Abstract
Genome-wide discovery efforts have identified more than 500 genetic loci associated with adiposity traits. The vast majority of these loci were found through large-scale meta-analyses for body mass index (BMI) and waist-to-hip ratio (WHR), and in European ancestry populations. However, alternative approaches, focusing on non-European ancestry populations, more refined adiposity measures, and low-frequency (minor allele frequency (MAF) < 5%) coding variants, identified additional novel loci which had not been identified before. Loci associated with overall obesity implicate pathways that act in the brain, whereas loci associated with fat distribution point to pathways involved in adipocyte biology. Pinpointing the causal gene within each locus remains challenging, but is a critical step towards translation of genome-wide association study (GWAS) loci into new biology. Ultimately, new genes may provide pharmacological targets for the development of weight loss drugs.
Introduction
Obesity is a major risk factor of disease, not only posing an enormous burden on people’s personal health [1], but also on societies as a whole [2,3]. Over the past four decades, the prevalence of obesity among adults has nearly quadrupled worldwide [4,5]. While in most high-income countries the rise in BMI seems to have slowed down as of late, albeit at a high level, in many low- and middle-income countries the increase continues. Particularly alarming is the global rise in obesity among children and adolescents [4–6].
Initiatives to prevent obesity or promote weight loss through lifestyle changes have limited success and are often short-lived, both at the community and individual levels [7,8], suggesting that innate mechanisms, encoded by the genome, also contribute to energy homeostasis [9]. Estimates of genetic contribution vary by study design and adiposity outcome, but are sufficiently high to warrant gene discovery studies (Table 1).
Table 1.
Heritability estimates for body mass index (BMI) and waist-to-hip ratio (WHR), by study design
In the past 10 years, genome-wide association studies (GWASs) have been particularly effective in the identification of genetic loci associated with adiposity outcomes. However, translation of these loci into new biology has been challenging. Here, I review recent progress and insights gained from these discoveries.
Conventional GWAS – Common (MAF ≥ 5%) variants for commonly studied adiposity phenotypes
In 2007, GWASs discovered the first genetic locus in FTO that showed robust association with BMI and obesity risk [10,11]. More than 500 genetic loci, for a range of adiposity traits, have since been identified (Figure 1). The vast majority of these (92%) were first identified for body mass index (BMI; n=341 loci), a proxy for overall adiposity, and for BMI-adjusted waist-to-hip ratio (WHRadjBMI; N=129), a proxy for body fat distribution. Because data on BMI and WHR are easily obtained, sample sizes have grown rapidly, resulting in a steep increase of new discoveries over the past 10 years. For example, the most recent GWAS meta-analyses by the GIANT (Genetic Investigation of Anthropometric Traits) Consortium included data from 339,224 individuals and 125 GWAS studies on BMI [12] and 210,088 individuals from 101 studies on WHRadjBMI [13]. In the latest GWAS for BMI, data from the Biobank Japan Project (N=173,430) was combined with the BMI summary statistics from the GIANT Consortium [14] for a total sample size of >512,000 individuals [15].
Figure 1.
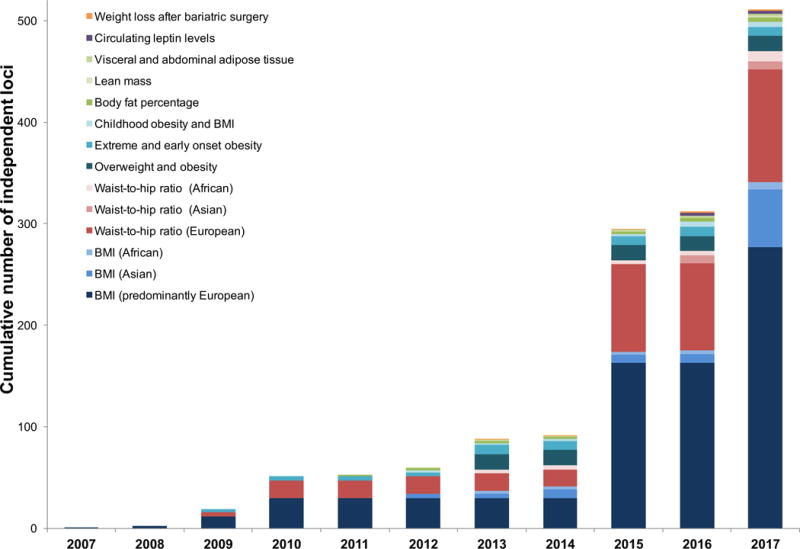
Cumulative number of loci identified since 2007. Color coding and shading corresponds to adiposity trait (and population ancestry) for which the initial discovery was made.
More than 80% of loci were first identified in populations that were exclusively or predominantly of European ancestry. However, despite much smaller sample sizes, GWASs of exclusively Asian or African ancestry populations have identified at least 64 loci for BMI and 18 for WHR that had not been identified in much larger European ancestry GWASs [12,13,15,16]. For most loci, associations are directionally consistent across ancestries, but allele frequencies and/or effect sizes may differ.
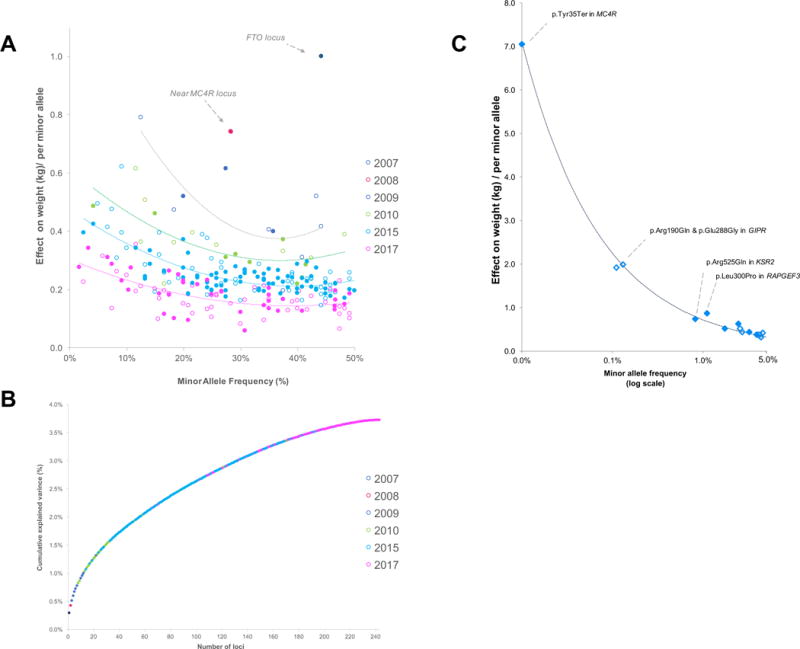
Loci discovered in the earliest, and thus smallest, meta-analyses tend to have the largest (albeit modest) effect sizes (Figure 2). As sample sizes increase with each new meta-analysis, the power to identify variants with smaller effect sizes and/or lower minor allele frequencies (MAFs) increases and the variance explained by each new locus becomes incremental (Figure 2). Current GWAS-identified loci combined explain ~4% of the phenotypic variation of BMI. For WHRadjBMI, effect sizes and variance explained tend to be larger for women (~2.7%) than for men (~1.4%).
Figure 2.
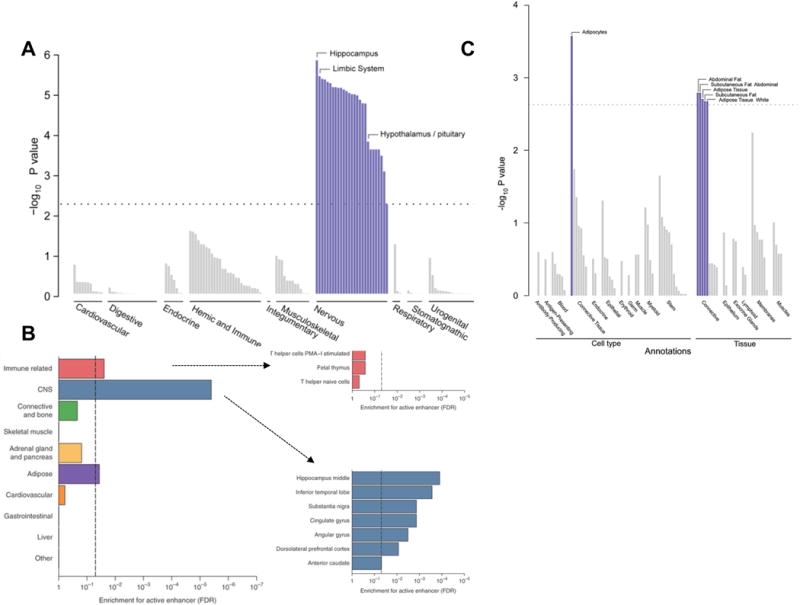
Tissue expression of genes at BMI-associated loci (Panel A) and WHRadjBMI-associated loci (Panel C) [Adapted from Locke et al. [12] and Shungin et al. [13], respectively). Enrichment of BMI-associated variants included in the 99% credible sets for active enhancer in 10 cell groups (Panel B) [Adapted from Akiyama et al. [15]].
GWASs have been successful in identifying numerous novel adiposity loci, but the ultimate goal is to elucidate the biology that these loci represent. Gene set, tissue, and functional enrichment analyses based on BMI-associated loci have implicated the central nervous system (CNS) as a key organ in the regulation of energy balance, highlighting not only the hypothalamus and pituitary gland (known appetite regulation sites), but also the hippocampus and limbic system (involved in learning, cognition, emotion and memory) [9,12,15,17] (Figure 3). Analyses that include the most recent BMI-associated loci have also provided support for a role of immune-related cells (lymphocytes, B cells) in the etiology of obesity [12,15]. Similar analyses based on the WHRadjBMI-associated loci have revealed a different biology, implicating adipogenesis, angiogenesis, and insulin resistance as processes affecting fat distribution [9,13].
Figure 3.
Effect size (in kg, assuming a 1.7m tall person) by MAF for GWAS-identified BMI loci (Panel A) and their cumulative explained variance (Panel B). Color coding corresponds with year of discovery and dotted line represent trends of effect sizes by year. Effect size (in kg, assuming a 1.7m tall person) by MAF for low-frequency and rare exomechip-identified loci (Panel C) [Adapted from Turcot et al. [42]]. Filled markers indicate that the minor allele is associated with higher BMI, and unfilled markers indicate that the minor allele is associated with lower BMI. The dotted line represents 80% power.
While some of these pathways overlap with the broad biology already established by human and animal models of extreme obesity and fat distribution, GWAS loci reveal genes that have not previously been implicated in known and novel pathways. Pinpointing the causal gene/variant in loci remains a major challenge. For example, over the past 10 years, the FTO locus has been studied in great depth [18], but the mechanisms through which it affects body weight are still not fully understood. Recent studies suggest that FTO’s BMI-associated variants mediate their effect not (only) through FTO, but also by influencing nearby genes. A study that used an extensive battery of tests, including epigenomic analyses, allelic activity, motif conservation, regulator expression, and gene coexpression patterns, suggested that rs1421085 in FTO is the causal variant that disrupts the ARID5B-mediated repression of IRX3 and IRX5 in preadipocytes, thereby suppressing white adipocyte browning, reducing thermogenesis and promoting lipid storage in a brain-independent way [19]. However, an earlier study, which used chromosome conformation capture, evolutionary conservation, tissue-specific gene-expression and a transgenic mouse model also proposed a role for IRX3 in the regulation of energy homeostasis and body composition, but through hypothalamic-mediated pathway [20]. Others found that the BMI-increasing alleles of FTO’s rs1421085/rs8050136 affect the binding affinity of transcription factor CUX1, suppressing the neuronal expression of FTO and nearby RPGRIP1L, which results in increased food intake and adiposity in mice [21,22]. Thus, current studies suggest that multiple pathways in multiple tissues may link the FTO-locus and body weight. Slowly, more GWAS-identified loci are undergoing in-depth analyses to elucidate their biology (TMEM18 [23,24], CADM2 [25,26], LYPLAL1 [27], ADCY3 [28]), but many more are waiting to be scrutinized.
Alternative approaches for gene discovery
Conventional GWAS, described above, have been very fruitful, not only because of the large sample, but also because studied variants are common (MAF>5%); both are contributors to increased statistical power for discovery. However, several other approaches have identified new loci through leveraging specific phenotypic and genotypic features.
Refined adiposity phenotypes
While BMI and WHR are easily obtained phenotypes, they are rather crude and heterogeneous indices to capture adiposity; e.g. individuals with the same BMI may still differ substantially in body composition and fat distribution. Therefore, recent gene discovery efforts have focused on more refined adiposity phenotypes, such as body fat percentage (BFP), lean mass, adipose tissue depots, and circulating leptin level, and have identified several new loci that point to new aspects of biology.
The most recent GWAS meta-analysis for BFP (Nmax~100,000) identified five novel loci that had not been reported by much larger GWAS for BMI or WHRadjBMI [29]. Most notable is a locus near IRS1, of which the BFP-increasing allele protects against type 2 diabetes and cardiovascular disease–an unexpected association mediated through an effect on fat deposition [29]. Specifically, the BFP-increasing allele favors fat deposition in subcutaneous fat depots, but not the metabolically harmful visceral depots [30]. These findings mirror Irs1 knockout mice that are lean but insulin resistant [31,32], and whose cell lines suggest a role in adipocyte differentiation [33,34]. Several other BFP-associated loci (in/near COBLL1, TOMM40, PLA2G6) stand out because of cross-trait associations similar to the near-IRS1 locus [29].
A GWAS meta-analysis of lean mass (Nmax~100,000) identified two novel loci; both of which are missense variants (p.Gly428Asp in VCAN, p.Gln283Arg in HSD17B11) in genes previously implicated in musculoskeletal health [35].
To more accurately assess fat distribution, a GWAS meta-analysis was performed on various adipose tissue depots, quantified using computed tomography and magnetic resonance imaging [36]. Despite a relatively small sample size (Nmax~18,000), seven new loci associated with various ectopic-adiposity traits were identified [36]. Functional analyses in mice showed that two loci (ATXN1, UBE2E2) play a role in adipogenesis [36].
Leptin is a hormone, predominantly secreted by adipocytes that plays a key role in food intake and energy-balance. A GWAS meta-analysis (Nmax=52,126) identified four loci (in/near LEP, SLC32A1, GCKR, CCNL1) associated with circulating leptin levels [37]. Functional follow-up of candidate genes in each locus, using an adipose tissue explant model in mice, showed that knockdown of Adig, located near Slc32a1, influences leptin release and secretion [37]. ADIG encodes adipogenin, known to be a potent regulator of adipogenesis [38,39], plays possibly also a role in leptin regulation [37].
Taken together, GWAS meta-analyses for more refined adiposity traits have identified loci that were not revealed in much larger BMI and WHRadjBMI discovery efforts. Furthermore, the interpretation of these loci may have been facilitated by the fact that phenotypes were less heterogeneous and more closely related to adiposity biology.
Low frequency (MAF:1<5%) and rare (MAF<1%) (coding) variants
GWAS-identified loci are typically common, non-coding and often intergenic, mainly because discovery has been limited by array design and available imputation reference panels. To further characterize the genetic architecture of adiposity traits, recent gene-discovery efforts have focused on low-frequency and rare variants. It had been speculated that such variants have larger effects and can therefore be identified with smaller sample sizes. However, recent whole genome and whole exome sequence based efforts did not identify novel low-frequency variants for adiposity traits [40,41], suggesting that their effects may not be as pronounced as expected and that larger sample sizes are needed for their discovery.
In a large-scale effort by the GIANT Consortium, BMI association summary statistics of 718,734 individuals from 125 studies with ExomeChip genotype data were combined, focusing on ~216,000 low-frequency (MAF:1<5%) and rare (MAF<1%) coding variants, which may alter gene function [42]. Four rare and 10 low-frequency variants in 13 genes were identified, eight of which were in genes (ZBTB7B, ACHE, RAPGEF3, RAB21, ZFHX3, ENTPD6, ZFR2, ZNF169) newly implicated in human obesity (Figure 2). Two rare variants (MC4R, KSR2) had been observed previously in extreme obesity [43–45], and two were found in GIPR, a gene previously implicated in common obesity and glycemic traits [12,46]. Effect sizes of low-frequency and, in particular, rare variants were larger than for common variants. Nevertheless, very large sample sizes were still needed for their discovery. The largest effect was observed for the MC4R stop-codon (p.Tyr35Ter); carriers (1 in 5,000 people) weigh on average ~7kg more than non-carriers. p.Tyr35Ter results in MC4R-deficiency and was one of the first mutations discovered in monogenic cases of obesity [43]. However, not all mutation carriers are obese; e.g. of 30 carriers in the UK Biobank, six were of normal weight, suggesting incomplete penetrance and compensation by other genetic or environmental factors [42]. Pathway analyses based on low-frequency and rare variants confirm a key role for the CNS in body weight regulation and provide new evidence for adipocyte and energy expenditure biology. Enrichment analyses based on low-frequency/rare variants provided more robust results than those based on common variants [42].
Interactions with lifestyle and demographics
Obesity is a multifactorial condition, resulting from an intricate interaction between genetic and non-genetic factors. Gene-by-environment (GxE) interaction studies show that some previously identified GWAS loci have indeed sex-specific and/or lifestyle-specific effects on adiposity outcomes. For example, effects of WHRadjBMI-associated loci are generally more pronounced in women than in men [13] and the genetic susceptibility to obesity, assessed by a genetic risk score of multiple BMI-associated loci, was higher among individuals who lived unhealthy lives [47–50].
However, genome-wide searches to discover novel loci that interact with lifestyle and demographic factors has proven to be challenging. A large-scale GWAS (Nmax~320,000) that examined the effect of age and sex on genetic associations with adiposity traits confirmed previously observed sex-specific effects for WHRadjBMI-associated loci (mostly larger in women), and newly reported age-specific effects for BMI-associated loci (mostly larger among younger adults (age<50yrs)), but found few new loci [51]. Two GWASs that aimed to identify loci of which the association depends on physical activity or smoking status found no new loci for BMI and WHRadjBMI [52,53]. Despite the large sample size, it seems that current genome-wide GxE interaction studies do not have sufficient statistical power, suggesting that interaction effects are likely small, and/or the precision and accuracy with which non-genetic (lifestyle) factors are assessed is low. In addition, non-genetic factors are often not measured in a uniform manner and, despite rigorous efforts to harmonize across studies, this may increase heterogeneity and further reduce statistical power.
Populations with specific genomic features
While increasing sample size has been a major driver for continued discovery in traditional GWAS, recent studies have taken advantage of specific demographic, evolutionary and/or genomic features of relatively small populations. For example, a recent GWAS of BMI in Samoans (Nmax=3,072), a unique founder population with a high prevalence of obesity, identified a coding variant (p.Arg457Gln) in CREBRF that is common among Samoans (MAF=26%) and other Pacific populations, but practically non-existent in other populations [54,55]. The Gln-allele is associated with a 1.4 kg/m2 higher BMI (equivalent to ~4kg for a 1.7m tall person) and was found to reduce energy use and promote fat storage in an adipocyte model [55]. Particularly intriguing is that the Gln-allele protects significantly against type 2 diabetes [55].
A study in a Greenlandic Inuit population (N~4,000) characterized by an extreme demographic history [56], identified a functional variant in ADCY3 (c.2433-1G>A) [57], a gene located in a locus previously identified by GWAS in which common variants were found to associated with BMI [58]. While relatively common in the Greenlandic population (~3%), the variant is monomorphic in other populations [57]. Other mutations in ADCY3 were found to cause monogenic obesity in a consanguineous population from Pakistan [59]. Functional follow-up analyses showed that ADCY3 co-localizes with MC4R at the primary cilia of a subset of hypothalamic neurons, previously implicated in body weight regulation [28].
While the identified variants were population-specific, the genes (and encoded proteins) may have a role in energy-homeostasis across all ancestries.
Genetic correlation and Mendelian randomization
Genetic correlation and Mendelian randomization are complementary approaches to assess shared etiology and causal relationships between adiposity and other traits.
Genetic correlation studies correlate SNP-association effects of one trait with those of another trait. SNP-association effects of BMI and WHRadjBMI were found to positively correlate with those of cardiometabolic traits, excessive daytime sleepiness, sleep duration [60–62], and negatively with those for anorexia nervosa, age at menarche, years of education, alcohol use, and self-rated health [60,63,64].
Mendelian randomization, also dubbed nature’s randomized trial, relies on the fact that SNPs, associated with a trait or biomarker, “randomize” a population – in an unbiased way – into low-to-highly exposed individuals. For example, the FTO genotype randomizes the population in three groups (by genotype); those with a low obesity risk (homozygous for the protective allele), intermediate risk (heterozygous) and high obesity risk (homozygous for the risk allele). This design allows testing whether BMI is causally associated with other traits. Consistent with findings from observational studies, Mendelian randomization studies have reported that increased BMI is causally related to higher risk of metabolic and cardiovascular diseases [65–70], higher risk of pancreatic, gastric, colorectal and breast cancer [71–76], lower risk of lung and skin cancer [76], higher risk of asthma [77], and increased bone mineral density [78]. Furthermore, they have also provided evidence of lesser-known causal effects; i.e. of increased BMI on higher risk of multiple sclerosis [79,80] and lower risk of Parkinson’s disease [81], and of increased childhood adiposity on higher risk of type 1 diabetes [82] and of disordered eating in adolescence [83].
Genetic information to predict obesity
Historically, genetic tests have been used to provide a genetic diagnosis to patients with rare forms of extreme and early-onset obesity that may be due to a single mutation [84]. For some patients, such a genetic diagnosis has been instrumental in their treatment [85,86]. As more variants are being discovered and genome sequencing is becoming mainstream, there is a growing expectation that genetic tests will help clinicians predict and diagnose patients’ risks of complex disease, such as common forms of obesity. However, unlike monogenic forms, common obesity is polygenic and multifactorial; i.e. numerous genetic variants, and also lifestyle and demographic factors contribute to a person’s obesity susceptibility. Therefore, it is no surprise that genetic prediction of obesity, even when based on nearly 100 GWAS-identified common variants, is poor (AUCROC~0.60) and unfit for use in clinical settings [12,87]. Because the heritability is modest, a test based solely on genetic variants will never accurately predict common obesity.
Conclusions and future perspectives
In the past decade, large-scale genome-wide discovery efforts have uncovered numerous new loci that harbor genetic variants associated with adiposity traits. With the advent of additional large population studies (e.g. UK Biobank, Million Veterans Project, All of Us), the number of loci will continue to increase rapidly in coming years. Preliminary analyses suggest that these loci highlight pathways that broadly overlap with the biology previously identified in extreme models of obesity in human and animals. However, the value of GWAS is that identified loci implicate new genes in both novel and known pathways. Pinpointing of these genes within each locus remains an important challenge, but may be sped up with the advent of novel approaches that integrate multiple sources of information [88,89]. The identification of the causal gene/variant is a critical step towards the translation of genetic loci into biology and requires close collaboration between geneticists and physiologists. Ultimately, new genes, even in known pathways, may provide new avenues for the development of obesity drugs, a field that had seen limited progress in the past four decades [90].
Acknowledgments
Ruth Loos is supported by the National Institutes of Health [R01 DK107786, R01 DK110113, U01HG007417].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement Nothing declared
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Collaborators GBDO, Afshin A, Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L, Mokdad AH, Moradi-Lakeh M, Naghavi M, et al. Health effects of overweight and obesity in 195 countries over 25 years. N Engl J Med. 2017;377(1):13–27. doi: 10.1056/NEJMoa1614362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang YC, McPherson K, Marsh T, Gortmaker SL, Brown M. Health and economic burden of the projected obesity trends in the USA and the uk. Lancet. 2011;378(9793):815–825. doi: 10.1016/S0140-6736(11)60814-3. [DOI] [PubMed] [Google Scholar]
- 3.Spieker EA, Pyzocha N. Economic impact of obesity. Prim Care. 2016;43(1):83–95. viii–ix. doi: 10.1016/j.pop.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 4.NCD Risk Factor Collaboration. Trends in adult body-mass index in 200 countries from 1975 to 2014: A pooled analysis of 1698 population-based measurement studies with 19.2 million participants. The Lancet. 2016;387(10026):1377–1396. doi: 10.1016/S0140-6736(16)30054-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fact sheet: Obesity and overweight. WHO; 2017. http://www.who.int/mediacentre/factsheets/fs311/en/ [Google Scholar]
- 6.NCD Risk Factor Collaboration. Abarca-Gómez L, Abdeen ZA, Hamid ZA, Abu-Rmeileh NM, Acosta-Cazares B, Acuin C, Adams RJ, Aekplakorn W, Afsana K, Aguilar-Salinas CA. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: A pooled analysis of 2416 population-based measurement studies in 1289 million children, adolescents, and adults. The Lancet. 2017 doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberto CA, Swinburn B, Hawkes C, Huang TT, Costa SA, Ashe M, Zwicker L, Cawley JH, Brownell KD. Patchy progress on obesity prevention: Emerging examples, entrenched barriers, and new thinking. Lancet. 2015;385(9985):2400–2409. doi: 10.1016/S0140-6736(14)61744-X. [DOI] [PubMed] [Google Scholar]
- 8.Ezzati M, Riboli E. Can noncommunicable diseases be prevented? Lessons from studies of populations and individuals. Science. 2012;337(6101):1482–1487. doi: 10.1126/science.1227001. [DOI] [PubMed] [Google Scholar]
- 9**.Ghosh S, Bouchard C. Convergence between biological, behavioural and genetic determinants of obesity. Nature reviews Genetics. 2017;18(12):731–748. doi: 10.1038/nrg.2017.72. This review provides a comprehensive overview of the multiple biological, behavioural and genetic determinants and correlates of obesity and how they converge. The authors performed additional analyses on existing data. [DOI] [PubMed] [Google Scholar]
- 10.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, Perry JR, Elliott KS, Lango H, Rayner NW, Shields B, et al. A common variant in the fto gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, et al. Genome-wide association scan shows genetic variants in the fto gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, Strawbridge RJ, Pers TH, Fischer K, Justice AE, Workalemahu T, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.http://www.broadinstitute.org/collaboration/giant/index.Php/main_page: (2017). http://www.broadinstitute.org/collaboration/giant/index.php/Main_Page
- 15*.Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, Iwasaki M, et al. Genome-wide association study identifies 112 new loci for body mass index in the japanese population. Nat Genet. 2017;49(10):1458–1467. doi: 10.1038/ng.3951. Largest meta-analysis of GWAS for BMI, combining data from the GIANT Consortium and the Biobank Japan Project (N >480,000 indiviudals), resulting in the discovery of 112 novel loci associated with BMI. Enrichement analysis confirm the role of central nervous system in body weight regulation, but also implicate immune related and adipose tissue. [DOI] [PubMed] [Google Scholar]
- 16.Ng MCY, Graff M, Lu Y, Justice AE, Mudgal P, Liu CT, Young K, Yanek LR, Feitosa MF, Wojczynski MK, Rand K, et al. Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of african ancestry: African ancestry anthropometry genetics consortium. PLoS Genet. 2017;13(4):e1006719. doi: 10.1371/journal.pgen.1006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, Ripke S, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung YC, Yeo GS, O’Rahilly S, Coll AP. Obesity and fto: Changing focus at a complex locus. Cell Metab. 2014;20(5):710–718. doi: 10.1016/j.cmet.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, et al. Fto obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, et al. Obesity-associated variants within fto form long-range functional connections with irx3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stratigopoulos G, Martin Carli JF, O’Day DR, Wang L, Leduc CA, Lanzano P, Chung WK, Rosenbaum M, Egli D, Doherty DA, Leibel RL. Hypomorphism for rpgrip1l, a ciliary gene vicinal to the fto locus, causes increased adiposity in mice. Cell Metab. 2014;19(5):767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stratigopoulos G, Burnett LC, Rausch R, Gill R, Penn DB, Skowronski AA, LeDuc CA, Lanzano AJ, Zhang P, Storm DR, Egli D, et al. Hypomorphism of fto and rpgrip1l causes obesity in mice. J Clin Invest. 2016;126(5):1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larder R, Sim MFM, Gulati P, Antrobus R, Tung YCL, Rimmington D, Ayuso E, Polex-Wolf J, Lam BYH, Dias C, Logan DW, et al. Obesity-associated gene tmem18 has a role in the central control of appetite and body weight regulation. Proc Natl Acad Sci U S A. 2017;114(35):9421–9426. doi: 10.1073/pnas.1707310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiemerslage L, Gohel PA, Maestri G, Hilmarsson TG, Mickael M, Fredriksson R, Williams MJ, Schioth HB. The drosophila ortholog of tmem18 regulates insulin and glucagon-like signaling. J Endocrinol. 2016;229(3):233–243. doi: 10.1530/JOE-16-0040. [DOI] [PubMed] [Google Scholar]
- 25.Rathjen T, Yan X, Kononenko NL, Ku MC, Song K, Ferrarese L, Tarallo V, Puchkov D, Kochlamazashvili G, Brachs S, Varela L, et al. Regulation of body weight and energy homeostasis by neuronal cell adhesion molecule 1. Nat Neurosci. 2017;20(8):1096–1103. doi: 10.1038/nn.4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan X, Wang Z, Schmidt V, Gauert A, Willnow TE, Heinig M, Poy MN. Cadm2 regulates body weight and energy homeostasis in mice. Mol Metab. 2017 doi: 10.1016/j.molmet.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watson RA, Gates AS, Wynn EH, Calvert FE, Girousse A, Lelliott CJ, Barroso I. Lyplal1 is dispensable for normal fat deposition in mice. Disease models & mechanisms. 2017 doi: 10.1242/dmm.031864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siljee JE, Wang Y, Bernard AA, Ersoy BA, Zhang S, Marley A, Von Zastrow M, Reiter JF, Vaisse C. Subcellular localization of mc4r with adcy3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018 doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, Cousminer DL, Dastani Z, Drong AW, Esko T, Evans DM, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nature communications. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kilpelainen TO, Zillikens MC, Stancakova A, Finucane FM, Ried JS, Langenberg C, Zhang W, Beckmann JS, Luan J, Vandenput L, Styrkarsdottir U, et al. Genetic variation near irs1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43(8):753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Araki E, Lipes MA, Patti ME, Bruning JC, Haag B, 3rd, Johnson RS, Kahn CR. Alternative pathway of insulin signalling in mice with targeted disruption of the irs-1 gene. Nature. 1994;372(6502):186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- 32.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372(6502):182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 33.Miki H, Yamauchi T, Suzuki R, Komeda K, Tsuchida A, Kubota N, Terauchi Y, Kamon J, Kaburagi Y, Matsui J, Akanuma Y, et al. Essential role of insulin receptor substrate 1 (irs-1) and irs-2 in adipocyte differentiation. Mol Cell Biol. 2001;21(7):2521–2532. doi: 10.1128/MCB.21.7.2521-2532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng YH, Kriauciunas KM, Kokkotou E, Kahn CR. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol. 2004;24(5):1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zillikens MC, Demissie S, Hsu YH, Yerges-Armstrong LM, Chou WC, Stolk L, Livshits G, Broer L, Johnson T, Koller DL, Kutalik Z, et al. Large meta-analysis of genome-wide association studies identifies five loci for lean body mass. Nature communications. 2017;8(1):80. doi: 10.1038/s41467-017-00031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu AY, Deng X, Fisher VA, Drong A, Zhang Y, Feitosa MF, Liu CT, Weeks O, Choh AC, Duan Q, Dyer TD, et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat Genet. 2017;49(1):125–130. doi: 10.1038/ng.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kilpelainen TO, Carli JF, Skowronski AA, Sun Q, Kriebel J, Feitosa MF, Hedman AK, Drong AW, Hayes JE, Zhao J, Pers TH, et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nature communications. 2016;7:10494. doi: 10.1038/ncomms10494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong YH, Hishikawa D, Miyahara H, Tsuzuki H, Nishimura Y, Gotoh C, Choi KC, Hokari Y, Takagi Y, Lee HG, Cho KK, et al. Up-regulation of adipogenin, an adipocyte plasma transmembrane protein, during adipogenesis. Mol Cell Biochem. 2005;276(1-2):133–141. doi: 10.1007/s11010-005-3673-0. [DOI] [PubMed] [Google Scholar]
- 39.Kim JY, Tillison K, Smas CM. Cloning, expression, and differentiation-dependent regulation of smaf1 in adipogenesis. Biochem Biophys Res Commun. 2005;326(1):36–44. doi: 10.1016/j.bbrc.2004.10.200. [DOI] [PubMed] [Google Scholar]
- 40.Tachmazidou I, Suveges D, Min JL, Ritchie GRS, Steinberg J, Walter K, Iotchkova V, Schwartzentruber J, Huang J, Memari Y, McCarthy S, et al. Whole-genome sequencing coupled to imputation discovers genetic signals for anthropometric traits. Am J Hum Genet. 2017;100(6):865–884. doi: 10.1016/j.ajhg.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hendricks AE, Bochukova EG, Marenne G, Keogh JM, Atanassova N, Bounds R, Wheeler E, Mistry V, Henning E, Korner A, Muddyman D, et al. Rare variant analysis of human and rodent obesity genes in individuals with severe childhood obesity. Sci Rep. 2017;7(1):4394. doi: 10.1038/s41598-017-03054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Turcot V, Lu Y, Highland HM, Schurmann C, Justice AE, Fine RS, Bradfield JP, Esko T, Giri A, Graff M, Guo X, et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat Genet. 2018;50(1):26–41. doi: 10.1038/s41588-017-0011-x. Largest exome-wide disocvery study for BMI, by the GIANT Consortium, including exome-chip association data of >700,000 indiviudals, disovering 14 low-frequency and rare variants in 13 genes, of 8 genes had not previously been implicated in body weight regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sina M, Hinney A, Ziegler A, Neupert T, Mayer H, Siegfried W, Blum WF, Remschmidt H, Hebebrand J. Phenotypes in three pedigrees with autosomal dominant obesity caused by haploinsufficiency mutations in the melanocortin-4 receptor gene. American Journal of Human Genetics. 1999;65(6):1501–1507. doi: 10.1086/302660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinney A, Schmidt A, Nottebom K, Heibult O, Becker I, Ziegler A, Gerber G, Sina M, Gorg T, Mayer H, Siegfried W, et al. Several mutations in the melanocortin-4 receptor gene including a nonsense and a frameshift mutation associated with dominantly inherited obesity in humans. Journal of Clinical Endocrinology Metabolism. 1999;84:1483–1486. doi: 10.1210/jcem.84.4.5728. [DOI] [PubMed] [Google Scholar]
- 45.Pearce LR, Atanassova N, Banton MC, Bottomley B, van der Klaauw AA, Revelli JP, Hendricks A, Keogh JM, Henning E, Doree D, Jeter-Jones S, et al. Ksr2 mutations are associated with obesity, insulin resistance, and impaired cellular fuel oxidation. Cell. 2013;155(4):765–777. doi: 10.1016/j.cell.2013.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU, Kao WH, et al. Genetic variation in gipr influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB, Chasman DI, et al. Sugar-sweetened beverages and genetic risk of obesity. New England Journal of Medicine. 2012;367(15):1387–1396. doi: 10.1056/NEJMoa1203039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Zhao JH, Luan Ja, Ekelund U, Luben RN, Khaw KT, Wareham NJ, Loos RJF. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from epic-norfolk prospective population study. PLoS Med. 2010;7(8):e1000332. doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyrrell J, Wood AR, Ames RM, Yaghootkar H, Beaumont RN, Jones SE, Tuke MA, Ruth KS, Freathy RM, Davey Smith G, Joost S, et al. Gene-obesogenic environment interactions in the uk biobank study. Int J Epidemiol. 2017;46(2):559–575. doi: 10.1093/ije/dyw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kilpelainen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, Ahmad T, Mora S, Kaakinen M, Sandholt CH, Holzapfel C, et al. Physical activity attenuates the influence of fto variants on obesity risk: A meta-analysis of 218,166 adults and 19,268 children. PLoS Med. 2011;8(11):e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winkler TW, Justice AE, Graff M, Barata L, Feitosa MF, Chu S, Czajkowski J, Esko T, Fall T, Kilpelainen TO, Lu Y, et al. The influence of age and sex on genetic associations with adult body size and shape: A large-scale genome-wide interaction study. PLoS Genet. 2015;11(10):e1005378. doi: 10.1371/journal.pgen.1005378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52*.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, Winkler TW, Chu AY, Mahajan A, Hadley D, Xue L, et al. Genome-wide physical activity interactions in adiposity - a meta-analysis of 200,452 adults. PLoS Genet. 2017;13(4):e1006528. doi: 10.1371/journal.pgen.1006528. Genome-wide search in Samoans, a founder population characterized by very high prevalence of obesity, identifies a coding variant in CREBRF that is associated with increased BMI and obesity risk, as well as reduced risk of type 2 diabetes. This variants is common among Samoans, but monomorphic among most other non-Polynesian populations. This study enphasises the value of studying populations with specific demographic, evolutionary, and or genetic features. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Justice AE, Winkler TW, Feitosa MF, Graff M, Fisher VA, Young K, Barata L, Deng X, Czajkowski J, Hadley D, Ngwa JS, et al. Genome-wide meta-analysis of 241,258 adults accounting for smoking behaviour identifies novel loci for obesity traits. Nature communications. 2017;8:14977. doi: 10.1038/ncomms14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berry SD, Walker CG, Ly K, Snell RG, Atatoa Carr PE, Bandara D, Mohal J, Castro TG, Marks EJ, Morton SMB, Grant CC. Widespread prevalence of a crebrf variant amongst maori and pacific children is associated with weight and height in early childhood. Int J Obes (Lond) 2017 doi: 10.1038/ijo.2017.230. [DOI] [PubMed] [Google Scholar]
- 55.Minster RL, Hawley NL, Su CT, Sun G, Kershaw EE, Cheng H, Buhule OD, Lin J, Reupena MS, Viali S, Tuitele J, et al. A thrifty variant in crebrf strongly influences body mass index in samoans. Nat Genet. 2016;48(9):1049–1054. doi: 10.1038/ng.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedersen CT, Lohmueller KE, Grarup N, Bjerregaard P, Hansen T, Siegismund HR, Moltke I, Albrechtsen A. The effect of an extreme and prolonged population bottleneck on patterns of deleterious variation: Insights from the greenlandic inuit. Genetics. 2017;205(2):787–801. doi: 10.1534/genetics.116.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Grarup N, Moltke I, Andersen MK, Dalby M, Vitting-Seerup K, Kern T, Mahendran Y, Jorsboe E, Larsen CVL, Dahl-Petersen IK, Gilly A, et al. Loss-of-function variants in adcy3 increase risk of obesity and type 2 diabetes. Nat Genet. 2018 doi: 10.1038/s41588-017-0022-7. Genome-wide search in Greenlandic Inuits, a population characterized by a harsh demographic history, identifies a coding variant in ADCY3 that is associated with increased BMI and obesity risk. This variants is common among Greenlandic Inuits, but monomorphic among most other populations. This study enphasises the value of studying populations with specific demographic, evolutionary, and or genetic features. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Allen HL, Lindgren CM, Luan Ja, Magi R, Randall JC, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saeed S, Bonnefond A, Tamanini F, Mirza MU, Manzoor J, Janjua QM, Din SM, Gaitan J, Milochau A, Durand E, Vaillant E, et al. Loss-of-function mutations in adcy3 cause monogenic severe obesity. Nat Genet. 2018 doi: 10.1038/s41588-017-0023-6. [DOI] [PubMed] [Google Scholar]
- 60.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C. Patterson N, Daly MJ, Price AL, Neale BM. Ld score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lane JM, Liang J, Vlasac I, Anderson SG, Bechtold DA, Bowden J, Emsley R, Gill S, Little MA, Luik AI, Loudon A, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, Yaghootkar H, Hu Y, Teder-Laving M, Hayward C, Roenneberg T, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills AG, Evans LM, Hopfer C. Phenotypic and genetic relationship between bmi and drinking in a sample of uk adults. Behav Genet. 2017;47(3):290–297. doi: 10.1007/s10519-017-9838-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harris SE, Hagenaars SP, Davies G, David Hill W, Liewald DCM, Ritchie SJ, Marioni RE, Metastroke Consortium ICfBPG-WAS, International Consortium for Blood Pressure Genome-Wide Association S, Aging CC, Longevity G et al. Molecular genetic contributions to self-rated health. Int J Epidemiol. 2017;46(3):994–1009. doi: 10.1093/ije/dyw219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lyall DM, Celis-Morales C, Ward J, Iliodromiti S, Anderson JJ, Gill JMR, Smith DJ, Ntuk UE, Mackay DF, Holmes MV, Sattar N, et al. Association of body mass index with cardiometabolic disease in the uk biobank: A mendelian randomization study. JAMA Cardiol. 2017;2(8):882–889. doi: 10.1001/jamacardio.2016.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emdin CA, Khera AV, Natarajan P, Klarin D, Zekavat SM, Hsiao AJ, Kathiresan S. Genetic association of waist-to-hip ratio with cardiometabolic traits, type 2 diabetes, and coronary heart disease. JAMA. 2017;317(6):626–634. doi: 10.1001/jama.2016.21042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hagg S, Fall T, Ploner A, Magi R, Fischer K, Draisma HH, Kals M, de Vries PS, Dehghan A, Willems SM, Sarin AP, et al. Adiposity as a cause of cardiovascular disease: A mendelian randomization study. Int J Epidemiol. 2015;44(2):578–586. doi: 10.1093/ije/dyv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lindstrom S, Germain M, Crous-Bou M, Smith EN, Morange PE, van Hylckama Vlieg A, de Haan HG, Chasman D, Ridker P, Brody J, de Andrade M, et al. Assessing the causal relationship between obesity and venous thromboembolism through a mendelian randomization study. Hum Genet. 2017;136(7):897–902. doi: 10.1007/s00439-017-1811-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dale CE, Fatemifar G, Palmer TM, White J, Prieto-Merino D, Zabaneh D, Engmann JEL, Shah T, Wong A, Warren HR, McLachlan S, et al. Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: A mendelian randomization analysis. Circulation. 2017;135(24):2373–2388. doi: 10.1161/CIRCULATIONAHA.116.026560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fall T, Hagg S, Ploner A, Magi R, Fischer K, Draisma HH, Sarin AP, Benyamin B, Ladenvall C, Akerlund M, Kals M, et al. Age- and sex-specific causal effects of adiposity on cardiovascular risk factors. Diabetes. 2015;64(5):1841–1852. doi: 10.2337/db14-0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carreras-Torres R, Johansson M, Gaborieau V, Haycock PC, Wade KH, Relton CL, Martin RM, Davey Smith G, Brennan P. The role of obesity, type 2 diabetes, and metabolic factors in pancreatic cancer: A mendelian randomization study. J Natl Cancer Inst. 2017;109(9) doi: 10.1093/jnci/djx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mao Y, Yan C, Lu Q, Zhu M, Yu F, Wang C, Dai J, Ma H, Hu Z, Shen H, Jin G. Genetically predicted high body mass index is associated with increased gastric cancer risk. Eur J Hum Genet. 2017;25(9):1061–1066. doi: 10.1038/ejhg.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thrift AP, Gong J, Peters U, Chang-Claude J, Rudolph A, Slattery ML, Chan AT, Locke AE, Kahali B, Justice AE, Pers TH, et al. Mendelian randomization study of body mass index and colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1024–1031. doi: 10.1158/1055-9965.EPI-14-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo Y, Warren Andersen S, Shu XO, Michailidou K, Bolla MK, Wang Q, Garcia-Closas M, Milne RL, Schmidt MK, Chang-Claude J, Dunning A, et al. Genetically predicted body mass index and breast cancer risk: Mendelian randomization analyses of data from 145,000 women of european descent. PLoS Med. 2016;13(8):e1002105. doi: 10.1371/journal.pmed.1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guo Q, Burgess S, Turman C, Bolla MK, Wang Q, Lush M, Abraham J, Aittomaki K, Andrulis IL, Apicella C, Arndt V, et al. Body mass index and breast cancer survival: A mendelian randomization analysis. Int J Epidemiol. 2017;46(6):1814–1822. doi: 10.1093/ije/dyx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Benn M, Tybjaerg-Hansen A, Smith GD, Nordestgaard BG. High body mass index and cancer risk-a mendelian randomisation study. Eur J Epidemiol. 2016;31(9):879–892. doi: 10.1007/s10654-016-0147-5. [DOI] [PubMed] [Google Scholar]
- 77.Skaaby T, Taylor AE, Thuesen BH, Jacobsen RK, Friedrich N, Mollehave LT, Hansen S, Larsen SC, Volker U, Nauck M, Volzke H, et al. Estimating the causal effect of body mass index on hay fever, asthma and lung function using mendelian randomization. Allergy. 2018;73(1):153–164. doi: 10.1111/all.13242. [DOI] [PubMed] [Google Scholar]
- 78.Kemp JP, Sayers A, Smith GD, Tobias JH, Evans DM. Using mendelian randomization to investigate a possible causal relationship between adiposity and increased bone mineral density at different skeletal sites in children. Int J Epidemiol. 2016;45(5):1560–1572. doi: 10.1093/ije/dyw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gianfrancesco MA, Glymour MM, Walter S, Rhead B, Shao X, Shen L, Quach H, Hubbard A, Jonsdottir I, Stefansson K, Strid P, et al. Causal effect of genetic variants associated with body mass index on multiple sclerosis susceptibility. Am J Epidemiol. 2017;185(3):162–171. doi: 10.1093/aje/kww120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mokry LE, Ross S, Timpson NJ, Sawcer S, Davey Smith G, Richards JB. Obesity and multiple sclerosis: A mendelian randomization study. PLoS Med. 2016;13(6):e1002053. doi: 10.1371/journal.pmed.1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Noyce AJ, Kia DA, Hemani G, Nicolas A, Price TR, De Pablo-Fernandez E, Haycock PC, Lewis PA, Foltynie T, Davey Smith G, International Parkinson Disease Genomics C et al. Estimating the causal influence of body mass index on risk of parkinson disease: A mendelian randomisation study. PLoS Med. 2017;14(6):e1002314. doi: 10.1371/journal.pmed.1002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Censin JC, Nowak C, Cooper N, Bergsten P, Todd JA, Fall T. Childhood adiposity and risk of type 1 diabetes: A mendelian randomization study. PLoS Med. 2017;14(8):e1002362. doi: 10.1371/journal.pmed.1002362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reed ZE, Micali N, Bulik CM, Davey Smith G, Wade KH. Assessing the causal role of adiposity on disordered eating in childhood, adolescence, and adulthood: A mendelian randomization analysis. Am J Clin Nutr. 2017;106(3):764–772. doi: 10.3945/ajcn.117.154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27(7):710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 85.Farooqi IS, O’Rahilly S. 20 years of leptin: Human disorders of leptin action. J Endocrinol. 2014;223(1):T63–70. doi: 10.1530/JOE-14-0480. [DOI] [PubMed] [Google Scholar]
- 86.Paz-Filho G, Mastronardi CA, Licinio J. Leptin treatment: Facts and expectations. Metabolism. 2015;64(1):146–156. doi: 10.1016/j.metabol.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 87*.Loos RJ, Janssens AC. Predicting polygenic obesity using genetic information. Cell Metab. 2017;25(3):535–543. doi: 10.1016/j.cmet.2017.02.013. Review that summarizes the value of common genetic variation to predict a person’s risk of obesity. Genetic variants identified through GWAS are generally poor predictors of future obesity. [DOI] [PubMed] [Google Scholar]
- 88.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, Jansen R, de Geus EJ, Boomsma DI, Wright FA, Sullivan PF, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mancuso N, Shi H, Goddard P, Kichaev G, Gusev A, Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am J Hum Genet. 2017;100(3):473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Srivastava G, Apovian CM. Current pharmacotherapy for obesity. Nat Rev Endocrinol. 2018;14(1):12–24. doi: 10.1038/nrendo.2017.122. [DOI] [PubMed] [Google Scholar]
- 91.Elks CE, den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, Ong KK. Variability in the heritability of body mass index: A systematic review and meta-regression. Frontiers in endocrinology. 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Silventoinen K, Jelenkovic A, Sund R, Yokoyama Y, Hur YM, Cozen W, Hwang AE, Mack TM, Honda C, Inui F, Iwatani Y, et al. Differences in genetic and environmental variation in adult body mass index by sex, age, time period, and region: An individual-based pooled analysis of 40 twin cohorts. Am J Clin Nutr. 2017 doi: 10.3945/ajcn.117.153643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rose KM, Newman B, Mayer-Davis EJ, Selby JV. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes Res. 1998;6(6):383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 94.Henneman P, Aulchenko YS, Frants RR, van Dijk KW, Oostra BA, van Duijn CM. Prevalence and heritability of the metabolic syndrome and its individual components in a dutch isolate: The erasmus rucphen family study. J Med Genet. 2008;45(9):572–577. doi: 10.1136/jmg.2008.058388. [DOI] [PubMed] [Google Scholar]
- 95.Robinson MR, English G, Moser G, Lloyd-Jones LR, Triplett MA, Zhu Z, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, LifeLines Cohort S. Esko T, et al. Genotype-covariate interaction effects and the heritability of adult body mass index. Nat Genet. 2017;49(8):1174–1181. doi: 10.1038/ng.3912. [DOI] [PubMed] [Google Scholar]
- 96.Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AA, Lee SH, Robinson MR, Perry JR, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, et al. Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet. 2015;47(10):1114–1120. doi: 10.1038/ng.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kutalik Z, Whittaker J, Waterworth D, consortium G. Beckmann JS, Bergmann S. Novel method to estimate the phenotypic variation explained by genome-wide association studies reveals large fraction of the missing heritability. Genet Epidemiol. 2011;35(5):341–349. doi: 10.1002/gepi.20582. [DOI] [PubMed] [Google Scholar]


