Abstract
Presenilins (Psen1 and Psen2 in mice) are polytopic transmembrane proteins that act in the γ-secretase complex to make intra-membrane cleavages of their substrates, including the well-studied Notch receptors. Such processing releases the Notch intracellular domain, allowing it to physically relocate from the cell membrane to the nucleus where it acts in a transcriptional activating complex to regulate downstream genes in the signal-receiving cell. Previous studies of Notch pathway mutants for Jagged1, Notch2, and Rbpj demonstrated that canonical signaling is a necessary component of normal mouse lens development. However, the central role of Psens within the γ-secretase complex has never been explored in any developing eye tissue or cell type. By directly comparing Psen single and double mutant phenotypes during mouse lens development, we found a stronger requirement for Psen1, although both genes are needed for progenitor cell growth and to prevent apoptosis. We also uncovered a novel genetic interaction between Psen1 and Jagged1. By quantifying protein and mRNA levels of key Notch pathway genes in Psen1/2 or Jagged1 mutant lenses, we identified multiple points in the overall signaling cascade where feedback regulation can occur. Our data are consistent with the loss of particular genes indirectly influencing the transcription level of another. However, we conclude that regulating Notch2 protein levels is particularly important during normal signaling, supporting the importance of post-translational regulatory mechanisms in this tissue.
Keywords: lens development, Notch signaling, Psen, gamma secretase (γ-secretase), Jagged1, fiber cell differentiation
Introduction
Cellular changes in growth, morphogenesis, and differentiation all contribute to tissue composition and shape, but ultimately, influence functionality. The vertebrate ocular lens is a prime example of this complex process. This spheroidal, transparent tissue must achieve precise size and curvature during development to focus images onto the retina for proper vision. Lens development initiates at the surface ectoderm in a region overlaying the optic vesicle, known as the presumptive lens ectoderm (PLE) (McAvoy et al., 1999). Fundamental experiments in frog embryos demonstrated that lens induction requires signals from the developing optic vesicle (Spemann, 1938). PLE cells respond by thickening into the lens placode, which folds into a pit shape that eventually pinches off from the surface ectoderm to give rise to the lens vesicle, a uniform, hollow structure comprised of proliferating progenitor cells. This is followed by two temporal waves of fibergenesis that both begin prenatally. Primary fibergenesis initiates when posterior lens vesicle cells elongate across the lumen and differentiate. At this time, anterior vesicle cells remain proliferative and coalesce anteriorly to create the anterior epithelial layer (AEL). During secondary fibergenesis, AEL cells move peripherally through the germinative zone where they become postmitotic and progress into the transition zone for terminal differentiation (Lovicu and Robinson, 2004).
A variety of intrinsic and extrinsic factors are essential for proper lens growth and fiber cell differentiation. Some key transcription factors include: Pax6, Sox2, FoxE3, Prox1, and c-Maf (Blixt et al., 2000; Chow et al., 1999; Hill et al., 1991; Kamachi et al., 1995; Kim et al., 1999; Wigle et al., 1999). These factors regulate lens growth, fiber cell differentiation, elongation, cell-cycle exit and organelle-clearing. Moreover, lens formation also relies on many signaling factors. Classic lens inversion experiments performed in chicks embryos offered the first clue that signals in the aqueous humor or vitreous regulate lens fiber elongation and polarity (Coulombre and Coulombre, 1963). Much later some of the signals were identified, including Fgfs, Wnts, Bmp, and Notch (Cain et al., 2008; Chamberlain and McAvoy, 1987; Faber et al., 2002; Garcia et al., 2011; Jia et al., 2007; Le et al., 2009; Rowan et al., 2008; Stump et al., 2003). One pathway with an essential role in secondary fibergenesis is Notch signaling (Jia et al., 2007; Rowan et al., 2008). Canonical cell-cell signaling enables ligands on the surface of one cell to engage with a Notch receptor expressed on the neighboring cell (Kopan and Ilagan, 2009; Kovall et al., 2017). There are two families of Notch ligands: Deltalike (Dll) and Jagged (Jag). Upon ligand binding, the Notch receptor protein undergoes a conformational change which exposes its negative regulatory region for cleavage by proteases. This fundamental activation step ultimately releases the Notch intracellular domain (NICD) (Schroeter et al., 1998). Receptor cleavage is sequentially mediated by two different protein complexes. First, ADAM secretase removes the large extracellular domain (Brou et al., 2000; Mumm et al., 2000) and then γ-secretase cuts within the transmembrane region to release NICD from the plasma membrane (De Strooper et al., 1999). The γ-secretase complex is comprised of four protein subunits: Nicastrin, PEN2, APH1, and the catalytic subunit Presenilin (Psen). The two mouse Psen genes, Psen1 and Psen2, encode proteins that catalyze substrate proteolysis, however each γ-secretase complex contains only one Psen paralog (Donoviel et al., 1999; Struhl and Greenwald, 1999). After cleavage and release, the NICD first binds the DNA-binding protein Rbpj and subsequently to a cofactor, Mastermind. This complex transcriptionally activates downstream genes, such as the Hes and Hey genes (Iso et al., 2003; Kovall, 2007; Kuroda et al., 1999; Tamura et al., 1995). Previously, we demonstrated that the activities of Notch pathway genes Jag1, Notch2, and Rbpj are required for lens development (Le et al., 2009; Rowan et al., 2008; Saravanamuthu et al., 2012). Given the pivotal role of the γ-secretase complex in canonical Notch signaling, we wished to explore the role(s) of Psen genes within the context of mouse lens development.
Psen1 mutations cause embryonic lethality, whereas Psen2 mutants are adult viable, illustrating the vastly different requirements for γ-secretase complexes containing either Psen protein (Herreman et al., 1999; Shen et al., 1997). There is also evidence of distinct subcellular distribution of Psen1- versus Psen2-containing γ-secretase complexes, which correlates with differential substrate specificity (Sannerud et al., 2016). For the Notch receptors, NICD generation is more heavily dependent on Psen1 than Psen2 (Zhang et al., 2000). However, for the lens, the extent to which Psen1- versus Psen2-containing γ-secretase complexes regulate Notch receptor activation is unknown. Moreover, given that γ-secretase can cleave up to 90 different proteins (Haapasalo and Kovacs, 2011), it is possible that Psen loss may have greater consequences during lens development, than do other Notch pathway mutants. To address these questions, we utilized the Le-Cre driver and Psen1CKO/CKO;Psen2−/− mouse stocks to generate an allelic series during lens formation that includes Psen1/2 double mutants, which block all Notch receptor activation. Here we report the lens phenotypes of this Psen1/2 allelic series. We found that although the Psen1/2 double mutant lenses complete primary fibergenesis, and initiate secondary fibergenesis, at E14.5 proliferation was dramatically reduced, along with FoxE3 downregulation, and there was significantly more apoptosis. Postnatally, Psen1/2 double mutants progressively lose lens tissue, exhibiting aphakia by P21. We also uncovered a genetic interaction between Psen1 and Jag1, wherein Le-Cre;Psen1CKO/+;Jag1CKO/+ adult mice display synergistic lens phenotypes. To understand the basis for this interaction, we measured the mRNA and protein levels of relevant Notch pathway genes in each other's mutant lens tissue. We found that at the onset of secondary fibergenesis, Notch2 protein levels are particularly sensitive to both Jag1 and Psen1/2 activity. Together, these data demonstrate Presenilins play a critical role in lens growth and homeostasis. We conclude that while Notch signaling is controlled at both the level of transcription and post-translation, in the lens it is the latter mechanism that is utilized for feedback regulation.
Materials and Methods
Animals
Psen1tm1Shn/tm1Shn;Psen2tm1Bdes/tm1Bdes (Psen1CKO/CKO;Psen2−/−) mice, Jagged1tm1JLew/tm1Lew mice (Jag1CKO/CKO), Notch1tm2Rko/tm2Rko (Notch1CKO/CKO) mice, Notch2tm3Grid/tm3Grid (Notch2CKO/CKO) mice and Notch1CKO/CKO; Notch2CKO/CKO mice were each maintained on a mixed 129-C57BL/6 background and genotyped as described (Beglopoulos et al., 2004; Brooker et al., 2006; McCright et al., 2006; Yang et al., 2004). Hes1tm1Hojo mice (Hes1CKO/CKO) were maintained on a CD-1 background and genotyped as described (Kita et al., 2007). Le-Cre Tg/+ mice were maintained on a FVB/N background and PCR genotyped as described (Ashery-Padan et al., 2000). MLR10-Cre Tg/+ mice were maintained on a FVB/N background and PCR genotyped as described (Zhao et al., 2004). Our Le-Cre Tg/+ line does not exhibit DNA damage pathway activation (Loonstra et al., 2001), shown by anti-H2A.X western blotting (Supplemental Fig. 2F). We also monitored Pax6 mRNA levels among all genotypes produced in different litters, and found no changes correlating with inheritance of the Le-Cre transgene alone.
Breeding schemes for all analyses mated one Cre Tg/+ mouse (Le- or MLR10-Cre) to one homozygous for a conditional allele (e.g. Psen1CKO/CKO or Psen1CKO/CKO;Psen2−/−) to create F1 Cre;gene of interestCKO/+ heterozygous mice. All embryonic or postnatal analyses used F2 generation animals generated by timed matings between Cre;gene of interestCKO/+ X gene of interestCKO/CKO mice. The resulting lens phenotypes occurred at expected Mendelian recessive ratios, completely correlating with the loss of gene of interest. Le-Cre;Jag1CKO/+;Psen1CKO/+ double heterozygote phenotypic analyses also used F2 mice, generated in timed matings between Le-Cre;Psen1CKO/+ X Jag1CKO/CKO mice, or Le-Cre;Jag1CKO/+ X Psen1CKO/CKO mice. Finally, we also analyzed F2 litters containing Le-Cre;Jag1CKO/+;Psen1CKO/+;Psen2+/− triple heterozygotes by timed matings of either Le-Cre;Psen1CKO/+;Psen2+/− F1 males and Jag1CKO/CKO females, or Le-Cre;Jag1CKO/+ F1 males to Psen1CKO/CKO;Psen2−/− females.
The embryonic age was based on vaginal plug detection at day E0.5. The heads of anesthetized adult mice were imaged with a Leica MZ8 dissecting microscope, DFC290 HD camera and Leica LAS V4.2 software. Standard H&E paraffin histology of P21 eyes was also performed and those data gathered using a Nikon eclipse E800 scope, Olympus DP74-CU camera and cellSens Dimension software (v1.17). All mice were housed and cared for in accordance with the guidelines provided by the National Institutes of Health, Bethesda, Maryland, and the Association for Research in Vision and Ophthalmology, and conducted with approval and oversight from the Cincinnati Childrens and UC Davis Institutional Animal Care and Use Committees.
Immunohistochemistry and Cell Counting
Embryonic tissue was fixed in 4% paraformaldehyde/PBS for 1hr on ice, processed by stepwise sucrose/PBS incubation and embedded in OCT, then 10 µm frozen sections were generated for marker analyses as described in (Brown et al., 1998). Anti-BrdU labeling was performed as in Le et al. (2006). The primary antibodies used were mouse anti-BrdU (1:100, Becton Dickinson Cat#:347580), rat anti-Ccnd2 (1:200, Santa Cruz Cat#:sc-452), rat anti- Cdh1 (1:500, Invitrogen Cat#:13-1900), rabbit anti-cleaved PARP (1:500, Cell Signaling Cat#:9544), rabbit anti-Cryba1 (1:5000, gift from Richard Lang), goat anti-FoxE3 (1:200, Santa Cruz Cat#:sc-48162-discontined), goat anti-Jag1 (1:200, Santa Cruz Cat#:sc-6011-discontinued), rabbit anti-Prox1 (1:5000, Millipore Cat#:AB5475), rabbit anti-Psen1 (1:100, Santa Cruz Cat#:sc-7860-discontinued). Sections were subsequently incubated with directly conjugated Alexafluor secondary antibodies (1:400, Jackson ImmunoResearch or Molecular Probes) or biotinylated secondary antibodies (1:500, Jackson ImmunoResearch or ThermoScientific) followed by Alexafluor conjugated streptavidin (1:500, Jackson ImmunoResearch). Nuclear staining was performed with DAPI (1:1000 dilution of a 1mg/ml solution, Sigma-Aldrich Cat#:28718-90-3).
Antibody labeled cryosections were imaged using either a Zeiss fluorescent microscope, Zeiss camera and Apotome deconvolution device or a Leica DM5500 microscope, equipped with a SPEII solid state confocal and processed using Axiovision (v5.0) or Leica LASAF and Adobe Photoshop (CS4) software programs, respectively. All images were equivalently adjusted among genotypes for brightness, contrast, and pseudo-coloring. For quantification of marker+ cells, 3 individuals per genotype were analyzed using at least 2 sections per individual. Cell counts were performed using the count tool in Adobe Photoshop CS4. Statistical significance was determined using one-way ANOVA analysis.
In Situ Hybridization
Dissected embryos were immediately embedded unfixed, then cryosectioned. Sections were fixed in 4% PFA/PBS for 10 minutes and hybridized with digoxygenin-labeled RNA probes as previously described (Brown et al., 1998). Full length Presenilin1 or Presenilin2 cDNA were obtained from Origene Technologies (Cat#:MC203369, Cat#:MC206272) and subcloned into pBluescript II KS+/−. Dig-labeled sense and antisense cRNA probes were used side-by-side in experiments and detected by anti-digoxigenin antibody coupled to alkaline phosphatase (Roche Cat#:11093274910). Images were captured on a Nikon eclipse E800 scope equipped with an Olympus DP74-CU camera and cellSens Dimension software (v1.17).
Western blotting
Individual pairs of mouse lenses were hand dissected away from other ocular tissues and harvested in cold PBS and snap frozen. Those of identical genotypes were pooled and lysed in RIPA buffer (150mM NaCl, 50mM Tris pH8, 1% NP40, 0.5% DOC, 0.1% SDS) plus cOmplete mini protease inhibitor tablet (Sigma-Aldrich Cat#:11836153001) for 2 hours using micro stir bars and vigorous stirring. The cell extracts were centrifuged at 15,800 rcf, the lysates collected, total protein quantified by Bradford assay (BioRad Protein Assay Cat#:500-0006), and loaded onto NuPage 4–12% Bis-Tris gel (Invitrogen Cat#:NP0322BOX). 25ug of total protein, equivalent to approximately five E14.5 lenses, was electrophoresed in MES running buffer (Invitrogen Cat#:NP0002-02) and transferred onto 0.2um nitrocellulose membranes (Invitrogen Cat#:LC2000). Blots were blocked in 5% milk/0.1M Tris (pH 7.4)/0.15M NaCl/0.1% Tween20. Protein detection was performed with subsequent primary antibodies: mouse anti-β-actin (1:3000, Sigma-Aldrich Cat#:A1978), goat anti-Jag1 (1:2000, Santa Cruz Cat#:sc-6011-discontinued), rabbit anti-Jag1 (1:1000, Santa Cruz Cat#:sc-8303-discontinued), rabbit anti-Hes1 (1:1000, Cell Signaling Cat#:11988), mouse anti-Notch1 (1:100, Sigma-Aldrich Cat#:N6786), rat anti-Notch2 (1:100, DSHB Cat#:C651.6DbHN), rabbit anti- Presenilin1 (1:2000, Santa Cruz Cat#:sc-7860-discontinued), rabbit anti-Phospho-Histone H2A.X (Ser139) (1:500, Cell Signaling Cat#:2577). Blots were incubated with HRP-conjugated secondary antibodies from Jackson ImmunoResearch. ECL kit (ThermoFisher Cat#:34078) was used for visualization as described by manufacturer. Signals were detected using the Konica Minolta SRX-101A medical film processor. Jag1-CTF protein levels were quantified from x-ray films by densitometry of serial exposures (ImageJ, NIH, Bethesda, MD), normalizing to control lenses and the β-actin loading control, and averaging signal intensity among n = 3 independent experiments. Statistical significance was determined using one-way ANOVA analysis.
RNA purification and Quantitative PCR Analysis
Total RNA from one pair of E14.5 lenses (single individual) was isolated using the Zymo Research Quick RNA miniprep kit (Cat#:R1055). RNA concentrations were measured with a Qubit 3.0 Fluorometer and Molecular Probes Qubit RNA HS Assay kit (Cat#:Q32852). 50ng of total lens RNA was reverse transcribed into cDNA via the Bio-Rad iScript cDNA Synthesis kit (Cat#:170-8891) and used for qPCR analysis using Applied BioSystems Fast Sybr Green Master Mix (Cat#:4385614) on an Applied Biosystems StepOnePlus machine. Relative Quantification (RQ) values were calculated using comparative CT method (Livak and Schmittgen, 2001) with β-actin as a normalization control. Statistical significance was determined by one-way ANOVA or a two-tailed Student’s t-Test using Microsoft Excel 2016 Analysis ToolPak, with p-values <0.05 considered significant.
Results
Presenilins expression in developing lens progenitor cells
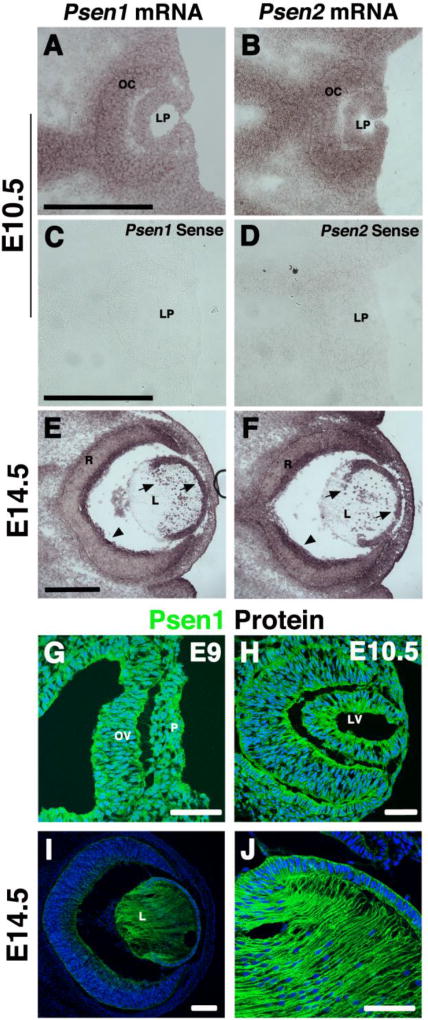
Although Presenilin expression has been reported in the adult primate and mouse lens (Frederikse and Zigler, 1998), we wished to visualize Psen1 and Psen2 localization during mouse primary (E10.5) and secondary (E14.5) fiber cell differentiation. Using full length Psen1 and Psen2 cDNA clones, we synthesized cRNA antisense and sense control probes and performed mRNA in situ hybridization (Fig. 1). Psen1 and Psen2 exhibit overlapping and essentially uniform expression patterns in the developing eye. We also used an antibody specific for Psen1 (Supplemental Fig. 1), to demonstrate protein localization throughout the early embryo at E9.5 (Fig 1G). At E10.5, Psen mRNAs and Psen1 protein are ubiquitously expressed in the lens pit, optic cup, forming pigment epithelium, surrounding mesenchyme, and ventral hypothalamus (Figs. 1A, B, H). By E14.5, Psen1 and Psen2 mRNA lens expression is restricted to AEL, transition zone cells, and fiber cell nuclei, with the rest of the fiber cell compartment devoid of signal. Psen1 and Psen2 expression is also detected in the corneal epithelium layer (Figs. 1E, F). By contrast, Psen1 protein localizes to the plasma membrane of all lens cells, including differentiated fiber cells (Figs. 1I, J). We conclude that both Psen genes are expressed at the right time and place to catalyze the proteolysis of multiple Notch receptors (Notch1, Notch2, and Notch3) co-expressed in AEL progenitors, plus overlap with two Notch ligands Deltalike1 and Jagged1 (Bao and Cepko, 1997; Le et al., 2009; Saravanamuthu et al., 2009; Weinmaster et al., 1991).
Figure 1. Presenilin expression during mouse lens development.
(A–D) In situ hybridization using Psen1 or Psen2 cRNA probes shows uniform expression throughout the developing eye at E10.5. Sense probes for both Psens illustrate background expression. (E, F) At E14.5, both genes are expressed by the anterior epithelial layer and lens fiber nuclei (arrows). Both genes are also expressed in the developing retina, particularly in the inner retinal ganglion layer (arrowheads). (G–J) Anti-Psen1 labeling of cryosections at three ages highlights ubiquitous expression in the lens placode, lens vesicle, and E14.5 anterior epithelial and fiber cells. n ≥ 3 at E9 and E10.5; n = 4 at E14.5. Anterior is right in all panels, L= lens, LP= lens pit, LV= lens vesicle, OV= optic vesicle, OC= optic cup, P= placode, R= retina. Bar in A, B, C= 500um, I= 100um and G, H, J= 50um.
Lens-specific loss of Presenilins causes microphthalmia and postnatal aphakia
To assess the consequences of removing Presenilins in the mammalian lens, we generated Psen single and double mutant mice. Although Psen2−/− germline mice are viable, analogous Psen1−/− mutants die during early embryogenesis (Donoviel et al., 1999; Shen et al., 1997). To circumvent this issue, we used a Psen1 conditional allele (Yu et al., 2001) and the Le-Cre driver (Ashery-Padan et al., 2000), to delete Psen1 exons 2 and 3 containing 5’ UTR, the ATG start codon, and a portion of an N-terminal cytoplasmic domain, just prior to the first transmembrane domain. We verified that this strategy produces a protein null mutation, using a specific antibody (Fig. 6A and Supplemental Fig. 1). We employed the Le-Cre driver for these studies so that the resulting phenotypes would be directly comparable to those previously described for multiple Notch pathway genes (Jia et al., 2007; Le et al., 2009; Rowan et al., 2008; Saravanamuthu et al., 2012), and because the developmental timing of conditional deletion of this pathway is critical. To support this idea, we also obtained another lens Cre driver, MLR10-Cre (Zhao et al., 2004), and assessed the P21 lens phenotypes of MLR10-Cre; Psen1CKO/CKO;Psen2−/− double mutants (Supplemental Fig. 2B), MLR10-Cre;Jagged1CKO/CKO single mutants (data not shown) and MLR10-Cre;Hes1CKO/CKO single mutants (Supplemental Fig. 2C). However, none displayed gross or histologic lens phenotypes (n = 3/ genotype). The MLR10-Cre driver removed nuclear Hes1 protein specifically from the E14.5 lens (Supplemental Figs. 2D, E).
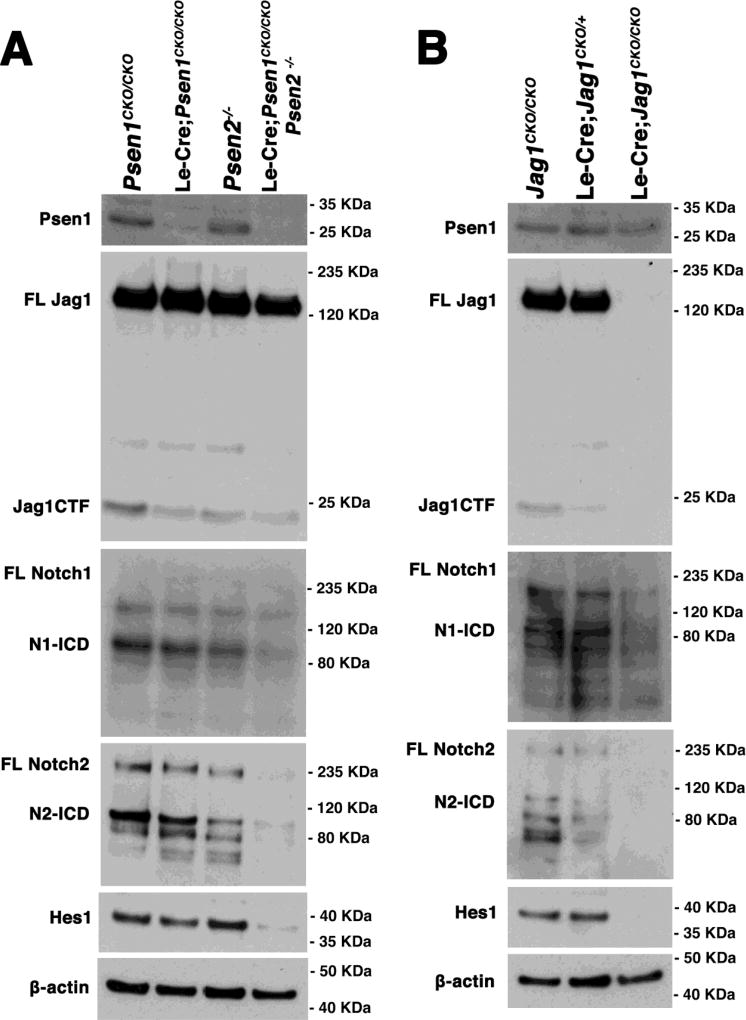
Figure 6. Notch2 protein levels are more sensitive to Psen1/2 redundancy and Jag1 activity in the E14.5 lens.
(A) Western analysis of protein lysates from Psen single or double mutant embryonic lenses. Near complete loss of Psen1 and partial loss of Jag1-CTF in Le-Cre;Psen1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− mutants, with no change in full length Jag1 protein levels. Psen1/2 double mutants have a dramatic loss of full length Notch2, as well as expected downregulation of N1-ICD and N2-ICD isoforms. Nearly complete loss of downstream Notch target, Hes1, confirms defects in canonical pathway signaling. (B) Western blot comparison with a Le-Cre;Jag1 allelic series. Psen1 expression was unaffected by loss of Jag1 and anti-Jag1 validates the conditional mutation. Although loss of Hes1 expression was expected (Le et al., 2009), there was essentially no Notch2 (full length, N2-NEXT, N2-ICD isoforms) in Jag1 mutants, with a less severe reduction in the N1-ICD isoform. In both A and B, anti-β-actin blot reprobing served as a loading control. Each blot is representative of three independent experiments, except anti-Notch1-ICD (n = 2).
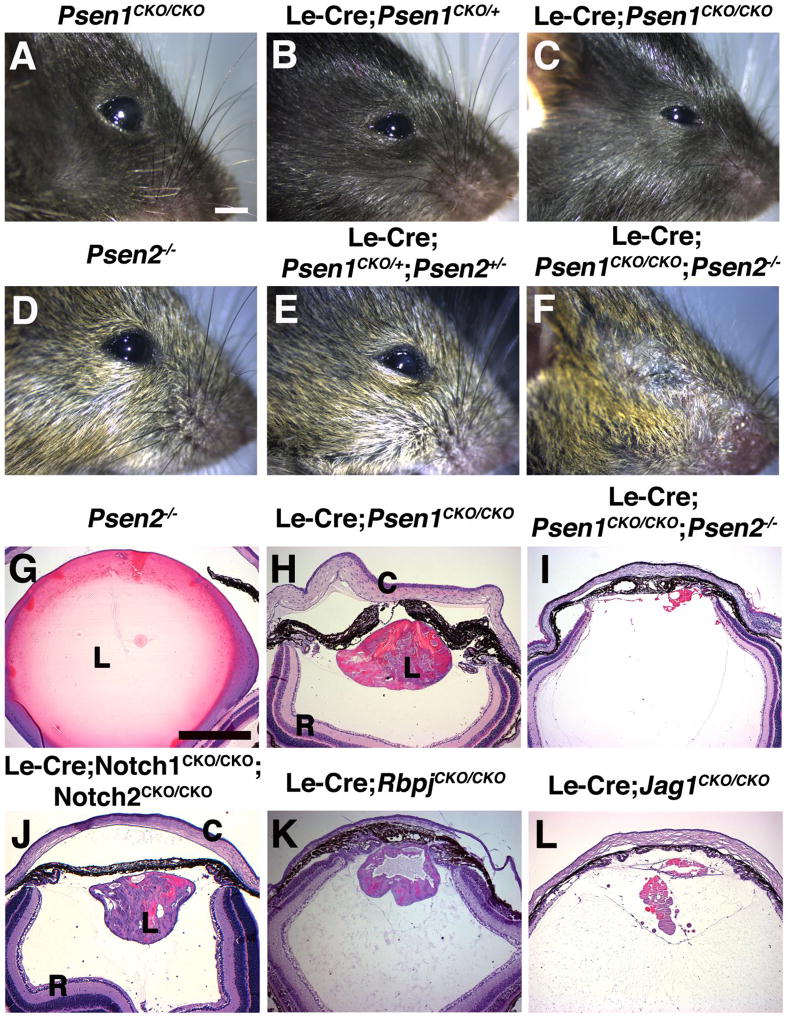
By contrast, P21 Le-Cre;Psen1CKO/CKO mice exhibited ocular microphthalmia, with reduced eye size that correlates with Psen1 gene dosage. For example, P21 Le-Cre;Psen1CKO/+ heterozygotes, have smaller-sized eyes than controls (Figs. 2A–C). Conversely, no eye phenotypes were found in Psen2−/− eyes (Fig. 2D). Histologic sections of P21 eyes highlight the obviously smaller lenses of Le-Cre;Psen1CKO/CKO eyes versus Psen2−/− (Figs. 2G, H). These phenotypes were fully penetrant for each genotype. We concluded that like other tissues, developing lens cells have a stronger requirement for Psen1. However, there is some level of redundancy between Psen1 and Psen2, since Le-Cre;Psen1CKO/CKO;Psen2−/− double mutants show more phenotypic severity (Figs. 2C, F n=3/3 double mutants). P21 Le-Cre;Psen1CKO/CKO;Psen2−/− eyes contained a near total loss of lens tissue, and lacked pupillary openings (Fig. 2I). This is in contrast to Le-Cre;Notch1CKO/CKO;Notch2CKO/CKO and Le-Cre;RbpjCKO/CKO P21 eyes, whose less severe lens phenotypes phenocopy one another (Rowan et al., 2008; Saravanamuthu et al., 2012) (Figs. 2I–K). Interestingly, the more severe defects in Le-Cre;Psen1CKO/CKO;Psen2−/− eyes strongly resemble those in Jag1 lens mutants (Le et al., 2009) (Figs. 2I, L), suggesting the possibility that during lens formation Psen1/2 and Jag1 also have activities independent from their roles in canonical Notch signaling.
Figure 2. Adult eye phenotypes of Presenilin single and double mutants.
Psen1 conditional deletion used the Le-Cre driver whereas Psen2 mutants are germline nulls. (A, D) P21 Psen1CKO/CKO (no Cre) and Psen2−/− eyes are normal. The severity of microphthalmia correlates with Psen1 gene dosage as Le-Cre;Psen1CKO/+ (B) and Psen1/2 double heterozygotes (E) have slightly smaller eyes, while Le-Cre;Psen1CKO/CKO mice (C) are obviously microphthalmic. (F) Le-Cre;Psen1CKO/CKO;Psen2−/− mice appear anophthalmic at this gross level. (G–L) P21 H&E stained sections at the level of the optic nerve, imaged at identical magnification. Le-Cre;Psen1CKO/CKO mutants (H) phenocopy Le-Cre;Notch1CKO/CKO;Notch2CKO/CKO (J) and Le-Cre;RbpjCKO/CKO (K) mutants. However, Le-Cre;Psen1CKO/CKO;Psen2−/− eyes (I) show lens aphakia, strongly resembling Le-Cre;Jag1CKO/CKO phenotype in (L). n ≥ 3/ genotype with all phenotypes showing complete penetrance. Anterior is up in panels G–L, C= cornea, L= lens, R= retina. Bar in A= 2mm and G= 500um.
Progressive AEL and transition zone thinning in prenatal Le-Cre;Psen1CKO/CKO;Psen2−/− lenses
To explore the chronology and molecular nature of the adult lens defects found in the Psen1/2 allelic series, we next turned our attention to embryonic lens formation. We selected several lens developmental markers to label E14.5 ocular cryosections of various genotypes, including Le-Cre;Psen1CKO/CKO;Psen2−/− double mutants. For fiber cell differentiation, we analyzed β-Crystallin (Cryba1) expression. Crystallins are abundant, water-soluble proteins found in lens fiber cells and aberrant Crystallins cause lens opacification seen in cataracts (Chambers and Russell, 1991). Although the fiber cell compartment was smaller in E14.5 Le-Cre;Psen1CKO/+;Psen2+/− and Le-Cre;Psen1CKO/CKO;Psen2−/− mice, we noted that Cryba1 was present in fiber cells indicating their differentiated status (Figs. 3A–C). Then, we examined FoxE3 expression within the AEL progenitor cells. FoxE3 encodes a winged-helix forkhead transcription factor that is expressed by lens progenitor cells, beginning in the lens placode stage and continuing into adulthood (Brownell et al., 2000). We noted E14.5 Le-Cre;Psen1CKO/+;Psen2+/− double heterozygotes had a normal FoxE3 pattern, despite the smaller size lens, but in Le-Cre;Psen1CKO/CKO;Psen2−/− double mutants there was essentially no FoxE3 expression (Figs. 3D–F). FoxE3 is necessary for lens vesicle separation from the surface ectoderm and AEL cell proliferation (Blixt et al., 2000; Medina-Martinez et al., 2005). The loss of FoxE3 by E14.5 in Le-Cre;Psen1CKO/CKO;Psen2−/− lenses is suggestive of reduced AEL proliferation. Previously we reported that Jag1 lens conditional mutants had significantly fewer FoxE3+ AEL cells at E14.5, but that Notch2 and Rbpj lens mutants only displayed a slight reduction (Le et al., 2009; Rowan et al., 2008; Saravanamuthu et al., 2012).
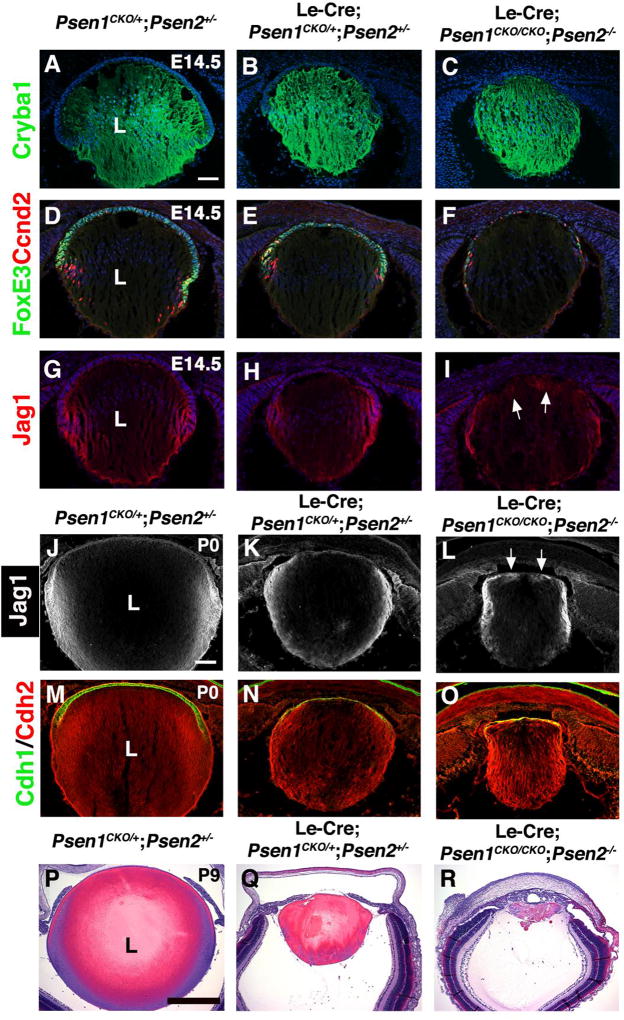
Figure 3. Progressive loss of the lens in Psen1/2 double mutants.
(A–C) At E14.5, Cryba1 expression in LeCre;Psen1CKO/CKO;Psen2−/− mice indicates establishment of secondary fibergenesis despite smaller lens size. (D–F) FoxE3/ CcnD2 double labeling of double mutants reveals thinner AEL (FoxE3+ cells) and malformed transition zone (CcnD2+ cells). (G–I) Jag1 protein domain, normally localized to the lens transition zone, is reduced and abnormally expressed at the anterior poles of fiber cells in double mutants (arrows in I). Note that double heterozygotes display milder defects (B, E, H). (J–L) At P0, Psen1/2 double mutants display obvious lens microphthalmia, and abnormal Jag1 expression in the anterior lens region (arrows in L), beyond its normal domain at the lens equator and in nascent fiber cells. (M–O) Also by P0, Cdh1/E-Cad is dramatically reduced from lens AEL cells, whereas Cdh2/N-Cad is abnormally localized similar to Jag1. (P–R) H&E stained sections of P9 eyes at the level of the optic nerve highlight the additional loss of lens tissue in both Le-Cre;Psen1CKO/+;Psen2+/− (Q) and Le-Cre;Psen1CKO/CKO;Psen2−/− (R) mutants. n ≥ 3/ genotype at E14.5 and P9, and n = 2/ genotype at P0. Anterior is up in all panels, L= lens. Bar in A= 50um, in J= 100um, and in P= 500um.
CyclinD2 (Ccnd2) is normally expressed by lens equatorial (transition zone) cells as they initiate fibergenesis (Reneker and Overbeek, 1996). In addition to fewer Ccnd2+ cells in E14.5 Le-Cre;Psen1CKO/CKO;Psen2−/− lens sections, we noted that their arrangement was sparser including abnormal localization to the AEL, compared to controls (Figs. 3D–F). This phenotype strongly resembles that of E14.5 Le-Cre;RbpjCKO/CKO lenses (Rowan et al., 2008), and is indicative of cell cycle exit defects. Another protein expressed by lens transition zone cells is the Notch ligand, Jag1. Although Jag1 is ubiquitous during lens pit and vesicle stages, at E14.5 it is specifically localized to the cell membrane of transition zone cells (Bao and Cepko, 1997; Le et al., 2009). We noticed that Le-Cre;Psen1CKO/CKO;Psen2−/− lenses also had abnormal Jag1 localization at the anterior pole of fiber cells (Figs. 3G–I), and that this mispatterning persisted to at least P0 (Fig. 3J–L), when the progressive nature of the lens double mutants became more pronounced. We also examined Cdh1 (E-Cadherin) and Cdh2 (N-Cadherin) expression, which further highlighted the abnormal morphology, size, and cell composition of P0 double mutant lenses (Figs. 3M–O). Similar to the Le-Cre;Rbpj conditional mutants (Rowan et al., 2008), we also noted mispatterning of Cdh2 expression at the anterior pole of P0 Psen1/2 double mutant lenses. Finally, the essentially continuous loss of lens tissue in Psen1/2 mutants was more pronounced at P9 (Figs. 3P–R). Overall, we conclude that the AEL and transition zone compartments are not maintained, as early as E14.5, and this likely contributes to the eventual aphakia found in adult Le-Cre;Psen1CKO/CKO;Psen2−/− mice.
Psen1/2 double mutants have both reduced proliferation and increased apoptosis
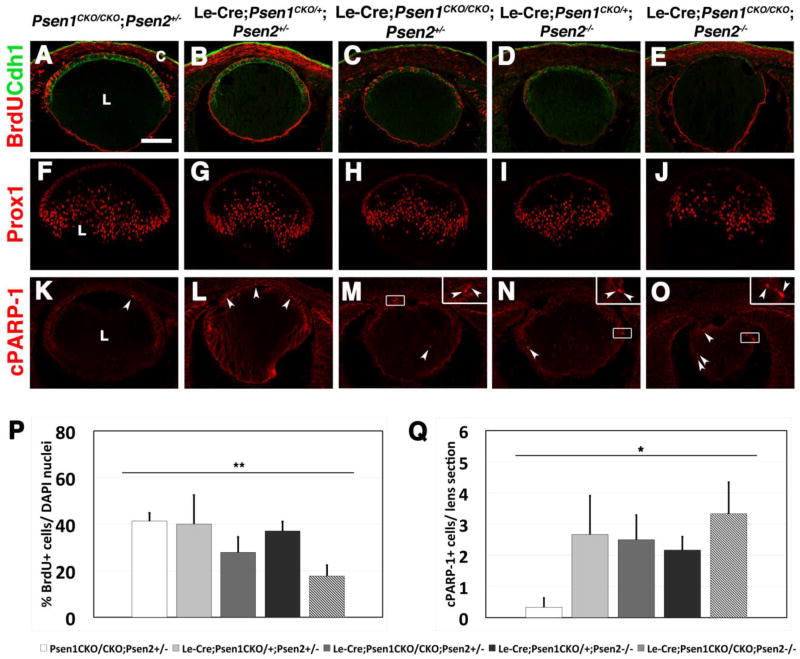
Both the smaller lens size and loss of FoxE3+ AEL cells at E14.5 implied potential defects in cell proliferation, premature fiber cell differentiation, increased apoptosis, or a combination thereof. For proliferation, we quantified the percentage of E14.5 S-phase (BrdU-pulse labeled in red) AEL cells co-stained with Cdh1 expression in green (Figs. 4A–E). By among all five genotypes analyzed (ANOVA), we noted a significant decrease in the percentage of BrdU+ AEL cells (p = 0.013), but individual 2-sample comparisons (T-test), between control and each mutant revealed that there was only significantly reduced percentages of S-phase cells in the AEL of Le-Cre;Psen1CKO/CKO;Psen2+/− (p = 0.032) and Le-Cre;Psen1CKO/CKO;Psen2−/− (p = 0.0002) eyes (Fig. 4P). From this, we conclude that removal of Psen1 had the greatest impact on proliferation, since the lenses of mice retaining just one wild type copy of Psen1 were comparable to controls. Next, to assess fiber cell differentiation, we looked at Prox1 expression, which encodes a transcription factor present in differentiated fiber cell nuclei. Prox1 promotes elongation and polarization of lens fiber cells (Wigle et al., 1999) and regulates a variety of lens genes (Audette et al., 2016). Although Prox1 is also expressed in the cytoplasm and nucleus of AEL and transition zone cells, it is confined to the nucleus upon fiber cell differentiation (Duncan et al., 2002). However, we did not detect any differences in the equatorial position or proportions of E14.5 Prox1+ fiber cells, across the Psen1/2 allelic series (Figs. 4F–J). To test for precocious fibergenesis, we also performed a Cryba1 labeling on younger E10–E12 ocular sections, but all genotypes showed normal onset of expression in the forming lens (data not shown). Finally, to test for apoptosis, we quantified the number of cleaved PARP-1 (cPARP-1) cells in E14.5 lens sections among the various genotypes (Figs. 4K–O). We found a significant increase in cPARP-1+ cells for Le-Cre;Psen1CKO/+;Psen2+/−, Le-Cre;Psen1CKO/CKO;Psen2+/−, Le-Cre;Psen1CKO/+;Psen2−/−, and Le-Cre;Psen1CKO/CKO;Psen2−/− mice compared to wildtype. The double mutants had the largest increase (Fig. 4Q). Overall, we conclude that a combination of reduced AEL proliferation and increased apoptosis across all lens compartments, can explain the smaller lens size of Le-Cre;Psen1CKO/CKO;Psen2−/− double mutants.
Figure 4. E14.5 Psen1/2 allelic series highlights defects in proliferation and apoptosis.
(A–E, P) BrdU+ cells are significantly reduced in Le-Cre;Psen1CKO/CKO;Psen2+/− and Le-Cre;Psen1CKO/CKO;Psen2−/− lenses at E14.5. (E) Cdh1 expression in the AEL in Le-Cre;Psen1CKO/CKO;Psen2−/− eyes is also obviously abnormal. (F–J) By comparison, the arrangement and proportion of Prox1+ fiber cells were relatively normal among the different Presenilin mutants. (K–O, Q) Double heterozygotes, and all embryos lacking three or four alleles, exhibit a significantly increased number of cPARP-1+ apoptotic cells (arrowheads) with greatest increase seen in double mutants. Inserts in (M–O) are zoomed views of the boxed regions to show the cPARP-1+ cells. n ≥ 3/ genotype. Anterior is up in all panels, C= cornea, L= lens. Bar in A= 50um. Error bars in P and Q represent the standard error of the mean (SEM); *p≤0.05, **p≤0.01, and ***p≤0.001.
Psen1 and Jag1 genetically interact
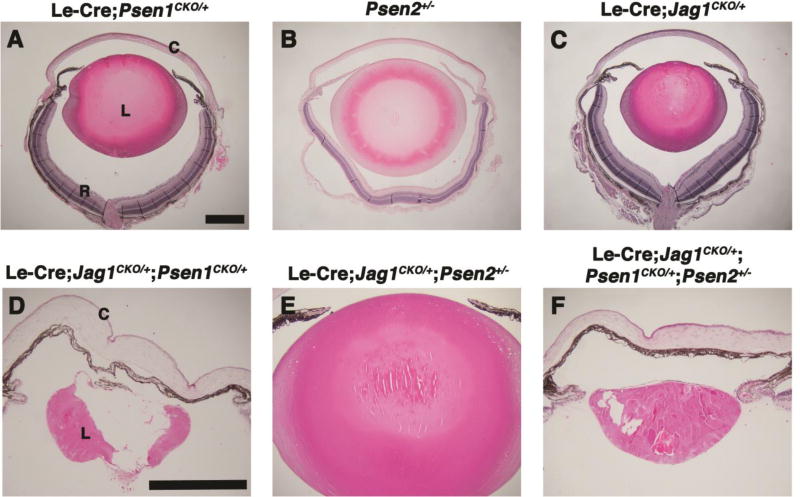
Since Le-Cre;Jag1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− P21 lens defects are more severe than those of other signaling pathway genes and appear to phenocopy one another (Figs. 2I, L), and owing to the precedent that Jag1 and Psen1 can physically interact in vitro (LaVoie and Selkoe, 2003), we formally tested whether Jag1 genetically interacts with Psen1 and/or Psen2 in vivo. P21 double and triple heterozygote mice were generated and their lens anatomy histologically compared to each other, and to relevant heterozygotes, with each phenotype fully penetrant. Among single heterozygotes, the Le-Cre;Psen1CKO/+ and Le-Cre;Jag1CKO/+ lenses were slightly smaller than Psen2+/−, but no other abnormalities were noted (Figs. 5A–C). As for the double heterozygotes, we saw a dramatic enhancement of lens defects in Le-Cre;Jag1CKO/+;Psen1CKO/+ but not in Le-Cre;Jag1CKO/+;Psen2+/− eyes (Figs. 5D, E). Importantly, the lens phenotypes of double Le-Cre;Jag1CKO/+;Psen1CKO/+ versus triple Le-Cre;Jag1CKO/+;Psen1CKO/+;Psen2+/− heterozygotes were essentially indistinguishable (Figs. 5D, F). Thus, we conclude that Jag1 and Psen1 genetically interact during mouse lens formation. While this synergy might be expected when reducing the levels of two components in the same signaling pathway, it is also consistent with the ability of these proteins to physically interact (LaVoie and Selkoe, 2003).
Figure 5. Psen1 and Jag1 genetically interact.
(A–F) H&E stained P21 sections. (A–C) Le-Cre;Psen1CKO/+ and Le-Cre;Jag1CKO/+ eyes have smaller lenses but with essentially normal lens structure, whereas Psen2+/− lenses were normal in both respects. Le-Cre;Jag1CKO/+;Psen1CKO/+ and Le-Cre;Jag1CKO/+;Psen1CKO/+;Psen2+/− eyes had obviously microphthalmic lenses and lack pupillary openings (D, F). By comparison, Le-Cre;Jag1CKO/+;Psen2+/− double heterozygous lenses phenocopied Le-Cre;Jag1CKO/+ single heterozygous eyes. n ≥ 3/ genotype, which each mutant showing completed penetrance. Anterior is up in all panels, C= cornea, L= lens, R= retina. Bars in A and D= 500um.
Loss of Presenilins feedbacks onto the Notch Pathway at both the protein and transcript levels
Because Jag1 expression becomes mispatterned in Psen1/2 double mutants, we were interested to quantify Jag1 protein levels in Psen mutant lenses, via western blotting of E14.5 single and double mutant lenses (Fig. 6A). First, we validated the loss of Psen1 in Le-Cre;Psen1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− mutant lenses, and also noted that Psen2−/− mutant lenses contain normal levels of Psen1. Next, we analyzed Jag1 expression, using an antibody raised against the C-terminus of Jag1 that recognizes most isoforms (LaVoie and Selkoe, 2003). Full-length Jag1 (FL-Jag1) was present at normal levels in Psen1/2 double mutants (Fig. 6A), despite the abnormal patterning found in vivo (Fig. 3K). However, Jag1-CTF levels were significantly reduced in E14.5 lenses of Le-Cre;Psen1CKO/CKO (0.61 ± 0.1) and Le-Cre;Psen1CKO/CKO;Psen2−/− (0.62 ± 0.04) mice (Supplemental Fig. 3). The Jag1-CTF is an isoform created by ADAM protease cleavage, which precedes a subsequent cleavage by γ-secretase in vitro (LaVoie and Selkoe, 2003). Therefore, we predicted that without γ-secretase activity in Le-Cre;Psen1CKO/CKO;Psen2−/− lenses, the Jag1-CTF may accumulate abnormally. However, this was not the case and suggests that Jag1 proteolytic processing in the developing mouse lens differs from what was reported in vitro. To verify that Psen1/2 double mutants block γ-secretase cleavage of the Notch1 and Notch2 receptor proteins, we also examined each receptor intracellular domain (ICD) in the Psen1/2 allelic series. Here we noted that while the 110KDa Notch1 intracellular domain (N1-ICD), was only reduced in Le-Cre;Psen1CKO/CKO;Psen2−/− lenses, by contrast two Notch2 protein isoforms (full length and N2-ICD) were completely missing (Fig. 6A). The greater reliance of Notch2 (over Notch1) on γ-secretase activity is highly correlative with the stronger genetic requirement for Notch2 in the lens (Saravanamuthu et al., 2012). Finally, we saw a nearly complete loss of Hes1 protein in Psen1/2 double mutant lenses, indicating that canonical intracellular signaling has been abolished. We conclude that losing γ-secretase activity not only blocks canonical signaling (loss of N-ICD and Hes1) but also produces signaling feedback at the level of Notch2 protein stability in E14.5 lenses.
Although Jag1 protein expression was unaffected by loss of Psen1 and Psen2, we wished to likewise compare Notch pathway component protein expression among E14.5 control, Le-Cre;Jag1CKO/+, and Le-Cre;Jag1CKO/CKO lenses. First, we noted that Psen1 protein levels were unaffected by the loss of Jag1, which was validated by a direct reprobing with an anti-Jag1 antibody (Fig. 6B). Somewhat surprisingly, we observed that although N1-ICD levels were reduced once again both Notch2 protein isoforms were missing in Le-Cre;Jag1CKO/CKO lenses (Fig. 6B). The downregulation of Notch2 protein was sensitive to Jag1 gene dosage, since Le-Cre;Jag1CKO/+ lenses expressed less Notch2 full length and N2-ICD than wild type controls, relative to actin loading controls. Finally, like Psen single and double mutants, we noted a complete absence of Hes1 expression in Le-Cre;Jag1CKO/CKO lenses. This further confirms Jag1 as the major Notch ligand for the developing mouse lens. We conclude that blocking canonical signaling, either by removing the major ligand or γ-secretase activity, not only causes the loss of a major target, Hes1, but also multiple Notch2 protein isoforms.
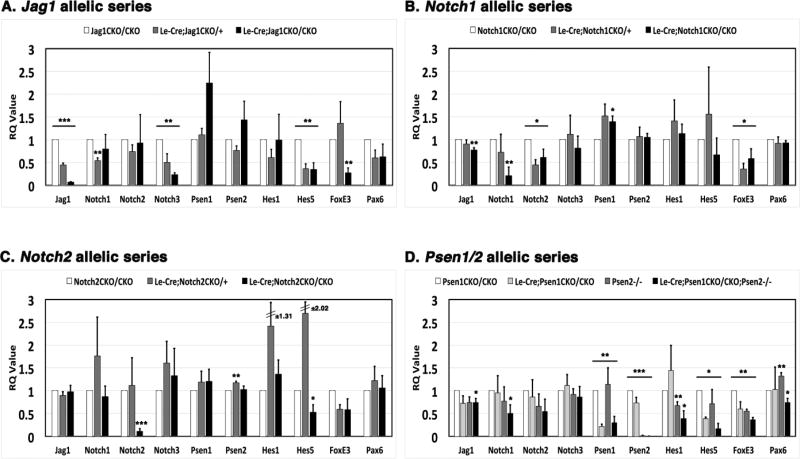
Although Notch receptor ICD levels should be affected by reduced ligand and γ-secretase activity via Psen1/2 gene dosage changes, the loss of full-length Notch2 protein in Jag1 and Psen1/2 lens mutants was unexpected. To further verify that Notch2 expression is specifically targeted, and whether such cross-regulation acts at the level of mRNA and/or protein expression, we collected E14.5 lenses and compared mRNA expression levels for multiple pathway genes, among different conditional mutants, namely Jag1, Notch1, Notch2 and the Psen1/2 allelic series. We also assayed two lens development genes, FoxE3, whose expression domain is dramatically reduced in both Jag1 and Psen1/2 mutant lenses (Fig. 3F) and Pax6, which was unaffected in Jag1 and Notch2 lens mutants (Le et al., 2009; Saravanamuthu et al., 2012). We also verified that each conditional mutation displayed gene dosage-sensitive loss of its endogenous mRNA (for example, in Fig. 7A there is a 50% loss in Le-Cre;Jag1CKO/+ and near complete loss in Le-Cre;Jag1CKO/CKO).
Figure 7. Notch pathway mRNA expression levels in various E14.5 conditionally mutant lenses.
Each graph portrays the relative mRNA levels of various Notch pathway genes, plus Foxe3 and Pax6, in (A) Jag1, (B) Notch1, (C) Notch2, and (D) single and double Psen mutants conditional lens mutants. The relative level of each gene was determined by qPCR, normalized to β-actin, and the respective wildtype control. RQ values were measured in three independent experiments, each performed in triplicate and graphed as the mean ± SEM. n = 3/ genotype. *p≤0.05, **p≤0.01, and ***p≤0.001.
First, we compared the mRNA levels of three Notch receptors (Notch1, Notch2, and Notch3) in the E14.5 lens. Notch1 mRNA was significantly lower only in Jag1 heterozygotes, returning to essentially wild type levels in Jag1 lens mutants (Fig. 7A). At the same time Notch2 mRNA levels were unaffected by loss of Jag1, yet, Notch3 was the most sensitive to changes in Jag1, displaying a 0.77-fold downregulation in Le-Cre;Jag1CKO/CKO lenses (Fig. 7A). By comparison, with the Psen1/2 allelic series, only Notch1 was significantly lower in Psen1/2 double mutant lenses (−0.49-fold change) (Fig. 7D). This is consistent with Notch1 protein downregulation in the same genotype (Fig. 6A), and with previous reports of Notch1 mRNA loss in the presomitic mesoderm of Psen1−/− embryos (Wong et al., 1997). As for Notch2, although mRNA levels exhibited a downward trend across the Psen1/2 allelic series, we only found a significant loss of this receptor in Notch1 mutant lenses (−0.39-fold) (Fig. 7B). We conclude that Psen1/2 double and Jag1 single mutants do not affect Notch2 transcription, suggesting that the dramatic loss of Notch2 protein (Figs. 6A, B) occurs via a post-transcriptional mechanism.
We were particularly interested in comparing receptor mRNA levels in Le-Cre;Notch1CKO/CKO or Le-Cre;Notch2CKO/CKO mutant lenses since Notch protein activities can compensate for one another in particular contexts (Fitzgerald et al., 1993; Liu et al., 2015; Liu et al., 2013). However, there are fewer examples of transcriptional cross-regulation. For instance, a couple studies have shown that deletion of Notch1 resulted in an increase in Notch2 expression (Graziani et al., 2008; Sweetwyne et al., 2015). In our study, we found that Notch1 levels are unaffected in Notch2 mutant lenses, and there was some measurable loss of Notch2 mRNA in Notch1 mutants (Figs. 7B, C), but Notch3 levels were unaffected in either Notch1 and Notch2 lens mutants. These data argue against Notch receptor compensation occurring in the context of the developing lens, at least at the transcriptional level.
As for Psen1 mRNA, we can still detect some transcripts in Le-Cre;Psen1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− lenses. This suggested the possibility that the Psen1 conditional deletion may not fully block transcription, at least in lens cells. However, such transcripts are presumably unstable since embryonic lenses with Psen1 deletion lack measurable protein levels by both western and IHC analysis (Fig. 6A, Supplemental Fig. 1). By comparison, the Psen mRNA levels were unaffected in other pathway mutants. However, we did detect a significant increase in Psen1 mRNA expression in Le-Cre;Notch1CKO/CKO lenses (Fig. 7B) and significantly higher levels of Psen2 mRNA in Le-Cre;Notch2CKO/+ heterozygotes (Fig. 7C).
To demonstrate the loss of canonical signaling in the different E14.5 lens mutants, we also compared Hes1 and Hes5 mRNA expression (Figs. 7A–D). Hes1 transcript levels were unaffected in Le-Cre;Jag1CKO/CKO lenses, which clearly lost all Hes1 protein expression (Fig. 6B), but also there was no significant changes in Hes1 mRNA in either receptor mutant (Figs. 7B,C). But, similar to its profound loss at the protein level in Psen1/2 double mutants, we observe around a 0.61-fold decrease in Hes1 at the mRNA level (Fig. 7D). However, the different conditional lens knockouts had a greater effect on the other Notch effector gene, Hes5. We detected around a 0.65-fold drop in Hes5 in both Le-Cre;Jag1CKO/+ and Le-Cre;Jag1CKO/CKO lenses (Fig. 7A), and a 0.47-fold drop in Le-Cre;Notch2CKO/CKO lenses (Fig. 7C). Hes5 expression went down 0.83-fold with an RQ value of 0.17 in Le-Cre;Psen1CKO/CKO;Psen2−/− lenses (Fig. 7D). Also, comparison of Psen single mutants revealed Psen1 contributes more to Hes5 mRNA expression than does Psen2. Since Hes proteins can co-repress one another (Hatakeyama et al., 2004; Riesenberg et al., 2018), it stands to reason that downregulation of Hes5 allows Hes1 levels to persist in these compromised Notch signaling backgrounds. These data also suggest that Hes5 may be the direct target of Notch pathway transcriptional regulation, whereas Hes1 seems to be more effectively regulated post-transcriptionally (Fig. 6).
Finally, the lens-specific factor FoxE3 is downregulated at the mRNA level in E14.5 Jag1, Notch1, Notch2 and Psen1/2, mutant lenses (Figs. 7A–D). We observed a 0.73-fold drop in Jag1 mutants, a 0.41-fold drop in Notch1 as well as in Notch2 mutants, and a 0.63-fold drop in Psen1/2 double mutant lenses. Particularly relevant here is that the dramatic loss of FoxE3 mRNA is in good agreement with the reduced ratio of FoxE3+ AEL cells in E14.5 Le-Cre;Psen1CKO/CKO;Psen2−/− eyes (Fig. 3F) and a previously reported loss of Foxe3+ cells in E14.5 Le-Cre;Jag1CKO/CKO lenses (Le et al., 2009). By comparison, Pax6 mRNA levels were comparable to wildtype in Jag1, Notch1, Notch2 and Psen mutant lenses (Figs. 7A–D), but specifically upregulated only in Psen2−/− germline mutant lenses as well as significantly downregulated (0.26-fold) in Le-Cre;Psen1CKO/CKO;Psen2−/− lenses.
In summary, our data demonstrate that loss of Jag1 or Psen1/2 results in dramatic loss of Notch2 protein isoforms in the E14.5 lens, and that this feeds back on receptor levels post-transcriptionally. We conclude that during mammalian lens formation, Notch receptor turnover is sensitive to levels of active Notch signaling. In addition, we found that Hes5 mRNA expression is dependent on Jag1-Notch2-Psen1 activity. At E14.5, Notch2 and Hes5 are normally expressed by AEL cells, and their downregulation in Psen1/2 double mutants completely correlates with a loss of AEL-specific markers like Foxe3, and progenitor cell proliferation (summarized in Fig. 8). Similar to removal of other Notch pathway genes, these changes at the onset of secondary fibergenesis contribute to an overall increase in apoptosis and the progressive loss of lens tissue and an eventual lens aphakia at P21.
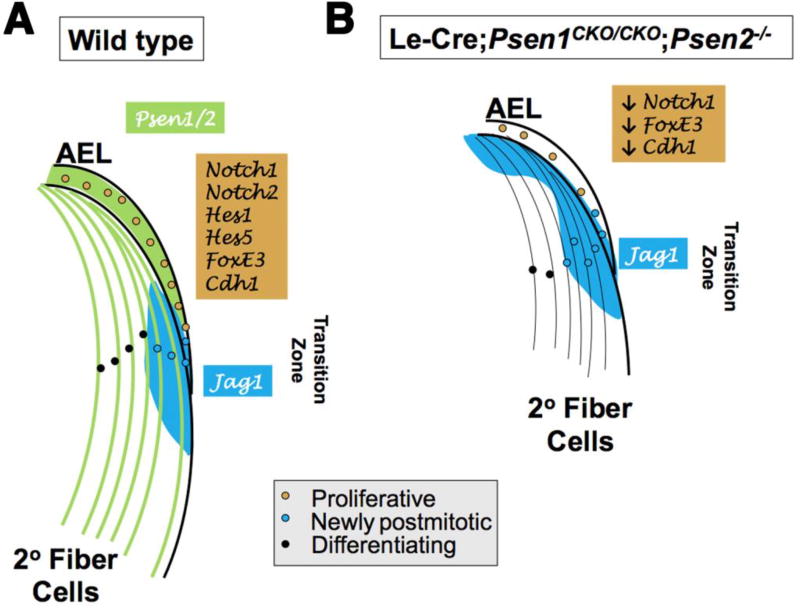
Figure 8. Summary diagram of Psen1/2 function in embryonic lens development.
(A) Normal protein expression for Psen1/2 (green) and Jag1 (blue) at E14.5. Presenilins and Jag1 (Le et al., 2009) are required for normal AEL marker expression (brown box). (B) Loss of both Presenilins causes a smaller lens, starting at E14.5, along with an inappropriate anterior expansion of Jag1, a reduction in AEL cell proliferation and marker expression. There is also a loss of Notch2 protein, Hes1 protein, and Hes5 mRNA expression that suggests Psen1/2 are components of a pathway signaling feedback, which normally maintains Notch protein expression levels. Anterior is up in both panels.
Discussion
In this study we found that removal of both Psen1 and Psen2 from the developing lens is overall consistent with the previously reported phenotypes of Jag1, Notch2 or Rbpj (Jia et al., 2007; Le et al., 2009; Rowan et al., 2008; Saravanamuthu et al., 2012). Although lens induction and primary fibergenesis were unaffected, at E14.5, as secondary fiber differentiation commences, Psen1/2 double mutants displayed an obvious reduction in lens size, molecular marker expression, proliferation, as well as increased apoptosis. Moreover, we anticipated the loss of the Notch ICD isoform and downstream effector Hes1 in Psen1/2 double mutant lenses, but our finding of substantial loss in full length Notch2 protein was surprising. It appears that perturbing the Notch pathway prior to receptor cleavage leads to loss of overall receptor protein expression. We also noted that the adult phenotypes of Psen1/2 double and Jag1 single mutant lenses strongly resemble one another and are each much more severe than the removal of Rbpj or Notch1/Notch2. Despite uncovering a synergistic genetic interaction between Psen1 and Jag1, we did not find evidence supporting their cross-regulation of each other, or that Jag1 protein is a substrate of γ-secretase activity in this context. Instead, we conclude that Notch2 in AEL cells specifically depends on Jag1 in the transition zone and nascent fiber cells to sustain pathway signaling and that the loss of Jag1 affects Notch protein expression similarly to its loss when all γ-secretase activity is missing. It is plausible that Psen1/2 and Jag1 influence the stability or turnover of the Notch2 receptor protein (and to a smaller extent Notch1). For example, multiple E3 ubiquitin ligases (Itch and Cbl in mammals) have been identified that target non-activated Notch receptors for degradation (Chastagner et al., 2008; Jehn et al., 2002), so characterizing their expression patterns and levels, as well as assessing Notch2 ubiquitylation states in Psen1/2 double mutant lenses would be interesting. Future studies involving mass spectrometry analysis of Psen1/2 double and Jag1 mutant lenses could also shed more light on the mechanism for Notch receptor turnover by identifying more factors involved in downregulating the Notch2 protein level.
One interesting finding is that Hes5 is more susceptible to Notch pathway hindrance than Hes1 in the lens. Our lab has recently shown that these two Notch effectors are both expressed by early lens progenitor cells, prior to lens vesicle separation from the surface ectoderm, and that Hes5 is potentially more dynamic in its expression than Hes1 (Riesenberg et al., 2018). Hes5 is subsequently present in both AEL and transition zone cells, while Hes1 expression is more restricted to AEL cells. These different spatial expression domains, along with our finding that Hes5 mRNA is more sensitive to overall changes in Notch pathway signaling, suggest different modes of regulating Hes1 versus Hes5 in the lens.
Although Psen1 has a stronger role than Psen2 in the context of the lens, there were no major differences at the transcriptional or translational level when comparing mRNA and proteins levels of the various Notch pathway components in Le-Cre;Psen1CKO/CKO and Psen2−/− single mutant lenses. Again, this suggests that Psen1 may have a separate, Notch-independent role in the lens. Presenilins are known to have a variety of substrates, most notably the Amyloid Precursor Protein (APP), so it is not surprising that γ-secretase would have additional substrates in this tissue (Haapasalo and Kovacs, 2011). Moreover, given the genetic interaction we found between Psen1 and Jag1, and the observation that Jag1 mutants exhibit a greater loss in lens tissue than seen in Rbpj mutants, both components would appear to be involved in some process independent of the canonical pathway. This would be consistent with multiple examples of Notch-independent roles for these genes. Presenilins and Jag1 have been implicated in remodeling adherens junctions and/or regulating Cdh1 levels, suggesting their Notch-independent function in the lens may involve this cell adhesion protein. Previous reports show that Psen1 physically interacts with adherens junction components and promotes cell-to-cell adhesion when over-expressed in human kidney cells (Georgakopoulos et al., 1999). Moreover, Psen1 was shown to cleave Cdh1, and thus, facilitate disassembly of adhesion complexes containing Cdh1 (Marambaud et al., 2002). In Drosophila, the Notch pathway was shown to be involved in remodeling adherens junctions necessary for stretched cell flattening (Grammont, 2007). The study showed that the Delta and Serrate mutant clones (Notch ligand homologues) had abnormally persistent junctions. Furthermore, Jag1-mediated Notch signaling in human breast epithelial cells also represses Cdh1 and promotes epithelial-to-mesenchymal transition (Leong et al., 2007). And interestingly, a study examining Jag1 and its processed isoforms thought to be generated by ADAM and γ-secretase activities, showed that FL-Jag1 and the ectodomain form of Jag1 can repress Cdh1 at the protein and mRNA level (Delury et al., 2013).
Moreover, Cdh1 has been shown to be necessary for proper lens development (Pontoriero et al., 2009). Le-Cre;Cdh1CKO/CKO mice have microphthalmic eyes and small, vacuolated lenses. Their lens defects are less severe than our Le-Cre;Psen1CKO/CKO;Psen2−/−, as well as LeCre;Jag1CKO/CKO, mice. Interestingly, our lab showed dramatic loss of Cdh1 expression by E14.5 in the AEL of Le-Cre;Psen1CKO/CKO;Psen2−/− mice, similar to what we saw in Le-Cre;Jag1CKO/CKO, both of which were greater than the loss seen in Le-Cre;RbpjCKO/CKO (Le et al., 2009; Rowan et al., 2008). It may be that Psen1 and Jag1 loss impairs both the Notch pathway and Cdh1 processing, leading to a more enhanced lens phenotype, especially in the AEL. This greater defect in AEL can also be seen by FoxE3 staining, which again, is markedly reduced in our Le-Cre;Psen1CKO/CKO;Psen2−/− and Le-Cre;Jag1CKO/CKO mice, but not in Le-Cre;RbpjCKO/CKO. It would be interesting for future studies to assess Cdh1 levels in the lenses of Le- Cre;Psen1CKO/CKO;Psen2−/− and Le-Cre;Jag1CKO/CKO mice, as well as Le-Cre;Jag1CKO/+;Psen1CKO/+ mice, to better understand the loss of epithelial cells in these tissues.
Although one explanation for Psen lens phenotypes in this study could be that the Le-Cre transgene, containing the Pax6 enhancer, may diminish overall Pax6 levels (Dora et al., 2014). We do not favor this explanation because Pax6 mRNA levels were normal in eleven of thirteen different genotypes analyzed here (Fig. 7). This is consistent with another recent study showing Pax6 acts upstream of the Notch pathway in the prenatal mouse lens (Thakurela et al., 2016). However, we did find a decrease in Pax6 mRNA levels in E14.5 Psen1/2 double mutants (−0.29-fold) (Fig. 7D). This is highly correlated with the dramatic loss of Foxe3 mRNA and protein expression and a reduced proliferation of AEL cells, with the latter activity dependent on Pax6 activity (Shaham et al., 2009). Moreover, to alleviate concerns that the Le-Cre transgene may be toxic to developing lens cells (Loonstra et al., 2001), we monitored levels of the DNA damage marker Phospho-Histone H2A.X (Ser139), which we find equivalently expressed between wildtype and Le-Cre transgenic P0 lenses (Supplemental Fig. 2F). We selected the Le-Cre transgene because it is the only Cre driver with appropriate early expression in the mouse lens. We also evaluated MLR10-Cre (Zhao et al., 2004) as a tool for this study, by using it to first delete Psen1/2, but then also Jag1 and Hes1. However, in all three cases, no adult lens defects were found (Supplemental Figs. 2B, 2C, and data not shown), although relevant protein expression was abolished by E14.5 (Supplemental Figs. 2D, 2E). Indeed, when these same two drivers were used to delete the Smoothened gene, only Le-Cre induced phenotypes, although Smoothened is also clearly expressed by lens progenitor cells from E9.5 onwards (Choi et al., 2014). These outcomes suggest that the staggered onset of these two Cre drivers (Le-Cre at E9 and MLR10-Cre E11) (Ashery-Padan et al., 2000; Zhao et al., 2004) imply that the critical window for Notch and Shh signaling occurs between these two ages. Certainly, all of the genes to which this test has been applied: Jag1, Hes1, Psen1 and Smoothened are expressed and active prior to the onset of MLR10 Cre expression and activity. Alternatively, Le-Cre expression is not lens-specific. At these embryonic ages, is also expressed in the surface ectoderm and other tissues that derive from it (Ashery-Padan et al., 2000). Thus, it is possible that disruption of these signaling pathways simultaneously in multiple tissues is more catastrophic than a lens-vesicle specific removal.
A long-standing question in the Notch field is to what extent different receptors carry out distinct functions. Many studies have shown Notch1- versus Notch2-mediated signaling induce different, even opposite, biologic outcomes (Chu et al., 2011; Fan et al., 2004; Graziani et al., 2008). Indeed, this was most clearly demonstrated by swapping particular protein domains in elegant chimeric receptor gene replacement studies (Aster et al., 2011; Fitzgerald et al., 1993; Liu et al., 2015). For the lens we found only a few differences between Le-Cre;Notch1CKO/CKO and Le-Cre;Notch2CKO/CKO eyes, namely that Psen1 mRNA was increased in Notch1 mutants, yet Psen2 mRNA was only slightly elevated in Notch2 heterozygotes. This may reflect a compensatory response to an overall diminishment of Notch signaling (increasing expression of an upstream component in the pathway to boost signal output). Although Notch1 and Notch2 mutants exhibit different phenotypes during lens formation, we find no support for cross regulation between these receptors, at least at the transcriptional level.
Supplementary Material
Supplemental Figure 1. Anti-Psen1 specificity. E14.5 Psen1CKO/+ (A) and Le-Cre;Psen1CKO/CKO (B) tissue sections labeled as in Figure 1 and imaged at 20× (A, B) and 40× (A’, B’) magnification. Le-Cre;Psen1CKO/CKO lenses are devoid of Psen1 protein while retinal expression is unaffected, verifying lens-specific removal of Psen1. Anterior is right in all panels, L= lens, R= retina. Bar in A= 100um, B= 50um.
Supplemental Figure 2. Data supporting use of Le-Cre driver in this study. (A–B) P21 H&E stained sections at the level of the optic nerve indicates eyes in MLR10-Cre;Psen1CKO/CKO;Psen2−/− mice are comparable to control (n = 3/genotype). (C) Another MLR10-Cre mutant of the Notch pathway, MLR10-Cre;Hes1CKO/CKO, exhibits no eye phenotype although complete removal of Hes1 protein can be detected by E14.5 (D, E). (F) DNA damage levels measured in P0 lens extracts by Western blotting using anti-H2A.X (Ser139) antibody. No difference in DNA damage marker was observed between wildtype and Le-Cre heterozygote samples. β-actin serves as loading control. Anterior is up in A-E, C= cornea, L= lens, R= retina. Bar in A= 500um, D= 50um.
Supplemental Figure 3. Quantification of Jag1-CTF protein levels in Figure 6. Histogram comparison of Jag1-CTF protein for E14.5 control, Psen single or double mutant lenses. Jag1-CTF was reduced in Le-Cre;Psen1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− mutants to 0.61 ± 0.1 and 0.62 ± 0.04, respectively. Values plotted are normalized relative densities, measured in three independent experiments and graphed as the mean ± SEM. n = 3/ genotype. *p≤0.05, **p≤0.01, and ***p≤0.001.
Supplemental Table 1. qPCR primer pair sequences. All primer pair sequences were obtained from the Harvard PrimerBank, with the exception of FoxE3 primers (Terrell et al., 2015).
Highlights.
Psen1 mutants are microphthalmic and Psen1/2 mutants are aphakic by adulthood
Psen1/2 lens mutants are more severe than canonical Notch pathway mutants
Psen1 and Jag1 genetically interact in the mouse lens
Feedback on overall Notch receptor protein level occurs in Psen1/2 mutants
Acknowledgments
The authors thank Jie Shen and Raphael Kopan for Psen1flox;Psen2KO mice; Tasuko Honjo for Rbpj flox mice; Raphael Kopan for Notch1 flox;Notch2 flox mice; Julien Lewis for Jagged1 flox mice; Ruth Ashery-Padan for Le-Cre transgenic mice; Michael Robinson for MLR-Cre mice; Richard Lang for antibody reagents and use of Zeiss Apotome imaging system; Brad Shibata and Paul FitzGerald for assistance with lens histology; April Bird and Kelly McCulloh for mouse husbandry and tissue collections; and Henry Ho for critical reading of this manuscript.
Abbreviations
- CKO
conditional knock-out
- ICD
intracellular domain
- NEXT
Notch extracellular truncation
- PLE
presumptive lens ectoderm
- AEL
anterior epithelial layer
- E0.5
embryonic day 0.5
- H&E
hematoxylin and eosin
- OCT
optimal cutting temperature
- RQ
relative quantification
- IHC
Immunohistochemistry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions:
M.A., T.T.L. and N.L.B. developed the concepts, experimental approach and complex mouse stocks, M.A. and T.T.L. performed the experiments, M.A. and N.L.B. analyzed the data, prepared and edited the manuscript.
References
- Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Bodnar N, Xu L, Karnell F, Milholland JM, Maillard I, Histen G, Nam Y, Blacklow SC, Pear WS. Notch ankyrin repeat domain variation influences leukemogenesis and Myc transactivation. PLoS One. 2011;6:e25645. doi: 10.1371/journal.pone.0025645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audette DS, Anand D, So T, Rubenstein TB, Lachke SA, Lovicu FJ, Duncan MK. Prox1 and fibroblast growth factor receptors form a novel regulatory loop controlling lens fiber differentiation and gene expression. Development. 2016;143:318–328. doi: 10.1242/dev.127860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao ZZ, Cepko CL. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglopoulos V, Sun X, Saura CA, Lemere CA, Kim RD, Shen J. Reduced beta-amyloid production and increased inflammatory responses in presenilin conditional knock-out mice. J Biol Chem. 2004;279:46907–46914. doi: 10.1074/jbc.M409544200. [DOI] [PubMed] [Google Scholar]
- Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14:245–254. [PMC free article] [PubMed] [Google Scholar]
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development. 2006;133:1277–1286. doi: 10.1242/dev.02284. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brownell I, Dirksen M, Jamrich M. Forkhead Foxe3 maps to the dysgenetic lens locus and is critical in lens development and differentiation. Genesis. 2000;27:81–93. doi: 10.1002/1526-968x(200006)27:2<81::aid-gene50>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Cain S, Martinez G, Kokkinos MI, Turner K, Richardson RJ, Abud HE, Huelsken J, Robinson ML, de Iongh RU. Differential requirement for beta-catenin in epithelial and fiber cells during lens development. Dev Biol. 2008;321:420–433. doi: 10.1016/j.ydbio.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Chamberlain CG, McAvoy JW. Evidence that fibroblast growth factor promotes lens fibre differentiation. Curr Eye Res. 1987;6:1165–1169. doi: 10.3109/02713688709034890. [DOI] [PubMed] [Google Scholar]
- Chambers C, Russell P. Deletion mutation in an eye lens beta-crystallin. An animal model for inherited cataracts. J Biol Chem. 1991;266:6742–6746. [PubMed] [Google Scholar]
- Chastagner P, Israel A, Brou C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One. 2008;3:e2735. doi: 10.1371/journal.pone.0002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JJ, Ting CT, Trogrlic L, Milevski SV, Familari M, Martinez G, de Iongh RU. A role for smoothened during murine lens and cornea development. PLoS One. 2014;9:e108037. doi: 10.1371/journal.pone.0108037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Chu D, Zhang Z, Zhou Y, Wang W, Li Y, Zhang H, Dong G, Zhao Q, Ji G. Notch1 and Notch2 have opposite prognostic effects on patients with colorectal cancer. Ann Oncol. 2011;22:2440–2447. doi: 10.1093/annonc/mdq776. [DOI] [PubMed] [Google Scholar]
- Coulombre JL, Coulombre AJ. Lens Development: Fiber Elongation and Lens Orientation. Science. 1963;142:1489–1490. doi: 10.1126/science.142.3598.1489. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Delury C, Tinker C, Rivers S, Hodges M, Broughton S, Parkin E. Differential regulation of E-cadherin expression by the soluble ectodomain and intracellular domain of Jagged1. International Journal of Biochemistry Research and Review. 2013;3:278–290. [Google Scholar]
- Donoviel DB, Hadjantonakis AK, Ikeda M, Zheng H, Hyslop PS, Bernstein A. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dora NJ, Collinson JM, Hill RE, West JD. Hemizygous Le-Cre transgenic mice have severe eye abnormalities on some genetic backgrounds in the absence of LoxP sites. PLoS One. 2014;9:e109193. doi: 10.1371/journal.pone.0109193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MK, Cui W, Oh DJ, Tomarev SI. Prox1 is differentially localized during lens development. Mech Dev. 2002;112:195–198. doi: 10.1016/s0925-4773(01)00645-1. [DOI] [PubMed] [Google Scholar]
- Faber SC, Robinson ML, Makarenkova HP, Lang RA. Bmp signaling is required for development of primary lens fiber cells. Development. 2002;129:3727–3737. doi: 10.1242/dev.129.15.3727. [DOI] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Wilkinson HA, Greenwald I. glp-1 can substitute for lin-12 in specifying cell fate decisions in Caenorhabditis elegans. Development. 1993;119:1019–1027. doi: 10.1242/dev.119.4.1019. [DOI] [PubMed] [Google Scholar]
- Frederikse PH, Zigler JS., Jr Presenilin expression in the ocular lens. Curr Eye Res. 1998;17:947–952. doi: 10.1076/ceyr.17.9.947.5135. [DOI] [PubMed] [Google Scholar]
- Garcia CM, Huang J, Madakashira BP, Liu Y, Rajagopal R, Dattilo L, Robinson ML, Beebe DC. The function of FGF signaling in the lens placode. Dev Biol. 2011;351:176–185. doi: 10.1016/j.ydbio.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos A, Marambaud P, Efthimiopoulos S, Shioi J, Cui W, Li HC, Schutte M, Gordon R, Holstein GR, Martinelli G, Mehta P, Friedrich VL, Jr, Robakis NK. Presenilin-1 forms complexes with the cadherin/catenin cell-cell adhesion system and is recruited to intercellular and synaptic contacts. Mol Cell. 1999;4:893–902. doi: 10.1016/s1097-2765(00)80219-1. [DOI] [PubMed] [Google Scholar]
- Grammont M. Adherens junction remodeling by the Notch pathway in Drosophila melanogaster oogenesis. J Cell Biol. 2007;177:139–150. doi: 10.1083/jcb.200609079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM, Strack PR, Miele L, Bocchetta M. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008;68:9678–9685. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Kovacs DM. The many substrates of presenilin/gamma-secretase. J Alzheimers Dis. 2011;25:3–28. doi: 10.3233/JAD-2011-101065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Bessho Y, Katoh K, Ookawara S, Fujioka M, Guillemot F, Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, Baekelandt V, Dressel R, Cupers P, Huylebroeck D, Zwijsen A, Van Leuven F, De Strooper B. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc Natl Acad Sci U S A. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Jehn BM, Dittert I, Beyer S, von der Mark K, Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J Biol Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- Jia J, Lin M, Zhang L, York JP, Zhang P. The Notch signaling pathway controls the size of the ocular lens by directly suppressing p57Kip2 expression. Mol Cell Biol. 2007;27:7236–7247. doi: 10.1128/MCB.00780-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi Y, Sockanathan S, Liu Q, Breitman M, Lovell-Badge R, Kondoh H. Involvement of SOX proteins in lens-specific activation of crystallin genes. EMBO J. 1995;14:3510–3519. doi: 10.1002/j.1460-2075.1995.tb07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 1999;96:3781–3785. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita A, Imayoshi I, Hojo M, Kitagawa M, Kokubu H, Ohsawa R, Ohtsuka T, Kageyama R, Hashimoto N. Hes1 and Hes5 control the progenitor pool, intermediate lobe specification, and posterior lobe formation in the pituitary development. Mol Endocrinol. 2007;21:1458–1466. doi: 10.1210/me.2007-0039. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol. 2007;17:117–127. doi: 10.1016/j.sbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Kovall RA, Gebelein B, Sprinzak D, Kopan R. The Canonical Notch Signaling Pathway: Structural and Biochemical Insights into Shape, Sugar, and Force. Dev Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- Le TT, Conley KW, Brown NL. Jagged 1 is necessary for normal mouse lens formation. Dev Biol. 2009;328:118–126. doi: 10.1016/j.ydbio.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Brunskill E, Varnum-Finney B, Zhang C, Zhang A, Jay PY, Bernstein I, Morimoto M, Kopan R. The intracellular domains of Notch1 and Notch2 are functionally equivalent during development and carcinogenesis. Development. 2015;142:2452–2463. doi: 10.1242/dev.125492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Chen S, Boyle S, Zhu Y, Zhang A, Piwnica-Worms DR, Ilagan MX, Kopan R. The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development. Dev Cell. 2013;25:585–598. doi: 10.1016/j.devcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, Robinson ML. Development of the ocular lens. Cambridge University Press; Cambridge, UK ; New York: 2004. [Google Scholar]
- Marambaud P, Shioi J, Serban G, Georgakopoulos A, Sarner S, Nagy V, Baki L, Wen P, Efthimiopoulos S, Shao Z, Wisniewski T, Robakis NK. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21:1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy JW, Chamberlain CG, de Iongh RU, Hales AM, Lovicu FJ. Lens development. Eye (Lond) 1999;13(Pt 3b):425–437. doi: 10.1038/eye.1999.117. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44:29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- Medina-Martinez O, Brownell I, Amaya-Manzanares F, Hu Q, Behringer RR, Jamrich M. Severe defects in proliferation and differentiation of lens cells in Foxe3 null mice. Mol Cell Biol. 2005;25:8854–8863. doi: 10.1128/MCB.25.20.8854-8863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian XL, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Molecular Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Pontoriero GF, Smith AN, Miller LA, Radice GL, West-Mays JA, Lang RA. Co-operative roles for E-cadherin and N-cadherin during lens vesicle separation and lens epithelial cell survival. Dev Biol. 2009;326:403–417. doi: 10.1016/j.ydbio.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneker LW, Overbeek PA. Lens-specific expression of PDGF-A alters lens growth and development. Dev Biol. 1996;180:554–565. doi: 10.1006/dbio.1996.0328. [DOI] [PubMed] [Google Scholar]
- Riesenberg AN, Conley KW, Le TT, Brown NL. Separate and coincident expression of Hes1 and Hes5 in the developing mouse eye. Dev Dyn. 2018;247:212–221. doi: 10.1002/dvdy.24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Conley KW, Le TT, Donner AL, Maas RL, Brown NL. Notch signaling regulates growth and differentiation in the mammalian lens. Dev Biol. 2008;321:111–122. doi: 10.1016/j.ydbio.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Esselens C, Ejsmont P, Mattera R, Rochin L, Tharkeshwar AK, De Baets G, De Wever V, Habets R, Baert V, Vermeire W, Michiels C, Groot AJ, Wouters R, Dillen K, Vints K, Baatsen P, Munck S, Derua R, Waelkens E, Basi GS, Mercken M, Vooijs M, Bollen M, Schymkowitz J, Rousseau F, Bonifacino JS, Van Niel G, De Strooper B, Annaert W. Restricted Location of PSEN2/gamma-Secretase Determines Substrate Specificity and Generates an Intracellular Abeta Pool. Cell. 2016;166:193–208. doi: 10.1016/j.cell.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanamuthu SS, Gao CY, Zelenka PS. Notch signaling is required for lateral induction of Jagged1 during FGF-induced lens fiber differentiation. Developmental Biology. 2009;332:166–176. doi: 10.1016/j.ydbio.2009.05.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanamuthu SS, Le TT, Gao CY, Cojocaru RI, Pandiyan P, Liu C, Zhang J, Zelenka PS, Brown NL. Conditional ablation of the Notch2 receptor in the ocular lens. Dev Biol. 2012;362:219–229. doi: 10.1016/j.ydbio.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- Shaham O, Smith AN, Robinson ML, Taketo MM, Lang RA, Ashery-Padan R. Pax6 is essential for lens fiber cell differentiation. Development. 2009;136:2567–2578. doi: 10.1242/dev.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Spemann H. Embryonic development and induction. Yale University Press; H. Milford, Oxford University Press; New Haven, London: 1938. [Google Scholar]
- Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- Stump RJ, Ang S, Chen Y, von Bahr T, Lovicu FJ, Pinson K, de Iongh RU, Yamaguchi TP, Sassoon DA, McAvoy JW. A role for Wnt/beta-catenin signaling in lens epithelial differentiation. Dev Biol. 2003;259:48–61. doi: 10.1016/s0012-1606(03)00179-9. [DOI] [PubMed] [Google Scholar]
- Sweetwyne MT, Gruenwald A, Niranjan T, Nishinakamura R, Strobl LJ, Susztak K. Notch1 and Notch2 in Podocytes Play Differential Roles During Diabetic Nephropathy Development. Diabetes. 2015;64:4099–4111. doi: 10.2337/db15-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Terrell AM, Anand D, Smith SF, Dang CA, Waters SM, Pathania M, Beebe DC, Lachke SA. Molecular characterization of mouse lens epithelial cell lines and their suitability to study RNA granules and cataract associated genes. Exp Eye Res. 2015;131:42–55. doi: 10.1016/j.exer.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S, Tiwari N, Schick S, Garding A, Ivanek R, Berninger B, Tiwari VK. Mapping gene regulatory circuitry of Pax6 during neurogenesis. Cell Discov. 2016;2:15045. doi: 10.1038/celldisc.2015.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmaster G, Roberts VJ, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wong PC, Zheng H, Chen H, Becher MW, Sirinathsinghji DJ, Trumbauer ME, Chen HY, Price DL, Van der Ploeg LH, Sisodia SS. Presenilin 1 is required for Notch1 and DII1 expression in the paraxial mesoderm. Nature. 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- Yang X, Klein R, Tian X, Cheng HT, Kopan R, Shen J. Notch activation induces apoptosis in neural progenitor cells through a p53-dependent pathway. Dev Biol. 2004;269:81–94. doi: 10.1016/j.ydbio.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Yu H, Saura CA, Choi SY, Sun LD, Yang X, Handler M, Kawarabayashi T, Younkin L, Fedeles B, Wilson MA, Younkin S, Kandel ER, Kirkwood A, Shen J. APP processing and synaptic plasticity in presenilin-1 conditional knockout mice. Neuron. 2001;31:713–726. doi: 10.1016/s0896-6273(01)00417-2. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Nadeau P, Song W, Donoviel D, Yuan M, Bernstein A, Yankner BA. Presenilins are required for gamma-secretase cleavage of beta-APP and transmembrane cleavage of Notch-1. Nat Cell Biol. 2000;2:463–465. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–1939. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Anti-Psen1 specificity. E14.5 Psen1CKO/+ (A) and Le-Cre;Psen1CKO/CKO (B) tissue sections labeled as in Figure 1 and imaged at 20× (A, B) and 40× (A’, B’) magnification. Le-Cre;Psen1CKO/CKO lenses are devoid of Psen1 protein while retinal expression is unaffected, verifying lens-specific removal of Psen1. Anterior is right in all panels, L= lens, R= retina. Bar in A= 100um, B= 50um.
Supplemental Figure 2. Data supporting use of Le-Cre driver in this study. (A–B) P21 H&E stained sections at the level of the optic nerve indicates eyes in MLR10-Cre;Psen1CKO/CKO;Psen2−/− mice are comparable to control (n = 3/genotype). (C) Another MLR10-Cre mutant of the Notch pathway, MLR10-Cre;Hes1CKO/CKO, exhibits no eye phenotype although complete removal of Hes1 protein can be detected by E14.5 (D, E). (F) DNA damage levels measured in P0 lens extracts by Western blotting using anti-H2A.X (Ser139) antibody. No difference in DNA damage marker was observed between wildtype and Le-Cre heterozygote samples. β-actin serves as loading control. Anterior is up in A-E, C= cornea, L= lens, R= retina. Bar in A= 500um, D= 50um.
Supplemental Figure 3. Quantification of Jag1-CTF protein levels in Figure 6. Histogram comparison of Jag1-CTF protein for E14.5 control, Psen single or double mutant lenses. Jag1-CTF was reduced in Le-Cre;Psen1CKO/CKO and Le-Cre;Psen1CKO/CKO;Psen2−/− mutants to 0.61 ± 0.1 and 0.62 ± 0.04, respectively. Values plotted are normalized relative densities, measured in three independent experiments and graphed as the mean ± SEM. n = 3/ genotype. *p≤0.05, **p≤0.01, and ***p≤0.001.
Supplemental Table 1. qPCR primer pair sequences. All primer pair sequences were obtained from the Harvard PrimerBank, with the exception of FoxE3 primers (Terrell et al., 2015).