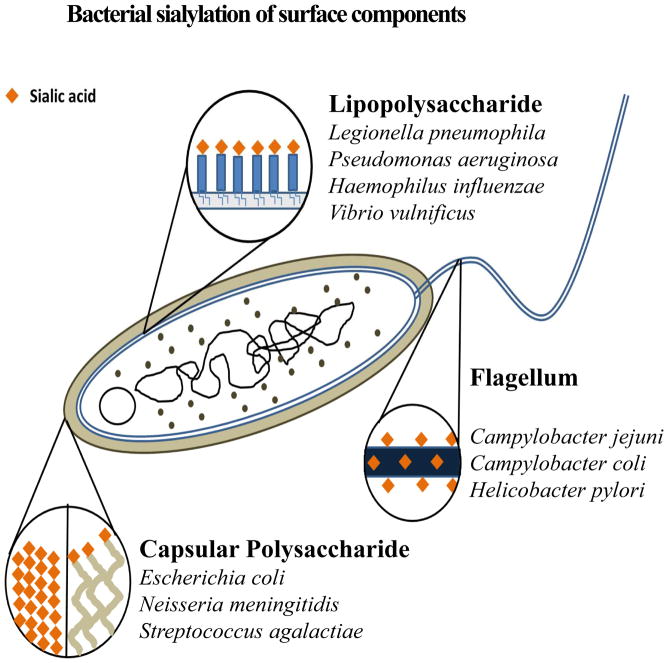
In humans, sialic acid is generated by the function of four primary genes. The precursor molecule in this pathway is the six carbon sugar UDP-N-acetylglucosamine which is converted by the bifunctional enzyme, UDP-N-acetylglucosamine-2-epimerase (NeuC)/N-acetylmannosamine kinase, to N-acetylmannosamine-6-phosphate (ManAc-6P). ManAc-6P, with the addition of phosphoenolpyruvate (PEP), is combined by the condensation action of N-acetylneuraminic-9-phosphate synthase (NeuB) to form N-acetylneuraminic-9-phosphate (Neu5Ac-9-P). Neu5Ac-9-P is converted to Neu5Ac by a phosphatase enzyme and the final 9-carbon product is activated by CMP-N-acetylneuraminic synthase (NeuA) generating CMP-Neu5Ac. This activated form is then recognized by sialylotransferases for ensuing glycosylation. Bacterial sialic acid synthesis follows a similar pathway with homologs of NeuC, NeuB, and NeuA required for de novo synthesis (119–121). Two sialic acid-like molecules, legionaminic and pseudaminic acid are exclusively synthesized by bacteria (60, 61). Bacteria have been observed to decorate three different surface structures depending on the species and the strain: lipopolysaccharide (LPS) in Gram-negative bacteria; the flagellum; and capsular polysaccharides (Fig. 1). It has been proposed that one of the roles of sialic acid surface decoration in bacteria is to mimic the eukaryotic host cells leading to a dampening of immune responses as described above (12, 59, 82, 90). In addition, it has been proposed that sialylation plays a role in biofilm formation (55, 109) (89, 118). The ability of bacteria to biosynthesize sialic acid was once thought to be limited to a few pathogenic and commensal species; however more recent phylogenetic analysis of sialic acid biosynthesis genes indicates that this ability is highly prevalent across a large number of diverse bacterial lineages (65).
FIGURE 1.
Bacterial sialylation of surface components. This diagram depicts the different surface structures in bacteria that are known to be decorated with nonulosonic acids (neuraminic, pseudaminic, or legionaminic). Also indicated are bacterial species demonstrated to have different surfaces sialylated.
Sialic Acid Catabolism
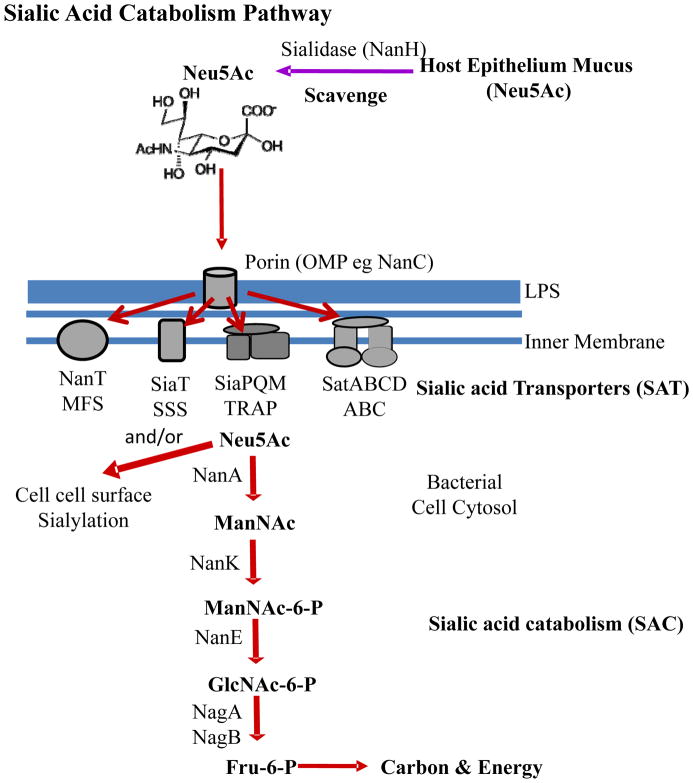
Conversely, the catabolism of Neu5Ac appears to be mainly the purview of bacterial species that associate with eukaryotic hosts either as a commensal or a pathogen (3, 5). In theory, all mucous covered epithelium surfaces are a potential food sources for sialic acid catabolizers since as stated previously in intestine mucin a key component of mucous over 65% of glycans contain sialic acid residues (97). In bacteria, Neu5Ac catabolism is dependent on the activity of N-acetylneuraminic acid lyase (NanA) (126). NanA cleaves pyruvate from Neu5Ac generating ManNAc. ManNAc is then converted to GlcNAc-6-P by the action of N-acetylmannosamine kinase (NanK) and N-acetylmannosamine 6-phosphate epimerase (NanE). GlcNAc-6-P is broken down further; first deacetylated by N-acetylglucosamine-6-phosphate deacetylase (NagA), and subsequently deaminated by glucosmine-6-phosphate deaminase (NagB) yielding fructose-6-phosphate, and ammonia (125, 126) (Fig. 2). Possessing Neu5Ac catabolic capabilities becomes significant when dealing with environments within eukaryotic hosts, where competition for space and nutrients is intense. Bacteria acquire sialic acid from their eukaryotic hosts either through the synthesis of a sialidase, a glycoside hydrolase, which cleaves terminal Neu5Ac residues from host glycoconjugates or simply by scavenging free Neu5Ac found in the environment released by other bacterial species (68). In Gram-negative bacteria, uptake of sialic acids require transport across the outer membrane by either a general porin such as OmpC/F or a sialic acid specific porin such as NanC (25) (Fig. 2). Transport across the inner membrane occurs by a diverse range of transporters. Four main systems have been shown to transport Neu5Ac: a major facilitator superfamily (MFS) permease designated as NanT, a tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM, a sodium solute symporter (SSS) SiaT, and an ATP-binding cassette (ABC) transport system SatABCD (2, 5, 75, 79, 96, 104–106, 124, 125) (Fig. 2). The first characterized bacterial Neu5Ac transporter was NanT from E. coli by the Vimr group, which contains a hydrophilic domain unique among sugar permeases, thought to facilitate the interaction with Neu5Ac (75, 125). The TRAP transporter systems, which were originally shown to be carboxylic acid transporters, were demonstrated to transport Neu5Ac in Haemophilus influenzae by the Apicella and Thomas groups (2, 79, 106). In Haemophilus ducreyi an ABC type transporter is proposed to be required for Neu5Ac transport (96). Thomas and colleagues have also demonstrated that Salmonella enterica encodes a SSS transporter along with NanT that both transport Neu5Ac into the cell (105).
FIGURE 2.
Schematic representation of the catabolism of sialic acid in Bacteria. The first step in catabolism is the uptake into the bacterial cell of free sialic acid molecules across the cell wall and cell membrane. Four type of transport systems have been described for transport across the cell membrane, MFS, SSS, TRAP, and ABC systems. NanH, Neuraminidase; Neu5Ac, N-acetylneuraminic acid. The sialic acid catabolic pathway involves several steps beginning with NanA. NanA, N-acetylneuraminic acid lyase; ManNAc, N-acetylmannosamine; NanK, N-acetylmannosamine kinase; ManNAc-6-P, N-acetyl-mannosamine-6-phosphate; NanE, N-acetylmannosamine-6-P epimerase; GlcNAc-6-P, N-acetylglucosamine-6-phosphate; NagA, N-acetylglucosamine-6-phosphate deacteyl-ase; GlcN-6-P, Glucosamine-6-phosphate; NagB, Glucosamine-6-phosphate deaminase; Fru-6-P, Fructose-6-phosphate; LPS, lipopolysaccharide.
Distribution of sialic acid catabolism gene clusters among sequenced bacteria
An investigation of the phylogenetic distribution of sialic acid catabolism among nearly 2000 finished and unfinished bacterial genomes was completed in 2009 (4). In that study, sialic acid gene clusters, consisting of NanA, NanE and NanK, were shown to be present in 46 species. These 46 species encompassed six bacterial families of Gamma-Proteobacteria (1. Enterobacteriaceae, 2. Pasteurellaceae, 3. Pseudoalteromonadaceae, 4. Psychromonadaceae, 5. Shewanellaceae, and 6. Vibrionaceae), one member of the genus Fusobacterium, and six Gram-positive families (1. Clostridiaceae, 2. Lactobacillaceae, 3. Lachnospiraceae, 4. Mycoplasmataceae, 5. Staphylococcaceae, 6. Streptococcaceae) encompassing both low and high GC representatives. A follow up study examined the distribution and phylogeny of NanA only and identified 86 species that contained this protein (3). Here we determined whether this distribution of sialic acid catabolism genes had expanded by examining the newly sequenced species from all completed and draft genomes published on NCBI (4497 genomes). The search was conducted using pBLAST with NanA of Vibrio cholerae N16961 (VC1776) and Staphylococcus aureus N315 (SA0304) as seeds. Candidate species were determined by the presence of NanA, and to eliminate false positives due to similarity to dihydropicolinate synthase (DapA), the presence of homologs of a sialic acid transporter as well as putative nanE/nanK genes or a sialidase was required. From this stringent analysis, we identified 265 species encompassing 9 phyla (Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, Proteobacteria, Planctomycetes Spirochaetes, Tenericutes, and Verrucomicrobia) with the potential to utilize sialic acid as a carbon and energy source. We speculate that the total number of species that can catabolize sialic acid is probably much larger if we just examined NanA distribution only. The distribution of sialic acid catabolizers among bacteria is composed primarily, but not exclusively, of commensal and pathogenic species of humans and animals. A major addition to the list of sialic acid catabolizers is 30 species from the phylum Actinobacteria, represented by 6 families: Arthrobacteraceae, Bifidobacteriaceae, Coriobacteriaceae, Corynebacteriaceae, Intrasporangiaceae and Streptomycetaceae. Many of the species within these families that contain sialic acid catabolism genes are associated with the oral cavity, the respiratory tract, the gastrointestinal tract, or the urogenital tract of humans. Examples include Bifidobacterium breve (commensal gut), Corynebacterium diphtheria (diphtheria), C. ulcerans (mastitis), C. pseudotuberculosis (lymphadenitis), Eggerthella lenta (commensal gut), Gardenella vaginalis (bacterial vaginosis), as well as soil dwellers such as any Streptomycetes spp and Kitasatospora setae. A greatly expanded group of potential sialic acid catabolizers is within the phylum Bacteroidetes, represented by 41 species from 8 families: Bacteroideteceae, Cytophagaceae, Cyclobacteriaceae, Chitinophagaceae, Flavobacteriaceae, Prevotellaceae, Porphyromonadaceae and Sphingobacteriaceae. Examples of species within this group that contain sialic acid catabolism genes include Bacteroides fragilis (commensal gut), B. dorei (commensal gut), B. vulgatus (commensal gut), Capnocytophaga ochracea (oral cavity), C. sputigena (periodontal disease), Cryptobacterium curtum (isolated from periodontal lesions), Tannerella forsythia (periodontal disease), Prevotella oralis (oral cavity), P. denticola (oral cavity), and P. salivae (oral cavity). Fusobacterium nucleatum from the phylum Fusobacteria (mucous membrane dwellers) was previously reported as a putative sialic acid utilizer, and now includes species F. varium, F. gonidiaformans, and F. necrophorum. The phyla Verrucomicrobia and Planctomycetes, are representative by 4 and 2 species respectively that have the potential to use sialic acid as a carbon source.
The phylum Firmicutes was also well represented with species capable of sialic acid catabolism; 76 species in total from 9 bacterial families, including 21 species from the family Streptococcaceae, 10 species from the family Staphylococcaceae, 11 species from Clostridiaceae, and 7 species from Lactobacillaceae. We identified 88 species from the phylum Proteobacteria that contain sialic acid catabolism genes, which included the well-studied pathogens E. coli, Salmonella enterica, H. influenzae, and Shigella spp. Members of potential sialic acid catabolizers within this phylum include several Citrobacter species, 9 Yersinia species, 8 species of Haemophilus, and Raoultella ornithinolytica (formally Klebsiella), Klebsiella pneumonia, and K. oxytoca. Within the family Vibrionaceae, 19 species contained sialic acid catabolism genes, including several pathogens of humans such as V. cholerae, V. mimicus and V. vulnificus.
The ability to catabolize sialic acid allows these species to utilize a ubiquitous carbon source found in the mucus rich surfaces of their eukaryotic hosts. In regards to these bacteria, the human body is an array of diverse ecosystems that houses and maintains a diverse consortium of bacterial species that compete for these nutritional resources. While the gastrointestinal tract is perhaps the best studied of these ecosystems, there exist a number of other locations that also contain a large amount of diverse species and, that can be invaded and colonized by pathogens that rely on sialic acid as an important carbon source. In the next sections, we will discuss the role of sialic acid catabolism in pathogenesis in a range of pathogens found in different regions of the human body (Table 1).
Table 1.
Bacterial species in which sialic acid catabolism was experimentally examined.
| Species | Transporter | Sialidase | Location | Reference |
|---|---|---|---|---|
| Bacteroides fragilis | NanT(MSF) | NanH | (14) | |
| Bifidobacterium longum | SAT (ABC) | NanH | Intestine | (131, 103) |
| Clostridium difficile | SSS | NanH | Intestine | (84) |
| Clostridium perfringens | SSS | NanH | Intestine | (83) |
| Corynebacterium glutamicum | SAT (ABC) | ? | (36) | |
| Cronobacter sakazakii | NanT(MSF) | - | Brain | (53, 54) |
| Escherichia coli | NanT(MSF) | - | Intestine | (22, 125) |
| Gardnerella vaginalis | NK | NanH | Urogenital tract | (68) |
| Haemophilus influenzae | SiaPQM (TRAP) | Respiratory tract | (106) | |
| Lactobacillus plantarum | SSS | (101) | ||
| Mycoplasma sinoviae | NanH | (76) | ||
| Pasteurella multocida | SiaPQM (TRAP) | Intestine | (113) | |
| Ruminococcus gnavus | SAT2 | NanH | Intestine | (27) |
| Salmonella enterica | NanT(MSF)SiaT (SSS) | - | Intestine | (4), (84, 105) |
| Staphylococcus aureus | Sym | Nose | (86) | |
| Streptococcus pneumoniae | SAT 2/3(ABC) | NanA,B,C | Respiratory tract | (74),(17) |
| Oralis group streptococci | NK | NK | Oral Cavity Endocarditis | (19, 20) |
| Milleri group streptococci | NK | NK | Oral Cavity Endocarditis | (19) |
| Group B streptococci | SAT (ABC) | - | Intestine Genital tract | (94) |
| Tannerella forsythiae | NanT(MSF) | NanH | Oral Cavity | (98) |
| Treponema denticola | NanT(MSF) | NanH | Oral Cavity | (62) |
| Vibrio cholerae N16961 | SiaPQM (TRAP) | NanH | Intestine | (5) |
| Vibrio mimicus | SiaPQM (TRAP) | NanH | Intestine | Boyd, unpublished |
| Vibrio fischeri ES114 | SSS | - | Light organ of squid | (5) |
| Vibrio vulnificus YJ016 | SiaPQM (TRAP) | - | Intestine/Blood | (50) |
| Yersinia enterocolitica | NanT(MSF) | - | Intestine | (4) |
| Yersinia pestis KIM | NanT(MSF) | - | Intestine Blood | (4) |
Oral Pathogens and sialic acid catabolism
The human oral microbiome is comprised of both eukaryotic and prokaryotic organisms, with approximately 1,000 species of bacteria comprising the dominant domain (128). Like other niches within the human body, the oral commensal microbiota plays an important role in preventing the colonization of harmful pathogenic bacteria in large part by outcompeting new comers for nutrients as proposed by Freter over 30 years ago (32, 33). Unfortunately, these commensals are not always successful in preventing harmful bacteria from taking root in the oral cavity, and a number of oral diseases are associated with bacterial pathogens. One such disease is periodontitis, which is the result of inflammation in the tissues that surround the teeth. This inflammation leads to degradation of the gum and bone and culminates with loss of the tooth. It is estimated that periodontitis affects 300 million individuals worldwide (98, 111). One of the culprits of periodontitis is the Gram-negative bacterium Tannerella forsythia, a Bacteroidetes, and a member of the family Porphyromonadaceae. This anaerobic bacterium is a dominant member of the subgingival plaque (biofilm), the presence of which is typically associated with periodontitis. Under in vitro culture conditions (liquid broth and plates), T. forsythia is fastidious and has traditionally been thought to only utilize the bacterial cell wall component N-acetylmuramic acid (NAM) as its sole carbon source (98). Additional analysis of the T. forsythia genome revealed the presence of a putative sialic acid utilization cluster (98, 111). Contained in this cluster are genes for sialic acid scavenging (including the sialidase nanH), inner and outer membrane transport, and genes for the conversion of Neu5Ac to the preferentially used carbon source NAM (98, 111). Furthermore, encoded elsewhere on the T. forsythia genome is the gene for a second sialadase, SiaHI. Similar to E. coli, T. forsythia contains the gene that encodes the enzyme (NanA) necessary for the conversion of sialic acid to ManNAc. While T. forsythia contains a copy of the nanE gene, it does not contain nanK. The NanE of T. forsythia is highly homologous (83% amino acid identity) to the NanE of Bacteroides fragilis, which also lacks the nanK gene. In B. fragilis, NanE has the ability to convert ManNAc to GlcNAc, after which it is then phosphorylated by the hexokinase RokA (14, 15, 98). The genome of T. forsythia contains a gene that is 82% homologous, on the amino acid level, to the RokA of B. fragilis, indicating that T. forsythia mostly likely utilizes sialic acid via the B. fragilis utilization pathway versus the canonical pathway observed in E. coli (98). Using bioinformatics, we identified this pathway in 8 species of the genus Prevotella from the phylum Bacteroidetes that were all isolated from the oral cavity suggesting that this is an important phenotypic trait in this environment.
Perplexingly, however, T. forsythia was shown to not utilize sialic acid as a nutritional source under growth on plates or in planktonic culture. In contrast sialic acid does appear to stimulate growth of T. forsythia under static growth conditions that is in biofilm (98). When statically grown cultures were supplemented with increasing concentrations of sialic acid, these cultures began to exceed the biomass of control cultures supplemented with NAM. It should be noted that the concentration of sialic acid required to exceed growth observed in cells utilizing NAM was much higher (6 mM vs 0.17 mM) (98).
Roy and colleagues also investigated the range of sialic acids that could promote the growth of T. forsythia in biofilm and found that growth was also supported by sialyllactose, which was a mixture of 2,3- and 2,6-sialyllactose, as well as glycolyl sialic acid (Neu5Gc). For both sialyllactose and Neu5Gc, growth of T. forsythia was comparable to levels reached using Neu5Ac as the sole carbon source. The addition of a sialidase inhibitor could abolish growth of T. forsythia in sialyllactose, indicating that the use of a sialidase was necessary for sialyllactose utilization (98). Given that sialyllactose is commonly found on the surface of host glycoproteins, this could indicate that the bacterial sialidase would also be important during colonization of the subgingival cavity.
In addition to playing roles in catabolism, the two known sialidases (NanH and SiaH1) of T. forsythia were also investigated for their role in the pathogenesis of this bacterium (40, 49, 115). Single mutants in both the nanH and siaH1genes were constructed and both strains were found to be defective in sialidase activity, though the defect was more severe for the nanH mutant than the siaH1 mutant (40). The two mutants and the wild-type strain were assayed for their ability to adhere to epithelial cells, and it was found that all three stains possessed the ability to adhere to and invade the cells. However, the nanH deletion mutant exhibited a significant defect in both adhesion and subsequent invasion of epithelial cells (40). Conversely the siaH1 deletion mutant demonstrated levels of adherence and invasion on par with the wild-type strain. Thus this group demonstrates that NanH is the principal sialidase while SiaH1 likely does not play a role in the cleavage of extracellular sialic acids (40). Another species associated with subgingival plaque is Treponema denticola, a Spirochaete, which has been shown to possess sialidase activity (62, 132). The sialidase of T. denticola cleaves both α2,3- and α2,6-linked sialic acid from glycoproteins. Additionally, a sialidase deletion mutant failed to grow in serum growth media, presumably due to its inability to cleave sialic acid from the serum proteins (62). This mutant was more susceptible to complement deposition and exhibited a defect in the mouse skin-infection model (62).
Many species of Streptococcus are associated with the oral cavity. Byers and colleagues demonstrated that S. oralis, S. sanguis, S. gordonii, S. mitis, S. intermedius, S. anginosus, S. constellatus and S. defectivus use Neu5Ac as a sole carbon source (19, 20). We identified in the genome sequence database a total of 10 species that contain sialic acid catabolism genes within the genus; S. gordonii, S. infantis, S. iniae, S. intermedius, S. mitis, S. oralis, S. parasanguinis, S. pneumonia, S. pyogenes, and S. sanguinis, and many of these species were isolated from the oral cavity and can cause diseases such as endocarditis.
Respiratory tract pathogens and sialic acid catabolism
The human lung has traditionally been considered a sterile environment in healthy individuals. However, recent metagenomic research indicates that the lungs do support their own consortium of microbiota, adding yet another microbial niche within the human body (7, 28, 46). Mucus production is important for lung tissue as it helps maintain optimum function by preventing the epithelium from becoming too dry. The lungs produce approximately 2 liters of mucus per day, which can serve as a rich nutrient source for pathogenic bacteria with the ability to cleave sugars from the glycoproteins found in host mucus. In cystic fibrosis patients, lung disease caused by polymicrobial infections is a serious concern. Traditionally the main players in CF lung infections were Proteobacteria such as Pseudomonas aeruginosa (NanH), Burkholderia cepacia complex, Haemophilus influenzae (NanA), Alcaligenes xylosoxidans, Stenotrophomonas maltophilia and the Firmicute Staphylococcus aureus (NanA). More recent metagenomic studies have shown that members of the phyla Actinobacteria, Bacteroidetes, Spirochaetes and Fusobacteria, are also present in the CF lung (7).
Haemophilus influenzae is a Gamma-Proteobacteria, a member of the family Pasteurellaceae and is an inhabitant of the human respiratory tract, associated with both upper and lower respiratory infections. Haemophilus influenzae strains can be subdivided into two groupings: encapsulated and non-encapsulated strains. Encapsulated strains are often associated with causing meningitis, while the non-encapsulated strains are commonly associated with mucosal diseases such as otitis media and bronchitis. Haemophilus influenzae strains have the ability to scavenge host sialic acid for both catabolism and sialylation of its LPS (2, 12, 41, 52, 70, 106, 123). Genes for the catabolism of sialic acid (nanEK nanA, siaR, nagBA) are present in the genome of H. influenzae within a single gene cluster, which has a different orientation than the cluster found in E. coli (2, 52, 106, 123). Located downstream of the catabolism gene cluster are the genes encoding for sialic acid TRAP transport (siaPQM) (2, 52, 106). Even though the catabolic pathway for sialic acid degradation is functional in H. influenzae, no defect was seen in vivo when comparing the infectious dose of the wild-type and the nanA deletion mutant strains using the intraperitoneal infant rat model (123). Additionally, when the wild-type and the nanA mutant were co-infected in the same mouse, the nanA mutant out-competed the wild-type strain, indicating that loss of sialic acid catabolism led to an increased fitness of the nanA deletion strain (123). Previous work demonstrated the importance of H. influenzae lipooligosaccharides (LOS) in vivo and that host derived sialic acid that is transported into the bacterial cell is incorporated into LPS, which is essential for resistance to host serum (2, 12, 41, 106). Interestingly, deletion of nanA appears to increase the amount of sialylation of H. influenzae LOS. It would stand to reason that hyper-sialylation could confer more resistance to host serum and lead to a competitive advantage in vivo. It is likely, based on the results observed above that sialic acid catabolism does not play a role in pathogenesis. However, all seven additional sequenced Haemophilus species: H. haemolyticus, H. parahaemolyticus, H. parainfluenzae, H. parasuis, H. pittmaniae, H. ducreyi, and H. sputorum, contain the Neu5Ac catabolism gene cluster similar to H. influenzae, which suggests that sialic acid is an important carbon source but not under the conditions examined in the above studies.
Streptococcus pneumoniae is a Gram-positive inhabitant of the human naso-oropharanx in healthy individuals. However, in some individuals the bacterium can progress from an asymptomatic resident to a pathogen, causing primarily pneumococcal pneumonia and an assortment of other diseases such as otitis media, bacteremia and meningitis. Strains of S. pneumoniae are especially adapted for life in an environment where sugar sources are bound to host proteins as they express up to 9 different glycosylases, 3 of which have been shown to have sialidase activity (NanA, NanB, and NanC) (58, 93). It was predicted from bioinformatics analysis that S. pneumoniae encodes the genes required for sialic acid catabolism and transport into the cell (4). Similar to H. influenzae, S. pneumonia can scavenge Neu5Ac for either catabolism and/or sialylation of their cell surface which serves as a key determinant of pathogenesis. And a recent study demonstrated that S. pneumoniae can utilize sialic acid as a carbon source and that an ABC type sialic acid transporter contributes to growth on a human glycoprotein and colonization in vivo (73).
Of the three sialidases identified in S. pneumoniae, nanA is present in all strains and is a surface associated sialidase. Studies have determined that NanA has activity common to other exosialidases in that it can cleave both α2,3- or α2,6-sialyllactose to release free sialic acid (58, 133). NanB is present in approximately 96% of S. pneumoniae strains and has been shown to act as a secreted sialidase (58, 133). And this sialidase has the ability to cleave α2,3-sialic acids to release 2,7-anhydro-sialic acid(58, 133). Lastly, NanC is present only in approximately 50% of S. pneumoniae isolates and is a trans-sialidase (133). NanC has been demonstrated to cleave α2,3-sialic acids to release 2,7-anhydro-sialic acid, and has been demonstrated to act as a sialidase inhibitor (133).
The roles of the S. pneumoniae sialidases in pathogenesis have been extensively investigated in vitro and in vivo, with a particular emphasis on the more widely distributed NanA and NanB enzymes that contribute to the bacterium’s ability to adhere to various epithelial cell lines (16). Overexpression of NanA also contributed to biofilm formation and growth on saliva-coated glass cover slips in S. pneumoniae (16). Lastly, nanA and nanB deletion mutants are defective for host colonization and the ability to cause sepsis following intranasal dosage (72, 87, 117).
Urogenital pathogens and sialic acid catabolism
The human vagina is another mucus covered surface that serves as a distinct ecosystem for the normal host microbiota. Vaginal mucus secretions are rich with sialic acids which serve as a nutrient source for the various bacterial species that inhabit this niche. Again, as in other locations on and in the human body, perturbations of the vaginal microbiota have been associated with the onset of disease. Specifically, bacterial vaginosis is a condition affecting the vaginal tract characterized by increased pH, thinning of vaginal secretions, and a fishy odor upon hydrogen peroxide treatment of vaginal samples. Women who have bacterial vaginosis are more at risk for pelvic inflammatory disease, sexually transmitted diseases, postsurgical complications, and pregnancy complications (110). The vaginal microbiota in healthy individuals is comprised mostly of lactobacilli and it is believed that bacterial vaginosis is caused by the loss of the normal microbiota (lactobacillus) and due to the overgrowth of a number of different anaerobic bacterial species (66, 68). Another hallmark of vaginosis is sialidase activity in vaginal secretions, which is not present in healthy individuals. Presumably, the presence of sialidases in vaginal secretion from individuals with vaginosis is due to the fluctuations in vaginal microbiota (66, 68).
A common bacterial species present in the microbiota of patients with bacterial vaginosis is Gardnerella vaginalis (110). Gardnerella vaginalis is an obligate anaerobe that belongs to the family Bifidobacteriaceae that produces sialidase and utilizes both Neu5Ac and Neu5Gc as carbon sources (68, 127). Recently, Lewis and colleagues characterized the role of sialidase in G. vaginalis in host vaginal colonization. They demonstrated that G. vaginalis strains that were sialidase positive could deplete sialic acid from bound sialoglycans in specialized culture media. However, sialic acid depletion and utilization by G. vaginalis was shown to be dependent on the host source of sialic acid: G. vaginalis was able to utilize human IgA as a source of sialic acid but was deficient in sialic acid utilization when grown using bovine submaxillary mucin(68). While free sialic acid is liberated from the bovine mucin, the amount of sialic acid utilized by G. vaginalis is much less than when grown using human mucin. Nonhuman mucin contains, in addition to Neu5Ac, a derivative Neu5Gc. The addition of Neu5Gc to media containing human IgA did not inhibit the increase of free Neu5Ac after bacterial inoculation (due to the presence of sialidase) but did appear to inhibit the bacteria from subsequently up-taking and utilizing the free Neu5Ac (68). Furthermore, inhibition of sialic acid catabolism by Neu5Gc was demonstrated to be at the level of transport and not by inhibiting sialidase or lyase enzymatic activity. It will be of interest to determine what type of sialic acid transporter is present in this species since NanT, TRAP and SSS systems were previously shown to transport Neu5Ac and Neu5Gc (43). Lastly, Lewis and colleagues demonstrated that G. vaginalis could free bound sialic acid in vivo using a vaginally infected mouse model, mimicking what is observed in human patients presenting with bacterial vaginosis (35, 68).
Group B streptococci (GBS) are a devastating pathogen of newborns and infants and transmission from mother to child is an enormous concern. A recent study by Pezzicoli and colleagues demonstrated that GBS utilize sialic acid as a carbon source and this ability was dependent upon an ABC sialic acid transporter (94). This group also demonstrated that the ability to catabolize sialic acid was important during in vivo mucosal colonization using an in vivo mouse model of intranasal and intravaginal GBS infection in which sialidase release of sialic acid was simulated by adding free Neu5Ac. The in vivo data demonstrated that exogenous sialic acid significantly increased the capacity of GBS to infect mice at the mucosal level.
Utilization of sialic acid as a carbon source by intestinal pathogens
The gastrointestinal tract is home to many commensals but pathogens also find their way into this environment, with the potential of causing disease. As with all epithelial cells, the intestinal tract is cover in a protective mucosal layer, which aids in preventing infection by the vast number of microorganisms that reside within a healthy human gut (77). As discussed previously, there is evidence which shows that both the secreted and cell surface intestinal mucus, the main component of which is mucin glycoproteins, act as mucosal barriers against potential pathogens (77). However, there are many enteric pathogens which have found ways to circumvent this mucosal barrier (77). Mucin glycoproteins, can serve as ligands for microbial adhesions and can also be utilized as an energy source by both commensal organisms as well as enteric pathogens (77).
There is a complex array of oligosaccharides present on the glycosylated domains of mucin and mucolytic bacteria release these mucin glycoproteins that can then be used as nutrient sources by commensals and pathogens alike. Some of the sugars available to enteric pathogens as food sources include fucose, galactose, galactosamine, glucosamine, Neu5Ac, mannose, glucose, glucuronate, gluconate and galacturonate (22, 63, 91). Neu5Ac is an excellent source of carbon and energy for intestinal commensals and pathogens, thus it is not surprising that there are a number of commensal species that encode the genes required to utilize Neu5Ac as a carbon source, such as, Bacteroides caccae, B. fragilis, B. ovatus, B. stercoris, B. uniformis, B. vulgatus, Parabacteroides distasonis,, B. breve, Escherichia coli, E. blattae, Edwardsiella tarda, Dorea formicigenerans, D. longicatena, Faecalibacterium prausnitzii, Fusobacterium nucleatum, Proteus mirabilis, Providencia rettgeri, Ruminococcus gnavus, Lactobacillus plantarum, L. sakei, L. salivarius, L. vadensis and Yokenella regensburgei. Among these species, only a handful has been experimentally shown to catabolize sialic acid. Examples of enteric pathogens which can utilize sialic acid as a carbon source include Citrobacter spp, Clostridium spp, Escherichia coli, E. hermannii, Enterobacter cloacae, E. cancerogenus, E. asburiae, E. aerogenes, Klebsiella pneumoniae, K. oxytoca, Salmonella enterica, Shigella spp., Vibrio cholerae, V. vulnificus, and 9 of 13 sequenced Yersinia spp; Y. pestis, Y. enterocolitica, Y. pseudotuberculosis, Y. bercovieri, Y. kristensenii, Y. mollaretii, Y. rohdei, Y. frederiksenii, and Y. ruckeri. And similar to commensal species, sialic acid metabolism has only been investigated in a handful of species (4, 104, 124).
Escherichia coli the most abundant Gram-negative facultative commensal in the intestinal tract can also be a pathogen, and utilizes many of the sugars present in the intestinal tract including sialic acid (22). Vimr and colleagues demonstrated the ability of E. coli K-12 to utilize sialic acid as a sole carbon source and identified an inducible catabolic system for sialic acids termed Nan (125). Since this initial work, it has been shown that sialic acid utilization is important for the in vivo survival of E. coli isolates (9, 22, 30). It was demonstrated that sialic acid catabolism was important for colonization initiation by E. coli MG1655 but not maintenance in the streptomycin treated mouse model of colonization and additionally, sialic acid was shown in vitro to be third in the order of preference of nutrients available within the intestinal tract (22).
In addition to looking at the ability of E. coli K-12 strains to utilize sialic acid, there have been studies comparing carbon metabolism between K-12 and pathogenic EDL933 O157:H7 strains (9, 30, 95). A carbon metabolism comparison between these two strains demonstrated a similar but not identical preference of nutrients in vitro, with both strains having Neu5Ac as their 4th most preferred nutrient (30). Catabolic genes for utilizing Neu5Ac were upregulated in the presence of sialic acid in E. coli EDL933, however, interestingly, mutation of the pathway for Neu5Ac catabolism caused colonization defects for E. coli MG1655 but not for E. coli EDL933, suggesting that sialic acid is not as important of a nutrient in vivo for E. coli EDL933 (30). It may be that the two strains utilize different carbon sources in order to occupy different intestinal niches and not compete for nutrients when both strains are present. A recent study using bovine small intestine contents as a growth media demonstrated that in E. coli O157:H7 the genes required for Neu5Ac catabolism were more highly expressed than in E. coli K12 and that Neu5Ac catabolism conferred a competitive growth advantage to the O157:H7 strain (9). Together these studies show the importance of sialic acid catabolism for E. coli colonization and demonstrate that there may be an interesting phenomenon of carbon preferences between commensal and pathogenic strains of the same species and in the same strain in different hosts.
It has also been shown that in addition to catabolizing Neu5Ac, the most prevalent sialic acid in nature, E. coli can also grow on alternative sialic acids such as 9-O-acetyl N-acetylneuraminic acid (112). The ability to utilize 9-O-acetyl N-acetylneuraminic requires YjhS (NanS), a 9-O-acetyl N-acetylneuraminic esterase, and has relevance to pathogenicity as this alternative sialic acid is commonly found in mammalian host mucosal sites (112). Additional alternative sialic acids that can be utilized and transported by E. coli K-12 include Neu5Gc and KDN, which are transported via NanT and catabolized using the NanA aldolase NanA (43). Hopkins and colleagues demonstrated that an E. coli nanT deletion strain could utilize Neu5Gc and KDN when expressing sialic acid transporters from two other human pathogens: the TRAP SiaPQM from H. influenzae and the SSS transporter from Salmonella enterica, demonstrating that potentially many human pathogens may be able to utilize KDN and Neu5Gc as carbon sources (43).
Another human enteric pathogen which utilizes sialic acid is S. enterica, which causes enterocolitis/diarrhea and infections have an incidence rate in the United States of 16.42 cases per 100,000 individuals (1, 24). It was shown in S. enterica serovar Typhimurium strain LT2 that this strain possesses a sialidase (NanH) that is absent from most other isolates (26, 45). NanH was shown to be homologous to clostridial sialidases (44). The function of NanH in vivo has not been studied. However, sialic acid was shown to be important for adherence of Salmonella to colonic cells (100). Sakarya and colleagues demonstrated that sialic acid is important for S. enterica serovar Typhi to adhere to Caco2 cells; when sialic acid was removed via sialidase treatment, adherence was reduced by 41% (100).
The Kingsley group identified genes important for host colonization via ChIP-seq and transcriptome analysis of the OmpR regulon and two of the operons subsequently identified (SL1068-71 and SL1066-67) were shown to be required for growth on sialic acid (92). They demonstrated that in a mixed inoculum experiment of the streptomycin treated mouse model of colitis that a deletion of operon SL1068-71 exhibited a significant reduction in the ability to colonize the cecum and ileum, and this operon exhibits sequence similarity to sialic acid uptake systems (92). In another recent paper examining expansion of enteric pathogens following antibiotic treatment and their ability to utilize microbiota-liberated host sugars, it was shown that S. enterica serovar Typhimurium utilized sialic acid and if this pathway was abolished through gene deletion, the competitiveness of the organism in vivo was reduced (84). The described studies demonstrate that Salmonella can utilize sialic acid and that it is an important nutrient in host colonization.
Vibrio cholerae, the causative agent of the profuse secretory diarrheal disease cholera and it is estimated, conservatively, that there are over a million cases of cholera worldwide, per year. Pathogenic isolates of this enteric extracellular pathogen are capable of utilizing sialic acid as a sole carbon source (5) (Fig. 3). V. cholerae contains a TRAP transporter SiaPQM (VC1777-VC1779), which is required for uptake of sialic acid (23, 80). A recent paper had suggested that an entirely different TRAP transporter (VC1927-VC1929) was the sole Neu5Ac transporter in V. cholerae (107). However, our bioinformatics, genomic and genetic analyses clearly demonstrated that this is not the case (23, 114). Our data show that VC1927-VC1929 encode a C4-dicarboxylate-specific TRAP transporter. We demonstrated that a deletion of VC1929 resulted in a defect in growth on C4-dicarboxylates but not Neu5Ac as the sole carbon source whereas deletion of siaP (VC1777) resulted in a mutant strain that was unable to support growth on Neu5Ac as the sole carbon source. These data unequivocally show that siaPQM (VC1777-1779) encoded a TRAP transporter and is the sole sialic acid transporter in V. cholerae (23).
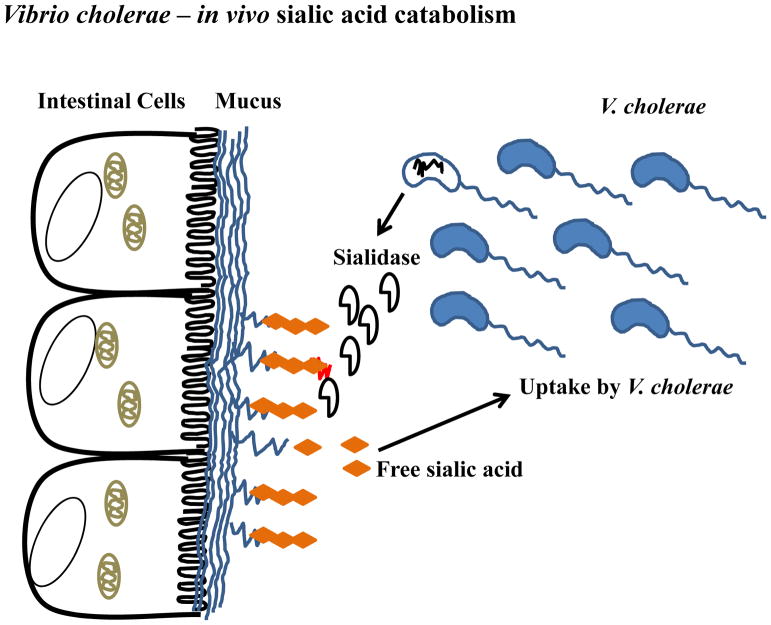
FIGURE 3.
Vibrio cholerae sialic acid metabolism. V. cholerae is an extracellular intestinal pathogen that colonizes the mucus layer of the small intestine and elaborates cholera toxin and sialidase. Sialidase cleaves sialic acid from high order gangliosides to release sialic acid and expose the GM1 ganglioside, the receptor for cholera toxin. Free sialic acid can be transported into the V. cholerae cell via the TRAP transporter SiaPQM contained on the pathogenicity island VPI-2, which also contains the genes for sialic acid catabolism.
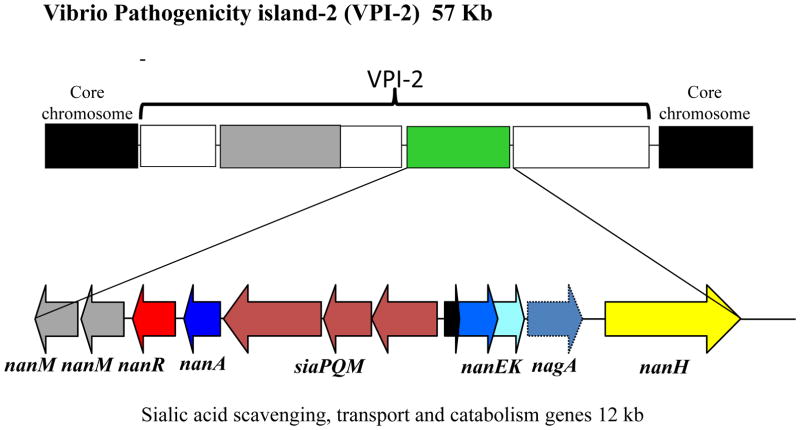
In addition to the catabolism and transporter genes, V. cholerae, also possesses a sialidase encoded by the nanH gene (51) (Fig. 4). It has been established that NanH removes two molecules of sialic acid from siaylated gangliosides found in intestinal epithelium, unmasking the GM1 ganglioside, the receptor for cholera toxin (34, 38, 39, 78). The sialidase along with the catabolism genes nanA, nanEK, and the TRAP transporter genes siaPQM are contained within a 57 kb pathogenicity island, named Vibrio Pathogenicity Island-2 (VPI-2) (Fig. 4) (51). This pathogenicity island is only present in pathogenic isolates and consists of 52 open reading frames (ORFs) in choleragenic V. cholerae isolates. Previously, we showed that VPI-2 displays all of the characteristics of a horizontally region; a lower G+C content compared to the entire genome; the presence of a tyrosine recombinase integrase; and a tRNA chromosomal insertion site. VPI-2 in choleragenic isolates encodes a type 1 restriction modification system, the sialic acid scavenging, transport and catabolism genes and a region containing several hypothetical proteins (51). In pathogenic V. cholerae isolates that cause inflammatory diarrhea, VPI-2 is also present but in these isolates the restriction modification system is replaced by a type three secretion system (81). Additionally, in V. mimicus isolates that cause inflammatory diarrhea, VPI-2 is present and also contains a type three secretion system. The key point is that the ability to catabolize sialic acid is retained and present in these enteric pathogens.
FIGURE 4.
Schematic diagram of the pathogenicity island VPI-2 in V. cholerae choleragenic isolates. ORFs of interest are depicted as arrows indicting the direction of transcription. The entire sialic acid catabolism cluster encompasses 12 kb on the 57 kb VPI-2 region.
A role for sialic acid catabolism in V. cholerae has been demonstrated in choleragenic isolates (5). V. cholerae colonizes the heavily sialylated mucus of the human gut and the ability to catabolize sialic acid as a carbon and energy source should confer the organism with a growth advantage compared to strains unable to utilize sialic acid. In single infection assays in the infant mouse model of cholera, which compared a wild type strain with a nanA mutant, it was found that in the early stages of infection (3 and 6 hr post infection) that the mutant was unable to maintain the same high cell density compared to the wild type strain. This suggests that the ability to catabolize sialic acid as a carbon source in the early stages of infection increases the fitness and likelihood of colonization in the highly populated environment of the human gut (5). Similarly, in in vivo co-infection competition assays, it was shown that the competitive index of the mutant versus wild-type was 0.06 indicating a decrease fitness of the mutant. In a recent study from our group, we demonstrated that sialic acid transport is important in in vivo fitness in a streptomycin treated adult mouse model of colonization. We found that a sialic acid transporter deficient strain was outcompeted by wild type in colonization persistence assays (13). These data show that the ability to utilize sialic acid as a carbon source by V. cholerae confers them with a competitive advantage in the sialic acid rich environment of the gut.
Another pathogenic Vibrio species which can utilize sialic acid as a sole carbon source is V. vulnificus (4). Phylogenetic analysis revealed that sialic acid transport and catabolism genes are present predominantly in clinical isolates and much less frequently in environmental isolates (71). Our group demonstrated that the TRAP transporter, SiaPQM that clusters with the catabolism genes, is essential for sialic acid uptake in this species (71). Interestingly, unlike V. cholerae, V. vulnificus is also capable of biosynthesis of nonulosonic acid a phenotype that is present in all V. vulnificus strains (67). However, our data suggest that V. vulnificus does not synthesize Neu5Ac but rather either of two sialic acid-like molecules, legionaminic or pseudaminic acid (67). This would indicate that transport of Neu5Ac into the cell is for the sole purpose of catabolism and not sialylation. More recently, we have shown that V. vulnificus decorates its LPS with nonulosonic acids and expression of these moclules is required for biofilm formation, motility and flagellar expression. In an in vivo mouse model of septicemia, we found that a sialylation defective mutant had a significant survival disadvantage compared wild type strain (Lubin, Boyd, and Lewis, unpublished data).
It has been proposed for V. vulnificus that catabolism of Neu5Ac is important for enteropathogenesis (50). A nanA mutant was shown to be incapable of utilizing Neu5Ac as a carbon source and was defective for intestinal colonization (50). This group also proposed that sialic acid catabolism is important in virulence as the nanA mutant exhibited decreased cytotoxicity towards INT-407 epithelial cells, decreased adherence in vitro, and also displayed decreases in histopathological damage in the jejunum and colon tissues from the mouse intestine (50). The Nan cluster of V. vulnificus has been shown to be transcriptionally repressed by NanR, and is induced by the presence of N-acetylmannosamine-6-phosphate (ManNAc-6P) which specifically binds to NanR (47, 57). The interaction between ManNAc-6P and NanR has been shown to be important in V. vulnificus pathogenesis. A mutant strain containing a NanR mutation that prevented binding of the ligand, ManNAc-6P, was shown to have growth impairment when sialic acid was the sole carbon source and the mutant strain was also less fit than the wild type strain in vivo, displaying decreased virulence (47). Together, these results demonstrate that catabolism of sialic acid is a feature of clinical isolates and is an important component of survival and virulence in vivo in V. vulnificus, a food-borne pathogen.
One of the first bacteria in which the ability to utilize sialic acid as a carbon source was demonstrated was Clostridium perfringens, a bacterium associated with food poisoning (31, 83). In C. perfringens, NanA, the sialic acid lyase, is necessary for growth in minimal medium supplemented with sialic acid as the sole carbon source and transcription of nanA is inducible by sialic acid (129). By bioinformatics, we identified putative sialic acid catabolism genes among 10 Clostridium species including C. difficile, a leading cause of antibiotic-associated diarrhea and colitis (11). A recent study of C. difficile demonstrated that post-antibiotic expansion of this organism is aided by the elevation of sialic acid levels in vivo (84). This study showed that not only does sialic acid levels have an impact on expansion of C. difficile but strains of C. difficile unable to utilize sialic acid exhibited reduced colonization following antibiotic treatment of mice, despite a spike in sialic acid levels (84). Taken together these results demonstrate that intestinal pathogenic species of Clostridia catabolize sialic acid and this is an important component of pathogenesis and host interaction of these bacteria.
Clostridia also produce a number of sialidases, the function of which was first examined in C. perfringens and was found to be variably present among strains (31, 83). A recent study examined C. perfringens Type D Strain CN3718, which encodes three sialidases, NanI, NanJ, and NanH, and demonstrated that secreted sialidiase, NanI, was important for enhanced binding to MDCK cells and cytotoxic effects of the episilon toxin, ETX (69) suggesting NanI may contribute to intestinal colonization and increased ETX action.
In summary, here we have highlighted some of the research demonstrating the importance of sialic acid as a carbon source for pathogens and that this ability can play a significant role in pathogenesis. One of the main challenges invading pathogens face is the limited availability of nutrient sources and in order to survive, they must be able to compete for these resources. This can be achieved through the use of alternative carbon sources such as amino sugars like sialic acids (22, 30, 32, 33, 42, 84). In conclusion these studies on sialic acid further our understanding of the biology of sialic acid and how it is exploited in a variety of ways, including as an alternative carbon nutrient source, by gut pathogens in vivo.
Acknowledgments
Research on Vibrio species in the Boyd group is supported by a National Science Foundation CAREER award DEB-0844409. We thank M.R. Carpenter, S. S. Kalburge, N. McDonald, and S.Y. Ongagna-Yhombi, for comments and discussion.
References
- 1.Incidence and trends of infection with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 1996–2012. Morb Mortal Wkly Rep. 2013;62:283–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA. Novel sialic acid transporter of Haemophilus influenzae. Infect Immun. 2005;73:5291–300. doi: 10.1128/IAI.73.9.5291-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almagro-Moreno S, Boyd EF. Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes. 2010;1:45–50. doi: 10.4161/gmic.1.1.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Almagro-Moreno S, Boyd EF. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–16. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chemical reviews. 2002;102:439–69. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 7.Beck JM, Young VB, Huffnagle GB. The microbiome of the lung. Transl Res. 2012;160:258–66. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett M, Schmid K. Immunosuppression by human plasma alpha 1-acid glycoprotein: importance of the carbohydrate moiety. Proc Natl Acad Sci U S A. 1980;77:6109–13. doi: 10.1073/pnas.77.10.6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environ Microbiol. 2013;15:610–22. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blix G. Concerning the carbohydrate groups of submaxillary mucin. Hoppe-Seylers Zeitschrift Fur Physiologische Chemie. 1936;240:43–54. [Google Scholar]
- 11.Borriello SP. Clostridial disease of the gut. Clin Infect Dis. 1995;20(Suppl 2):S242–50. doi: 10.1093/clinids/20.supplement_2.s242. [DOI] [PubMed] [Google Scholar]
- 12.Bouchet V, Hood DW, Li J, Brisson JR, Randle GA, Martin A, Li Z, Goldstein R, Schweda EKH, Pelton SI, Richards JC, Moxon ER. Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc Natl Acad Sci U S A. 2003;100:8898–8903. doi: 10.1073/pnas.1432026100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd EF, Chowdhury N, McDonald ND, Lubin JB. Host sialic acids are an important bacterial nutrient source that increase fitness of intestinal pathogens in vivo. 114th General Meeting of the American Society for Microbiology; 2014. ASM Abstracts. [Google Scholar]
- 14.Brigham C, Caughlan R, Gallegos R, Dallas MB, Godoy VG, Malamy MH. Sialic acid (N-acetyl neuraminic acid) utilization by Bacteroides fragilis requires a novel N-acetyl mannosamine epimerase. J Bacteriol. 2009;191:3629–38. doi: 10.1128/JB.00811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brigham CJ, Malamy MH. Characterization of the RokA and HexA broad-substrate-specificity hexokinases from Bacteroides fragilis and their role in hexose and N-acetylglucosamine utilization. J Bacteriol. 2005;187:890–901. doi: 10.1128/JB.187.3.890-901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brittan JL, Buckeridge TJ, Finn A, Kadioglu A, Jenkinson HF. Pneumococcal neuraminidase A: an essential upper airway colonization factor for Streptococcus pneumoniae. Mol Oral Microbiol. 2012;27:270–83. doi: 10.1111/j.2041-1014.2012.00658.x. [DOI] [PubMed] [Google Scholar]
- 17.Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol. 2008;190:221–30. doi: 10.1128/JB.01251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butor C, Diaz S, Varki A. High level O-acetylation of sialic acids on N-linked oligosaccharides of rat liver membranes. Differential subcellular distribution of 7- and 9-O-acetyl groups and of enzymes involved in their regulation. J Biol Chem. 1993;268:10197–206. [PubMed] [Google Scholar]
- 19.Byers HL, Homer KA, Beighton D. Utilization of sialic acid by viridans streptococci. J Dent Res. 1996;75:1564–71. doi: 10.1177/00220345960750080701. [DOI] [PubMed] [Google Scholar]
- 20.Byers HL, Tarelli E, Homer KA, Hambley H, Beighton D. Growth of Viridans streptococci on human serum alpha1-acid glycoprotein. J Dent Res. 1999;78:1370–80. doi: 10.1177/00220345990780071201. [DOI] [PubMed] [Google Scholar]
- 21.Cameron DJ, Churchill WH. Specificity of macrophage mediated cytotoxicity: role of target cell sialic acid. Jpn J Exp Med. 1982;52:9–16. [PubMed] [Google Scholar]
- 22.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101:7427–32. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury N, Norris J, McAlister E, Lau SY, Thomas GH, Boyd EF. The VC1777-VC1779 proteins are members of a sialic acid-specific subfamily of TRAP transporters (SiaPQM) and constitute the sole route of sialic acid uptake in the human pathogen Vibrio cholerae. Microbiology. 2012;158:2158–67. doi: 10.1099/mic.0.059659-0. [DOI] [PubMed] [Google Scholar]
- 24.Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85:112–8. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 25.Condemine G, Berrier C, Plumbridge J, Ghazi A. Function and Expression of an N-Acetylneuraminic Acid-Inducible Outer Membrane Channel in Escherichia coli. J Bacteriol. 2005;187:1959–1965. doi: 10.1128/JB.187.6.1959-1965.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crennell SJ, Garman EF, Philippon C, Vasella A, Laver WG, Vimr ER, Taylor GL. The structures of Salmonella typhimurium LT2 neuraminidase and its complexes with three inhibitors at high resolution. J Mol Biol. 1996;259:264–80. doi: 10.1006/jmbi.1996.0318. [DOI] [PubMed] [Google Scholar]
- 27.Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of Mucin Glycans by the Human Gut Symbiont Ruminococcus gnavus Is Strain-Dependent. PLoS One. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Culling CFA, Reid PE, Clay MG, Dunn WL. The histochemical demonstration of O-acylated sialic acid in gastrointestinal mucins. Their association with the potassium hydroxide-periodic acid-schiff effect. J Histochem Cytochem. 1974;22:826–831. doi: 10.1177/22.8.826. [DOI] [PubMed] [Google Scholar]
- 30.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, McHargue JW, Hightower GA, Smith JT, Autieri SM, Leatham MP, Lins JJ, Allen RL, Laux DC, Cohen PS, Conway T. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect Immun. 2008;76:1143–52. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fraser AG, Collee JG. The production of neuraminidase by food poisoning strains of Clostridium welchii (C. perfringens) J Med Microbiol. 1975;8:251–63. doi: 10.1099/00222615-8-2-251. [DOI] [PubMed] [Google Scholar]
- 32.Freter R. Virulence mechanisms of bacterial pathogens. American Society for Microbiology; Washington, DC: 1988. Mechanisms of bacterial colonization of the mucosal surfaces of the gut; pp. 45–60. [Google Scholar]
- 33.Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges DJ, editor. Human intestinal microflora in health and disease. Academic Press, Inc; New York: 1983. pp. 33–54. [Google Scholar]
- 34.Galen JE, Ketley JM, Fasano A, Richardson SH, Wasserman SS, Kaper JB. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60:406–15. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert NM, Lewis WG, Lewis AL. Clinical features of bacterial vaginosis in a murine model of vaginal infection with Gardnerella vaginalis. PLoS One. 2013;8:e59539. doi: 10.1371/journal.pone.0059539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruteser N, Marin K, Krämer R, Thomas GH. Sialic acid utilization by the soil bacterium Corynebacterium glutamicum. FEMS Microbiol Lett. 2012;336:131–138. doi: 10.1111/j.1574-6968.2012.02663.x. [DOI] [PubMed] [Google Scholar]
- 37.Ho JJ, Cheng S, Kim YS. Access to peptide regions of a surface mucin (MUC1) is reduced by sialic acids. Biochem Biophys Res Commun. 1995;210:866–73. doi: 10.1006/bbrc.1995.1738. [DOI] [PubMed] [Google Scholar]
- 38.Holmgren J, Lonnroth I, Mansson J, Svennerholm L. Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proc Natl Acad Sci U S A. 1975;72:2520–4. doi: 10.1073/pnas.72.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holmgren J, Lonnroth I, Svennerholm L. Fixation and inactivation of cholera toxin by GM1 ganglioside. Scand J Infect Dis. 1973;5:77–8. doi: 10.3109/inf.1973.5.issue-1.15. [DOI] [PubMed] [Google Scholar]
- 40.Honma K, Mishima E, Sharma A. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect Immun. 2011;79:393–401. doi: 10.1128/IAI.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hood DW, Makepeace K, Deadman ME, Rest RF, Thibault P, Martin A, Richards JC, Moxon ER. Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol Microbiol. 1999;33:679–92. doi: 10.1046/j.1365-2958.1999.01509.x. [DOI] [PubMed] [Google Scholar]
- 42.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 43.Hopkins AP, Hawkhead JA, Thomas GH. Transport and catabolism of the sialic acids N-glycolylneuraminic acid and 3-keto-3-deoxy-D-glycero-D-galactonononic acid by Escherichia coli K-12. FEMS Microbiol Lett. 2013;347:14–22. doi: 10.1111/1574-6968.12213. [DOI] [PubMed] [Google Scholar]
- 44.Hoyer LL, Hamilton AC, Steenbergen SM, Vimr ER. Cloning, sequencing and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol Microbiol. 1992;6:873–84. doi: 10.1111/j.1365-2958.1992.tb01538.x. [DOI] [PubMed] [Google Scholar]
- 45.Hoyer LL, Roggentin P, Schauer R, Vimr ER. Purification and properties of cloned Salmonella typhimurium LT2 sialidase with virus-typical kinetic preference for sialyl alpha 2----3 linkages. J Biochem. 1991;110:462–7. doi: 10.1093/oxfordjournals.jbchem.a123603. [DOI] [PubMed] [Google Scholar]
- 46.Huang YJ, Lynch SV. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med. 2011;5:809–21. doi: 10.1586/ers.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang J, Kim BS, Jang SY, Lim JG, You DJ, Jung HS, Oh TK, Lee JO, Choi SH, Kim MH. Structural insights into the regulation of sialic acid catabolism by the Vibrio vulnificus transcriptional repressor NanR. Proc Natl Acad Sci U S A. 2013;110:E2829–37. doi: 10.1073/pnas.1302859110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue S, Kitajima K. KDN (deaminated neuraminic acid): dreamful past and exciting future of the newest member of the sialic acid family. Glycoconj J. 2006;23:277–90. doi: 10.1007/s10719-006-6484-y. [DOI] [PubMed] [Google Scholar]
- 49.Ishikura H, Arakawa S, Nakajima T, Tsuchida N, Ishikawa I. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J Med Microbiol. 2003;52:1101–7. doi: 10.1099/jmm.0.05349-0. [DOI] [PubMed] [Google Scholar]
- 50.Jeong HG, Oh MH, Kim BS, Lee MY, Han HJ, Choi SH. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect Immun. 2009;77:3209–17. doi: 10.1128/IAI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jermyn WS, Boyd EF. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology. 2002;148:3681–93. doi: 10.1099/00221287-148-11-3681. [DOI] [PubMed] [Google Scholar]
- 52.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, Gibson BW, Apicella MA, Munson RS., Jr Regulation of sialic acid transport and catabolism in Haemophilus influenzae. Mol Microbiol. 2007;66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 53.Joseph S, Desai P, Ji Y, Cummings CA, Shih R, Degoricija L, Rico A, Brzoska P, Hamby SE, Masood N, Hariri S, Sonbol H, Chuzhanova N, McClelland M, Furtado MR, Forsythe SJ. Comparative analysis of genome sequences covering the seven cronobacter species. PLoS One. 2012;7:e49455. doi: 10.1371/journal.pone.0049455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joseph S, Hariri S, Masood N, Forsythe S. Sialic acid utilization by Cronobacter sakazakii. Microb Inform Exp. 2013;3:3. doi: 10.1186/2042-5783-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–8. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kazatchkine MD, Fearon DT, Austen KF. Human alternative complement pathway: membrane-associated sialic acid regulates the competition between B and beta1 H for cell-bound C3b. J Immunol. 1979;122:75–81. [PubMed] [Google Scholar]
- 57.Kim BS, Hwang J, Kim MH, Choi SH. Cooperative regulation of the Vibrio vulnificus nan gene cluster by NanR protein, cAMP receptor protein, and N-acetylmannosamine 6-phosphate. J Biol Chem. 2011;286:40889–99. doi: 10.1074/jbc.M111.300988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.King SJ. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol Oral Microbiol. 2010;25:15–24. doi: 10.1111/j.2041-1014.2009.00564.x. [DOI] [PubMed] [Google Scholar]
- 59.Kline KA, Schwartz DJ, Lewis WG, Hultgren SJ, Lewis AL. Immune activation and suppression by group B streptococcus in a murine model of urinary tract infection. Infect Immun. 2011;79:3588–95. doi: 10.1128/IAI.00122-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knirel YA, Rietschel ET, Marre R, Zähringer U. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. European Journal of Biochemistry. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- 61.Knirel YA, Vinogradov EV, L’Vov LV, Kocharova NA, Shashkov AS, Dmitriev BA, Kochetkov NK. Sialic acids of a new type from the lipopolysaccharides of Pseudomonas aeruginosa and Shigella boydii. Carbohydr Res. 1984;133:C5–8. doi: 10.1016/0008-6215(84)85213-1. [DOI] [PubMed] [Google Scholar]
- 62.Kurniyati K, Zhang W, Zhang K, Li C. A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola. Mol Microbiol. 2013;89:842–56. doi: 10.1111/mmi.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LaMont JT, Ventola AS. Purification and composition of colonic epithelial mucin. Biochim Biophys Acta. 1980;626:234–43. doi: 10.1016/0005-2795(80)90214-7. [DOI] [PubMed] [Google Scholar]
- 64.Lanoue A, Batista FD, Stewart M, Neuberger MS. Interaction of CD22 with alpha2,6-linked sialoglycoconjugates: innate recognition of self to dampen B cell autoreactivity? Eur J Immunol. 2002;32:348–55. doi: 10.1002/1521-4141(200202)32:2<348::AID-IMMU348>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 65.Lewis AL, Desa N, Hansen EE, Knirel YA, Gordon JI, Gagneux P, Nizet V, Varki A. Innovations in host and microbial sialic acid biosynthesis revealed by phylogenomic prediction of nonulosonic acid structure. Proc Natl Acad Sci U S A. 2009;106:13552–7. doi: 10.1073/pnas.0902431106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol. 2012;14:1174–82. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 67.Lewis AL, Lubin JB, Argade S, Naidu N, Choudhury B, Boyd EF. Genomic and metabolic profiling of nonulosonic acids in Vibrionaceae reveal biochemical phenotypes of allelic divergence in Vibrio vulnificus. Appl Environ Microbiol. 2011;77:5782–93. doi: 10.1128/AEM.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted Actinobacterium Gardnerella vaginalis. J Biol Chem. 2013;288:12067–79. doi: 10.1074/jbc.M113.453654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li J, Sayeed S, Robertson S, Chen J, McClane BA. Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog. 2011;7:e1002429. doi: 10.1371/journal.ppat.1002429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lilley GG, Barbosa JA, Pearce LA. Expression in Escherichia coli of the putative N-acetylneuraminate lyase gene (nanA) from Haemophilus influenzae: overproduction, purification, and crystallization. Protein Expr Purif. 1998;12:295–304. doi: 10.1006/prep.1997.0841. [DOI] [PubMed] [Google Scholar]
- 71.Lubin JB, Kingston JJ, Chowdhury N, Boyd EF. Sialic acid catabolism and transport gene clusters are lineage specific in Vibrio vulnificus. Appl Environ Microbiol. 2012;78:3407–15. doi: 10.1128/AEM.07395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manco S, Hernon F, Yesilkaya H, Paton JC, Andrew PW, Kadioglu A. Pneumococcal neuraminidases A and B both have essential roles during infection of the respiratory tract and sepsis. Infect Immun. 2006;74:4014–20. doi: 10.1128/IAI.01237-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marion C, Aten AE, Woodiga SA, King SJ. Identification of an ATPase, MsmK, which energizes multiple carbohydrate ABC transporters in Streptococcus pneumoniae. Infect Immun. 2011;79:4193–200. doi: 10.1128/IAI.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marion C, Burnaugh AM, Woodiga SA, King SJ. Sialic acid transport contributes to pneumococcal colonization. Infect Immun. 2011;79:1262–9. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez J, Steenbergen S, Vimr E. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J Bacteriol. 1995;177:6005–10. doi: 10.1128/jb.177.20.6005-6010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.May M, Brown DR. Genetic variation in sialidase and linkage to N-acetylneuraminate catabolism in Mycoplasma synoviae. Microb Pathog. 2008;45:38–44. doi: 10.1016/j.micpath.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–78. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 78.Moustafa I, Connaris H, Taylor M, Zaitsev V, Wilson JC, Kiefel MJ, Von Itzstein M, Taylor G. Sialic acid recognition by Vibrio cholerae neuraminidase. J Biol Chem. 2004;279:40819–26. doi: 10.1074/jbc.M404965200. [DOI] [PubMed] [Google Scholar]
- 79.Mulligan C, Geertsma ER, Severi E, Kelly DJ, Poolman B, Thomas GH. The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc Natl Acad Sci U S A. 2009;106:1778–83. doi: 10.1073/pnas.0809979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mulligan C, Leech AP, Kelly DJ, Thomas GH. The membrane proteins SiaQ and SiaM form an essential stoichiometric complex in the sialic acid tripartite ATP-independent periplasmic (TRAP) transporter SiaPQM (VC1777-1779) from Vibrio cholerae. J Biol Chem. 2012;287:3598–608. doi: 10.1074/jbc.M111.281030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murphy RA, Boyd EF. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol. 2008;190:636–47. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mushtaq N, Redpath MB, Luzio JP, Taylor PW. Prevention and Cure of Systemic Escherichia coli K1 Infection by Modification of the Bacterial Phenotype. Antimicrobial Agents and Chemotherapy. 2004;48:1503–1508. doi: 10.1128/AAC.48.5.1503-1508.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nees S, Schauer R, Mayer F. Purification and characterization of N-acetylneuraminate lyase from Clostridium perfringens. Hoppe Seylers Z Physiol Chem. 1976;357:839–53. doi: 10.1515/bchm2.1976.357.1.839. [DOI] [PubMed] [Google Scholar]
- 84.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature. 2013;502:96–9. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ogasawara Y, Namai T, Yoshino F, Lee MC, Ishii K. Sialic acid is an essential moiety of mucin as a hydroxyl radical scavenger. FEBS Letters. 2007;581:2473–2477. doi: 10.1016/j.febslet.2007.04.062. [DOI] [PubMed] [Google Scholar]
- 86.Olson ME, King JM, Yahr TL, Horswill AR. Sialic acid catabolism in Staphylococcus aureus. J Bacteriol. 2013;195:1779–88. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190:1661–9. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 88.Ozkan AN, Ninnemann JL. Suppression of in vitro lymphocyte and neutrophil responses by a low molecular weight suppressor active peptide from burn-patient sera. J Clin Immunol. 1985;5:172–9. doi: 10.1007/BF00915508. [DOI] [PubMed] [Google Scholar]
- 89.Parker D, Soong G, Planet P, Brower J, Ratner AJ, Prince A. The NanA neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun. 2009;77:3722–30. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parsons NJ, Patel PV, Tan EL, Andrade JR, Nairn CA, Goldner M, Cole JA, Smith H. Cytidine 5′-monophospho-N-acetyl neuraminic acid and a low molecular weight factor from human blood cells induce lipopolysaccharide alteration in gonococci when conferring resistance to killing by human serum. Microb Pathog. 1988;5:303–9. doi: 10.1016/0882-4010(88)90103-9. [DOI] [PubMed] [Google Scholar]
- 91.Peekhaus N, Conway T. What’s for dinner?: Entner-Doudoroff metabolism in Escherichia coli. J Bacteriol. 1998;180:3495–502. doi: 10.1128/jb.180.14.3495-3502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Perkins TT, Davies MR, Klemm EJ, Rowley G, Wileman T, James K, Keane T, Maskell D, Hinton JC, Dougan G, Kingsley RA. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol Microbiol. 2013;87:526–38. doi: 10.1111/mmi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pettigrew MM, Fennie KP, York MP, Daniels J, Ghaffar F. Variation in the presence of neuraminidase genes among Streptococcus pneumoniae isolates with identical sequence types. Infect Immun. 2006;74:3360–5. doi: 10.1128/IAI.01442-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pezzicoli A, Ruggiero P, Amerighi F, Telford JL, Soriani M. Exogenous sialic acid transport contributes to group B streptococcus infection of mucosal surfaces. J Infect Dis. 2012;206:924–31. doi: 10.1093/infdis/jis451. [DOI] [PubMed] [Google Scholar]
- 95.Polzin S, Huber C, Eylert E, Elsenhans I, Eisenreich W, Schmidt H. Growth media simulating ileal and colonic environments affect the intracellular proteome and carbon fluxes of enterohemorrhagic Escherichia coli O157:H7 strain EDL933. Appl Environ Microbiol. 2013;79:3703–15. doi: 10.1128/AEM.00062-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Post DM, Mungur R, Gibson BW, Munson RS., Jr Identification of a novel sialic acid transporter in Haemophilus ducreyi. Infect Immun. 2005;73:6727–35. doi: 10.1128/IAI.73.10.6727-6735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Robbe C, Capon C, Coddeville B, Michalski JC. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. Biochem J. 2004;384:307–16. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roy S, Douglas CW, Stafford GP. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J Bacteriol. 2010;192:2285–93. doi: 10.1128/JB.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rutishauser U. Polysialic acid in the plasticity of the developing and adult vertebrate nervous system. Nat Rev Neurosci. 2008;9:26–35. doi: 10.1038/nrn2285. [DOI] [PubMed] [Google Scholar]
- 100.Sakarya S, Gokturk C, Ozturk T, Ertugrul MB. Sialic acid is required for nonspecific adherence of Salmonella enterica ssp. enterica serovar Typhi on Caco-2 cells. FEMS Immunol Med Microbiol. 2010;58:330–5. doi: 10.1111/j.1574-695X.2010.00650.x. [DOI] [PubMed] [Google Scholar]
- 101.Sanchez-Carron G, Garcia-Garcia MI, Lopez-Rodriguez AB, Jimenez-Garcia S, Sola-Carvajal A, Garcia-Carmona F, Sanchez-Ferrer A. Molecular characterization of a novel N-acetylneuraminate lyase from Lactobacillus plantarum WCFS1. Appl Environ Microbiol. 2011;77:2471–8. doi: 10.1128/AEM.02927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Scudder PR, Chantler EN. Control of human cervical mucin glycosylation by endogenous fucosyl and sialyltransferases. Adv Exp Med Biol. 1982;144:265–7. doi: 10.1007/978-1-4615-9254-9_40. [DOI] [PubMed] [Google Scholar]
- 103.Sela DA, Mills DA. Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 2010;18:298–307. doi: 10.1016/j.tim.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–22. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 105.Severi E, Hosie AH, Hawkhead JA, Thomas GH. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol Lett. 2010;304:47–54. doi: 10.1111/j.1574-6968.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 106.Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, Kelly D, Hood D, Thomas GH. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol. 2005;58:1173–85. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 107.Sharma SK, Moe TS, Srivastava R, Chandra D, Srivastava BS. Functional characterization of VC1929 of Vibrio cholerae El Tor: role in mannose-sensitive haemagglutination, virulence and utilization of sialic acid. Microbiology. 2011;157:3180–6. doi: 10.1099/mic.0.050245-0. [DOI] [PubMed] [Google Scholar]
- 108.Slomiany BL, Murty VLN, Piotrowski J, Slomiany A. Salivary mucins in oral mucosal defense. General Pharmacology: The Vascular System. 1996;27:761–771. doi: 10.1016/0306-3623(95)02050-0. [DOI] [PubMed] [Google Scholar]
- 109.Soong G, Muir A, Gomez MI, Waks J, Reddy B, Planet P, Singh PK, Kaneko Y, Wolfgang MC, Hsiao YS, Tong L, Prince A. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J Clin Invest. 2006;116:2297–2305. doi: 10.1172/JCI27920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Srinivasan S, Hoffman NG, Morgan MT, Matsen FA, Fiedler TL, Hall RW, Ross FJ, McCoy CO, Bumgarner R, Marrazzo JM, Fredricks DN. Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One. 2012;7:e37818. doi: 10.1371/journal.pone.0037818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stafford G, Roy S, Honma K, Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol Oral Microbiol. 2012;27:11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Steenbergen SM, Jirik JL, Vimr ER. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J Bacteriol. 2009;191:7134–9. doi: 10.1128/JB.01000-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Steenbergen SM, Lichtensteiger CA, Caughlan R, Garfinkle J, Fuller TE, Vimr ER. Sialic Acid metabolism and systemic pasteurellosis. Infect Immun. 2005;73:1284–94. doi: 10.1128/IAI.73.3.1284-1294.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas GH, Boyd EF. On sialic acid transport and utilization by Vibrio cholerae. Microbiology. 2011;157:3253–4. doi: 10.1099/mic.0.054692-0. discussion 3254–5. [DOI] [PubMed] [Google Scholar]
- 115.Thompson H, Homer KA, Rao S, Booth V, Hosie AH. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J Bacteriol. 2009;191:3623–8. doi: 10.1128/JB.01618-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thornton DJ, Carlstedt I, Howard M, Devine PL, Price MR, Sheehan JK. Respiratory mucins: identification of core proteins and glycoforms. Biochem J. 1996;316(Pt 3):967–75. doi: 10.1042/bj3160967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tong HH, Blue LE, James MA, DeMaria TF. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect Immun. 2000;68:921–4. doi: 10.1128/iai.68.2.921-924.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J Infect Dis. 2009;199:1497–505. doi: 10.1086/598483. [DOI] [PubMed] [Google Scholar]
- 119.Vann WF, Daines DA, Murkin AS, Tanner ME, Chaffin DO, Rubens CE, Vionnet J, Silver RP. The NeuC protein of Escherichia coli K1 is a UDP N-acetylglucosamine 2-epimerase. J Bacteriol. 2004;186:706–12. doi: 10.1128/JB.186.3.706-712.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vann WF, Silver RP, Abeijon C, Chang K, Aaronson W, Sutton A, Finn CW, Lindner W, Kotsatos M. Purification, properties, and genetic location of Escherichia coli cytidine 5′-monophosphate N-acetylneuraminic acid synthetase. J Biol Chem. 1987;262:17556–62. [PubMed] [Google Scholar]
- 121.Vann WF, Tavarez JJ, Crowley J, Vimr E, Silver RP. Purification and characterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology. 1997;7:697–701. doi: 10.1093/glycob/7.5.697. [DOI] [PubMed] [Google Scholar]
- 122.Varki A. Loss of N-glycolylneuraminic acid in humans: Mechanisms, consequences, and implications for hominid evolution. Am J Phys Anthropol Suppl. 2001;33:54–69. doi: 10.1002/ajpa.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vimr E, Lichtensteiger C, Steenbergen S. Sialic acid metabolism’s dual function in Haemophilus influenzae. Mol Microbiol. 2000;36:1113–23. doi: 10.1046/j.1365-2958.2000.01925.x. [DOI] [PubMed] [Google Scholar]
- 124.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–53. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vimr ER, Troy FA. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J Bacteriol. 1985;164:845–53. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vimr ER, Troy FA. Regulation of sialic acid metabolism in Escherichia coli: role of N-acylneuraminate pyruvate-lyase. J Bacteriol. 1985;164:854–60. doi: 10.1128/jb.164.2.854-860.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.von Nicolai H, Hammann R, Salehnia S, Zilliken F. A newly discovered sialidase from Gardnerella vaginalis. Zentralbl Bakteriol Mikrobiol Hyg A. 1984;258:20–6. doi: 10.1016/s0176-6724(84)80003-6. [DOI] [PubMed] [Google Scholar]
- 128.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–43. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 129.Walters DM, Stirewalt VL, Melville SB. Cloning, sequence, and transcriptional regulation of the operon encoding a putative N-acetylmannosamine-6-phosphate epimerase (nanE) and sialic acid lyase (nanA) in Clostridium perfringens. J Bacteriol. 1999;181:4526–32. doi: 10.1128/jb.181.15.4526-4532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang B. Molecular mechanism underlying sialic acid as an essential nutrient for brain development and cognition. Adv Nutr. 2012;3:465S–72S. doi: 10.3945/an.112.001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res. 2007;51:1398–405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 132.Wyss C, Moter A, Choi BK, Dewhirst FE, Xue Y, Schupbach P, Gobel UB, Paster BJ, Guggenheim B. Treponema putidum sp. nov., a medium-sized proteolytic spirochaete isolated from lesions of human periodontitis and acute necrotizing ulcerative gingivitis. Int J Syst Evol Microbiol. 2004;54:1117–22. doi: 10.1099/ijs.0.02806-0. [DOI] [PubMed] [Google Scholar]
- 133.Xu G, Kiefel MJ, Wilson JC, Andrew PW, Oggioni MR, Taylor GL. Three Streptococcus pneumoniae sialidases: three different products. J Am Chem Soc. 2011;133:1718–21. doi: 10.1021/ja110733q. [DOI] [PubMed] [Google Scholar]