Abstract
Osteosarcoma is an aggressive cancer of the skeletal system, which is associated with a poor prognosis due to the high recurrence rate. Although previous studies have revealed that competitive endogenous RNAs (ceRNAs) are involved in various biological processes in the physiology and development of osteosarcoma, the roles of ceRNAs in osteosarcoma recurrence remain largely unexplored. The present study constructed a ceRNA-ceRNA network for osteosarcoma by systematically integrating matched expression profiles for microRNAs (miRNAs/miRs) and mRNAs, and identified two ceRNA-mediated modules that were associated with recurrence in patients with osteosarcoma. A multivariate Cox regression analysis demonstrated that the recurrence-free prognosis associated with the expression of the two modules was independent of other clinical factors. In addition, hsa-miR-335-3p was identified as an upstream regulating factor for both modules. In conclusion, the results of the present study suggested that ceRNAs may act as potential therapeutic biomarkers for predicting the recurrence of osteosarcoma, and may help to identify patients with osteosarcoma at a high risk of recurrence, who may benefit from adjuvant therapy.
Keywords: competitive endogenous RNAs, microRNA, regulation, network, prognostic modules, osteosarcoma
Introduction
As the most common primary bone malignancy, osteosarcoma is one of the major causes of cancer-associated mortality in children and young adults (1,2). In recent years, with the development of comprehensive treatments, the 5-year survival rate for patients with osteosarcoma has significantly improved (3–6). However, patients that experience osteosarcoma recurrence still have a low 5-year survival rate, as few clinical prognostic biomarkers for recurrence have been identified (7,8). Previous studies have characterized the cellular mechanisms underlying osteosarcoma and have identified numerous potential therapeutic targets (9–11). Wang et al reported that a competitive endogenous RNA (ceRNA) may be a therapeutic target for the molecular therapy of osteosarcoma (12). Furthermore, a growing number of studies have demonstrated that ceRNAs regulate the processes of osteosarcoma initiation and progression, including tumor cell differentiation, proliferation, apoptosis and metastasis (13,14). These studies have implicated ceRNAs in various biological processes in normal physiology and the development of osteosarcoma; however, the prognostic role of ceRNAs in recurrent osteosarcoma remains unclear. In particular, there are currently no satisfactory biomarkers for the recurrence of osteosarcoma following standard treatment. Therefore, it is important to identify potential molecular markers associated with the recurrence of osteosarcoma, which may provide valuable information for the diagnosis and treatment of this disease.
The present study constructed a ceRNA-ceRNA network for osteosarcoma (OSceNet) by repurposing publicly available matched expression profiles for microRNA (miRNA/miR) and mRNA from the Gene Expression Omnibus (GEO) database. The present analysis identified two recurrence-free survival-associated modules that were regulated by hsa-miR-335-5p, and determined that the expression levels of the ceRNAs in each module could distinguish patients with good and poor survival outcomes based on the risk score model method. These findings demonstrated the association between ceRNA modules and tumor recurrence in osteosarcoma, potentially allowing for the detection of ceRNA biomarkers associated with osteosarcoma recurrence.
Materials and methods
Data collection
miRNA and mRNA expression profiles for osteosarcoma prior to and following treatment were obtained from the GEO database (accession nos. GSE39040 and GSE39055, respectively; https://www.ncbi.nlm.nih.gov/geo/) (15). Probes were mapped to genes; a gene with numerous probes was represented by the mean of the expression values of the probes. Matching miRNA and mRNA expression profiles were obtained for further analysis. Clinical data for the patients with osteosarcoma were also obtained for the two datasets, including age, sex, recurrence status and other clinical information.
Human miRNA-mRNA target data was collected from TarBase (16), miRTarBase (17) and miRecords (18), which manually curate data regarding high-quality, experimentally validated miRNA-mRNA target interactions from published experiments. By integrating data from these three databases, a total of 44,181 non-redundant miRNA-mRNA target interactions were obtained.
Construction of the OSceNet
The OSceNet was constructed using R software (v3.2.1; http://mirrors.tuna.tsinghua.edu.cn/CRAN/) according to two principles: The two mRNAs were regulated by the same miRNA, and the two mRNAs followed the same expression pattern. Firstly, correlations between the previously validated miRNA-mRNA target pairs in the matched miRNA and mRNA expression profiles were evaluated by Pearson correlation coefficients. The pairs with a significant negative correlation were regarded as relevant miRNA-mRNA pairs in the context of osteosarcoma (R< −0.4, adjusted P<0.05). Secondly, all mRNA-mRNA pairs that were both targeted by at least one miRNA were listed as candidate ceRNA pairs. Thirdly, for each candidate ceRNA pair, the Pearson correlation coefficient between them was calculated. All candidate ceRNA pairs with R>0.4 and adjusted P<0.05 were considered ceRNA-ceRNA interactions. By assembling all the identified ceRNA pairs, the OSceNet was constructed. Within the OSceNet, each node represents an mRNA, and two nodes are connected if they are co-regulated by at least one miRNA and are co-expressed in osteosarcoma.
Network visualization
The networks were visualized using Cytoscape 3.3.0 (19), including the OSceNet and the modules view.
Recurrence analysis using the risk score model
The present study assessed the impact on the recurrence-free prognosis for each module of the OSceNet using the risk score model. The risk score model was constructed by considering the power of each of the ceRNAs in the module, as evaluated by univariate Cox regression analysis, and the relative expression of each ceRNA, as follows:
where n is the number of ceRNAs in the module, expi is the expression level of ceRNA i, and coefi is the estimated regression coefficient for ceRNA i from the univariate Cox regression analysis. The median risk score value was selected as a cutoff to classify patients into high- and low-risk groups. Recurrence-free survival analyses were performed to assess the difference in recurrence probability between the high- and low-risk groups, and the statistical significance was determined by a log-rank test using the R package ‘survival’ (https://github.com/therneau/survival).
Statistical analysis
Random networks with the same architecture in OSceNet were generated using R package ‘igraph’ (http://igraph.org). Modules in OSceNet were identified using cFinder software (20). Multivariate Cox regression analyses were performed using Cox proportional hazards regression model to determine whether the prognostic models were independent of other clinical variables, adjusting for age, sex, primary disease site and chemosensitivity as covariates. The time-dependent receiver operating characteristic (ROC) curve analysis was performed using R software. All statistical analyses were performed using R software (v3.2.1), including Fisher’s exact test and Wilcoxon sum rank test. In the present study, P<0.1 was considered to indicate a statistically significant difference due to the small sample size (37 patients).
Results
Characterization of the topological features and competitive interactions in the OSceNet
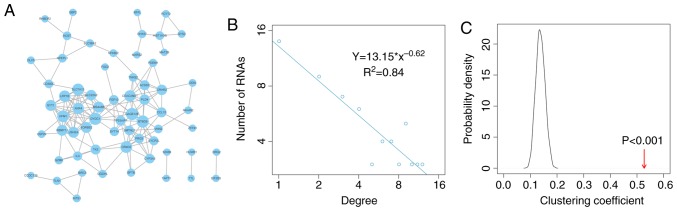
The matched miRNA and mRNA expression profiles of 37 patients with osteosarcoma were obtained from the GEO database, after which the OSceNet was constructed. Three steps were followed to generate the network: i) A total of 130 miRNA-mRNA pairs were determined between 28 miRNAs and 122 mRNAs, which were significantly negatively correlated in osteosarcoma (R< -0.4, adjusted P<0.05). ii) A total of 1,826 candidate ceRNA-ceRNA pairs were identified, which were co-regulated by at least one miRNA. iii) A total of 156 candidate ceRNA-ceRNA pairs were identified as positively correlated with each other (R>0.4, adjusted P<0.05), forming a network of 64 mRNAs (Fig. 1A).
Figure 1.
OSceNet and topological features. (A) Global view of the OSceNet. Larger nodes denote a higher degree in the OSceNet. (B) Degree distribution of OSceNet. (C) Clustering coefficient of the OSceNet was higher than that of random networks. Red arrow represents the clustering coefficient in the OSceNet. ceRNA, competing endogenous RNA; OSceNet, ceRNA-ceRNA network for osteosarcoma.
To explore the structure and features of the OSceNet, degree distribution analysis of the nodes in the network was performed. The present study identified that most nodes had relatively few interactions, whereas a small proportion of nodes had several interactions, which fits with a power-law distribution, thus suggesting that the network was scale-free and different from randomly generated networks (Fig. 1B). Subsequently, 1,000 random networks were generated that preserved the same number of nodes and edges, and the same degree for each node as in OSceNet using the R package ‘igraph’. Comparing the clustering coefficient between the OSceNet and the random networks revealed strong, non-random competitive interactions between the OSceNet ceRNA-ceRNA pairs (Fig. 1C; P<0.001).
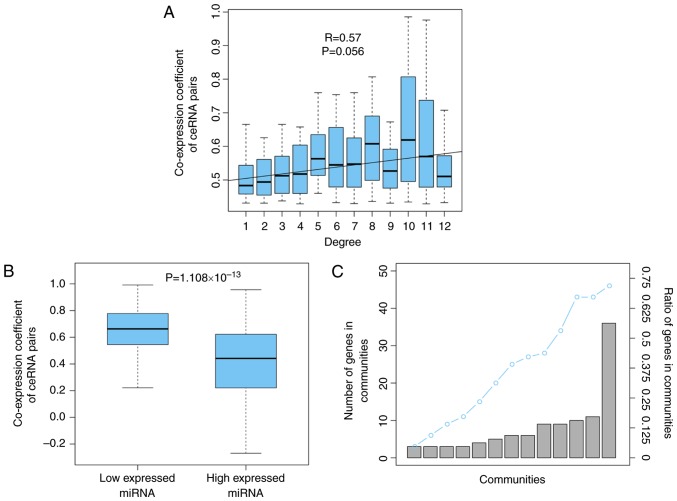
To analyze the competitive intensity among the ceRNA interaction pairs in OSceNet, the present study demonstrated that the nodes with a higher degree, indicating more competitors, were more strongly regulated by their interacting neighbors (R=0.57, P=0.056; Fig. 2A), which was consistent with a previous study (21). In addition, the present study investigated the effects of miRNA expression on ceRNA-ceRNA interactions. For each miRNA shared by a ceRNA pair, the tumor expression profiles were divided into high and low miRNA expression, using the median value for miRNA expression as the cutoff. By comparing the co-expression of the ceRNA pairs between these two groups of tumor samples, it was revealed that the ceRNA pairs displayed significantly increased competition in samples with low miRNA expression (P=1.108×10−13; Fig. 2B). This is understandable, as mRNAs will be fully suppressed when the interacting miRNA is abundantly expressed; however, when the miRNA molecules are relatively sparse, the targeted mRNAs will only be partially suppressed, and the upregulation of one mRNA will further weaken the miRNA suppression for its partner, resulting in strong co-expression of the ceRNA pairs. Subsequently, the present study analyzed the modular structure of the OSceNet. A total of 13 modules were identified using cFinder software (22). Analysis of the genes in the modules showed that 72% of the nodes from the OSceNet were part of at least one module, thus indicating that the ceRNA pairs tend to interact with other pairs, as a module, in osteosarcoma (Fig. 2C).
Figure 2.
Competitive interactions of the OSceNet. (A) ceRNAs with higher degree appeared to be more co-expressed with neighbors. (B) Co-expression of ceRNA pairs is stronger in the samples with low miRNA expression. (C) Most genes in the OSceNet were involved in at least one module. ceRNA, competing endogenous RNA; miRNA, microRNA; OSceNet, ceRNA-ceRNA network for osteosarcoma.
Identification of two recurrence-free survival-associated modules
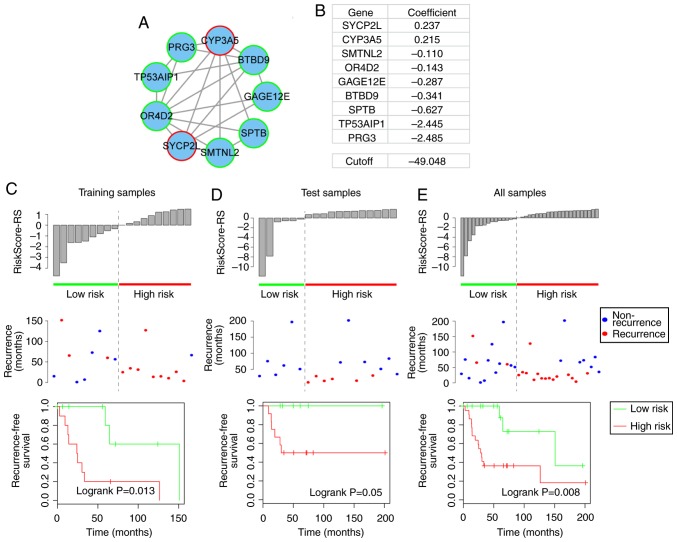
To identify the modules with the most robust ability to predict recurrence-free survival for patients with osteosarcoma, the present study randomly divided the 37 samples into two groups: The training dataset, which contained 19 samples, and the test dataset, which consisted of 18 samples. To avoid the prognostic noise of clinical factors, the division was balanced to obtain two datasets with similar clinical information (Table I). For each of the 13 modules identified, the associated recurrence-free survival prognosis was evaluated using a module-specific risk score model (23,24). The risk score model was constructed according to the linear combination of the expression values of genes in the module weighted by the regression coefficients derived from univariate Cox regression analysis (25,26). For example, module 1 included two risk-associated genes, which had positive regression coefficients, and seven protective genes, which had negative regression coefficients, as identified by univariate Cox regression analysis, and the risk score model for module 1 was as follows: Risk score=[0.237× expression value of synaptonemal complex protein 2-like (SYCP2L)]+[0.215× expression value of cytochrome P450 family 3 subfamily A member 5 (CYP3A5)]+[−0.11× expression value of smoothelin-like 2 (SMTNL2)]+[−0.143× expression value of olfactory receptor family 4 subfamily D member 2 (OR4D2)]+[−0.287× expression value of G antigen 12E (GAGE12E)]+[−0.341× expression value of BTB domain-containing 9 (BTBD9)]+[−0.627× expression value of spectrin β, erythrocytic (SPTB)]+[−2.445× expression value of tumor protein p53-regulated apoptosis-inducing protein 1 (TP53AIP1)]+[−2.485× expression value of proteoglycan 3, pro eosinophil major basic protein 2 (PRG3)]. The present study then calculated the risk score of each module for each sample in the training dataset, and divided the samples into a low-risk group (e.g. n=9 in module 1) and a high-risk group (e.g. n=10 in module 1) with the median risk score (e.g. risk score=−49.048 in module 1) as the cutoff. The prognosis of each module was then validated in the test dataset and complete datasets, using the same procedure with an unchanged risk score model and cutoff. The two modules that most robustly predicted the recurrence-free survival prognosis were then selected, as shown in Figs. 3 and 4.
Table I.
Clinical factors of patients in the training and test datasets.
| Variable | Training set (n=19) |
Test set (n=18) |
P-value |
|---|---|---|---|
| Sex | |||
| Female | 8 | 9 | 0.746a |
| Male | 11 | 9 | |
| Site | |||
| Left | 11 | 13 | 0.495a |
| Right | 8 | 5 | |
| Chemosensitivity | |||
| Favorable | 7 | 7 | 1a |
| Unfavorable | 12 | 11 | |
| Recurrence | |||
| Yes | 12 | 6 | 0.103a |
| No | 7 | 12 | |
| Age (mean ± SD) | 13.32±6.87 | 13.67±14.91 | 0.445b |
| Recurrence time (mean ± SD) | 46.89±45.11 | 59.28±55.55 | 0.328b |
P-values were calculated by Fisher’s exact test;
P-values were calculated by Wilcoxon rank sum test. The statistical analyses were performed using R software. SD, standard deviation.
Figure 3.
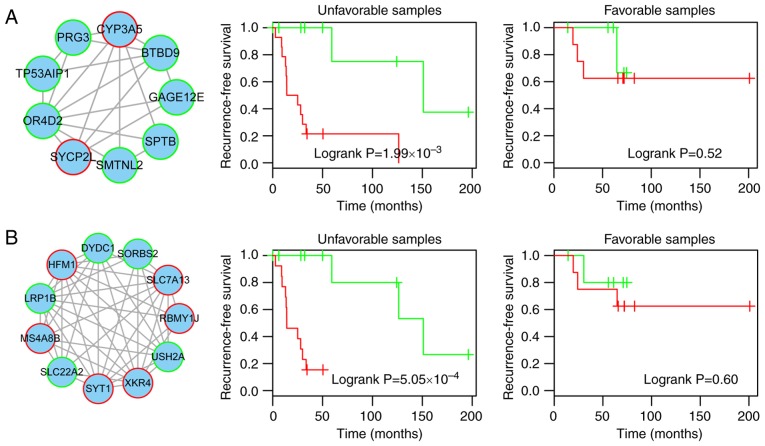
Recurrence-free prognosis of module 1. (A) Module 1. Red-bordered nodes represent risk genes, whereas green-bordered nodes represent protective genes for osteosarcoma. (B) Risk coefficients of genes in module 1, as calculated by the univariate Cox regression model and the cutoff (median value of training samples) of the risk score for high-risk and low-risk patients. (C–E) Risk score distribution derived from module 1; recurrence status and recurrence-free survival of high-risk and low-risk patients in the training, test and complete datasets.
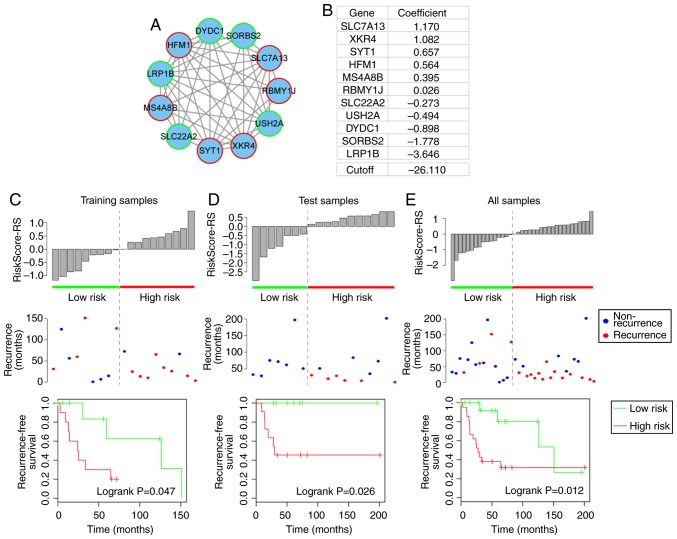
Figure 4.
Recurrence-free prognosis of module 2. (A) Module 2. Red-bordered nodes represent risk genes, whereas green-bordered nodes represent protective genes for osteosarcoma. (B) Risk coefficients of genes in module 2, as calculated by the univariate Cox regression model and the cutoff (median value of training samples) of the risk score for high-risk and low-risk patients. (C-E) Risk score distribution derived from module 2; recurrence status and recurrence-free survival of high-risk and low-risk patients in the training, test and complete datasets.
Prognostic module 1
Prognostic module 1 consisted of two risk genes (SYCP2L and CYP3A5) and seven protective genes (SMTNL2, OR4D2, GAGE12E, BTBD9, SPTB, TP53AIP1 and PRG3) (Fig. 3A and B). Of these, previous studies also revealed CYP3A5 to be a risk gene in osteosarcoma. For example, Dhaini et al reported that the expression levels of CYP3A5 were significantly increased in primary biopsies from patients who developed distant metastatic disease compared with biopsies from patients with nonmetastatic disease (27). Furthermore, Trujillo-Paolillo et al also reported that CYP genes serve an important role in osteosarcoma tumorigenesis at primary and metastatic sites, as well as in treatment response (28). The present study revealed a novel potential mechanism by which CYP3A5 may promote osteosarcoma tumorigenesis via its ceRNA activity.
Compared with the low-risk samples in the training, test and complete datasets, the high-risk samples, as defined by module 1 exhibited a higher rate of recurrence (training, 33.3 vs. 90%; test, 0 vs. 50%; total, 20 vs. 68.2%) and a significantly reduced recurrence-free survival time (log-rank P=0.013, 0.05 and 0.008, respectively; Fig. 3C–E).
Prognostic module 2
Prognostic module 2, as shown in Fig. 4A, contained 11 genes, including six risk genes [solute carrier family 7 member 13 (SLC7A13), XK-related 4 (XKR4), synaptotagmin 1, HFM1, ATP-dependent DNA helicase homolog (HFM1), membrane spanning 4-domains A8 and RNA-binding motif protein, Y-linked, family 1, member J] and five protective genes [solute carrier family 22 member 2, usherin (USH2A), DPY30 domain-containing 1 (DYDC1), sorbin and SH3 domain-containing 2 and low-density lipoprotein (LDL) receptor-related protein 1B (LRP1B)]. From this module, LRP1B, which is a member of the LDL receptor family, is particularly notable, as another member of the LDL receptor family (LRP1/LRP1A) is significant in osteoblast behavior (29). In addition, the present study revealed that LRP1B is a protective gene in osteosarcoma (Fig. 4B). The high- and low-risk samples defined by module 2 also exhibited significantly different associated recurrence-free survival prognosis in the training, test and complete datasets (log-rank P=0.047, 0.026 and 0.012, respectively) (Fig. 4C–E).
Effects of the two identified modules on recurrence-free prognosis are independent of clinical factors
To determine whether the association of the two modules with recurrence-free prognosis was independent and robust, a multivariate Cox regression analysis was conducted, including risk score defined by the modules, age, sex, primary disease site and chemosensitivity as covariates. The results indicated that risk score was independently significant and the most valuable prognostic factor (HR=9.03, P=0.0045 for module 1; HR=13.42, P=0.0014 for module 2; Table II). In addition, chemosensitivity of the samples was also associated with recurrence-free prognosis (P=0.03 for module 1, P=0.0068 for module 2; Table II). Therefore, the present study subsequently inspected the ability of the risk score to predict recurrence-free prognosis in the samples stratified by chemosensitivity into favorable (n=14) and unfavorable (n=23) strata. The patients within each chemosensitivity stratum were then classified into high and low risk scores based on modules 1 and 2, respectively. For the unfavorable samples, the results demonstrated that the risk score variables derived from modules 1 or 2 had significant prognostic value for recurrence-free survival (log-rank P=1.99×10−3 and 5.05×10−4, respectively, Fig. 5). For the favorable chemosensitivity stratum, the prognostic impact of the modules was reduced (Fig. 5). These results indicated that the prognostic power of the module-based risk score was also chemosensitivity-independent, particularly for patients with unfavorable chemosensitivity. For patients with unfavorable chemosensitivity, early diagnosis may be possible using the risk score. In addition, combining chemosensitivity and the risk score may provide more accurate prognostic information for patients.
Table II.
Multivariate Cox regression analysis of the risk score defined by the two modules and recurrence-free survival for patients with osteosarcoma.
| Variable | Module 1
|
Module 2
|
||||
|---|---|---|---|---|---|---|
| HR | 95% CI of HR | P-value | HR | 95% CI of HR | P-value | |
| Risk score | 9.03 | (1.98–41.25) | 4.5×10−3 | 13.42 | (2.73–66.07) | 1.4×10−3 |
| Age | 1.03 | (0.98–1.08) | 0.22 | 1.00 | (0.96–1.06) | 0.76 |
| Sex | ||||||
| Female | 1 (reference) | |||||
| Male | 1.84 | (0.68–4.98) | 0.23 | 3.34 | (1.13–9.90) | 0.03 |
| Site | ||||||
| Left | 1 (reference) | |||||
| Right | 2.76 | (0.95–8.03) | 0.06 | 1.47 | (0.52–4.19) | 0.47 |
| Chemosensitivity | ||||||
| Favorable | 1 (reference) | |||||
| Unfavorable | 3.73 | (1.14–12.17) | 0.03 | 6.04 | (1.64–22.20) | 6.8×10−3 |
CI, confidence interval; HR, hazard ratio.
Figure 5.
Recurrence-free prognosis of modules 1 and 2 in the samples stratified by chemosensitivity. (A) Recurrence-free survival of high- and low-risk patients derived from module 1 classified into unfavorable and favorable chemosensitivity strata. (B) Recurrence-free survival of high- and low-risk patients derived from module 2 classified into unfavorable and favorable chemosensitivity strata.
Two prognostic modules are regulated by hsa-miR-335-5p
The present study revealed that the two modules were independent prognostic factors for recurrence-free survival in osteosarcoma. No common genes existed common between the two modules. When considering upstream miRNAs that regulate the two modules, the same miRNA, hsa-miR-335-3p, was shared by all nodes in modules 1 and 2 (Fig. 6A). Using the median relative expression value for hsa-miR-335-3p across all samples, the samples were divided into high and low hsa-miR-335-3p expression groups. In the two modules, the co-expression coefficients for ceRNA pairs in the low group were significantly higher than those in the high group (P=0.002, <2.2×10−16, respectively), thus suggesting that the two modules are highly regulated by hsa-miR-335-3p (Fig. 6B).
Figure 6.
hsa-miR-335-5p regulates ceRNA modules and recurrence-free prognosis. (A) Modules 1 and 2 were all regulated by hsa-miR-335-5p. (B) ceRNA pairs in both modules exhibit stronger co-expression in the low hsa-miR-335-5p expression samples. (C) Genomic location of hsa-miR-335-5p and ceRNAs from both modules. (D) Recurrence-free prognosis for the combination of the modules with hsa-miR-335-5p expression. ceRNA, competing endogenous RNA; miR, microRNA.
When analyzing the genomic location of the ceRNAs in the modules, it was revealed that the majority of the ceRNA pairs regulated each other across chromosomes, as only three out of 21 and two out of 45 ceRNA pairs were located on the same chromosome for module 1 and module 2 (SYCP2L and BTBD9 on chromosome 6, TP53AIP1 and PRG3 on chromosome 11, OR4D2 and SMTNL2 on chromosome 17 for module 1; HFM1 and USH2A on chromosome 1, XKR4 and SLC7A13 on chromosome 8 for module 2), respectively, which is consistent with previous studies (21,30). In addition, the miRNA-mRNA regulation had a similar result, as only one miRNA-mRNA pair (hsa-miR-335-5p and CYP3A5) was located on the same chromosome in module 1 (Fig. 6C). By combining the expression levels of the genes from the two modules and hsa-miR-335-5p, a novel prognostic model for recurrence-free survival was generated with enhanced power. In this novel risk score model, samples in the high-risk group had a significantly reduced recurrence-free survival time compared with those in the low-risk group (median recurrence-free survival, 28.9 vs. 151.0 months, log-rank P=0.008; Fig. 6D).
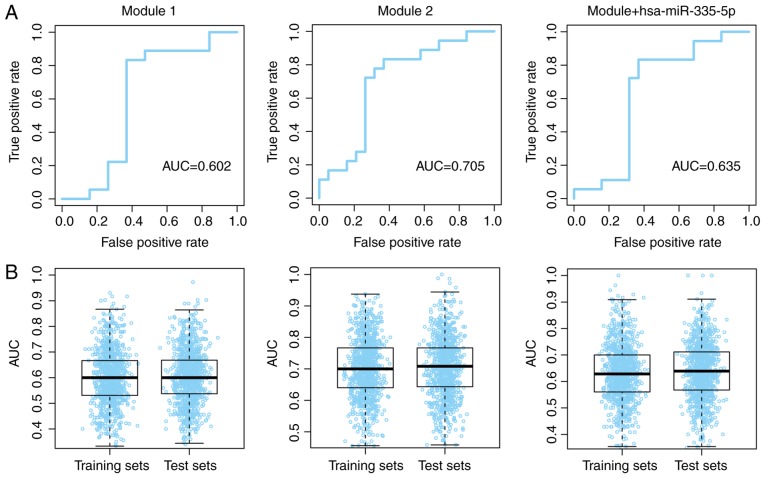
The prognostic value for modules 1 and 2, and the combination of the modules with hsa-miR-335-5p, was further validated using ROC curves. The patients with and without recurrence were used as true positive and true negative data-sets, respectively. Good area under the curve (AUC) values were obtained (Fig. 7A). To avoid model overfitting to the training and test datasets, the group division process was repeated with 1,000 iterations, and the AUC values for the three models were evaluated in each training and test dataset. The median AUC values ranged between 0.60 and 0.71 across the different dataset divisions (Fig. 7B). These results indicated that the modules may possess a robust ability to predict the recurrence-free survival of patients with osteosarcoma.
Figure 7.
Prognostic value of modules 1 and 2, and combination of the modules with hsa-miR-335-5p. (A) Prognostic value of both modules for all patients with osteosarcoma. (B) Prognostic value of both modules for random dataset divisions. AUC, area under the curve.
Discussion
Osteosarcoma is the most commonly occurring primary bone malignancy in children and young adults; however diagnostic accuracy and the effectiveness of treatment are far from satisfactory, particularly for patients experiencing osteosarcoma recurrence (31,32). Previous studies have demonstrated that ceRNAs serve a crucial role in the development of osteosarcoma (12,31,33); however, it remains unclear whether ceRNAs are implicated in tumor recurrence. Therefore, the present study aimed to investigate the potential role of ceRNAs in osteosarcoma recurrence, and to identify potential biomarkers.
The present study identified two modules (modules 1 and 2), with no common ceRNAs between them, with prognostic power for predicting the recurrence-free survival outcome for patients with osteosarcoma. Either module 1 or 2 could effectively divide patients with osteosarcoma into high- and low-risk groups, as defined by recurrence-free survival, through the relative expression levels of genes in each module. When comparing the results of the patients with osteosarcoma between the two modules, it was revealed that 32 out of 37 patients were classified into the same group by modules 1 and 2. Considering the mutually exclusive ceRNAs in the two modules, these consistent predictive results indicated the accuracy of modules 1 and 2 in predicting the prognosis for patients with osteosarcoma.
Numerous studies have identified osteosarcoma-associated ceRNAs, and have demonstrated their value in predicting prognosis for survival or recurrence, including small nucleolar RNA host gene 1 and X inactive specific transcript (non-protein coding) (10,34,35). However, the majority of these studies focus on just one ceRNA-ceRNA pair, instead of taking a global view of the ceRNA-ceRNA network and modules. In the prognostic modules 1 and 2 identified in the present study, no single gene exhibited a significant prognostic ability for the recurrence-free survival of patients with osteosarcoma, with the exception of TP53AIP1, which was somewhat associated with prognosis (P=0.08). This may be the reason why these ceRNAs have not been identified in previous studies; therefore, it may be hypothesized that the ceRNAs in the modules affect osteosarcoma development as a whole, rather than by acting individually. In addition, the results revealed that hsa-miR-335-5p may influence the recurrence of osteosarcoma by regulating these two ceRNA interaction modules, rather than by itself (P=0.717, univariate Cox regression analysis). These results indicated that hsa-miR-335-5p may be considered a potential novel therapeutic target, as the regulator of these two modules. The present identification of recurrence-free survival-associated factors at the level of modules opens novel avenues for leveraging publicly available data to identify prognostic biomarkers for patients with osteosarcoma.
In conclusion, the present study integrated confirmed experimental interactions between miRNAs and mRNAs, and used gene expression data for osteosarcoma from the GEO database to construct a ceRNA-ceRNA interaction network. Two modules were subsequently identified, and further survival analysis demonstrated that these modules were significantly associated with the recurrence-free survival of patients with osteosarcoma. The ability of these two modules to predict recurrence-free prognosis was independent of other clinical factors. Furthermore, hsa-miR-335-3p regulated all of the ceRNAs from the two modules, and the vast majority of the ceRNA pairs regulated each other across chromosomes, as determined by analyzing their genomic locations. These findings may provide additional useful data for the development of clinical therapeutic targets against osteosarcoma recurrence.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The full network containing 64 mRNAs is available in the bilab repository (http://shiyabshi230.j2eeall.com).
Authors’ contributions
ZB and YC conceived and designed the experiments. YC, QC, JZ and YZ analyzed the data. ZB and YC wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Conflicts of interest
The authors declare that they have no conflicts of interest.
References
- 1.Lee L, Fei L, Pope J, Wagner LM. Early lymphocyte recovery and outcome in osteosarcoma. J Pediatr Hematol Oncol. 2017;39:179–183. doi: 10.1097/MPH.0000000000000717. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cam M, Gardner HL, Roberts RD, Fenger JM, Guttridge DC, London CA, Cam H. ∆Np63 mediates cellular survival and metastasis in canine osteosarcoma. Oncotarget. 2016;7:48533–48546. doi: 10.18632/oncotarget.10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao L, Wang J, Wang PQ. MiR-326 is a diagnostic biomarker and regulates cell survival and apoptosis by targeting Bcl-2 in osteosarcoma. Biomed Pharmacother. 2016;84:828–835. doi: 10.1016/j.biopha.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 5.Tu B, Zhu J, Liu S, Wang L, Fan Q, Hao Y, Fan C, Tang TT. Mesenchymal stem cells promote osteosarcoma cell survival and drug resistance through activation of STAT3. Oncotarget. 2016;7:48296–48308. doi: 10.18632/oncotarget.10219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y, Lu Q, Xu J, Yan R, Zhu J, Xu J, Jiang X, Li J, Wu F. The effect of pathological fractures on the prognosis of patients with osteosarcoma: A meta-analysis of 14 studies. Oncotarget. 2017;8:73037–73049. doi: 10.18632/oncotarget.20375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li YJ, Yao K, Lu MX, Zhang WB, Xiao C, Tu CQ. Prognostic value of the C-reactive protein to albumin ratio: A novel inflammation-based prognostic indicator in osteosarcoma. Onco Targets Ther. 2017;10:5255–5261. doi: 10.2147/OTT.S140560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XX, Liao Y, Hong L, Zeng Z, Yuan TB, Xia X, Qin J. Tissue microarray staining reveals PLD1 and Sp1 have a collaborative, pro-tumoral effect in patients with osteosarcomas. Oncotarget. 2017;8:74340–74347. doi: 10.18632/oncotarget.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YM, Shen QC, Gokavarapu S, Ong HS, Cao W, Ji T. Osteosarcoma of the Mandible: A site-specific study on survival and prognostic factors. J Craniofac Surg. 2016;27:1929–1933. doi: 10.1097/SCS.0000000000002968. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Dou P, Liu T, He S. Application of long Noncoding RNAs in osteosarcoma: Biomarkers and therapeutic targets. Cell Physiol Biochem. 2017;42:1407–1419. doi: 10.1159/000479205. [DOI] [PubMed] [Google Scholar]
- 11.Simpson S, Dunning MD, de Brot S, Grau-Roma L, Mongan NP, Rutland CS. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet Scand. 2017;59:71. doi: 10.1186/s13028-017-0341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Zhang Y, Yang T, Zhao W, Wang N, Li P, Zeng X, Zhang W. Long non-coding RNA MALAT1 for promoting metastasis and proliferation by acting as a ceRNA of miR-144 3p in osteosarcoma cells. Oncotarget. 2017;8:59417–59434. doi: 10.18632/oncotarget.19727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao S, Cheng C, Chen H, Li M, Liu K, Wang G. IGF1 3’UTR functions as a ceRNA in promoting angiogenesis by sponging miR-29 family in osteosarcoma. J Mol Histol. 2016;47:135–143. doi: 10.1007/s10735-016-9659-2. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly AD, Haibe-Kains B, Janeway KA, Hill KE, Howe E, Goldsmith J, Kurek K, Perez-Atayde AR, Francoeur N, Fan JB, et al. MicroRNA paraffin-based studies in osteosarcoma reveal reproducible independent prognostic profiles at 14q32. Genome Med. 2013;5:2. doi: 10.1186/gm406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(Database issue):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: An integrated resource for microRNA-target interactions. Nucleic acids Res. 2009;37(Database issue):D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palla G, Derényi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- 21.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adamcsek B, Palla G, Farkas IJ, Derényi I, Vicsek T. CFinder: Locating cliques and overlapping modules in biological networks. Bioinformatics. 2006;22:1021–1023. doi: 10.1093/bioinformatics/btl039. [DOI] [PubMed] [Google Scholar]
- 23.Alizadeh AA, Gentles AJ, Alencar AJ, Liu CL, Kohrt HE, Houot R, Goldstein MJ, Zhao S, Natkunam Y, Advani RH, et al. Prediction of survival in diffuse large B-cell lymphoma based on the expression of 2 genes reflecting tumor and microenvironment. Blood. 2011;118:1350–1358. doi: 10.1182/blood-2011-03-345272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 25.Zhou M, Wang X, Shi H, Cheng L, Wang Z, Zhao H, Yang L, Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi H, Cheng L, Sun J. Relapse-related long non-coding RNA signature to improve prognosis prediction of lung adenocarcinoma. Oncotarget. 2016;7:29720–29738. doi: 10.18632/oncotarget.8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dhaini HR, Thomas DG, Giordano TJ, Johnson TD, Biermann JS, Leu K, Hollenberg PF, Baker LH. Cytochrome P450 CYP3A4/5 expression as a biomarker of outcome in osteosarcoma. J Clin Oncol. 2003;21:2481–2485. doi: 10.1200/JCO.2003.06.015. [DOI] [PubMed] [Google Scholar]
- 28.Trujillo-Paolillo A, Tesser-Gamba F, Petrilli AS, de Seixas Alves MT, Garcia Filho RJ, de Oliveira R, de Toledo SRC. CYP genes in osteosarcoma: Their role in tumorigenesis, pulmonary metastatic microenvironment and treatment response. Oncotarget. 2017;8:38530–38540. doi: 10.18632/oncotarget.15869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemeier A, Kassem M, Toedter K, Wendt D, Ruether W, Beisiegel U, Heeren J. Expression of LRP1 by human osteoblasts: A mechanism for the delivery of lipoproteins and vitamin K1 to bone. J Bone Miner Res. 2005;20:283–293. doi: 10.1359/JBMR.041102. [DOI] [PubMed] [Google Scholar]
- 30.Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell. 2011;147:344–357. doi: 10.1016/j.cell.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cong C, Wang W, Tian J, Gao T, Zheng W, Zhou C. Identification of serum miR-124 as a biomarker for diagnosis and prognosis in osteosarcoma. Cancer Biomark. 2018;21:449–454. doi: 10.3233/CBM-170672. [DOI] [PubMed] [Google Scholar]
- 32.Jin H, Luo S, Wang Y, Liu C, Piao Z, Xu M, Guan W, Li Q, Zou H, Tan QY, et al. miR-135b stimulates osteosarcoma recurrence and lung metastasis via Notch and Wnt/β-catenin signaling. Mol Ther Nucleic Acids. 2017;8:111–122. doi: 10.1016/j.omtn.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sha B, Gao W, Han Y, Wang S, Wu J, Xu F, Lu T. Potential application of titanium dioxide nanoparticles in the prevention of osteosarcoma and chondrosarcoma recurrence. J Nanosci Nanotechnol. 2013;13:1208–1211. doi: 10.1166/jnn.2013.6081. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Cao L, Wu J, Wang Q. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52:77–88. doi: 10.3892/ijo.2017.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang R, Xia T. Long non-coding RNA XIST regulates PDCD4 expression by interacting with miR-21-5p and inhibits osteosarcoma cell growth and metastasis. Int J Oncol. 2017;51:1460–1470. doi: 10.3892/ijo.2017.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request. The full network containing 64 mRNAs is available in the bilab repository (http://shiyabshi230.j2eeall.com).