Abstract
Gly missense mutations in type I collagen, which replace a conserved Gly in the repeating (Gly-Xaa-Yaa)n sequence with a larger residue, are known to cause Osteogenesis Imperfecta (OI). The clinical consequences of such mutations range from mild to lethal, with more serious clinical severity associated with larger Gly replacement residues. Here, we investigate the influence of the identity of the residue replacing Gly within and adjacent to the integrin binding 502GFPGER507 sequence on triple-helix structure, stability and integrin binding using a recombinant bacterial collagen system. Recombinant collagens were constructed with Gly substituted by Ala, Ser or Val at four positions within the integrin binding region. All constructs formed a stable triple-helix structure with a small decrease in melting temperature. Trypsin was used to probe local disruption of the triple helix, and Gly to Val replacements made the triple helix trypsin sensitive at three of the four sites. Any mutation at Gly505, eliminated integrin binding, while decreased integrin binding affinity was observed in the replacement of Gly residues at Gly502 following the order Val > Ser > Ala. Molecular dynamics simulations indicated that all Gly replacements led to transient disruption of triple-helix interchain hydrogen bonds in the region of the Gly replacement. These computational and experimental results lend insight into the complex molecular basis of the varying clinical severity of OI.
Keywords: Osteogenesis Imperfecta, collagen, missense mutation, integrin binding sites, molecular dynamics, recombinant protein expression, triple-helix
1. Introduction
Collagen, the most abundant protein in vertebrates, is crucial to the structural integrity and mechanical stability of connective tissues, including skin, tendon, cartilage, cornea and bone. The collagen triple-helical domain, a common structural motif of all types of collagen, is composed of a repetitive (Gly-Xaa-Yaa)n amino acid sequence, where the Xaa and Yaa positions are often occupied by imino acid residues, proline and hydroxyproline (Brodsky and Persikov, 2005). As the smallest amino acid residue, Gly is required at every third position in the polypeptide because it is the only residue capable of fitting into the core of the triple helix (Ramachandran and Kartha, 1955; Rich and Crick, 1961). Up to 28 collagen types have been identified in human and can be categorized into two general groups, fibrillar (e.g. types I, II, III, V and XI) and non-fibrillar collagens. Among the 28 types of human collagen, type I is the most abundant and is the major protein in bone. Type I collagen is natively heterotrimeric and consists of two α1(I) chains and one α2(I) chain, encoded by the COL1A1 and COL1A2 genes, respectively.
Owing to the importance and abundance of collagen, mutations in collagen disrupt extracellular matrix (ECM) structure and function, causing a diverse set of diseases. A prime example is Osteogenesis Imperfecta (OI), a hereditary disorder characterized by bone malformations and fractures (Marini et al., 2007). The majority of OI cases are autosomal dominant and result from mutations in the α1(I) or α2(I) chain of type I collagen, with the phenotypes ranging from mild disease to a perinatal lethality. More recently, autosomal recessive forms of OI have been reported which are caused by mutations in genes encoding proteins involved in collagen processing (Forlino and Marini, 2016).
Among mutations in type I collagen that lead to autosomal dominant OI, missense mutations resulting in substitutions of a single Gly residue by a bulkier residue in the triple-helical domain of type I collagen are the most common. Such replacements break the Gly-Xaa-Yaa repeats and lead to pathological conditions (Royce and Steinmann, 2003). A singlebase substitution in a Gly codon can lead to any of eight different residues (Ser, Cys, Arg, Val, Glu, Asp, Ala and Trp). The identity of the residue replacing Gly appears to be closely related to the degree of clinical severity of OI cases. Gly→ Ser substitutions are the most frequently observed OI missense mutation, and seen to lead to a wide range of lethal and non-lethal phenotypes. Substitutions of Gly by Ala, the smallest replacement residue, are often mild and fewer are observed than predicted on the basis of a mutation rate analysis residue replacing Gly (Persikov et al., 2004). In contrast, substitutions of Gly by Val, one of the largest replacement residues, result in a lethal phenotype in 73% of the cases reported (Marini et al., 2007). Moreover, a similar pattern of the differential effects of amino acid identity on severity was observed in distinct OI cases with various mutations at the same Gly residue. For example, at position Gly844, replacement by Ala leads to the moderate type of OI5 (type I/IV), replacement by Ser leads to a severe form (type III OI), and replacement by Val leads to a lethal phenotype (type II OI) (COL1A1 Database of Osteogenesis Imperfecta Variants (www.le.ac.uk/ge/collagen/ updated September 29, 2017).
The mechanism by which a Gly missense mutation leads to bone fragility in OI has been investigated, but the way in which the identity of the residue replacing Gly affects the clinical phenotype remains elusive. A Gly replacement by any larger residue distorts the triple-helix conformation (Bella et al., 1994), leads to a small loss of stability of the collagen triple helix (Makareeva et al., 2008), delays collagen folding (Raghunath et al., 1994), can interfere with secretion (Forlino et al., 2007), and can affect procollagen processing and assembly (Lightfoot et al., 1994; Lightfoot et al., 1992). Previous studies of a collagen-like model peptide showed that the degree of destabilization in short (Gly-Pro-Hyp)8 triple- helical peptides depended on the identity of the residue replacing Gly, following the order Ala ≤ Ser < Cys < Arg < Val < Glu ≤ Asp, which correlated well with the severity of OI for a given Gly replacement residue in the α1(I) chain (Beck et al., 2000). Besides structural destabilization, another model to explain the genotype-phenotype relationship of OI is that some mutations in collagen may interfere with collagen’s interactions with various cell receptors, chaperones and other ECM proteins (Marini et al., 2007). It is possible that Gly replacements by residues with very large side chains, e.g. Val, lead to a greater perturbation of triple-helix structure or greater interference with interactions, which could explain the high percentage of lethal OI cases (e.g. 73% lethal for Gly to Val mutations).
Using a bacterial collagen system, the effects of Gly→Ser mutations on collagen structure and specific collagen-protein interactions were previously reported (Chhum et al., 2016; Yigit et al., 2016). Flere, these studies are extended to further investigate the effect of the identity of the residues replacing Gly on both collagen structure and interactions. Specifically, the effect of the replacement of Gly residues by Ala, Ser, or Val on collagen stability and integrin binding activity is reported and used to explore the relationship between altered biophysical properties and OI pathology. The structure and stability of these recombinant proteins were characterized by CD, DSC and trypsin sensitivity, and their integrin binding affinity was examined by solid-state binding assay. Based on the experimental analyses, molecular dynamic simulations were further performed to better visualize the structural changes at the molecular level. The results indicated the importance of the identity of the Gly replacement residue and its position on both structural and interaction parameters.
2. Materials and methods
2.1. Construction and preparation of recombinant bacterial collagen proteins
All Gly missense mutant plasmids were derived from the previously constructed plasmid pCold-VCL-Int (Yigit et al., 2016). DNA sequence of the original bacterial collagen VCL was optimized for expression in E. coli based on the colla- gen-like Scl2 protein from Streptococcus pyogenes (An et al., 2014). The VCL construct harbored the DNA sequence for the V trimerization domain and the (Gly-Xaa-Yaa)79 CL domain with Flis6 tag at N-terminus for purification purpose, while its derivative VCL-Int was constructed by insertion of the integrin-binding region G496AR-G499ER-G502FP-G505ER- G508VQ-G511PP in the human αl(I) chain after triplet number 30 in the CL domain (Yigit et al., 2016). Gly missense substitutions were generated by replacing Gly residues at 4 different Gly sites (G502, G505, G508, and G511) with Ser, Ala and Val using Q5 Site-Directed Mutagenesis Kit (NEB), respectively. The recombinant plasmids were transformed into E. coli DF15a competent cells, extracted and verified by DNA sequencing.
All constructs in the pCold III vector were transformed and expressed in E. coli BL21 cells. Ampicillin-resistant colonies were picked and inoculated into 20 ml LB medium containing 100 μg/ml of Ampicillin. After incubation at 37°C overnight in a rotary shaker (250 rpm), 10 ml of pre-culture was transferred into 500 ml of LB-Ampicillin medium in a 2-liter flask. Cells was cultured at 37°C with shaking at 250 rpm, until reached an OD600 of 0.8–1.0. Cultures was induced by adding IPTG to the final concentration of 1 mM, then grown at 20°C with shaking overnight. Purification of recombinant bacterial collagens was performed on an AECTA pure system (GE Healthcare) as described previously (An et al., 2016). Briefly, the cells were harvested by centrifugation at 8,000 g for 20 min at 4°C, resuspended in 10 ml of binding buffer (20 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole, pH 7.4) and lysed by sonication. The crude lysate was centrifuged at 8,000 g for 30 min at 4°C to remove cellular debris. The supernatant was loaded onto a pre-equilibrated Ni-NTA column and washed with 5 column volumes of binding buffer, 3 column volumes of binding buffer plus 50 mM imidazole and binding buffer plus 100 mM of imidazole, sequentially. The protein bound to the Ni-NTA resin was eluted by elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). Purity of the elution fractions was detected by SDS-PAGE. Elution fractions containing target protein were collected and dialyzed against lx PBS buffer (lOx PBS, pH 7.4; Fisher Scientific). Molecular weight of purified proteins was determined by MALDI-TOF mass spectrometry on a Microflex LT system (Bruker Corporation, Billerica, MA). Protein concentrations were measured by UV-Vis spectrophotometer (Aviv Biomedical Inc., Lake- wood, NJ) with an extinction coefficient of ε280 =9970 M1 cm-l
2.2. Circular dichroism (CD) analysis
CD spectra of recombinant collagens were obtained on an AVIV Model 420 CD spectrometer (AVIV Biomedical Inc.). Wavelength scans were collected at 0°C from 260 to 190 nm recording points at every 0.5 nm for 4 s using a bandwidth of 1 nm, averaging three scans for each sample.
Temperature scans were monitored by measuring MRE at 220 nm from 0 to 70°C with a 10-s averaging time and 1.5- nm bandwidth. Samples were equilibrated for 2 min at each temperature, and the temperature was increased at an average rate of 0.1°C/min. The melting temperature (Tm) is defined as the temperature at which the fraction-folded F(Tm) is equal to 0.5 as described previously (Bryan et al., 2011).
2.3. Differential scanning calorimetry (DSC) analysis
DSC profiles of recombinant collagens were obtained on a NANO DSC II model 6100 (Calorimetry Sciences Corp, Lindon, UT). Each sample was dialyzed against PBS overnight before measurement. Dialysis buffer was collected and used as reference for the corresponding sample. Samples were loaded into the cells at 0°C and heated at a rate of 1°C /mm till 70°C.
2.4. Trypsin and chymotrypsin cleavage assay
Purified protein samples (0.1 mg/ml in PBS) were treated with 0.01 mg/ml of trypsin (Sigma-Aldrich) or 5.0 pg/ml of chymotrypsin (Sigma-Aldrich) at 20°C for up to overnight. The reaction was terminated by adding phenylmethylsulfonyl fluoride (Sigma-Aldrich) to a final concentration of 1 mM, and the digests were further analyzed by SDS-PAGE (NuPAGE Bis-Tris 4 −12%, Thermo Fisher Scientific).
2.5. Solid-state binding assay
Binding of integrin α2 I domain to the recombinant bacterial collagen proteins was carried out according to the method reported previously (Knight et al., 2000; Tuckwell et al., 1995). The high-binding 96-well plate (R&D Systems, Minneapolis, MN) was coated with 100 μl of recombinant collagen (20 μg/ml) per well for 1 h at room temperature, blocked with 200 μ1 of 50 mg/mL BSA (Sigma-Aldrich) in PBS for 1 h, and washed four times with washing buffer (PBS containing 1 mg/ml BSA and 2 mM MgCl2). Then 100 μl of recombinant GST-tagged integrin α2 I domain (provided by Professor S. Hamaia and Professor R. W. Famdale) was added at a concentration of 20 μg/ml in wash buffer. Plates were incubated at room temperature for 1 h, and washed with washing buffer for four times. 100 μl of anti-GST F1RP antibody (1:10,000 dilution in washing buffer) was applied to wells and incubated for 1 h at room temperature (Sigma-Aldrich). After washing, binding was assessed through colorimetric analysis using a TMB substrate kit (Pierce). Absorbance was measured at 450 nm with SpectraMax M2 microplate reader (Molecular Devices Corp., Sunnyvale, CA). VCL-Int protein was used as the positive control for all the binding assays. For dose-response assays, serial concentrations of I domain at 0.10, 1.56, 6.25, 25, 50, 100, 200 and 400 ng/μl were used, corresponding to 0.002, 0.03, 0.125, 0.5, 1, 2, 4 and 8 μM I domain in molar concentration. Data as shown are representative of at least three repeat experiments.
2.6. Molecular dynamics simulations of collagen-like triple helices
To investigate the effects of Gly to Ala, Ser, and Val substitutions, respectively, on the triple-helical structure of type I collagen containing the integrin-binding sequence, molecular dynamics (MD) simulations were performed for the model α1(I)3 homotrimer collagen peptide and its mutants. The sequences of 6 triplets (residues 496–513) containing the 52GFP-GER507 integrin-binding triplets (underlined in the sequence below) were retrieved from the UniProt database from the human α1(I) chain (P02452) (Apweiler et al., 2004), and flanked by additional triplets of the bacterial construct sequence (shown in grey) and three GPO stabilizing triplets at both ends. The wildtype (WT) sequence used in the simulations is:
(GPO)3VGPAGPOGPRGEOGPO-GAR-GER-GFP-GER-GVQ-GPP-GLPGKDGEA-(GPO)3
The initial triple-helical structures for the WT collagen peptides were built using the Triple-Helical collagen Building Script (THeBuScr) (Rainey and Goh, 2004). The α1(I) chains were capped with an acetyl group and NH2 group in the Aland C-termini, respectively. In addition to the wildtype system, mutant collagen model peptides were also simulated by using the UCSF Chimera package (Pettersen et al., 2004) to make residue substitutions.
All simulations were performed with GROMACS 4.6.7 (Hess et al., 2008) using the GROMOS 54a7 force field (Schmid et al., 2011) and SPC water model (Berendsen et al., 1981). For each collagen triple helix, the starting structure was energy- minimized in vacuum using the steepest descent algorithm for a maximum of 2,000 steps. All heavy atoms of the triple helix were position restrained with a force constant of 1,000 kJ/mol/nm2. The vacuum-minimized structure was then solvated in a rectangular water box such that no collagen atom was closer than 1.5 nm to the edges of the box and the long axis of the collagen triple helix was parallel to the z-axis of the box. The system was then further energy-minimized using the steepest descent algorithm for a maximum of 5,000 steps. In this step, the periodic boundary condition was applied, and a cut-off value of 0.8 nm was used for both the Coulombic and the van der Waals interactions. The long- range Coulombic interaction was treated with the Particle- Mesh Ewald (PME) algorithm (Essmann et al., 1995). The Fourier spacing for PME was set to 0.12 nm and the cubic interpolation was also implemented. Long-range dispersion correction for energy was applied to account for the truncation of van der Waals interactions. During the minimization, the heavy atoms of the triple helix remained restrained.
The well-minimized system was then subjected to a two- stage equilibration process. In the first stage of equilibration, the system was annealed from 5 K to 300 K over 20 ps and was then equilibrated at 300 K for 30 ps in an NVT ensemble. The temperature of the system was maintained using the v-rescale thermostat with a time constant of 0.1 ps (Bussi et al., 2007). The system then underwent the second stage equilibration for 500 ps in an NPT ensemble. The pressure of the system was maintained at 1.0 bar using the isotropic Berendsen barostat with a time constant of 2.0 ps and an isothermal compressibility of 4.5xl0−5 bar−1 (Berendsen et al., 1984). The temperature was maintained at 300 K using the Nose-Hoover thermostat with a time constant of 1.0 ps (Hoover, 1985; Nose, 1984). To alleviate the “hot solvent- code solute” problem (Cheng and Merz, 1996; Lingenheil et al., 2008), separate thermostats were applied to the collagen and the solvent individually. During this two-stage equilibration, all the heavy atoms of the peptide remain restrained with a force constant of 1000 kJ/mol/nm2. The bond lengths for the peptide was constrained using the LINCS algorithm (Hess et al., 1997). The geometry of the water was constrained using the SETTLE algorithm (Miyamoto and Kollman, 1992). All the equilibrations were performed with a 2-fs time step using the leap-frog algorithm (Hockney et al., 1974). After equilibration, the 150-ns production run at 1.0 bar and 300 K was performed. During the production, only the first and last Cot atoms on each chain of the triple helix (six atoms in total) were restrained with a force constant of 10 kJ/mol/nm2.
3. Results
3.1. Design and expression of recombinant constructs with Gly mutations
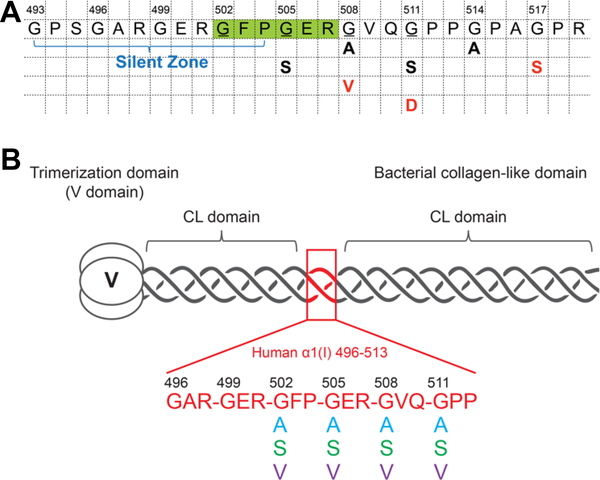
The recombinant bacterial system used here is based on the S. pyogenes Scl2 collagen-like protein, and contains an N- terminal trimerization domain (V domain) and a triple-helix domain (Gly-Xaa-Yaa)79 denoted as CL (Chhum et al., 2016; Yoshizumi et al., 2009). This VCL construct is expressed in E. coli, and the purified VCL protein forms a homotrimeric triple helix with stability similar to that of human collagens (Tm ~ 36–37°C), even though hydroxyproline is not formed in this system. A modified recombinant bacterial collagen VCL- Int was constructed with insertion of six triplets from the α1(I) chain of type I human collagen (496GAR-GER-GFP- GER-GVQ-GPP513) containing the integrin-binding site 502GFPGER507, and this homotrimeric protein showed a strong and specific binding for the integrin α2 I domain (Yigit et al., 2016). The effects of Gly to Ser substitutions on structure and cell adhesion were examined previously (Yigit et al., 2016), and here, recombinant constructs were extended to explore the impact of the identity of the residue replacing Gly on structural and functional properties of collagen. Ala, Ser and Val were selected to represent the smallest residue replacement (Ala), the most common replacement (Ser), and a replacement which generally leads to a lethal phenotype (Val). G502 and G505 were mutated within the essential integrin binding motif 502GFPGER507. Additionally, mutations at G508 and G511, C-terminal to the integrin binding site, were also studied because multiple mutations with different Gly replacement residues, resulting in OI cases of differing severity, were reported at these sites (Figure 1A).
Figure 1.
A. Reported OI fonns within and adjacent to the essential integrin binding sequence 502GFPGER507 (Galicka et al., 2003; Lindahl et al., 2015; Marini et al., 2007; Venturi et al., 2006; Wang et al., 2009; Wu et al., 2015). The essential integrin binding motif GFPGER was highlighted in green; the Gly residues studied in this work were underlined; the residue replacing Gly is shown below, together with the phenotypic severity of the OI (nonlethal OI case: black; lethal OI case: red; G505S: OI-IV; G508A: OI-III; G508V: OI-II; G511S: OI-III; G511D: OI-II; G514A: OI-III; G517S: OI-II). B. Schematic diagram of the recombinant bacterial constructs VCL-Int and Gly missense mutations, highlighting the insertion of (Gly-Xaa-Yaa)6 triplets from human α1(I) collagen chain.
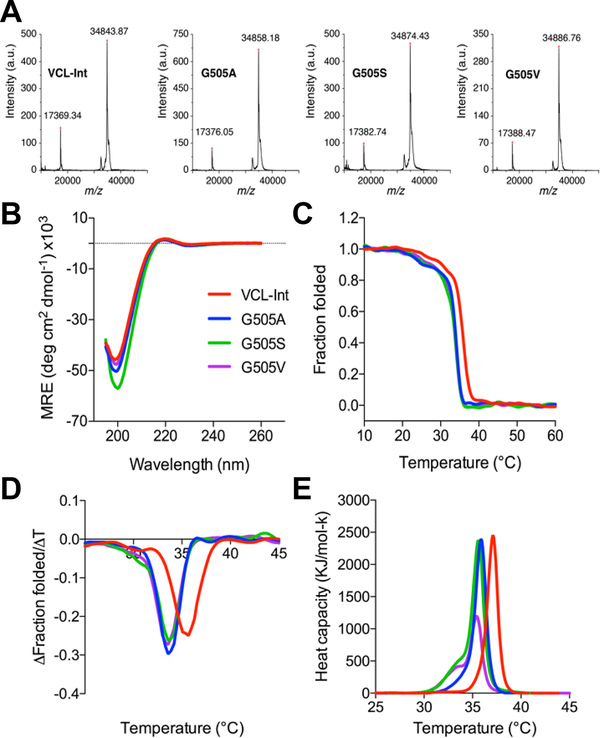
Recombinant bacterial constructs were generated where Gly at four positions within and adjacent to the integrin binding site (502, 505, 508 and 511) were replaced by Ala, Ser and Val, respectively (Figure 1B). The twelve proteins generated were denoted as G502A, G502S, G502V, G505A, G505S, G505V, G508A, G508S, G508V, G511A, G511S and G511V. The purity and identity of the proteins were confirmed by SDS-PAGE (data not shown) and mass spectrometry. Figure 2A shows the MALDI-TOF results for the wildtype VCL-Int construct, along with the G505A, G505S, and G505V mutants.
Figure 2.
Characterization of the recombinant collagen proteins with insertion of the six-triplet integrin binding sequence, and a representative example of Gly substitution at Gly505. Color code is as follows: VCL-Int (red); Gly→Ala substitution (blue); Gly→Ser substitution (green); Gly→Val substitution (purple). A. MALDI-TOF mass spectrometry of VCL-Int, G505A, G505S and G505V, showing the molecular mass of the proteins is in good agreement with that expected, a.u., arbitrary units; B. Circular dichroism (CD) wavelength spectra of the recombinant collagens showing features characteristic of triple helix formation; C. CD temperature scans showing the unfolding profiles; D. First derivative of the CD melting curves showing lower stability of all Gly505 mutants compared to the control VCL-Int protein; E. Differential scanning calorimetry (DSC) of Gly505 mutants showing heat capacity as a function of temperature.
3.2. Characterization of structure and stability of collagens with Gly substitutions
The triple-helical conformation and stability of the recombinant collagens were examined to investigate the structural consequences of the mutations. CD spectra of all proteins with Gly substitutions behaved similarly to that of the control VCL-Int, with a characteristic maximum near 220 nm and a minimum near 198 nm, indicating a triple-helical conformation (Figure 2B). The CD melting curve for the control protein VCL-Int showed a single sharp thermal transition from the trimer to monomer state, with Tm of 35.4°C (Figure 2C-D). All Gly substitutions led to a small decrease of 1.5– 2.5°C in thermal stability compared to the VCL-Int control. The thermal-transition values obtained by DSC followed the same trend (Figure 2E), with slightly higher Tm values than those measured by CD due to the faster heating rate under nonequilibrium conditions. Along with the decreased Tm values, a small shoulder at a lower temperature (30.1–30.7°C) was observed in mutant with Gly substitutions at position 502, suggesting the accumulation of a small amount of a lower-stability species (Figure S1). The DSC melting profiles for G502A, G502S and G502V mutants also exhibited a second lower-stability thermal transition, consistent with the CD results (Figure S1).
3.3. Trypsin sensitivity of collagens with Gly substitutions
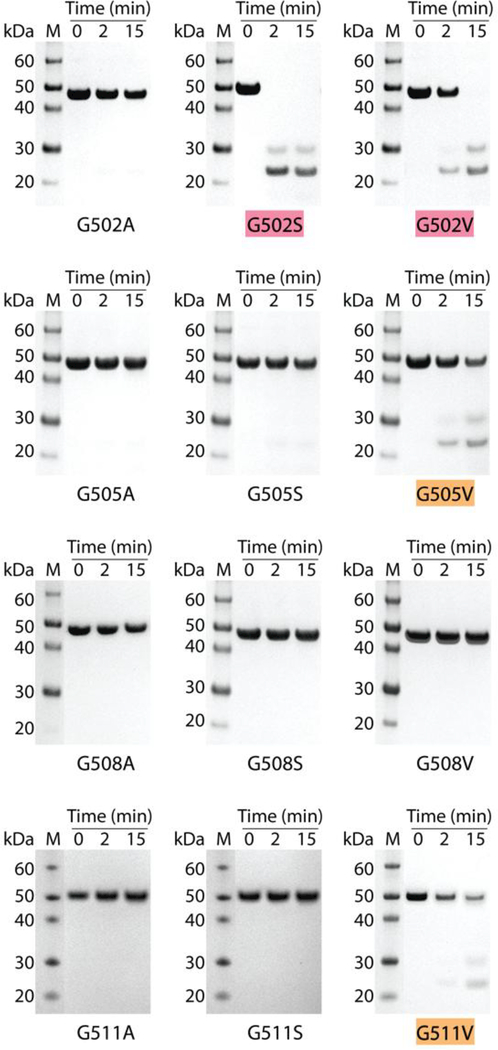
The tightly packed triple helix confers resistance to general proteases (Bruckner and Prockop, 1981; Yu et al., 2014), and the recombinant constructs with Gly missense mutants were treated with trypsin to assess disruption of the native triple helix. After a 15-min digestion at 20°C, the control protein VCL-Int remained resistant to trypsin digestion, as demonstrated by its intact size on SDS-PAGE (Figure S2A). Under the same conditions, some recombinant constructs with mutations became sensitive to trypsin, showing a dependence both on the position of the mutation and identity of the residue replacing Gly. In general, the replacement of Gly by the smaller Ala or Ser residues maintained the trypsin resistance seen for the VCL-Int control. Such resistant constructs include G502A, G505A/S, G508A/S and G511A/S. The G502S was an exception in being very susceptible to trypsin digestion and being totally degraded within 15 min (highlighted in red in Figure 3). The replacement of Gly by Val appeared to cause more disruption, leading to trypsin susceptibility for G502V, G505V and G511V, with detectable digestion after 15 min (highlighted in red/orange in Figure 3). Although G502V, G505V, and G511V were all digested by trypsin, G508V was resistant to digestion, even after an overnight incubation (Figure S2B). To confirm the unusual nature of this Val replacement, chymotrypsin digestion was also performed, and showed that G508V, like the VCL-Int control, was resistant to digestion, while G505V was susceptible (Figure S2C).
Figure 3.
Trypsin digestion of the recombinant bacterial protein with Gly to Ala, Gly to Ser and Gly to Val mutations at positions 502, 505, 508 and 511 in the 6-triplet insertion. SDS- PAGE of all recombinant collagens after trypsin digestion for time t=0, 2, and 15 min at 20°C. G502S, G502V, G505V and G511V showed various levels of trypsin susceptibility. Mutants completely degraded within 15 min were highlighted with red; mutants susceptible to trypsin but incompletely degraded within 15 min were highlighted with orange. Lane M, Novex® Sharp protein standard (Invitrogen); collagen monomer chains run slower than expected, as reported previously; molecular mass indicated in kDa.
Since the VCL-Int control is resistant to trypsin digestion, it is likely that the mutations lead to a local unwinding within the inserted human sequence, and mass spectrometry was carried out to confirm the location of the initial cleavage. Detection of tryptic digests using mass spectroscopy confirmed that trypsin cleaved at the C-terminal sides of residues Arg501 and Arg507 in G502S and G502V (Figure S2D), while hydrolysis occurred after Arg507 in G505V and G511V.
3.4. Molecular dynamics simulations of triple helices with Gly substitutions
To investigate the effect of Gly to Ser substitutions on the triple-helical structure of type I human collagen containing the integrin-binding sequence (502GFPGER507 ), computational studies were carried out for model collagen-like triple helices. Molecular dynamics (MD) simulations at 300 K were performed for the wildtype (WT) system and its mutants, which were Gly → Ala, Ser, or Val, respectively at positions 502, 505, 508, and 511, resulting in a total of twelve mutant systems (Figure S3).
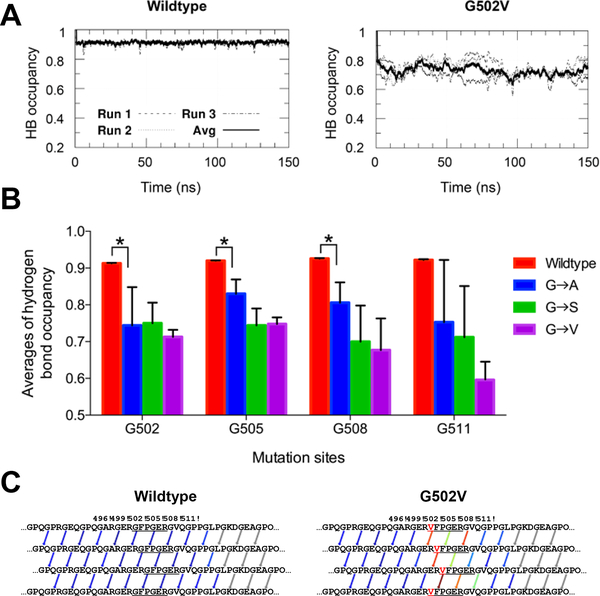
To characterize the structure stability of the collagen molecules, formation of each interchain hydrogen bond was monitored during the MD simulations. A hydrogen bond was considered formed if both the N···O distance was smaller than A and the H-N···O angle was less than 30°. To further quantify the effects of Gly substitutions, the occupancies of the interchain hydrogen bonds that are within five triplets N- terminal and four triplets C-terminal to the substitution site in the wildtype and mutant triple helices were averaged. Average hydrogen bond occupancy over time was first plotted to determine when all systems were equilibrated, indicated by relatively leveled hydrogen bond occupancy and occurred in the last 50 ns of all the MD runs (Figure 4A). Consequently, the occupancies for each interchain hydrogen bond were averaged over the last 50 ns of simulation time. The wildtype system showed a high average hydrogen bond occupancy of above 0.90, indicating a stable triple helix (Figure 4B). In the mutant systems, all three substitutions (Gly to Ala, Ser or Val) resulted in broken interchain hydrogen bonds localized near the mutation, indicated by the significantly lower average hydrogen bond occupancy. Visualization of HB patterns indicated that the hydrogen bonds near the mutation site were weaker in the representative mutant G502V (Figure 4C). The large error bars for the average hydrogen bond occupancies of the systems are likely reflective of the small number of MD runs performed for each system. As a result, statistical analysis is quite difficult to perform for such systems.
Figure 4.
A. Graph depicting average hydrogen bond occupancy over time for wildtype and G502V model collagen-like triple helices. A hydrogen bond is defined as donor-acceptor distance ≤ A and hydrogen-donor-acceptor angle ≤ 30°. B. Averages of hydrogen bond occupancy of the last 50 ns of the three production runs of each system. Sample standard deviation of the three runs was used to calculate error bars. The significance level was determined by p value using paired Student’s t test between the means of two samples. Statistically significant levels were marked with asterisks: *, P ≤ 0.05. C. Visualization of the hydrogen bond forming/breaking pattern of the wildtype and G502V mutant with hydrogen bonds colored based on their occupancies, ranging from red (no hydrogen bond formed) to blue (hydrogen bond formed the whole time in the production time. Hydrogen bond occupancies are averaged over the last 50 ns of production time.
3.5. Integrin binding to recombinant constructs with Gly substitutions
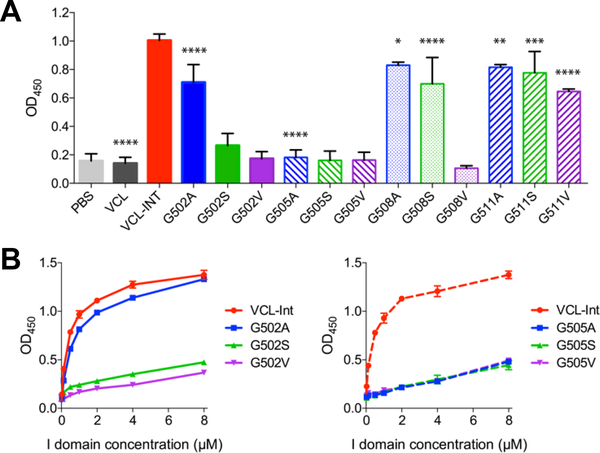
Solid-state binding assays were performed to investigate if the identity of residue replacing Gly within/in the C-terminal region of the essential integrin binding 502GFPGER507 sequence affected its specific binding affinity to integrin. As expected, the original VCL construct without the GFPGER insertion did not bind the integrin I domain, whereas the positive control VCL-Int containing the integrin binding motif GFPGER showed a high affinity to the I domain (Figure 5A), consistent with the previous work (Yigit et al., 2016). As assessed at a fixed integrin concentration (c=20 μg/ml), an obvious decrease in the binding signals for constructs G502A and G502S was observed when compared to the control VCL-Int, while G502V showed essentially no binding. In contrast, all Gly substitutions at position 505 showed essentially no binding. Gly replacement C-terminal to the 502GFPGER507 motif at G508 and G511 showed a noticeable decrease in binding for G508V.
Figure 5.
Solid-state binding assay of integrin α2 I domain binding to all recombinant bacterial collagens. A. VCL-Int and all Gly substitution constructs were immobilized on 96-well microplates and incubated with 20 μg/ml (0.4 μM) of I domain. Bound proteins were detected with anti-GST HRP antibodies and measured as absorbance at 450 mn. Experiments were carried out in triplicates, with results based on the averages of data points and standard deviations presented as error bars. The significance level was determined by p value using paired Student’s t test between the means of VCL-Int and other samples. Statistically significant levels were marked with asterisks: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. B. Dose response of I domain to recombinant collagens with Gly substitution at Gly-502 and Gly- 505 positions adsorbed onto 96-well microplates. To cover all samples on the same 96-well microplate, the experiments for the dose-response curves consisted of duplicate or triplicate measurements and two independent dose response curves of I domain to VCL-Int were performed.
Due to the importance of 202GFPGER507 sequence in the interaction with the integrin I domain (Emsley et al., 2000), G502 and G505 mutants were selected for further analysis of integrin-recombinant collagen interaction by assessing dose- response curves over a range of integrin concentrations (Figure 5B). These titration curves confirmed the trend seen for binding at c=20 μg/ml (0.4 μM) in Figure 5A. For example, the left panel of Figure 5B shows that compared to VCL-Int, G502A showed a slight decrease in binding and G502S exhibited a marked loss of binding affinity, while the binding activity was totally abolished when Gly502 was replaced with Val. In contrast, the right panel of Figure 5B shows that no binding of integrin was detected to a recombinant protein with any Gly substitution at position 505 at all integrin concentrations, pointing to the essential nature of Gly505. Thus, the results obtained at one fixed integrin concentration is a reasonable measure of relative affinity. It is not possible to calculate Km binding values because of the qualitative nature of solid-state binding assays due to the high non-linearity of the detection signals in solid-state binding assays (Tangemann and Engel, 1995).
4. Discussion
Analysis of the OI database shows a general correlation between the identity of the residue replacing Gly and the degree of clinical severity, yet the way in which the identity of a specific residue leads to different OI pathology is still not well understood. Different approaches have been developed to relate the mutations in the collagen chain with clinical outcomes. Direct studies on OI collagens have been challenging because missense mutations within heterotrimeric type I collagen are linked to the dominant form of OI and result in a mixture of molecular species that are hard to separate (e.g. a Gly substitution in the α1(I) chain will lead to 25% normal trimers, 50% molecules containing one mutant chain and 25% molecules with two mutant chains). In this work, in vitro studies on a recombinant system were used to assess the effect of the identity of the residue replacing Gly on several factors, including global thermal stability, local triple-helix disruptions, and integrin binding, as well as interchain hydrogen bonding through MD simulations. A major advantage of the recombinant bacterial system is the production of pure well defined homotrimers useful for structural and functional studies.
4.1. Effect of mutation on collagen stability
Previous studies on relatively short collagen-like peptides indicated the Gly substitutions dramatically reduced triple- helix global stability, with the degree of disruption dependent on the residue replacing Gly (Beck et al., 2000; Persikov et al., 2004). In contrast, examination of 41 fibroblast OI collagens with Gly missense mutations showed only a small decrease in melting temperature compared with controls (0– 4.6°C). No correlation was seen between the identity of the substituting residue and triple helix destabilization, but the amount of the decrease in Tm showed regional variations (Makareeva et al., 2008). As seen here, the VCL recombinant bacterial proteins are similar to OI collagens in exhibiting a small but reproducible decrease in Tm, regardless of the identity of the residue replacing Gly. Mutations at four adjacent positions (Gly502, 505, 508 and 511) studied here had similar Tm decreases, consistent with Makareeva’s regional hypothesis (Makareeva et al., 2008).
In contrast to the global stability probed by CD melting curves and DSC profiles, trypsin digestion probes the presence of any local disruptions in the triple helix. Using a short digestion time (15 min), three Gly to Val constructs (at G502, G505 and G511) and one Gly to Ser construct at Gly502 became highly susceptible to trypsin, while after overnight trypsin treatment, most mutants were degraded, regardless of the position and identity of the mutation. These results suggest that replacement of Gly by the large Val residue causes a significant local unfolding of the triple helix in general, but the degree of trypsin sensitivity is modulated by the local sequence environment of the Gly, as well as its position relative to a susceptible Arg/Lys residue. The trypsin and chymo- trypsin resistance of the construct where Gly508 in the VG508VQ triplet was replaced by Val (RV508VQ) was unusual, and it is possible that the hydrophobic core formed by the two adjacent Val residues could make Arg507 inaccessible to trypsin or chymotrypsin.
MD simulations showed that Gly→Ala, Gly→ Ser and Gly-→Val substitutions significantly reduced the average occupancy of the interchain hydrogen bonds in the collagen triple helix, with the majority of the hydrogen bonds disrupted in immediate proximity to the mutation site. Furthermore, the hydrogen bonds were broken largely C-terminal to the mutation. Destabilized triplets that are C-terminal to OI mutations have been associated with some lethal OI phenotypes (Bodian et al., 2008), so this asymmetry in triple-helix disruption relative to the mutation may play a role in clinical severity. Both identity and location of the mutation did not have a deterministic effect on the influence of broken hydrogen bonds, as there was no statistically significant difference in the average hydrogen bond occupancy between Ala, Ser, and Val substitutions at any locations (Figure 4B). The similar disruptive effects of all mutations are consistent with the almost complete cleavage of all mutant constructs, except for G508V, after an overnight trypsin digestion (Figure S2B), but do not reflect the distinctions observed between residues after a short trypsin digest; it is possible that an increased number of longer MD runs might enable better statistics and better model the different behaviors with shorter trypsin digests.
In agreement with the studies of short peptides (Beck et al., 2000), previous computational studies using model collagen- like (Gly-Pro-Hyp)n sequences, in which a Gly in the middle GPO triplet was mutated, suggested that different residue replacements resulted in various changes in free energy (Lee et al., 2011) and in a decreased Young’s modulus of the triple helix (Gautieri et al., 2009). In our MD simulations, even the large Val residue did not lead to a significantly larger hydrogen bond disruption than Ala or Ser. This discrepancy may relate to the use of a real human collagen sequence in this study, compared with a (Gly-Pro-Hyp)n sequence in the previous reports.
4.2. Effect of mutation on integrin binding
Collagen-integrin interactions play important roles in fibril assembly, remodeling and angiogenesis, as well as cell growth, adhesion, migration and differentiation (Marini et al., 2007). The crystal structure of the GFOGER peptide-integrin α2 I domain complex, shows that the Glu residue within the GER triplet coordinates the metal in the I domain’s MIDAS motif directly and the Arg residue makes a salt bridge to D219 of the I domain (Emsley et al., 2000). The aromatic ring of the Phe residue is also seen to bind to a hydrophobic pocket in the I domain (Emsley et al., 2000). It has been suggested that Gly missense mutations within ligand binding regions of collagen may interfere with collagen interactions and lead to the dominant form of OI (Forlino and Marini, 2016). Here we found that the decrease in integrin binding affinity showed a clear correlation with the identity of the residue replacing Gly at the site G502, following the order G-→V > G-→S > G-→A. Any changes to G505 completely abolished integrin binding, regardless of the identity of the replacement residues, suggesting the essential role of G505 in integrin binding.
4.3. Correlation between identity of the residue replacing Gly and OI clinical severity
The in vitro and in silico studies lend insight into possible factors related to observed OI cases at the two Gly positions in the essential GFPGER sequence. (1) Gly502: There are no reported OI mutations at position Gly502; it falls into a “silent zone” of four triplets with no OI cases. Since Gly502S and Gly502V show a strikingly high sensitivity to trypsin, it is possible that any mutation would lead to general proteolytic susceptibility and early embryonic lethality, as proposed previously (Chhum et al., 2016). (2) Gly505: Our in vitro studies indicate that any mutation at this position completely eliminates integrin binding in our homotrimer recombinant constructs. Yet there is a Gly to Ser mutation reported at this position leading to moderate OI (OI IV) (Venturi et al., 2006). The impact of a Gly substitution in all three chains on integrin binding could be greater than that of the same substitution in just one or two chains. In addition, the α1ß1, α10ß1 and α11ß1 integrins are also known to recognize the triple- helical GFOGER sequence (Barczyk et al., 2010), and the presence of other GxOGER integrin binding motifs in fibrillar collagen may mitigate the consequences of losing a functional GFOGER (Hamaia and Famdale, 2014). The Gly residues in triplets immediately C-terminal to the GFOGER site, G508 and G511, are both sites of multiple OI mutations that result in varying clinical severity (Figure 1A). Our results show that the consequences of Gly replacement by the large Val residue differ from replacement by the smaller Ala or Ser residues; however, it remains unclear still how these perturbations to binding or local triple-helix structure relate to clinical consequences.
Our results define the effects of replacing Gly by different residues at four nearby positions within the collagen triple helix using purified recombinant molecules. Limitations of this system include its formation of homotrimers with mutations in all 3 chains, in contrast to the mutations in 1 or 2 chains found in OI type I collagen, and the possibility that recombinant integrin I domain binding to VCL protein on a plate may differ from physiological integrin-collagen interactions. The effects reported here of mutations leading to proteolysis and interfering with binding to integrin illustrate the need to look for a subtle and complex interplay of factors in understanding the clinical severity for OI.
Supplementary Material
Highlights:
The severity of the bone disorder Osteogenesis Imperfecta is influenced by the specific residue which replaces Gly in the repeating (Gly-Xaa-Yaa)n tripeptide sequence of type I collagen. A recombinant bacterial collagen system and molecular dynamics simulations were applied here to investigate the influence of Ala, Ser and Val replacing Gly within the integrin binding collagen region. The degree of local distortion of the triplehelix and the disruption of integrin binding were affected by the replacement residue identity at some but not all sites, and the implications for collagen diseases are discussed.
Acknowledgements
This work was supported by NIH grants #EB011620 (to BB and DLK) and #GM60048 (to BB) and by the Tufts start-up fund (to Y.-S. L.). We thank Dr. David Wilbur from Tufts University Chemistry Department for allowing us to access the MALDI-TOF MS equipment. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the NIH.
Abbreviations
- OI
osteogenesis imperfecta
- MD
molecular dynamics
- HB
hydrogen bonding
- CL
collagen-like
- Tm
melting temperature
- CD
circular dichroism
- DSC
Differential Scanning Calorimetry
- MRE
mean residue ellipticity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version.
The Sillence classification of the phenotypic severity of the OI: OI type II, perinatal lethal; OI type III, severe; OI type IV, moderate; OI type I, mild) (Marini et al., 2007)
References
- An B, Kaplan DL, Brodsky B, 2014. Engineered recombinant bacterial collagen as an alternative collagen-based biomaterial for tissue engineering. Frontiers in Chemistry 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Abbonante V, Xu H, Gavriilidou D, Yoshizumi A, Bihan D, Famdale RW, Kaplan DL, Balduini A, Leitinger B, Brodsky B, 2016. Recombinant collagen engineered to bind to discoidin domain receptor functions as a receptor inhibitor. Journal of Biological Chemistry 291, 4343–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Natale DA, O'Donovan C, Redaschi N, Yeh LS, 2004. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32, D115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczyk M, Carracedo S, Gullberg D, 2010. Integrins. Cell Tissue Res 339, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JA, Brodsky B, 2000. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc Natl Acad Sci U S A 97, 4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bella J, Eaton M, Brodsky B, Berman HM, 1994. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science 266, 75–81. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J, 1981. Interaction Models for Water in Relation to Protein Hydration, p. 331–342, in: Pullman B, (Ed.), Intermolecular Forces: Proceedings of the Fourteenth Jerusalem Symposium on Quantum Chemistry and Biochemistry Held in Jerusalem, Israel, April 13–16, 1981, Springer Netherlands, Dordrecht. [Google Scholar]
- Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR, 1984. Molecular-Dynamics with Coupling to an External Bath. Journal of Chemical Physics 81, 3684–3690. [Google Scholar]
- Bodian DE, Madhan B, Brodsky B, Klein TE, 2008. Predicting the clinical lethality of osteogenesis imperfecta from collagen glycine mutations. Biochemistry 47, 5424–5432. [DOI] [PubMed] [Google Scholar]
- Brodsky B, Persikov AV, 2005. Molecular structure of the collagen triple helix. Adv Protein Chem 70, 301–339. [DOI] [PubMed] [Google Scholar]
- Bruckner P, Prockop DJ, 1981. Proteolytic enzymes as probes for the triple-helical conformation of procollagen. Anal Biochem 110, 360–368. [DOI] [PubMed] [Google Scholar]
- Bryan MA, Cheng H, Brodsky B, 2011. Sequence environment of mutation affects stability and folding in collagen model peptides of osteogenesis imperfecta. Biopolymers 96, 4–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M, 2007. Canonical sampling through velocity rescaling. J Chem Phys 126, 014101. [DOI] [PubMed] [Google Scholar]
- Cheng AE, Merz KM, 1996. Application of the Nose-Hoover chain algorithm to the study of protein dynamics. Journal of Physical Chemistry 100, 1927–1937. [Google Scholar]
- Chhum P, Yu H, An B, Doyon BR, Fin Y-S, Brodsky B, 2016. Consequences of Glycine mutations in the fibronectin binding sequence of collagen. J. Biol. Chem 291, 27073–27086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley J, Knight CG, Famdale RW, Barnes MJ, Fiddington RC, 2000. Structural basis of collagen recognition by integrin α2β1. Cell 101,47–56. [DOI] [PubMed] [Google Scholar]
- Essmann U, Perera F, Berkowitz MF, Darden T, Lee H, Pedersen FG, 1995. A Smooth Particle Mesh Ewald Method. J Chem Phys 103, 8577–8593. [Google Scholar]
- Forlino A, Marini JC, 2016. Osteogenesis imperfecta. The Fancet 387, 1657–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlino A, Kuznetsova NV, Marini JC, Feikin S, 2007. Selective retention and degradation of molecules with a single mutant α1(I) chain in the Brtl IV mouse model of OI. Matrix Biology 26, 604–614. [DOI] [PubMed] [Google Scholar]
- Galicka A, Wolczynski S, Gindzienski A, Surazynski A, Palka J, 2003. Gly511 to Ser substitution in the COF1A1 gene in osteogenesis imperfecta type III patient with increased turnover of collagen. Mol Cell Biochem 248, 49–56. [DOI] [PubMed] [Google Scholar]
- Gautieri A, Vesentini S, Redaelli A, Buehler MJ, 2009. Single molecule effects of osteogenesis imperfecta mutations in tropocollagen protein domains. Protein Sci 18, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaia S, Famdale RW, 2014. Integrin recognition motifs in the human collagens. Adv Exp Med Biol 819, 127–142. [DOI] [PubMed] [Google Scholar]
- Hess B, Bekker H, Berendsen HJC, Fraaije JGEM, 1997. FINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry 18, 1463–1472. [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Findahl E, 2008. GROMACS 4: Algorithms for Highly Efficient, Foad-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput 4, 435–447. [DOI] [PubMed] [Google Scholar]
- Hockney RW, Goel SP, Eastwood JW, 1974. Quiet high-resolution computer models of a plasma. Journal of Computational Physics 14, 148–158. [Google Scholar]
- Hoover WG, 1985. Canonical Dynamics - Equilibrium Phase-Space Distributions. Phys Rev A 31, 1695–1697. [DOI] [PubMed] [Google Scholar]
- Knight CG, Morton EF, Peachey AR, Tuckwell DS, Famdale RW, Bames MJ, 2000. The Collagen-binding A-domains of Integrins α1β1 and α2β1 Recognize the Same Specific Amino Acid Sequence, GFOGER, in Native (Triple-helical) Collagens. Journal of Biological Chemistry 275, 35–40. [DOI] [PubMed] [Google Scholar]
- Lee KH, Kuczera K, Holl MMB, 2011. The Severity of Osteogenesis Imperfecta: A Comparison to the Relative Free Energy Differences of Collagen Model Peptides. Biopolymers 95, 182–193. [DOI] [PubMed] [Google Scholar]
- Lightfoot SJ, Atkinson MS, Murphy G, Byers PH, Kadler KE, 1994. Substitution of serine for glycine 883 in the triple helix of the pro alpha 1 (I) chain of type I procollagen produces osteogenesis imperfecta type IV and introduces a structural change in the triple helix that does not alter cleavage of the molecule by procollagen N- proteinase. Journal of Biological Chemistry 269, 30352–30357. [PubMed] [Google Scholar]
- Lightfoot SJ, Holmes DF, Brass A, Grant ME, Byers PH, Kadler KE, 1992. Type I procollagens containing substitutions of aspartate, arginine, and cysteine for glycine in the pro alpha 1 (I) chain are cleaved slowly by N-proteinase, but only the cysteine substitution introduces a kink in the molecule. J Biol Chem 267, 25521–25528. [PubMed] [Google Scholar]
- Lindahl K, Astrom E, Rubin CJ, Grigelioniene G, Malmgren B, Fjunggren O, Kindmark A, 2015. Genetic epidemiology, prevalence, and genotype-phenotype correlations in the Swedish population with osteogenesis imperfecta. Eur J Hum Genet 23, 1042–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingenheil M, Denschlag R, Reichold R, Tavan P, 2008. The “hot- solvent/cold-solute” problem revisited. J. Chem. Theory. Comput. 4, 1293–1306. [DOI] [PubMed] [Google Scholar]
- Makareeva E, Mertz EL, Kuznetsova NV, Sutter MB, DeRidder AM, Cabral WA, Bames AM, McBride DJ, Marini JC, Leikin S, 2008. Structural heterogeneity of type I collagen triple helix and its role in osteogenesis imperfecta. J Biol Chem 283, 4787–4798. [DOI] [PubMed] [Google Scholar]
- Marini JC, Forlino A, Cabral WA, Bames AM, San Antonio JD, Milgrom S, Hyland JC, Korkko J, Prockop DJ, De Paepe A, Coucke P, Symoens S, Glorieux FH, Roughley PJ, Lund AM, Kuurila-Svahn K, Hartikka H, Cohn DH, Krakow D, Mottes M, Schwarze U, Chen D, Yang K, Kuslich C, Troendle J, Dalgleish R, Byers PH, 2007. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum Mutat 28, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Kollman PA, 1992. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. Journal of Computational Chemistry 13, 952–962. [Google Scholar]
- Nosé S, 1984. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52, 255–268. [Google Scholar]
- Persikov AV, Pillitteri RJ, Amin P, Schwarze U, Byers PH, Brodsky B, 2004. Stability related bias in residues replacing glycines within the collagen triple helix (Gly-Xaa-Yaa) in inherited connective tissue disorders. Hum Mutat 24, 330–337. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, 2004. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Raghunath M, Bruckner P, Steinmann B, 1994. Delayed triple helix formation of mutant collagen from patients with osteogenesis imperfecta. Journal of Molecular Biology 236, 940–949. [DOI] [PubMed] [Google Scholar]
- Rainey JK, Goh MC, 2004. An interactive triple-helical collagen builder. Bioinformatics (Oxford, England) 20, 2458–2459. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Kartha G, 1955. Structure of collagen. Nature 176, 593–595. [DOI] [PubMed] [Google Scholar]
- Rich A, Crick FH, 1961. The molecular structure of collagen. J Mol Biol 3, 483–506. [DOI] [PubMed] [Google Scholar]
- Royce PM, Steinmann B, 2003. Connective tissue and its heritable disorders: molecular, genetic, and medical aspects John Wiley & Sons. [Google Scholar]
- Schmid N, Eichenberger AP, Choutko A, Riniker S, Winger M, Mark AE, van Gunsteren WF, 2011. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur Biophys J 40, 843–856. [DOI] [PubMed] [Google Scholar]
- Tangemann K, Engel J, 1995. Demonstration of non-linear detection in ELISA resulting in up to 1000-fold too high affinities of fibronogen binding to integrin αIIbβ3. FEBS Letters 358, 179–181. [DOI] [PubMed] [Google Scholar]
- Tuckwell D, Calderwood DA, Green LJ, Humphries MJ, 1995. Integrin α2 I-domain is a binding site for collagens. Journal of Cell Science 108, 1629–1637. [DOI] [PubMed] [Google Scholar]
- Venturi G, Tedeschi E, Mottes M, Valli M, Camilot M, Viglio S, Antoniazzi F, Tato L, 2006. Osteogenesis imperfecta: clinical, biochemical and molecular findings. Clinical Genetics 70, 131–139. [DOI] [PubMed] [Google Scholar]
- Wang Z, Yang Z, Ke Z, Yang S, Shi H, Wang L, 2009. Mutations in COL1A1 of type I collagen genes in Chinese patients with osteogenesis imperfecta. J Investig Med 57, 662–667. [DOI] [PubMed] [Google Scholar]
- Wu Q, Wang W, Cao L, Sun L, Xu Y, Zhong X, 2015. Diagnosis of fetal osteogenesis imperfecta by multidisciplinary assessment: a retrospective study of 10 cases. Fetal Pediatr Pathol 34, 57–64. [DOI] [PubMed] [Google Scholar]
- Yigit S, Yu H, An B, Hamaia S, Famdale RW, Kaplan DL, Lin Y-S, Brodsky B, 2016. Mapping the effect of Gly mutations in collagen on α2β1integrin binding. J. Biol. Chem. 291, 19196–19207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizumi A, Yu Z, Silva T, Thiagarajan G, Ramshaw JA, Inouye M, Brodsky B, 2009. Self-association of streptococcus pyogenes collagen-like constructs into higher order structures. Protein Sci 18, 1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, An B, Ramshaw JAM, Brodsky B, 2014. Bacterial collagenlike proteins that form triple-helical structures. Journal of Structural Biology 186, 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.