Abstract
Innate lymphoid cells (ILCs) that are capable of producing effector cytokines reminiscent of CD4+ T helper (Th) cells during infections and tissue inflammations have drawn much attention in the immunology field in recent years. Within the ILCs, the lymphoid tissue inducer (LTi) cells that play a critical role in lymphoid organogenesis were identified long before the establishment of the ILC concept. LTi cells, developed and functioning mainly at the fetal stage, and LTi-like cells, presumably generated during the adulthood, are regarded as a subset of type 3 ILCs (ILC3s) because they express the ILC3 lineage-defining transcription factor RORγt, and like other ILC3s, can produce an ILC3 signature cytokine IL-22 and initiate protective immune responses against extracellular bacteria. However, LTi/LTi-like cells have a unique gene expression pattern, and they develop from a progenitor that is distinct from the progenitor of all other ILCs and the progenitor of conventional natural killer (cNK) cells. There are also several other unique features of LTi/LTi-like cells comparing to non-LTi ILC3s. In addition to their classical function in lymphoid organogenesis, LTi/LTi-like cells also have specialized functions in association with the adaptive immune system, which include their effects on T and B cell development, activation and function. In this review, we summarize these specific features of LTi/LTi-like cells and propose that these cells should be considered as a separated innate lymphoid lineage in parallel with other non-LTi ILCs and cNK cells.
Graphical Abstract
1. Introduction
Our knowledge on the innate immune system has greatly expanded in the past few years due to the identification and defining of several new innate lymphoid populations, now known as innate lymphoid cells (ILCs)1. These new members of the innate immune system are featured by their functional similarity to CD4+ T helper (Th) cells in the adaptive immune system2. Based on their lineage-defining transcription factor (also called master regulator) expression and signature cytokine production, mature ILCs can be divided into three major subsets (ILC1s, ILC2s and ILC3s) mirroring distinct CD4+ T effector cells (Th1, Th2 and Th17 cells). ILC2s express the Th2 cell master regulator GATA-3, and secrete IL-5 and IL-13, but low levels of IL-43, 4, 5, 6. ILC3s express the Th17/Th22 cell master regulator RORγt and their effector cytokines, IL-22, IL-17A, and IL-17F7, 8, 9.
The defining of ILC1s was established later than other ILC subsets. The conventional natural killer (cNK) cells had been considered as an ILC1 subset1, however, the most recent view in the field prefers to classify cNK cells as the innate counterpart of cytotoxic CD8+ T cells since they both exhibit cytolytic activities and express transcription factor Eomes2, 10. Up to date, several non-NK ILC1s residing in various tissues have been discovered by different research groups11, 12, 13. Both ILC1s and cNK cells express the Th1 master regulator T-bet and several NK cell surface markers such as NK1.1 and NKp46 in C57BL/6 mice. However, Eomes expression may distinguish cNK cells from ILC1s13. Similar to Th1 cells, ILC1s do not express Eomes, but they can secrete effector cytokine IFN-γ and TNF-α upon stimulation by IL-12, IL-15, and/or IL-1811, 12, 13.
All these ILC subsets and cNK cells constitutively express an important transcriptional regulator, inhibitor of DNA-binding protein 2 (Id2), which is also required for an overall fate determination of innate lymphoid cell lineage through antagonizing the functions of E-box proteins13, 14, 15. Recent studies have also found that these terminally developed ILC subsets may retain certain plasticity to convert to each other16, 17, 18, a phenomenon that has been well described for differentiated CD4+ T helper cell subsets19, 20, 21.
Compared to ILC1s and ILC2s, ILC3s are much more complicated since they can be further divided into two distinct lineages, lymphoid tissue inducer (LTi) or LTi-like cells, and the remaining ILC3s some of which express natural cytotoxicity receptors (NCRs)22. LTi cells, named after their function in organogenesis of secondary lymphoid structures at fetal stage, are the ILC population that was first discovered23, 24, 25, 26, 27, 28. In the adult stage, there is also a group of cells that are phenotypically similar to LTi cells but unable to facilitate the generation of secondary lymphoid organs29, 30, 31, 32. Thus, they are referred to as LTi-like cells. LTi and LTi-like cells are similar in their gene expression profiles, except that LTi-like cells express the T cell survival molecules OX40L33, 34, 35 and CD30L36 (Figure 1). In mice, LTi cells or LTi-like cells can be distinguished from other ILC3s by their expression of a chemokine receptor, CCR622. A proportion of LTi/LTi-like cells can also express CD4, and these cells are considered as the most mature LTi cells. However, a gene expression comparison did not reveal substantial differences between the CD4+ and CD4− LTi-like cells37.
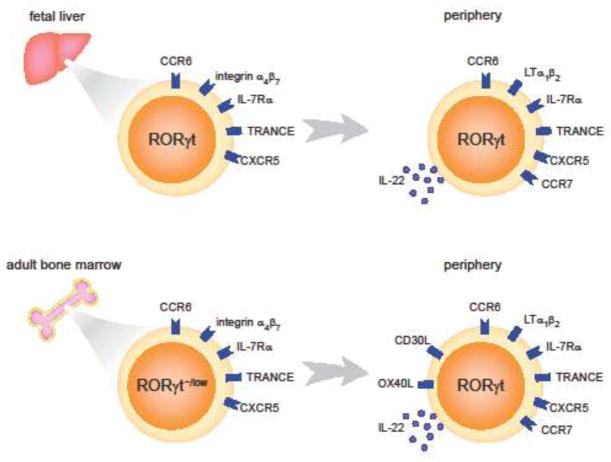
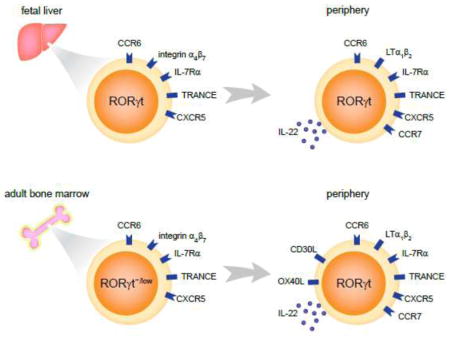
Figure 1. Development and differences of fetal and adult LTi cells.
In the fetal stage, LTi cells are developed from their progenitors in the fetal liver. LTi progenitor cells are featured by their high expression of integrin α4β7, IL-7Rα and RORγt. Other molecules such as CCR6, TRANCE and CXCR5 are also expressed by these progenitors. The progenitor cells then migrate to other organs such as fetal spleen, small intestine and thymus, where they further mature and start to express lymphtoxin α1β2 (LTα1β2) and IL-22. In the adult stage, LTi cells (or LTi-like cells) are developed from the progenitors in the bone marrow. These progenitors still express high levels of integrin α4β7 and IL-7Rα, but relatively low levels of RORγt. Gene expression in mature adult LTi-like cells are very similar to that in fetal LTi cells, except that adult LTi cells additionally express OX40L and CD30L.
The CCR6− ILC3s belong to another lineage22; some mouse CCR6− ILC3s express NKp467. The development of NKp46-expressing ILC3s depends on the master regulator T-bet22, 38, 39. Some of these NKp46+ ILC3s can further convert into ILC1s after losing RORγt expression16, 22, 40. In human, however, it is still unclear how to distinguish LTi/LTi-like cells from the NCR+ ILC3s since all the human RORγt+ ILC3s reported so far express CCR641. ILC3s in human are divided into different subsets mainly based on CD56 and NKp44 expression41, 42, 43, 44. The CD56− human ILC3s, regardless of NKp44 expression, are most likely the counterpart of the CCR6+ mouse LTi/LTi-like cells41. Effector cytokine production is also somewhat different between human and mouse ILC3 subsets. While both ILC3 lineages in mice are capable of producing IL-22, the IL-22 production is restricted to the NKp44+ populations, CD56+NKp44+ ILC3s in particular, in human42. Thus, the human ILC3s may have a greater diversity. In this review, we will discuss LTi/LTi-like cells mainly based on mice studies, if not specifically mentioned.
Although LTi/LTi-like cells express the ILC3 master regulator RORγt28 and effector cytokines IL-22, IL-17A and IL-17F9, their development is distinct from all other ILCs and cNK cells45, 46, 47, indicating that this population is a divergent member of the ILC family. Here, we summarize the current knowledge on LTi/LTi-like cells with respect to their characterization, development, and particular functions hoping to introduce a novel view on the innate lymphoid system.
2. Developmental difference between LTi cells and other ILCs
LTi cells were discovered much earlier than other ILCs23. Thus, how they developed had already been studied through in vitro and in vivo analyses even before the establishment of ILC concept24, 48, 49, 50, 51, 52, 53. However, only after the discovery of several common innate lymphoid cell progenitors45, 46, we started to have a comprehensive understanding of the innate lymphoid lineages generation including the development of LTi cells. The major argument for LTi/LTi-like cells being divergent from other ILCs is that they develop through distinct pathways.
The development of all innate lymphocytes is affected by deficiency of certain transcriptional regulators such as Id2, GATA-347, TCF-154, 55, 56, TOX57, 58, and Nfil-359, 60, indicating that a common ILC progenitor may exist. Indeed, an Id2-expressing common helper-like innate lymphoid progenitor (ChILP) population was discovered; these cells are able to generate all ILC subsets including LTi cells46. However, ChILP cells are not a homogeneous population as some of them can transiently express PLZF and these cells can generate all other ILCs except for LTi cells45. Although a unique progenitor for LTi-like cells in adult bone marrow has not been identified, it is clear that LTi progenitors in fetal liver express RORγt and integrin α4β761. A single cell gene expression analysis of LTi progenitors and PLZF+ ILC progenitors in the fetal liver confirmed that these were two distinct lineages with different transcriptional regulatory network62 (Figure 2). Thus, although in adult stage, LTi-like cells and NCR+ ILC3s show a great similarity and redundancy at their immune effector functions63, their development is quite different.
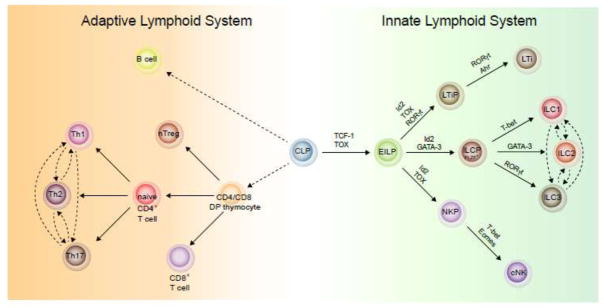
Figure 2. Similarities and differences between the development of innate and adaptive lymphoid system.
The development of innate lymphocytes and T cells shows certain similarities. At mature stage, CD4+ T cells and ILCs produce similar sets of effector cytokines, while CD8+ T cells and cNK cells exhibit cytotoxic activities. In the innate lymphoid system, the development of LTi cells is distinct from other ILCs and cNK cells though they all generated from an early innate lymphocyte progenitor (EILP). The non-LTi ILCs are generated from an PLZF-expressing common progenitor, ILCP, while LTi cells developed from their own progenitor which depends on the master transcription factor RORγt. At mature stage, while non-LTi ILCs show plasticity under certain circumstances similar to what have been observed in differentiated CD4+ T cells, LTi cells are relatively stable.
Even after development, some ILC subsets still exhibit certain degree of plasticity under some circumstances. For example, the NCR+ ILC3s can further develop into RORγt− ILC1s46, and conversely, some ILC1s are able to convert back to the NCR+ ILC3s16. In patient with chronic obstructive pulmonary disease (COPD), the ILC2s in inflamed lungs can convert into ILC1s which may contribute to the pathogenesis of the disease17, 18. However, the lineage conversion between LTi cells and other ILCs has not been reported (Figure 2). This is probably because the “boundary” between LTi lineage and other ILC lineages is much harder to cross than the “boundary” between non-LTi ILCs.
3. Molecular features of LTi/LTi-like cells
Although LTi cells and NCR+ ILC3s show great redundancy during immune responses, they also exhibit remarkable differences in gene expression and cellular function. Using a T-bet and RORγt double reporter mouse strain, we have isolated highly pure CCR6+T-bet− LTi/LTi-like cells and T-bet+NKp46+ ILC3s, and compared their gene expression differences through RNA-Seq analysis64. The result has revealed several classes of genes that are differently expressed by these two lineages. The genes that highly enriched in LTi/LTi-like cells include transcription regulators such as Egr2, Id1 and Id3, cytokines, chemokines and growth factors such as IL-17F, CCL21, Egfl7, BMP-2, antigen presenting molecules such as CD83 and H2-Oa, and other surface molecules such as CCR6, CXCR5, DLL1, Ly6C, Nrp-1, PD-1, and S1PR164. Many of these LTi/LTi-like cell-specific genes have already been reported to be associated with the unique functions of LTi cells and the rest of genes may also indicate other special functions of LTi/LTi-like cells which need further investigation.
As discussed above, the majority of LTi/LTi-like cells express CCR6, which can distinguish these cells from other RORγt+ ILC3s in mice22. CCL20, the chemokine ligand for CCR6, is constitutively expressed in the gut. Although Ccr6 deficiency does not alter the development and expansion of LTi cells in the gut, it seems to affect the production of effector cytokine IL-22. However, two studies from different labs using Ccr6-deficient mice showed opposite results. Sawa et al. observed an increase in IL-22 production by LTi cells in Ccr6-deficient mice65, while Lin and colleagues reported a decreased IL-22 production by Ccr6−/− ILC3s66. It is not clear why this contradiction occurs. One possibility is that the housing conditions of the animal facilities may affect IL-22 production by LTi cells.
Besides CCR6, there are also certain functionally related molecules uniquely expressed by LTi cells. The neurotrophic factor receptor RET is a tyrosine kinase receptor that is expressed mainly by the nervous system, kidney and hematopoietic progenitors to sense the environmental cues such as glial-derived neurontrophic factor (GDNF) family ligands (GFL)67. Interestingly, ILC3s in gut lamina propria also express RET68. We and others found that RET+ cells were highly enriched in the LTi/LTi-like population but not in the NCR+ ILC3s. In response to the neurotrophic factors from glial cells in the gut, the RET+ ILC3s produce more IL-22 through the p38 MAPK-ERK AKT cascade and STAT3 activation to control the activity of epithelial cells. Thus, RET expression on ILC3s forms a glial-ILC3-epithelial cell axis to orchestrate the homeostasis of the gut68.
4. The role of LTi/LTi-like cells in lymphoid structure generation
The naming of LTi cells is because of their critical role in secondary lymphoid organogenesis during the fetal stage69. In the fetal liver, all innate lymphocyte progenitors including that for LTi cells express integrin α4β746, 70, 71, which allows migration of LTi cells towards the lymph nodes or the Peyer’s patch anlagen through interacting with MAdCAM-1 expressed by high endothelial venules (HEVs)72, 73. At the sites of lymph node or the Peyer’s patch anlagen, a group of specialized stromal cells called lymphoid tissue organizer (LTo) cells50, 74 produce chemokine CXCL13, which recruits/attracts LTi cells that selectively express its receptor CXCR550, 75, 76, resulting in the formation of an initial hematopoietic cell cluster. LTi cells then start to express lymphotoxin α1β2 (LTα1β2), which will engage with its receptor LTβR on stromal cells48, 77, 78, 79. The signals through LTβR activate the classical and an alternative NF-κB signaling in stromal cells, leading to their further maturation and the expression of several genes related with secondary lymphoid structure organization such as TNF-related activation-induced cytokine (TRANCE)49, homeostatic chemokine CCL19, CCL21, and CXCL1380, as well as adhesion molecules MAdCAM-1, VCAM-1, and ICAM-181, 82. The IL-7R and TRANCE feedback on LTi cells to enhance their LTα1β2 expression, and thus sustained the activation of stromal cells48. The homeostatic chemokines recruit more lymphocytes including LTi cells and can also function in maintaining the LTα1β2 expression on LTi cells80, 81, 83. The adhesion molecules will also attract/retain LTi cells to expand the initial cluster. Once B and T cells start to be recruited to the developing lymph nodes or Peyer’s patches, they will take over the role of LTi cells in expressing LTα1β2 (Figure 3). The importance of the molecules mentioned above during this process has been demonstrated by the causative impairment in lymphoid structure generation in mice with deficiency in these molecules49, 52, 75, 84, 85.
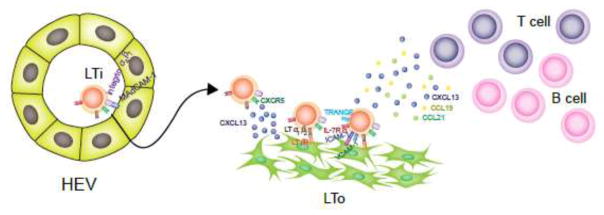
Figure 3. Role of LTi cells in secondary lymphoid structure organogenesis.
In the fetal stage, LTi cells will entry the lymph nodes or Peyer’s patches anlagen through the interaction between integrin α4β7 and MAdCAM-1 on the HEVs. CXCL13 secreted by LTo cells will also help to recruit LTi cells through interact with CXCR5 on them. Then LTi cells start to express lymphotoxin α1β2 (LTα1β2) and engage with the stromal cells expressing its receptor LTβR. This interaction will stimulate the stromal cells to upregulate TRANCE, CXCL13, CCL19, CCL21, MAdCAM-1, VCAM-1, and ICAM-1, which feedback on LTi cells to enhance its LTα1β2 expression and recruit more LTi as well as T and B cells in to the anlagen site.
5. LTi/LTi-like cells in association with T cells
5.1 Effect of thymic LTi cells on T cell development
At the embryo stage, LTi cells appear in the thymus starting from E14 and sustain throughout the adulthood31. These thymic LTi cells are phenotypically similar to LTi cells in other lymphoid tissues. These LTi cells also express TRANCE as other LTi cells, which enables the interaction between LTi cells and epithelial cells in thymic medulla that express the TRANCE receptor, receptor activator of NF-κB (RANK). The signaling initiated by TRANCE/RNAK interaction promotes the expression of AIRE in medullary epithelial cells31, resulting in subsequent expression of self-tissue-restricted antigens that are required for negative selection during T cell development86, 87. Thus, the thymic LTi cells are involved in regulating the central tolerance of T cells to self-antigens through its interaction with medullary epithelial cells in the thymus (Figure 4).
Figure 4. Interactions between LTi cell and T or B cells in the adaptive immune system.
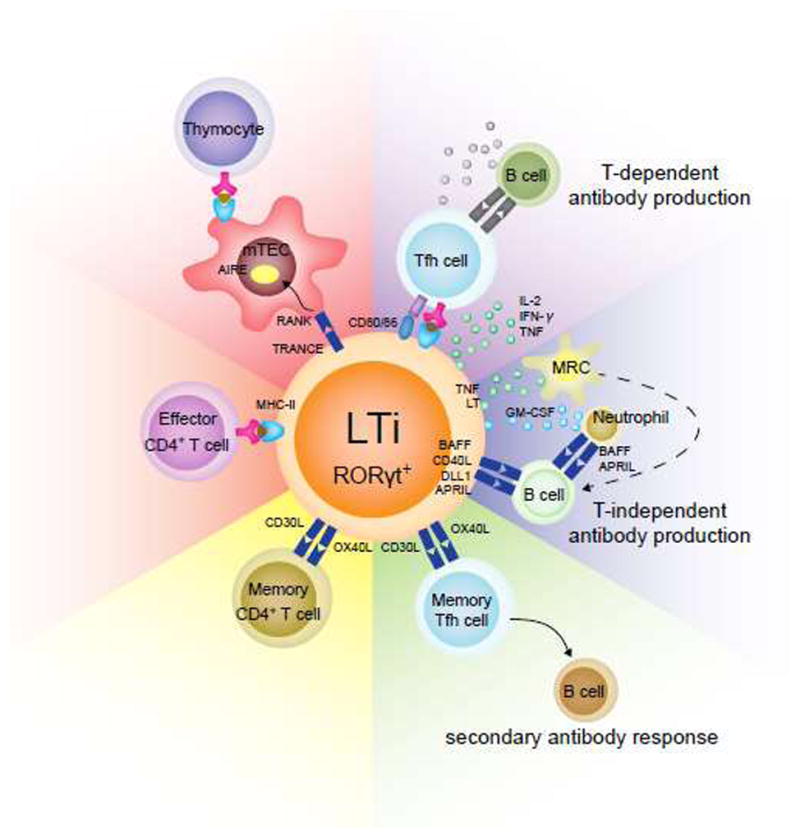
LTi cells can affect development and functions of T and B cells in the adaptive immune system through multiple mechanisms. They interact with the mTECs through RNAK/TRANCE interaction and thus induce the expression of AIRE, which is critical for T cell central tolerance. LTi cells can also directly regulate the effector T cell responses through antigen presentation by MHC-II. The adult LTi-like cells specifically express OX40L and CD30L, which may promote the survival of memory CD4+ T cells. The same mechanism may be responsible for the survival of memory follicular T helper (Tfh) cells and maintenance of memory antibody responses. In addition, LTi cells may promote antibody production by B cells through both T-dependent and -independent manners. Antigen presentation and cytokine secretion to Tfh cells from LTi cells can enhance B cell responses. Alternatively, LTi cells may directly activate B cell through BAFF, CD40L, DLL1 and APRIL expression. TNF, lymphotoxin (LT), and GM-CSF expression by LTi can also activate B cells through marginal reticular cells (MRC) and neutrophils.
5.2 LTi effect on CD4+ T cell memory maintenance
The initial knowledge about the function of LTi-like cells in facilitating memory CD4+ T cell maintenance is from the observation that memory cells do not survive well in Rorc−/− mice in which LTi-like cells are absent88. Further investigation confirmed that LTi-like cells in lymphoid organs are sufficient to mediate the survival of CD4+ memory T cells but not for the CD8+ memory T cells88. LTi-like cells in lymphoid organs locate at the sites of memory T cell recirculation. LTi-like cells in adult stage express two additional T cell survival molecules, OX40L and CD30L, comparing to LTi cells in fetal stage32, 89. While the memory CD4+ T cells express their receptors OX40 and CD30 on the surface. Thus, the interaction between LTi and memory CD4+ T cells will transduce the survival signaling to T cells and facilitate the maintenance of the CD4+ T cell memory (Figure 4).
5.3 Suppression of commensal antigen specific T cells by LTi cells
Besides the involvement in the central tolerance establishment during T cell development, LTi/LTi-like cells were also found to suppress the commensal antigen-specific T effector cells to maintain the homeostasis of gut environment. This role of LTi/LTi-like cells was initially realized from the phenomenon that Rorc-deficient mice harbored more effector/memory-like CD4+ T cells as well as elevated levels of commensal bacteria-specific IgG in serum, indicating that the commensal bacteria are involved in this abnormal activation of CD4+ T cells90. In consistent with this finding, antibiotic administration restores the percentage of effector/memory-like CD4+ T cells and commensal bacteria-specific IgG level in the Rorc-deficient mice to levels comparable to those in wild type mice. Further investigation indicates that expression of major histocompatibility complex-II (MHC-II) on LTi cells is required for limiting the effector/memory CD4+ T cell number, and conditional depletion of the MHC-II from ILC3s recapitulates the phenotype in the Rorc-deficient mice91. Similar as antigen presenting cells (APCs), MHC-II-expressing LTi/LTi-like cells are capable of presenting antigens to CD4+ T cells. However, unlike classical APCs, LTi/LTi-like cells do not express co-stimulatory molecules CD40, CD80 and CD86. Thus, such an antigen presentation by LTi/LTi-like cells in the absence of co-stimulation can induce T cell death and thus limit CD4+ T cell responses90 (Figure 4). Since LTi cells are abundant in intestinal lamina propria, such tolerance mechanism is crucial in maintaining local gut homeostasis. Dysregulated MHC-II expression on LTi cells is found to be linked to patients with inflammatory bowel diseases such as pediatric Crohn’s disease.
6. LTi/LTi-like cells and B cell responses
Increasing evidence indicates that ILC3s are involved in facilitating the antibody production by B cells in either a T cell-dependent or -independent manner92, 93. Though it is not fully clarified in these studies, from cell location or functional gene expression, these ILC3s are presumably LTi/LTi-like cells.
6.1 LTi/LTi-like cells facilitate systemic antibody production
The majority of ILC3s in lymphoid organs such as spleen and lymph nodes are CCR6+ LTi/LTi-like cells. Similar to the gut–resident LTi/LTi-like cells, the ILC3s in the lymphoid organs can also internalize antigens and present antigen peptides to CD4+ T cells, which triggering the T cell dependent antibody production. Different from the situation in the gut, the ILC3s in lymphoid organs start expressing co-stimulatory molecules CD80 and CD86 in the presence of IL-1β, and producing cytokines such as IL-2, IFN-γ and TNF-α towards fully activate T cells93 (Figure 4).
Besides, LTi/LTi-like cells can also promote T cell independent antibody generation against carbohydrate antigens92. LTi/LTi-like cells locate at the marginal zone and perifollicular zone of spleen and lymph nodes94, and can activate the marginal reticular cells (MRC) through the production of TNF and lymphotoxin94. They can also be induced to express BAFF, CD40L, DLL1 and APRIL to promote B cells to produce antibodies against T-independent antigens92. ILC3s in lymphoid organs can also indirectly help B cell responses by secreting GM-CSF which in turn acts on neutrophils to express BAFF and APRIL92 (Figure 4).
Furthermore, LTi/LTi-like cells may facilitate memory B cell responses owing to their role in promoting the T cell memory. The expression of OX40L and CD30L on the surface of LTi/LTi-like cells provides a survival signal to CD4+ T cells and thus enhances the generation of memory T cells including memory T follicular helper (Tfh) cells. As a result, in a secondary antibody response, B cells can respond and mature more quickly to generate high-affinity antibodies95 (Figure 4).
6.2 LTi/LTi-like cells promote mucosal antibody responses
IgA produced by B cells in mucosal tissues such as the intestine mucosa is critical for controlling the composition of commensal bacteria. This IgA production happens in either a T cell-dependent manner mainly in Peyer’s patches or a T cell-independent manner in the lamina propria or the isolated lymphoid follicles (ILFs) of the gut96, 97. In the lamina propria, ILC3s secrete soluble LTα3 which promotes the homing of CD40L-expressing T cells to the gut and thus induce IgA production by B cells.
As we mentioned earlier, LTi/LTi-like cells also express transmembrane LTα1β2 as well as TNF. These molecules are capable of activating the scavenger cells (SCs) in the gut to release CXCL13, CCL19 and CCL20 that are important for recruiting B cells and dendritic cells97. Then the interaction between dendritic cells and LTi/LTi-like cells, in addition to the stimulation of dendritic cells by microbial TLR ligands, will induce the generation of active TGF-β1, which is required for the IgA production97, 98. LTi/LTi-like cells can also promote SCs, dendritic cells and macrophages to produce BAFF through LTα1β2, TRANCE, and LIGHT97.
7. Concluding remarks
LTi cell population is the first identified innate lymphoid cells. They have been considered as a subgroup within the T helper-like ILC family due to their similarity to the NCR+ ILC3s in terms of their immune functions. However, several clues also indicate that LTi cells have their unique features that may have been overlooked. Obviously, LTi cells are associated with the secondary lymphoid organogenesis. Moreover, as we discussed above, LTi cells are also associated with the adaptive immune system in multiple aspects, including facilitating the central tolerance during the generation of T cells, repressing the activation of commensal antigen-specific CD4+ T effector cells in the gut, promoting the survival of CD4+ memory T cells, and supporting the production of T-dependent or -independent antibodies in lymphoid or mucosal tissues. Thus, regarding their roles in modulating adaptive immune system, it is possible that LTi cells and the generation of adaptive immune system during evolution may have a close relationship. All innate lymphocytes are supposed to originate from a common progenitor. Some recent studies indeed support this hypothesis. However, unlike the functional similarity between LTi and other ILC3s, the lineage development of LTi is nevertheless distinct from all other ILC subsets, as well as the cytotoxic conventional NK cells. Taking together, we should consider LTi cells as the third major group of innate lymphocytes, in parallel with cNK cells and T helper-like ILCs that are derived from the PLZF+ progenitors.
Summary.
Lymphoid tissue inducers (LTi cells) are developmentally distinct from other innate lymphoid cells (ILCs).
LTi cells have a unique transcriptome which is associated with their specialized functions.
LTi cells are associated with the adaptive immune system through multiple mechanisms.
LTi cells represent a third innate lymphocyte population which is in parallel with NK cells and other non-LTi ILCs.
Acknowledgments
C.Z. is supported by the One Hundred Talents Program of Peking University Health Science Center (BMU20160551) and the National Natural Science Foundation of China (No. 31770957). M.Z. and J.Z. are supported by the Intramural Research Program of the NIH, NIAID.
Biographies

Chao Zhong received his Ph.D. in biochemistry and molecular biology from Institute of Biophysics, Chinese Academy of Sciences in Beijing, China. He completed his postdoctoral training at the National Institutes of Health in Maryland, U.S., under the mentorship of Dr. Jinfang Zhu, where he studied extensively on the transcriptional regulations underlying innate lymphoid cell development, maturation and function. He is currently a principle investigator of Peking University in Beijing, China, and is continuing to focus his research on the development and functions of various innate/innate-like immunocytes in mucosal tissues.

Mingzhu Zheng received her Ph.D. in immunology from Zhejiang University School of Medicine at Hangzhou, Zhejiang, China. She is currently a postdoctoral fellow in the Laboratory of Immune System Biology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, under the mentorship of Dr. Jinfang Zhu, investigating the network relationship of multiple transcription factors during the development of innate lymphoid cell subsets.

Jinfang Zhu received his Ph.D. in biochemistry and molecular biology from the Shanghai Institute of Biochemistry, Chinese Academy of Sciences. He completed his postdoctoral training at the Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, with late Dr. William E. Paul, studying CD4 T helper cell differentiation. He started his own group in the Laboratory of Immunology as an Earl Stadtman investigator, and is now the section chief of the Molecular and Cellular Immunoregulation Section in the Laboratory of Immune System Biology. His lab studies functional heterogeneity and plasticity of CD4 T helper and innate lymphoid cell subsets in the steady state and during infection or inflammation.
Footnotes
Conflicts of interest
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 2.Eberl G, Di Santo JP, Vivier E. The brave new world of innate lymphoid cells. Nat Immunol. 2015;16:1–5. doi: 10.1038/ni.3059. [DOI] [PubMed] [Google Scholar]
- 3.Fallon PG, et al. Identification of an interleukin (IL)-25-dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. The Journal of experimental medicine. 2006;203:1105–1116. doi: 10.1084/jem.20051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moro K, et al. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- 5.Neill DR, et al. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price AE, et al. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11489–11494. doi: 10.1073/pnas.1003988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satoh-Takayama N, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. The Journal of experimental medicine. 2017;214:1861–1876. doi: 10.1084/jem.20170494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs A, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sojka DK, et al. Tissue-resident natural killer (NK) cells are cell lineages distinct from thymic and conventional splenic NK cells. eLife. 2014;3:e01659. doi: 10.7554/eLife.01659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delconte RB, et al. The Helix-Loop-Helix Protein ID2 Governs NK Cell Fate by Tuning Their Sensitivity to Interleukin-15. Immunity. 2016;44:103–115. doi: 10.1016/j.immuni.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Miyazaki M, et al. The E-Id Protein Axis Specifies Adaptive Lymphoid Cell Identity and Suppresses Thymic Innate Lymphoid Cell Development. Immunity. 2017;46:818–834 e814. doi: 10.1016/j.immuni.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernink JH, et al. Interleukin-12 and -23 Control Plasticity of CD127(+) Group 1 and Group 3 Innate Lymphoid Cells in the Intestinal Lamina Propria. Immunity. 2015;43:146–160. doi: 10.1016/j.immuni.2015.06.019. [DOI] [PubMed] [Google Scholar]
- 17.Ohne Y, et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat Immunol. 2016;17:646–655. doi: 10.1038/ni.3447. [DOI] [PubMed] [Google Scholar]
- 18.Lim AI, et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med. 2016;213:569–583. doi: 10.1084/jem.20151750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakayamada S, Takahashi H, Kanno Y, O’Shea JJ. Helper T cell diversity and plasticity. Curr Opin Immunol. 2012;24:297–302. doi: 10.1016/j.coi.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Paul WE. Heterogeneity and plasticity of T helper cells. Cell research. 2010;20:4–12. doi: 10.1038/cr.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 22.Klose CS, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 23.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3− LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 24.Fukuyama S, et al. Initiation of NALT organogenesis is independent of the IL-7R, LTbetaR, and NIK signaling pathways but requires the Id2 gene and CD3(−)CD4(+)CD45(+) cells. Immunity. 2002;17:31–40. doi: 10.1016/s1074-7613(02)00339-4. [DOI] [PubMed] [Google Scholar]
- 25.Mebius RE. Organogenesis of lymphoid tissues. Nature reviews. 2003;3:292–303. doi: 10.1038/nri1054. [DOI] [PubMed] [Google Scholar]
- 26.van de Pavert SA, Mebius RE. New insights into the development of lymphoid tissues. Nat Rev Immunol. 2010;10:664–674. doi: 10.1038/nri2832. [DOI] [PubMed] [Google Scholar]
- 27.Randall TD, Carragher DM, Rangel-Moreno J. Development of secondary lymphoid organs. Annu Rev Immunol. 2008;26:627–650. doi: 10.1146/annurev.immunol.26.021607.090257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eberl G, et al. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- 29.Kim MY. Roles of embryonic and adult lymphoid tissue inducer cells in primary and secondary lymphoid tissues. Yonsei Med J. 2008;49:352–356. doi: 10.3349/ymj.2008.49.3.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane P, et al. Lymphoid tissue inducer cells in adaptive CD4 T cell dependent responses. Semin Immunol. 2008;20:159–163. doi: 10.1016/j.smim.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Rossi SW, et al. RANK signals from CD4(+)3(−) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–1272. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim MY, et al. CD4(+)CD3(−) accessory cells costimulate primed CD4 T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity. 2003;18:643–654. doi: 10.1016/s1074-7613(03)00110-9. [DOI] [PubMed] [Google Scholar]
- 33.Soroosh P, Ine S, Sugamura K, Ishii N. OX40-OX40 ligand interaction through T cell-T cell contact contributes to CD4 T cell longevity. J Immunol. 2006;176:5975–5987. doi: 10.4049/jimmunol.176.10.5975. [DOI] [PubMed] [Google Scholar]
- 34.Rogers PR, Song J, Gramaglia I, Killeen N, Croft M. OX40 promotes Bcl-xL and Bcl-2 expression and is essential for long-term survival of CD4 T cells. Immunity. 2001;15:445–455. doi: 10.1016/s1074-7613(01)00191-1. [DOI] [PubMed] [Google Scholar]
- 35.Bansal-Pakala P, Halteman BS, Cheng MH, Croft M. Costimulation of CD8 T cell responses by OX40. J Immunol. 2004;172:4821–4825. doi: 10.4049/jimmunol.172.8.4821. [DOI] [PubMed] [Google Scholar]
- 36.Gaspal FM, et al. Mice deficient in OX40 and CD30 signals lack memory antibody responses because of deficient CD4 T cell memory. J Immunol. 2005;174:3891–3896. doi: 10.4049/jimmunol.174.7.3891. [DOI] [PubMed] [Google Scholar]
- 37.Robinette ML, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nature immunology. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rankin LC, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sciume G, et al. Distinct requirements for T-bet in gut innate lymphoid cells. The Journal of experimental medicine. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vonarbourg C, et al. Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cupedo T, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- 42.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoorweg K, et al. Functional Differences between Human NKp44(−) and NKp44(+) RORC(+) Innate Lymphoid Cells. Front Immunol. 2012;3:72. doi: 10.3389/fimmu.2012.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glatzer T, et al. RORgammat(+) innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity. 2013;38:1223–1235. doi: 10.1016/j.immuni.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508:397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klose CSN, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- 47.Yagi R, et al. The transcription factor GATA3 is critical for the development of all IL-7Ralpha-expressing innate lymphoid cells. Immunity. 2014;40:378–388. doi: 10.1016/j.immuni.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshida H, et al. Different cytokines induce surface lymphotoxin-alphabeta on IL-7 receptor-alpha cells that differentially engender lymph nodes and Peyer’s patches. Immunity. 2002;17:823–833. doi: 10.1016/s1074-7613(02)00479-x. [DOI] [PubMed] [Google Scholar]
- 49.Kim D, et al. Regulation of peripheral lymph node genesis by the tumor necrosis factor family member TRANCE. J Exp Med. 2000;192:1467–1478. doi: 10.1084/jem.192.10.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finke D, Acha-Orbea H, Mattis A, Lipp M, Kraehenbuhl J. CD4+CD3− cells induce Peyer’s patch development: role of alpha4beta1 integrin activation by CXCR5. Immunity. 2002;17:363–373. doi: 10.1016/s1074-7613(02)00395-3. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida H, et al. IL-7 receptor alpha+ CD3(−) cells in the embryonic intestine induces the organizing center of Peyer’s patches. Int Immunol. 1999;11:643–655. doi: 10.1093/intimm/11.5.643. [DOI] [PubMed] [Google Scholar]
- 52.Adachi S, et al. Essential role of IL-7 receptor alpha in the formation of Peyer’s patch anlage. Int Immunol. 1998;10:1–6. doi: 10.1093/intimm/10.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Hashi H, et al. Compartmentalization of Peyer’s patch anlagen before lymphocyte entry. J Immunol. 2001;166:3702–3709. doi: 10.4049/jimmunol.166.6.3702. [DOI] [PubMed] [Google Scholar]
- 54.Mielke LA, et al. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol. 2013;191:4383–4391. doi: 10.4049/jimmunol.1301228. [DOI] [PubMed] [Google Scholar]
- 55.Yang Q, et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 2013;38:694–704. doi: 10.1016/j.immuni.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang Q, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nature immunology. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seehus CR, et al. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nature immunology. 2015;16:599–608. doi: 10.1038/ni.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aliahmad P, de la Torre B, Kaye J. Shared dependence on the DNA-binding factor TOX for the development of lymphoid tissue-inducer cell and NK cell lineages. Nat Immunol. 2010;11:945–952. doi: 10.1038/ni.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geiger TL, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. The Journal of experimental medicine. 2014;211:1723–1731. doi: 10.1084/jem.20140212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W, et al. NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep. 2015;10:2043–2054. doi: 10.1016/j.celrep.2015.02.057. [DOI] [PubMed] [Google Scholar]
- 61.Possot C, et al. Notch signaling is necessary for adult, but not fetal, development of RORgammat(+) innate lymphoid cells. Nat Immunol. 2011;12:949–958. doi: 10.1038/ni.2105. [DOI] [PubMed] [Google Scholar]
- 62.Ishizuka IE, et al. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nature immunology. 2016;17:269–276. doi: 10.1038/ni.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song C, et al. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. The Journal of experimental medicine. 2015;212:1869–1882. doi: 10.1084/jem.20151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong C, et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nature immunology. 2016;17:169–178. doi: 10.1038/ni.3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sawa S, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 66.Lin YL, Ip PP, Liao F. CCR6 Deficiency Impairs IgA Production and Dysregulates Antimicrobial Peptide Production, Altering the Intestinal Flora. Front Immunol. 2017;8:805. doi: 10.3389/fimmu.2017.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Costantini F. GDNF/Ret signaling and renal branching morphogenesis: From mesenchymal signals to epithelial cell behaviors. Organogenesis. 2010;6:252–262. doi: 10.4161/org.6.4.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ibiza S, et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature. 2016;535:440–443. doi: 10.1038/nature18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fu YX, Chaplin DD. Development and maturation of secondary lymphoid tissues. Annu Rev Immunol. 1999;17:399–433. doi: 10.1146/annurev.immunol.17.1.399. [DOI] [PubMed] [Google Scholar]
- 70.Mebius RE, et al. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3− cells, as well as macrophages. J Immunol. 2001;166:6593–6601. doi: 10.4049/jimmunol.166.11.6593. [DOI] [PubMed] [Google Scholar]
- 71.Yoshida H, et al. Expression of alpha(4)beta(7) integrin defines a distinct pathway of lymphoid progenitors committed to T cells, fetal intestinal lymphotoxin producer, NK, and dendritic cells. J Immunol. 2001;167:2511–2521. doi: 10.4049/jimmunol.167.5.2511. [DOI] [PubMed] [Google Scholar]
- 72.Iizuka T, et al. Stage-specific expression of mucosal addressin cell adhesion molecule-1 during embryogenesis in rats. J Immunol. 2000;164:2463–2471. doi: 10.4049/jimmunol.164.5.2463. [DOI] [PubMed] [Google Scholar]
- 73.Mebius RE, Schadee-Eestermans IL, Weissman IL. MAdCAM-1 dependent colonization of developing lymph nodes involves a unique subset of CD4+CD3− hematolymphoid cells. Cell Adhes Commun. 1998;6:97–103. doi: 10.3109/15419069809004464. [DOI] [PubMed] [Google Scholar]
- 74.Onder L, et al. Lymphatic Endothelial Cells Control Initiation of Lymph Node Organogenesis. Immunity. 2017;47:80–92 e84. doi: 10.1016/j.immuni.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 75.Ansel KM, et al. A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature. 2000;406:309–314. doi: 10.1038/35018581. [DOI] [PubMed] [Google Scholar]
- 76.McDonald KG, McDonough JS, Dieckgraefe BK, Newberry RD. Dendritic cells produce CXCL13 and participate in the development of murine small intestine lymphoid tissues. Am J Pathol. 2010;176:2367–2377. doi: 10.2353/ajpath.2010.090723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Futterer A, Mink K, Luz A, Kosco-Vilbois MH, Pfeffer K. The lymphotoxin beta receptor controls organogenesis and affinity maturation in peripheral lymphoid tissues. Immunity. 1998;9:59–70. doi: 10.1016/s1074-7613(00)80588-9. [DOI] [PubMed] [Google Scholar]
- 78.Cupedo T, et al. Presumptive lymph node organizers are differentially represented in developing mesenteric and peripheral nodes. J Immunol. 2004;173:2968–2975. doi: 10.4049/jimmunol.173.5.2968. [DOI] [PubMed] [Google Scholar]
- 79.Rennert PD, James D, Mackay F, Browning JL, Hochman PS. Lymph node genesis is induced by signaling through the lymphotoxin beta receptor. Immunity. 1998;9:71–79. doi: 10.1016/s1074-7613(00)80589-0. [DOI] [PubMed] [Google Scholar]
- 80.Honda K, et al. Molecular basis for hematopoietic/mesenchymal interaction during initiation of Peyer’s patch organogenesis. J Exp Med. 2001;193:621–630. doi: 10.1084/jem.193.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dejardin E, et al. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 82.Drayton DL, Ying X, Lee J, Lesslauer W, Ruddle NH. Ectopic LT alpha beta directs lymphoid organ neogenesis with concomitant expression of peripheral node addressin and a HEV-restricted sulfotransferase. J Exp Med. 2003;197:1153–1163. doi: 10.1084/jem.20021761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cupedo T, Mebius RE. Cellular interactions in lymph node development. J Immunol. 2005;174:21–25. doi: 10.4049/jimmunol.174.1.21. [DOI] [PubMed] [Google Scholar]
- 84.Dougall WC, et al. RANK is essential for osteoclast and lymph node development. Genes Dev. 1999;13:2412–2424. doi: 10.1101/gad.13.18.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2:223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 86.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 87.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 88.Withers DR, et al. Cutting edge: lymphoid tissue inducer cells maintain memory CD4 T cells within secondary lymphoid tissue. J Immunol. 2012;189:2094–2098. doi: 10.4049/jimmunol.1201639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim MY, et al. OX40 ligand and CD30 ligand are expressed on adult but not neonatal CD4+CD3− inducer cells: evidence that IL-7 signals regulate CD30 ligand but not OX40 ligand expression. J Immunol. 2005;174:6686–6691. doi: 10.4049/jimmunol.174.11.6686. [DOI] [PubMed] [Google Scholar]
- 90.Hepworth MR, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hepworth MR, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4(+) T cells. Science (New York, NY) 2015;348:1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magri G, et al. Innate lymphoid cells integrate stromal and immunological signals to enhance antibody production by splenic marginal zone B cells. Nat Immunol. 2014;15:354–364. doi: 10.1038/ni.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.von Burg N, et al. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc Natl Acad Sci U S A. 2014;111:12835–12840. doi: 10.1073/pnas.1406908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim MY, et al. Function of CD4+CD3− cells in relation to B- and T-zone stroma in spleen. Blood. 2007;109:1602–1610. doi: 10.1182/blood-2006-04-018465. [DOI] [PubMed] [Google Scholar]
- 95.Lane PJ, Gaspal FM, Kim MY. Two sides of a cellular coin: CD4(+)CD3− cells regulate memory responses and lymph-node organization. Nat Rev Immunol. 2005;5:655–660. doi: 10.1038/nri1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fagarasan S, Kawamoto S, Kanagawa O, Suzuki K. Adaptive immune regulation in the gut: T cell-dependent and T cell-independent IgA synthesis. Annual review of immunology. 2010;28:243–273. doi: 10.1146/annurev-immunol-030409-101314. [DOI] [PubMed] [Google Scholar]
- 97.Tsuji M, et al. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 2008;29:261–271. doi: 10.1016/j.immuni.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 98.Cazac BB, Roes J. TGF-beta receptor controls B cell responsiveness and induction of IgA in vivo. Immunity. 2000;13:443–451. doi: 10.1016/s1074-7613(00)00044-3. [DOI] [PubMed] [Google Scholar]