Abstract
Respiratory syncytial virus (RSV) is a single-stranded negative sense RNA virus that belongs to the paramyxovirus family. RSV infections lead to a variety of clinical outcomes ranging from a mild “cold-like disease” to death. Infection is usually more severe in infants and the elderly. RSV is associated with the development and exacerbation of chronic lung conditions including asthma, and it is a major cause of hospitalizations in infants. Because of its clinical relevance, experimental animal models to study RSV in vivo are needed. The most common and accessible animal model in research laboratories is the mouse. However, commonly use RSV strains poorly establish infection in mice and thus titration of the virus from mouse lungs to confirm infection is not sensitive enough to detect early viral infection. Here we discuss in detail how to infect BALB/c mice with RSV and how to detect RSV genomes in the lung using reverse transcription quantitative PCR (RT-qPCR). This method allows detection of viral genomes as early as day 1 post-infection (shown in Figure 2), whereas traditional TCID50 fails to detect significant virus until after day 2 post-infection. Of note, despite of higher sensitivity, genome RT-qPCR only shows the production of viral genomes and thus positive results for this assay are not proof of production of infectious viral particles.
Materials and Reagents
Vacuum-driven Sterilcup® 500 ml Millipore ExpressPLUS 0.22 μm PES (Merck Millipore Corporation, catalog number: SCGPU05RE)
FastPrep® tubes (MP Biomedicals, catalog number: 5076-400)
¼” ceramic sphere (MP Biomedicals LLC, catalog number: 116540412)
CryoTubes™ vials for freezing viruses (Thermo Fisher Scientific, catalog number: 377267)
Surgical scissors (Roboz Surgical Instrument Co)
Tweezers (Roboz Surgical Instrument Co)
384-well PCR plate (VWR International, catalog number: 82051-470)
PCR strip tubes, 0.2 ml (Bioexpress, catalog number: T-3035-2)
0.22 μm filter used in the pipette aid (VWR International, catalog number: 28145-481)
BSL2 personal protective equipment (PPE)
Mice (Balb/c, 6-8 weeks old, gender-mixed) (Taconic)
Human RSV strain A2 (ATCC, catalog number: VR-1540)
SYBR green master mix (Applied Biosystems, catalog number: 4368708)
- qPCR primer sets
Genes Forward primer Reverse primer RSV g 5’AACATACCTGCCCAGAATC3’ 5’GGTCTTGACTGTTGTAGATTGCA3’ Rsp11 5’CGTGACGAAGATGAAGATGC3’ 5’ GCACATTGAATCGCACAGTC3’ α-tubulin 5’ TGCCTTTGTGCACTGGTATG3’ 5’ CTGGAGCAGTTTGACGACAC3’ RSV genome RT primer: 5’GATAAATATAGGCATGGGGAAAGTG3’
TRIzol reagent (Thermo Fisher Scientific, Ambion™, catalog number: 15596018)
Chloroform (Sigma-Aldrich, catalog number: C2432-25 ml)
-
Isopropanol (Sigma-Aldrich, catalog number: 437522)
Note: It is also named “2-propanol” on Sigma-Aldrich website.
Ultra-Pure nuclease-free water
SuperScript® III First-Strand Synthesis System (Thermo Fisher Scientific, Invitrogen™, catalog number: 18080-051)
High capacity RNA-to-cDNA™ kit (Thermo Fisher Scientific, Applied Biosystems™, catalog number: 4387406)
Ketamine (Henry Schein)
Xylazine (U.S. Food and Drug Administration, AnaSed, catalog number: 139-236)
Phosphate buffered saline (PBS) (Thermo Fisher Scientific, Gibco™, catalog number: 10010-023)
Ketamine/Xylazine solution (see Recipes)
Primer preparation (see Recipes)
Equipment
Biosafety hood in biosafety level 2 facility
-
Applied Biosystem ViiA™ 7 lightcycler (Life Technologies, catalog number: 4453536)
Note: Currently, it is “Thermo Fisher Scientific, catalog number: 4453536”.
-
Eppendorf centrifuge 5415R (Eppendorf AG, catalog number: 22636570)
Note: This product has been discontinued by Eppendorf AG.
NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, catalog number: ND-1000)
BioRad C1000 thermal cycler (Bio-Rad Laboratories, catalog number: 1851148EDU)
MP Fastprep-24 homogenizer (MP Biomedicals, catalog number: 116004500)
Procedure
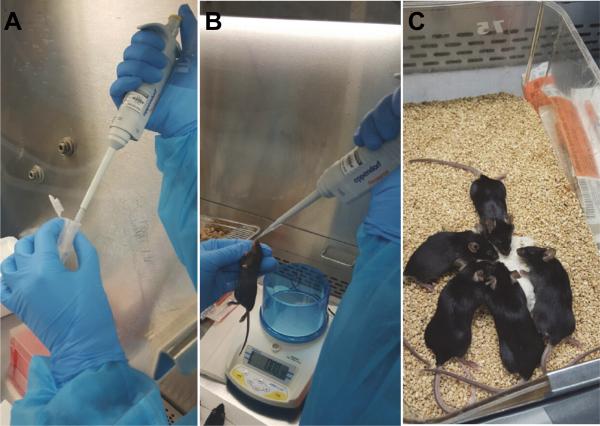
A. Infection (Figure 1)
Figure 1. Procedure for intranasal immunization of mice.
A. Use a P100 pipette to administer 35 μl of virus solution per mouse. B. Hand-hold a fully-anesthetized mouse vertically and place the virus solution into its nostril drop by drop. C. Place the mouse back into its cage and monitor it until it awakens.
Note: This section describes the infection of Balb/C mice with RSV using the intranasal route.
Dilute RSV stock [for preparation of RSV stocks see Sun and López (2016)] in sterile PBS to a concentration of 5 × 106 TCID50 / 35 μl. Keep the diluted virus on ice.
Anesthetize the mice with 40 μl/20 g of weight of Ketamine/Xylazine solution. Make sure each mouse is completely asleep and unresponsive to a toe pinch stimulus.
Hold the mouse vertically with the head tilted back and slowly deliver 35 μl of virus dilution into its nostrils using a 100 μl pipette. The solution should be completely aspirated into the lung and not be swallowed through the mouth.
Place the mouse back into its cage and monitor it until it wakes from anesthesia.
B. Lung tissue collection
Note: This section describes the collection of infected tissue from mice to later examine viral load.
-
5.
Prepare collection tubes: For each sample, add 1 ml TRIzol into one MP Fastprep tube containing one ceramic sphere. Other similar homogenizing techniques can be used.
-
6.
Euthanize the mice at the desired time point post infection (usually 6 h, 10 h, 24 h, and daily for up to one week) using CO2 or other approved method.
-
7.
Place a dead mouse on polystyrene foam and pin each limb with needles, belly up.
-
8.
Spray the fur of the mouse using 70% ethanol.
-
9.
Use scissors to cut open the abdominal and thoracic cavities. Opening the abdominal cavity exposing the underside of the diaphragm. Use scissors to pierce through the diaphragm. Cut away the ribcage of the mouse and fully expose the heart and lungs. Collect two of the left lobes and place into a Fastprep tube containing TRIzol. Always take the same lobes of the lung to maintain consistency. Other lobes can be used for other purposes.
-
10.
Homogenize the lung on the FastPrep-24 homogenizer or similar equipment. Homogenize for 20 sec. Repeat three times.
-
11.
Aliquot the homogenized samples into 500 μl aliquots. Place them at −80 °C for storage.
C. Total RNA extraction
Note: This section describes the extraction of RNA from infected tissue, which will be later used for detection of viral genomes.
-
12.
Thaw one aliquot and add 500 μl of fresh TRIzol for a 1 ml total volume.
-
13.
Add 200 μl of chloroform and shake vigorously for 15 sec.
-
14.
Leave for 3 min at room temperature to allow for phase separation. Centrifuge for 15 min at 12,800 x g in a microcentrifuge at 4 °C.
-
15.
Carefully collect 3/4 of the upper aqueous phase without disturbing or touching the interface or organic layer. Combine the aqueous phase with an equal volume of isopropanol. Gently invert to mix.
-
16.
Leave for 10-20 min at −20 °C for precipitation.
-
17.
Centrifuge for 15 min at 12,800 x g in a microcentrifuge at 4 °C.
-
18.
Discard the supernatant with care not to lose the pellet.
-
19.
Wash the pellet with 1 ml of 75% ethanol. Vortex briefly or pipette up and down the sample multiple times.
-
20.
Centrifuge for 10 min at 12,800 x g in a microcentrifuge at 4 °C.
-
21.
Remove the supernatant and invert the tubes. Allow the pellet to air-dry until clear.
-
22.
Resuspend the pellet in 30-40 μl of ultra-pure nuclease-free dH2O.
-
23.
Incubate at 65 °C for 5 min.
-
24.
Measure RNA concentration in a Nanodrop 1000 or equivalent.
D. Reverse transcription (RT)
Note: This section describes the conversion of total RNA prepared in section C into cDNA. In brief, mRNAs are converted into total cDNA (T-cDNA) using oilgo-dT. This cDNA will be then used to amplify housekeeping genes as controls. RSV genomes are converted into viral genome cDNA (V-cDNA) using a RSV genome RT primer that specifically binds to the 5’ end of the genome. V-cDNA will be used for the amplification of RSV viral genomes.
D1. RT of total mRNA (T-cDNA)
-
25.
Use 2 μg of RNA for reverse transcription.
-
26.
Prepare the RT reaction mix in PCR tubes on ice using the High Capacity RNA-cDNA kit as follows (per sample): 2x RT buffer 5 μl, 20x RT Enzyme mix 0.5 μl, RNA 2 μg, nuclease-free H2O quantity sufficient to 10 μl.
Note: The oligo-dT primers are included in the RT buffer.
-
27.
Gently mix and briefly centrifuge to collect all the components at the bottom of the tube.
-
28.
Place into a BioRad C1000 Thermal Cycler and run program to synthesize cDNA as follow: 37 °C for 60 min, 95 °C for 5 min, 4 °C hold.
-
29.
Store total cDNA (T-cDNA) at −20 °C until ready for qPCR analysis.
D2. RT of viral genomes (V-cDNA)
-
30.
Use 2 μg of RNA for reverse transcription.
-
31.
Mix the following ingredients per sample in one tube: 1 μl of the genome RT primer (50 μM), 1 μl of dNTPs (10 mM), 2 μg of RNA, nuclease-free H2O (add up to 10 μl).
-
32.
Incubate 10 min at 65 °C.
-
33.
Prepare the RT reaction mix in the PCR tubes on ice using SuperScript® III First-Strand Synthesis System as follows (per sample): 5x RT buffer 2 μl, 25 mM MgCl2 4 μl, 0.1 M DTT 2 μl, RNase OUT 1 μl, SS III 1 μl.
-
34.
After incubation, add 10 μl of RT mix to each sample. Mix well.
-
35.
Incubate at 50 °C for 50 min.
-
36.
Heat at 85 °C for 5 min.
-
37.
Add 1 μl of RNase H per sample (from SuperScript III reverse transcriptase kit) and incubate at 37 °C for 20 min.
-
38.
Keep the viral genome cDNA (V-cDNA) at −20 °C until ready for the qPCR analysis.
E. Quantitative PCR (qPCR)
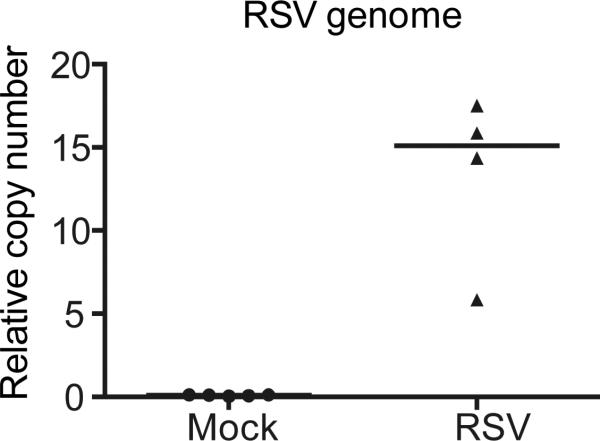
Note: This section describes the quantification of viral genomes presented in the infected tissue. T-cDNA and V-cDNA generated from section D are used as templates for housekeeping genes and RSV genomes, respectively. The copy numbers of two housekeeping genes are used to calculate the housekeeping index (hki) that is then used to calculate the relative copy number of viral genomes. An example of the results from viral genome qPCR at day 1 post-infection in vivo is shown in Figure 2.
Figure 2. RSV viral genome qPCR.
Mice were infected with PBS (mock) or RSV. Lungs were harvest on day 1 post-infection and analyzed using a RSV genome RT-qPCR assay. Each dot represents RSV genome quantification per mouse.
-
39.
qPCR reactions are performed in triplicate using specific primers and the Power SYBR® Green PCR Master Mixture in a Viia7 Applied Biosystem Lightcycler or equivalent.
-
40.
Thaw both T- and V-cDNA and spin down before use.
-
41.
Dilute cDNA 1:40 using dH2O.
-
42.
Load the plate with 4 μl of the diluted cDNA/well in triplicate.
-
43.
Prepare Master Mix: SYBR green (5 μl) with 1 μl of qPCR primer mix. Load the plate with Master Mix 6 μl/well. For housekeeping genes, Rsp11 and α-tubulin are examined as reference using T-cDNAs as template. For RSV genome, RSV g primer pair is used and V-cDNAs serve as template.
-
44.
Run the qPCR using the following program (Table 1):
-
45.
Viral genome copy numbers are normalized based on levels of Rsp11 and α-tubulin for mouse samples using the formula: relative copy number=2,500*1.93^ (Ctgenome - hki), hki=median of CtRsp11 and Ctα-tubulin. Sequences of primers used for qPCR can be found in the materials under qPCR primers. Results see Figure 2.
Table 1.
DI-PCR program
| Program | Temperature (°C) | Time | |
|---|---|---|---|
| 50 | 2 min | Hold | |
| Denaturation | 95 | 10 min | Hold |
| Denaturation | 95 | 5 sec | 40 cycles |
| Annealing | 55 | 25 sec | |
| Extension | 60 | 35 sec | |
| Denaturation | 95 | 15 sec | Melting curve |
| Annealing | 65 | 1 min | |
| Denaturation | 95 | 15 sec | |
Recipes
- Ketamine/Xylazine solution (2 ml)
- 1 ml ketamine
- 0.14 ml xylazine
- 1 ml PBS
- Mixed in BSL2 hood and store at room temperature
- Primer preparation
- Primers are diluted into 100 μM as stock solution.
- For reverse transcription (RT primer), dilute the stock solution 1:2 using nuclease-free
- H2O for a working concentration of 50 μM.
- For qPCR primers, reverse and forward primers are used as a pair. Prepare 750 μl of primer mix as a working stock as described below:
- 37.5 μl stock solution of forward primer
- 37.5 μl stock solution of reverse primer
- 675 μl nuclease-free H2O
Acknowledgments
This work was supported by grant AI083284 from the National Institute of Health (CBL). The protocol described herein was based on the following paper: Sun et al. (2015); Sun and López (2016).
References
- 1.Sun Y, López CB. Preparation of respiratory syncytial virus with high or low content of defective viral particles and their purification from viral stocks. Bio-protocol. 2016;6(9):e1820. doi: 10.21769/BioProtoc.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y, Jain D, Koziol-White CJ, Genoyer E, Gilbert M, Tapia K, Panettieri RA, Jr., Hodinka RL, Lopez CB. Immunostimulatory defective viral genomes from respiratory syncytial virus promote a strong innate antiviral response during infection in mice and humans. PLoS Pathog. 2015;11(9):e1005122. doi: 10.1371/journal.ppat.1005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tapia K, Kim WK, Sun Y, Mercado-Lopez X, Dunay E, Wise M, Adu M, Lopez CB. Defective viral genomes arising in vivo provide critical danger signals for the triggering of lung antiviral immunity. PLoS Pathog. 2013;9(10):e1003703. doi: 10.1371/journal.ppat.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]