Abstract
Aims
To investigate the impact of glucuronidation enzyme (UGT1A4*3 142T>G, UGT1A4*2 70C>A, UGT2B7 –161C>T) and transporter (MDR1/ABCB1 1236C>T, ABCG2 421C>A) polymorphisms on steady‐state disposition of lamotrigine and on the lamotrigine–valproate interaction.
Methods
Adults with epilepsy on lamotrigine monotherapy (n = 131) or lamotrigine + valproate treatment (n = 74) were genotyped and steady‐state lamotrigine and valproate morning troughs were determined as a part of routine therapeutic drug monitoring.
Results
No effect of UGT and MDR1/ABCB1 polymorphisms was observed. In the entire cohort, ABCG2 421A allele had no effect however an interaction between the variant allele and valproate was observed: (i) in lamotrigine‐only patients, variant allele (vs. wild type homozygosity) was independently (adjustments: age, sex, body mass index, lamotrigine dose, other polymorphisms) associated with mildly lower lamotrigine troughs [geometric means ratio (GMR) = 0.76, 95% confidence interval (CI) 0.59–0.98], whereas in lamotrigine + valproate patients it was associated with higher troughs (GMR = 1.72, 95%CI 1.14–2.62); (ii) valproate cotreatment was overall associated with markedly higher troughs vs. lamotrigine monotherapy (GMR = 3.49, 95%CI 2.73–4.44), but more so in variant allele carriers (GMR = 5.24, 95%CI 3.38–8.15) than in wild type homozygotes (GMR = 2.32, 95%CI 1.89–2.83); (iii) variant allele effects in two treatment subsets and valproate effects in two genotype subsets differed by 2.36‐fold (95%CI 1.39–3.67); (iv) increase in lamotrigine troughs associated with increasing valproate troughs was greater in variant allele carriers than in wild type homozygotes, i.e. variant allele effect increased with increasing valproate troughs.
Conclusion
This study is first to indicate a potentially relevant interaction between ABCG2 421C>A polymorphism and valproate in their effects on lamotrigine disposition.
Keywords: Clinical Pharmacology, Drug Interactions, Drug Transporter, Genetics And Pharmacogenetic, Pharmacogenomics, Pharmacokinetics
What is Already Known about this Subject
Pharmacokinetic variability of lamotrigine is largely due to comedication. Specifically, valproate increases exposure to lamotrigine around 2‐fold by inhibiting its metabolism (inhibits UGTs).
Lamotrigine is a substrate for ABCG2 transporter. The single nucleotide polymorphism ABCG2 421C>A has been suggested to affect disposition of lamotrigine, but data are scarce and contradictory.
What this Study Adds
Variant allele at ABCG2 421C>A and valproate interact in their effects on lamotrigine disposition.
Lamotrigine mono‐treated variant allele carriers (vs. wild type homozygotes) have 25% lower troughs, but when valproate cotreated, they have 70% higher lamotrigine troughs.
Valproate increases lamotrigine troughs by 2.3‐fold in wild type homozygotes and by 5.2‐fold in variant allele carriers.
Introduction
http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=2622 is a second‐generation antiepileptic drug (AED). In adults and adolescents aged >13 years it is approved as a mono‐ or an adjunctive treatment for partial or generalized seizures and seizures associated with the Lennox–Gastaut syndrome 1. Due to its rather variable pharmacokinetics it is considered that therapeutic drug monitoring (TDM; based on trough concentrations, as recommended for AEDs in general 2) could improve seizure control with lamotrigine: the likely range of therapeutic troughs has been suggested to be between 3.9 and 51 μmol l–1 with rapidly increasing risk of serious adverse events with concentrations >58 μmol l–1 2, 3. Elimination of lamotrigine is practically exclusively by metabolism: it undergoes N‐glucuronidation primarily via uridine‐diphosphate‐glucuronidase (UGT) isoform 1A4 (http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=988) with a contribution of UGT1A1 and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=988 1, 2, 3. In vitro studies suggest that UGT2B7 is particularly important at low lamotrigine concentrations 4. Pharmacokinetic variability of lamotrigine is due to variability of the elimination process and is largely explained by comedication, since a number of commonly coadministered AEDs and non‐AEDs are known UGT inducers 1, 2, 3. However, lamotrigine is commonly used with http://www.guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=7009, a classical AED and a known inhibitor of lamotrigine metabolism: exposure to lamotrigine increases around 2‐fold 5 as valproate inhibits UGT1A4 6 and UGT2B7 4.
Reports on a potential role of UGT polymorphisms in lamotrigine disposition variability have been somewhat conflicting with uncertain clinical relevance. Chinese patients homozygous for the variant allele at UTG1A4*3 had higher lamotrigine serum levels associated with a greater efficacy 7, while in Northern European 8 and Turkish patients 9 variant homozygosity or variant allele carriage, respectively, were associated with lower lamotrigine concentrations. In Chinese children cotreated with lamotrigine and valproate, both higher 10 and lower 11 lamotrigine concentrations were reported associated with this polymorphism. Regarding other UGT polymorphisms, one study in Northern Europeans suggested a trend of increasing lamotrigine concentrations in variant allele carriers at UGT1A4*2 8, while a study in Central‐Southern European subjects 12 found no association between UGT1A4*2 and lamotrigine pharmacokinetics. Studies investigating the impact of polymorphisms on activity of UGT2B7 were largely focused on a nonsynonymous polymorphism 802C>T [UGT2B7*2 (His268Tyr), rs7439366], which is rather frequent in different ethnic groups and is in a complete linkage disequilibrium with a promoter polymorphism UGT2B7 –161C>T 13, 14, 15. The latter has been suggested to affect the UGT2B7 transcription resulting in an increased glucuronidation activity for morphine 13, 16 and was also reported associated with valproate concentrations in epilepsy patients 17. In Central‐Southern European subjects 12, variant allele carriage at UGT2B7 –161C>T was associated with a 20% lower lamotrigine clearance, which is similar to results in Spanish 18 and Thai patients 19.
Little is known about the impact of ABC transporters on the disposition of lamotrigine and its variability. In vitro studies suggest that lamotrigine is a substrate for http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=768&familyId=152&familyType=TRANSPORTER transporter 20, 21. We found an association between a variant allele at MDR1/ABCB1 1236C>T and lower lamotrigine troughs 22, while a lack of effect of this polymorphism on lamotrigine pharmacokinetics was also reported 12. One in vitro study excluded lamotrigine as a potential http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=792 transporter substrate 23, but a subsequent one identified it as a substrate for human ABCG2 at therapeutic concentrations 21. Two studies in Chinese patients reported on the impact of ABCG2 polymorphisms on lamotrigine disposition – variant allele carriage at ABCG2 421 C>A was associated with somewhat lower troughs vs. wild type genotype in one study 24, while in another one, variant allele homozygotes had higher lamotrigine concentrations than wild type homozygotes and heterozygotes 25. In vitro, valproic acid was shown to concentration‐dependently increase expression of ABCG2 in human trophoblast BeWo cell line 26, making it plausible to hypothesize about ABCG2 transporter as an additional potential site of lamotrigine–valproate interaction.
Objective
Considering relative paucity of data and conflicting reports, which might be ethnicity‐specific, the primary objective of the present work was to evaluate the impact of the suggested UGT, ABCG2 and MDR1/ABCB1 polymorphisms on steady‐state disposition of lamotrigine as assessed by routine TDM morning trough concentrations 2 in a sample of Central‐Southern European adults and adolescents with epilepsy, and to explore their possible involvement in lamotrigine‐valproate interaction. For this purpose, lamotrigine‐treated and lamotrigine + valproate‐treated patients were genotyped for the following polymorphisms: UGT1A4*3 142T>G (rs2011425), UGT1A4*2 70C>A (rs6755571), UGT2B7 –161C>T (rs7668258), MDR1/ABCB1 1236C>T (rs1128503) and ABCG2 421C>A (rs2231142). Since a subset of patients cotreated with valproate was to be obtained, the secondary objective was to evaluate possible impact of these polymorphisms on valproate steady‐state disposition. This subset of patients (lamotrigine + valproate‐treated) was also genotyped for http://www.guidetopharmacology.org/GRAC/ObjectDisplayForward?objectId=1326 [CYP2C9*2 (c.430C>T, rs1799853), CYP2C9*3 (c.1075A>C, rs1057910)] and http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=262#1328 polymorphisms [CYP2C19*2 (c.681G>A, rs4244285), CYP2C19*17 (c.806C>T, rs12248560)] since the two CYPs are involved in valproic acid metabolism 27.
Methods
This prospective observational study based on routine TDM was conducted between 2012 and 2016 in line with the Declaration of Helsinki (the 2008 version) and was approved by the Institutional Ethics Committee (approval class: 8.1.‐14/78–2, registration number: 02/21/JG). Informed consent was obtained from all participants included in the study (parents/guardians for underage adolescents).
Subjects
Inclusion criteria were: (i) age > 16 years; (ii) informed consent; (iii) verified epilepsy with indication for treatment with lamotrigine (immediate‐release, Lamictal, GlaxoSmithKline) or with lamotrigine and valproate (extended‐release, Depakine Chrono, Sanofi‐Aventis; no other AEDs), and no absolute contraindication for either drug; (iv) preserved renal and liver function based on routine laboratory indicators (serum urea, creatinine, albuminuria, liver function tests, lactate dehydrogenase); (v) no use of known inducers of lamotrigine glucuronidation or drugs that affect valproate pharmacokinetics (as per approved labels) within 4 weeks prior to blood sampling for the present analysis. Excluded were patients with unregulated diabetes mellitus, hyper‐ or hypothyroidism, patients with chronic heart failure, history of or an ongoing malignant disease, and HIV‐positive subjects.
Drug dosing
Both drugs were dosed as per approved labels with gradual up‐titration based on response and tolerability. For monotherapy, lamotrigine was started at 25 mg once daily (weeks 1 and 2), increased to 50 mg once daily (weeks 3 and 4) and was then up‐titrated by 50 mg day–1 in (bi)weekly intervals. For combined treatment, it was started at 25 mg every other day (weeks 1 and 2), increased to 25 mg once daily (weeks 3 and 4) and was then up‐titrated by 25–50 mg day–1 in (bi) weekly intervals. When maintenance dosing was achieved, lamotrigine was delivered twice daily. Valproate was dosed twice daily starting with 300 mg day–1 in week 1 and was then up‐titrated by 300 mg day–1 in weekly intervals.
Blood sampling and analytical assays for lamotrigine and valproate concentrations
Morning troughs were taken between 07:00 and 09:00 h [24 ± 1 (lamotrigine titration) or 12 ± 1 h after the previous dose] after at least 21 days of regular (co)treatment. This period was sufficient for both lamotrigine and valproate to achieve steady‐state and to bridge the period of the initial UGT induction by lamotrigine 28. Serum lamotrigine was quantified using a validated high‐performance liquid chromatography method with a diode‐array detector (Shimadzu, Japan) as described previously 22. Serum valproate was measured by immunoassay (PETINIA) on a Dimension Expand analyser (Siemens; calibrator and control samples by Siemens, Germany). Both analytes are included in external quality control schemes (DGKL RfB and UK NEQAS).
Genotyping procedures
Genomic DNA was extracted from 3 ml of whole blood using the FlexiGene DNA Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. Genotyping of MDR1/ABCB1 1236C>T, ABCG2 421C>A, UGT1A4*2, UGT2B7 –161C>T CYP2C9*2, CYP2C9*3, CYP2C19*2 and CYP2C19*17 was performed using TaqMan Drug Metabolism Genotyping assays ID C_7586662_10, ID C_15854163_70, ID C_25957120_10 C_27827970_40, C_25625805_10, C_27104892_10, C_25986767_70, and C_469857_10, respectively, while genotyping of UGT1A4*3 was performed using Custom TaqMan SNP Genotyping assay (Applied Biosystems, Foster City, CA, USA) by real‐time polymerase chain reaction (PCR) genotyping method on the 7500 Real‐Time PCR System (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's instructions. Genotyping of UGT1A4*3 was confirmed by a PCR‐RFLP method on the Gene Amp PCR System 9700 (Applied Biosystems, Foster City, CA, USA) 29.
Sample size and power considerations
Based on previous investigations in the target population 22, 30, we expected frequency of the variant allele at MDR1/ABCB1 1236C>T to be around 40% and frequency of intermediate or slow CYP2C9 or CYP2C19 metabolizers to be around 20–30%. Since we had no insight into the frequency of variant alleles at other genotyped loci, which might be ethnically specific, we opted not to define the sample size on an a priori power calculation but reasoned that subjects should be enrolled until the smallest treatment‐by‐variant allele carriage subset included at least 10 subjects. At a two‐sided 5% α level, a comparison between two groups of 10 subjects each provides 80% power to detect a difference expressed as a geometric means ratio of 1.55 (55% higher) or 0.64 (36% lower) assuming a coefficient of variation of 35%. We considered differences of this size (and larger) to be potentially practically relevant.
Statistics
Log‐transformed lamotrigine and valproate morning troughs, as measured and also adjusted per mg dose, were analysed by fitting general linear models and effects were expressed as geometric means ratios (GMRs).
In respect to the primary objective, the main analysis was done in three steps: (i) For initial screening of a potential effect of the UGT and ABC transporter polymorphisms on lamotrigine disposition and/or valproate–lamotrigine interaction, a separate unadjusted model for each polymorphism was fitted to lamotrigine troughs with treatment (lamotrigine only or lamotrigine + valproate), genotype (variant allele carriage vs. wild type homozygosity), their interaction and lamotrigine dose in the case of measured troughs. Since four treatment‐polymorphism interactions were tested, α level for a significant interaction term was set at 0.0125 to prevent false‐positive findings. (ii) To evaluate whether effects were independent, adjusted models were fitted: (a) a main effects model included age, sex, body mass index, treatment, lamotrigine dose (for measured troughs) and genotypes at all UGT and transporter loci; (b) a series of models with interactions, a separate one for each polymorphism, with all the adjustments as in the main effects model plus a treatment–genotype interaction (for the specific polymorphism). Alpha level for the interaction terms was corrected as in the unadjusted analysis. (iii) For any polymorphism with an independent main effect or treatment–genotype interaction, a model including only adjustments with P < 0.1 (parsimonious) was fitted. In the case of a significant treatment–genotype interaction, two supportive analyses were performed. The first one aimed to explore a possibility that the moderating effect of a polymorphism on valproate–lamotrigine interaction could vary with the extent of exposure to valproate. The described models were re‐fitted, but treatment was not considered as a binary variable, rather the combination treatment patients were represented by their morning valproate troughs, while lamotrigine‐only patients were considered to have valproate 0. In the second one, adjusted main effects models (without the treatment effect) were fitted separately for lamotrigine‐only and lamotrigine + valproate patients: numerically different variant allele effects were illustrative of a treatment–genotype interaction.
Regarding the secondary objective and considering a limited number of valproate‐(co)treated patients, a separate model for each evaluated UGT, ABC transporter and CYP polymorphism was fitted to valproate troughs with adjustment for age, sex, body mass index and valproate dose (for measured troughs). We used SAS 9.4 for Windows (SAS Inc., Cary, NC, USA) for data analysis and CubeX 31 to test Hardy–Weinberg equilibrium and linkage disequilibrium.
Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY 32, and are permanently archived in the Concise Guide to PHARMACOLOGY 2017/18 33, 34.
Results
Patient characteristics
A total of 205 patients were enrolled, 131 treated with lamotrigine (LAM) and 74 treated with lamotrigine + valproate (LAM + VAL), fairly comparable regarding age, sex and body mass index (Table 1) with measured and dose‐adjusted lamotrigine troughs considerably higher in the latter (Table 1). Variant alleles were common at MDR1/ABCB1 1236C>T and at UGT2B7 –161C>T (40–50%; Table 2), rare at ABCG2 421 C>A and at UGT1A4*3 (10–13%; Table 2) and sporadic at UGT1A4*2 (1%; Table 2). All UGT/transporter variant allele carriage‐by‐treatment subsets included at least 10 subjects except for UGT1A4*2 – we considered further enrolment unfeasible (low prevalence), particularly regarding the reported lack of effect of these polymorphisms on lamotrigine pharmacokinetics 12. Consequently, it was not included in further analyses. In the LAM + VAL‐treated patients, CYP2C9*1 and CYP2C19*1 alleles and homozygotes prevailed (Table 2). No departure from Hardy–Weinberg equilibrium was observed at any locus, neither overall nor by treatment subset (not shown). Linkage disequilibrium between the investigated ABCG2 and UGT2B7 loci (long arm chromosome 4) could not be excluded (D' = 0.408, r 2 = 0.017, χ2 = 3.55, P = 0.059).
Table 1.
Patient demographics and lamotrigine (LAM) and valproate (VAL) doses and measured and dose‐adjusted trough concentrations. Data are median (range) or count (%)
| All patients | LAM only | LAM + VAL | |
|---|---|---|---|
| n | 205 | 131 | 74 |
| Men | 69 (33.6) | 40 (30.5) | 29 (39.2) |
| Age (years) | 34 (14–77) | 35 (16–74) | 32.5 (14–77) |
| Body mass index (kg m–2) | 24 (12–40.5) | 24.1 (18.7–40.5) | 23.3 (12–35.4) |
| LAM daily dose (mg) | 125 (12.5–400) | 100 (12.5–400) | 150 (25–350) |
| LAM trough (μmol l–1) | 10.5 (0.5–102) | 6.7 (0.5–37.6) | 26 (2.3–102) |
| LAM trough/mg dose (nmol l–1) | 92 (6.5–464) | 68 (6.5–318) | 172.3 (30.7–464) |
| VAL daily dose (mg) | ‐ | ‐ | 1000 (250–2000) |
| VAL trough (μmol l–1) | ‐ | ‐ | 376 (98.5–813.2) |
| VAL trough/mg dose (nmol l–1) | ‐ | ‐ | 412.9 (129.5–1295) |
Table 2.
Prevalence [n (%)] of genotypes, overall and by treatment – lamotrigine (LAM) or lamotrigine + valproate (VAL) CYP2C9 and CYP2C19 data only for LAM + VAL patients
| All patients | LAM only (n = 131) | LAM + VAL (n = 74) | |
|---|---|---|---|
| n | 205 | 131 | 74 |
| UGT1A4*3 | |||
| TT | 155 (75.6) | 99 (75.6) | 56 (75.6) |
| TG | 48 (23.4) | 31 (23.6) | 17 (23.0) |
| GG | 2 (1.0) | 1 (0.8) | 1 (1.4) |
| Variant allele frequency (%) | 12.7 | 12.6 | 12.8 |
| UGT1A4*2 | |||
| CC | 199 (97.1) | 128 (97.7) | 71 (95.9) |
| CA | 6 (2.9) | 3 (2.3) | 3 (4.1) |
| AA | 0 | 0 | 0 |
| Variant allele frequency (%) | 1.4 | 1.1 | 2.0 |
| UGT2B7 –161C>T | |||
| CC | 51 (24.9) | 26 (19.9) | 25 (33.8) |
| CT | 98 (47.8) | 65 (49.6) | 33 (44.6) |
| TT | 56 (27.3) | 40 (30.5) | 16 (21.6) |
| Variant allele frequency (%) | 51.2 | 55.3 | 43.9 |
| ABCG2 421C>A | |||
| CC | 167 (81.5) | 103 (78.6) | 64 (86.5) |
| CA | 36 (17.5) | 26 (19.9) | 10 (13.5) |
| AA | 2 (1.0) | 2 (1.5) | 0 |
| Variant allele frequency (%) | 9.7 | 11.5 | 6.8 |
| MDR1/ABCB1 1236C>T | |||
| CC | 75 (36.6) | 50 (38.2) | 25 (33.8) |
| CT | 93 (44.3) | 60 (45.8) | 33 (44.6) |
| TT | 37 (18.0) | 21 (16.0) | 16 (21.6) |
| Variant allele frequency (%) | 40.7 | 38.9 | 43.9 |
| CYP2C9 (*2,*3) | |||
| *1/*1 | ‐ | ‐ | 42 (56.8) |
| *1/*2 | ‐ | ‐ | 18 (24.3) |
| *1/*3 | ‐ | ‐ | 13 (17.6) |
| *2/*3 | ‐ | ‐ | 1 (1.3) |
| Allele frequencies *1/ *2/ *3 (%) | ‐ | ‐ | 77.7/12.8/9.5 |
| CYP2C19 (*2,*17) | |||
| *1/*1 | ‐ | ‐ | 28 (37.8) |
| *1/*17 | ‐ | ‐ | 23 (31.1) |
| *17/*17 | ‐ | ‐ | 2 (2.7) |
| *1/*2 | ‐ | ‐ | 13 (17.6) |
| *2/*17 | ‐ | ‐ | 5 (6.8) |
| *2/*2 | ‐ | ‐ | 3 (4.0) |
| Allele frequencies *1/ *2/ *17 (%) | ‐ | ‐ | 62.2 /16.2/21.6 |
Effects of UGT and ABC transporter variant alleles on steady‐state lamotrigine troughs and lamotrigine‐valproate interaction
Figure 1 shows measured and dose‐adjusted lamotrigine troughs by treatment‐by‐genotype (except for UGT1A4*2). Table 3 summarizes the main analysis (see Supporting Information Tables S1 and S2 for details). Models fitted to lamotrigine troughs in the entire cohort (unadjusted, adjusted and adjusted with treatment–genotype interactions) consistently indicted no main effect of any of the analysed polymorphisms, 2.5 to 3.5‐times higher lamotrigine troughs in patients cotreated with valproate (vs. lamotrigine only) and no significant treatment–genotype interaction, except for ABCG2 421C>A (Table 3). The results of the unadjusted, adjusted and parsimonious models (only adjustments with P < 0.1) pertaining to ABCG2 421C>A were consistent (Table 4): (i) measured (with lamotrigine dose as a covariate) and dose‐adjusted troughs were around 3.5‐times and around 3.0‐times higher, respectively, in LAM + VAL vs. LAM‐treated patients (valproate effect); (ii) there was no main effect of the variant allele; (iii) treatment–genotype interaction was significant (P < 0.0125) – (a) in wild type homozygotes, valproate cotreatment (vs. lamotrigine only) was associated with around 2.4‐times (measured) and around 2.2‐times (dose‐adjusted) higher troughs, whereas in variant allele carriers valproate effect was more pronounced and cotreatment was associated with around 5.3‐times (measured) and around 4.3‐times (dose‐adjusted) higher troughs; (b) conversely, in LAM‐only patients, variant allele (vs. wild type) was associated with around 24% (measured) and around 18% lower troughs, i.e. in this subset of patients it had an effect without the presence of valproate, whereas in LAM + VAL patients direction of the effect has changed, and variant allele was associated with around 70% (measured) and around 60% (dose‐adjusted) higher troughs; (c) geometric means ratios for the interaction terms indicated that the valproate effect in variant allele carriers was around 2.2–2.3‐times (measured troughs) and around 1.9–2.0‐times (dose‐adjusted troughs) higher than in wild type homozygotes, just as was the effect of the variant allele in LAM + VAL‐treated patients compared to that in LAM‐only patients. Figure 2A illustrates the interaction. Adjusted geometric means and geometric means ratios suggest that the extent of interaction is mainly defined by enhancement of the valproate effect by the variant allele, less so by its effect in the absence of valproate. The interaction was evidenced also by the supportive analyses (Table 5; see Supporting Information Table S4 for details). When the parsimonious model from Table 4 was refitted in a way that valproate cotreatment was represented by valproate troughs (in LAM‐only patients this value was set to zero), valproate trough–genotype interaction was again significant (Table 5): increase in valproate troughs was associated with a greater increase in lamotrigine troughs in variant allele carriers than in wild type homozygotes. Figure 2B illustrates the interaction from the alternative viewpoint: at zero valproate, adjusted geometric mean lamotrigine troughs were lower in variant allele carriers than in wild type homozygotes (by 25% and 19%, measured and dose‐adjusted, respectively), but increased to 33% /29% higher values at 100 μmol l–1 (changed direction of the variant allele effect); lamotrigine troughs continuously increased across valproate troughs in both variant allele carriers and wild type homozygotes, but apparently more so in the former hence the variant allele – wild type difference increased to 58%/49% at 400 μmol l–1 and to 72%/60% at 800 μmol l–1 (Figure 2B) indicating a greater variant allele effect at higher valproate troughs. Finally, in a model accounting for age, sex, body mass index, lamotrigine dose (for measured troughs) and variant allele carriage at all analysed loci fitted separately to LAM‐only and LAM + VAL‐treated patients, variant allele at ABCG2 421C>A was associated with around 20–25% lower troughs in the former (an effect in absence of valproate) and with around 55–65% higher troughs in the latter (Table 5). Across all models (Table 4, Supporting Information tables), measured lamotrigine troughs were by 13–20% higher with 25 mg day–1 increase in lamotrigine dose. Higher body mass index tended to be associated with lower troughs (by 3–5% for each 2 kg m–2 increase) across all models (Table 4, Supporting Information tables).
Figure 1.
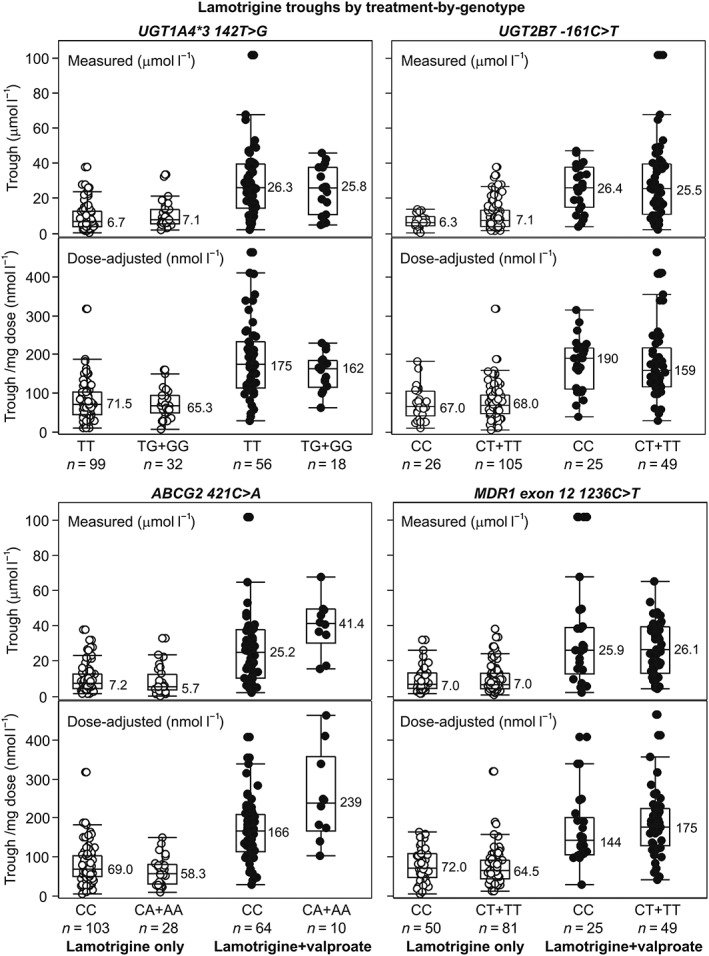
Measured and dose‐adjusted (per mg dose) steady‐state lamotrigine troughs by treatment‐by‐genotype – wild type homozygosity vs. variant allele carriage at UGT and ABC transporter loci, except for UGT1A4*2. Depicted are individual values (circles), medians (horizontal lines and numerical values), quartiles (boxes) and inner fences (bars). Values outside fences are outliers. For the lamotrigine‐treated wild type subjects at UGT1A4*2 (n = 128) measured lamotrigine troughs ranged between 0.5 and 37.6 μmol l–1 (median 6.6) and dose‐adjusted troughs ranged between 6.5 and 318 nmol l–1 (median 68.0). For the three variant allele carriers, the respective values were 5.2, 12.5 and 23.8 μmol l–1 (measured) and 79.3, 104.0 and 125 nmol l–1 (dose‐adjusted), i.e. within the wild type range. For the lamotrigine + valproate‐treated wild type subjects (n = 71), measured lamotrigine troughs ranged between 2.3 and 102 μmol l–1 (median 25.9) and dose‐adjusted troughs ranged between 30.7 and 464 nmol l–1 (median171.6). For the three variant allele carriers, the respective values were 6.5, 49.3 and 67.8 μmol l–1 (measured) and 130, 247 and 339 nmol l–1 (dose‐adjusted), i.e. within the wild type range. Data illustrate the valproate effect and do not indicate any effect of the variant allele carriage
Table 3.
Effects of variant allele carriage (vs. wild type homozygosity) at genotyped UGT and ABC transporter loci on steady‐state measured and dose‐adjusted lamotrigine troughs: summary of the main analysis (see Supporting Information Tables S1 and S2 for details). A significant treatment–genotype interaction indicates differences in the variant allele effects between patients treated with lamotrigine (LAM) + valproate (VAL) and patients treated with LAM only; conversely, it indicates differences in treatment effect (effect of VAL) between variant allele carriers and wild type homozygotes. Alpha level for the interaction term was set at 0.0125 to prevent false‐positive findings (four treatment–polymorphism tests were performed in unadjusted and adjusted analyses)
| Model type, purpose | Model effects | Main effect of the variant allele | Main treatment effect | Treatment*genotype interaction |
|---|---|---|---|---|
| Unadjusted; screening. Separate model for each polymorphism |
Treatment (LAM + VAL vs. LAM) Genotype (variant allele vs. wild type) Treatment*genotype interaction LAM daily dose (measured troughs) |
UGT1A4*3 142T>G – no effect UGT2B7 –161C>T – no effect MDR1 1236C>T – no effect ABCG2 421C>A – no effect |
In all models measured and dose‐adjusted troughs 2.5–3.5‐times higher with LAM + VAL vs. LAM |
UGT1A4*3 142T>G – insignificant UGT2B7 –161C>T – insignificant MDR1 1236C>T – insignificant ABCG2 421C>A –significant |
| Adjusted, main effects only; independent polymorphism effects |
Treatment Genotypes at all four loci LAM daily dose (measured troughs) Age, sex, body mass index |
UGT1A4*3 142T>G – no effect UGT2B7 –161C>T – no effect MDR1 1236C>T – no effect ABCG2 421C>A – no effect |
Measured and dose‐adjusted troughs around 2.5‐times higher with LAM + VAL vs. LAM | ‐ |
| Adjusted + interaction treatment*genotype; independent interaction effects. Separate model for each interaction |
Treatment Genotypes at all four loci Treatment*genotype interaction LAM daily dose (measured troughs) Age, sex, body mass index |
No variant allele effect at any locus was observed in any of the models. | In all models measured and dose‐adjusted troughs 2.5–3.5‐times higher with LAM + VAL vs. LAM |
UGT1A4*3 142T>G – insignificant UGT2B7 –161C>T – insignificant MDR1 1236C>T – insignificant ABCG2 421C>A – significant |
Table 4.
Effects of variant allele at ABCG2 421C>A (vs. wild type homozygosity) on measured and dose‐adjusted steady‐state lamotrigine troughs: main analysis. Modelsa were fitted to log‐transformed troughs: unadjusted, fully adjusted with treatment–ABCG2 genotype interaction and a parsimonious model (only adjustments with P < 0.1). Effects are geometric means ratios (GMR) with 95% confidence intervals (CI). For the interaction term, GMR is a relative difference in the effect of variant allele between lamotrigine (LAM) + valproate (VAL)‐treated patients and LAM‐only patients, i.e. a difference in the effect of VAL between variant allele carriers and wild type homozygotes
| Measured troughs | Dose‐adjusted troughs | |
|---|---|---|
| Models and effects | GMR (95% CI) | GMR (95% CI) |
| Unadjusted model | ||
| Treatment (LAM + VAL vs. LAM) | 3.57 (2.80–4.54) | 3.10 (2.45–3.93) |
| ABCG2 genotype (variant allele vs. wild type) | 1.13 (0.89–1.43) | 1.13 (0.89–1.42) |
| Treatment*ABCG2 genotype interaction | 2.19 (1.35–3.54)b | 1.89 (1.18–3.03)c |
| LAM + VAL vs. LAM at wild type | 2.41 (1.99–2.92) | 2.26 (1.87–2.73) |
| LAM + VAL vs. LAM at variant allele | 5.28 (3.39–8.20) | 4.26 (2.76–6.57) |
| Variant allele vs. wild type at LAM | 0.76 (0.59–0.98) | 0.82 (0.64–1.05) |
| Variant allele vs. wild type at LAM + VAL | 1.67 (1.11–2.52) | 1.55 (1.04–2.31) |
| LAM daily dose (by 25 mg) | 1.16 (1.13–1.19) | ‐ |
| Fully adjusted + treatment*ABCG2 interaction | ||
| Age (by 5 years) | 0.99 (0.95–1.02) | 0.99 (0.96–1.03) |
| Sex (men vs. women) | 0.94 (0.77–1.13) | 1.03 (0.85–1.23) |
| Body mass index (by 2 kg m b) | 0.96 (0.91–1.00) | 0.95 (0.91–0.99) |
| Treatment (LAM + VAL vs. LAM) | 3.49 (2.73–4.44) | 3.02 (2.38–3.83) |
| UGT1A4*3 142T>G (variant allele vs. wild type) | 0.94 (0.79–1.14) | 0.90 (0.74–1.10) |
| UGT2B7 –161C>T (variant allele vs. wild type) | 0.97 (0.80–1.19) | 0.97 (0.80–1.19) |
| MDR1 exon 12 1236C>T (variant vs. wild type) | 1.06 (0.89–1.27) | 1.03 (0.86–1.22) |
| ABCG2 421C>A (variant allele vs. wild type) | 1.14 (0.89–1.45) | 1.14 (0.90–1.45) |
| Treatment*ABCG2 421C>A interaction | 2.36 (1.39–3.64)b | 1.97 (1.22–3.18)c |
| LAM + VAL vs. LAM at wild type | 2.32 (1.89–2.83) | 2.15 (1.77–2.62) |
| LAM + VAL vs. LAM at variant allele | 5.24 (3.38–8.15) | 4.24 (2.75–6.54) |
| Variant allele vs. wild type at LAM | 0.76 (0.59–0.98) | 0.81 (0.63–1.04) |
| Variant allele vs. wild type at LAM + VAL | 1.72 (1.14–2.62) | 1.60 (1.07–2.62) |
| LAM daily dose (by 25 mg) | 1.16 (1.13–1.19) | ‐ |
| Parsimonious model | ||
| Body mass index (by 2 kg m b) | 0.95 (0.91–0.99) | 0.95 (0.92–0.99) |
| Treatment (LAM + VAL vs. LAM) | 3.50 (2.75–4.44) | 3.05 (2.41–3.85) |
| ABCG2 genotype (variant allele vs. wild type) | 1.13 (0.89–1.43) | 1.13 (0.89–1.42) |
| Treatment*ABCG2 genotype interaction | 2.29 (1.43–3.70)b | 1.98 (1.24–3.16)c |
| LAM + VAL vs. LAM at wild type | 2.31 (1.90–2.80) | 2.17 (1.80–2.62) |
| LAM + VAL vs. LAM at variant allele | 5.30 (3.43–8.20) | 4.29 (2.79–6.58) |
| Variant allele vs. wild type at LAM | 0.74 (0.58–0.96) | 0.80 (0.62–1.03) |
| Variant allele vs. wild type at LAM + VAL | 1.71 (1.14–2.55) | 1.58 (1.06–2.35) |
| LAM daily dose (by 25 mg) | 1.16 (1.13–1.19) | ‐ |
Unadjusted and fully adjusted with treatment–genotype interaction are models described in Table 3 used for the analysis of other polymorphisms
P = 0.001 in the unadjusted, P = 0.001 in the fully adjusted and P < 0.001 in the parsimonious model
P = 0.009 in the unadjusted, P = 0.006 in the fully adjusted and P = 0.005 in the parsimonious model
Figure 2.
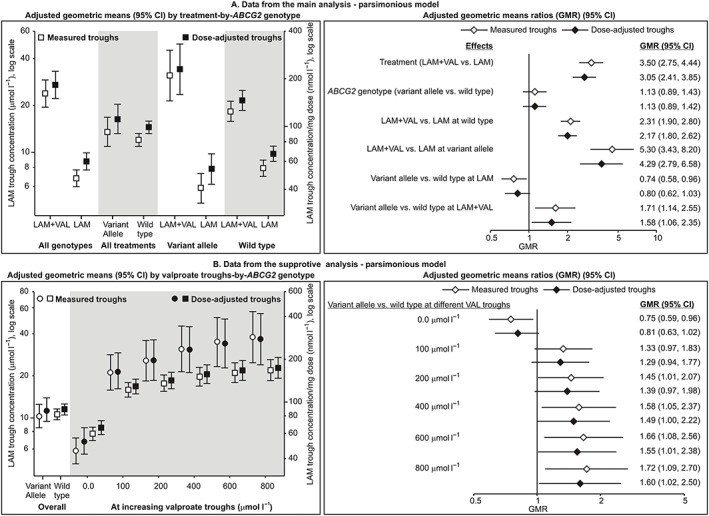
Illustration of the interaction between the variant allele at ABCG2 421C>A and valproate (VAL) in their effects on steady‐state lamotrigine (LAM) troughs. (A) Adjusted geometric means and geometric means ratios from the main analysis – parsimonious model in Table 4. (B) Adjusted geometric means (circles for variant allele carriers, squares for wild type homozygotes) and geometric means ratios from the supportive analysis – parsimonious model in Table 5
Table 5.
Effects of variant allele at ABCG2 421C>A (vs. wild type homozygosity) on measured and dose‐adjusted steady‐state lamotrigine troughs: summary of supportive analyses (see Supporting Information Tables S3 and S4 for details). General linear models were fitted to log‐transformed troughs, so effects are geometric means ratios (GMR) with 95% confidence intervals (CI). Model 1 is the parsimonious model from Table 4, refitted so that treatment was not considered as a binary variable but lamotrigine + valproate‐treated patients were represented by their valproate troughs and in lamotrigine‐only patients this value was set to 0. Valproate troughs are log‐transformed. Treatment, ABCG2 genotype and interaction effects are shown. GMR for the interaction term is a relative difference in the effect of a 2.71‐fold increase in valproate trough between variant allele carriers and wild type homozygotes, i.e. a relative increase in the effect of the variant allele with a 2.71‐fold increase in valproate concentration. Model 2 is the adjusted main effects model from Table 3 (but without effect of treatment) fitted only in lamotrigine‐treated patients. Model 3 is the same model fitted in lamotrigine + valproate‐treated patients. Only variant allele effects are shown
| Models | Measured troughs | Dose‐adjusted troughs |
|---|---|---|
| GMR (95% CI) | GMR (95% CI) | |
| Model 1 | ||
| Valproate trough (by 2.71‐fold) | 1.24 (1.19–1.29) | 1.22 (1.17–1.26) |
| ABCG2 421C>A (variant allele vs. wild type) | 0.98 (0.78–1.20) | 0.99 (0.81–1.27) |
| Valproate trough–ABCG2 421C>A interaction | 1.13 (1.05–1.22)a | 1.11 (1.02–1.20)b |
| Valproate trough at wild type | 1.17 (1.13–1.21) | 1.16 (1.12–1.19) |
| Valproate trough at variant allele | 1.32 (1.23–1.42) | 1.28 (1.19–1.37) |
| Model 2 (lamotrigine‐only patients) | ||
| ABCG2 421C>A (variant allele vs. wild type) | 0.75 (0.57–0.98) | 0.79 (0.60–1.03) |
| Model 3 (lamotrigine + valproate patients) | ||
| ABCG2 421C>A (variant allele vs. wild type) | 1.64 (1.14–2.36) | 1.56 (1.10–2.22) |
P = 0.005
P = 0.011
Effects of CYP2C9 and CYP2C19 polymorphisms and of UGT and ABC transporter variant alleles on steady‐state valproate troughs
A general linear model (age, sex, body mass index, valproate dose for measured troughs, CYP2C9 and CYP2C19 genotype) fitted to measured or dose‐adjusted valproate troughs in LAM + VAL‐treated patients indicated comparable concentrations in CYP2C9*1 homozygotes and *2/*3 allele‐carriers; and comparable concentrations in CYP2C19*1 homozygotes, combined *1/*17 and *17/*17 genotypes and *2 allele‐carriers (Supporting Information Figure S1). Similar models (age, sex, body mass index, valproate dose for measured troughs), a separate one for each of the UGT and ABC transporter polymorphisms, indicated no effect of variant alleles at any locus on valproate troughs (Supporting Information Figure S2).
Discussion
Pharmacokinetics of lamotrigine shows considerable interindividual variability that reflects on its efficacy and safety and is therefore subject to TDM 2. It is susceptible to effects of physiological (e.g. pregnancy) and diseased conditions (e.g. liver failure) and drugs 1. Several AEDs and non‐AEDs strongly induce glucuronidation of lamotrigine and may reduce its efficacy, but the most profound is the effect of a glucuronidation inhibitor valproic acid 1, 2, 3. Polymorphisms of UGT enzymes, most commonly UGT1A4*3, UGT1A4*2 and UGT2B7 –161C>T have also been investigated as potential sources of lamotrigine variability, but data have been rather sparse and contradictory 7, 8, 9, 10, 11, 12, 19. In vitro, lamotrigine is a substrate for MDR1/ABCB1 and ABCG2 transporters 20, 21 and sporadic studies have suggested that MDR1/ABCB1 exon 12 1236C>T polymorphism might be associated with somewhat lower lamotrigine troughs 22 or have no effect on lamotrigine pharmacokinetics 12 in Southern‐Central European adult patients, while ABCG2 421C>A polymorphism appeared associated with somewhat lower 24 or higher 25 concentrations in Chinese patients. Considering relative paucity of data and apparent contradictions, our primary objective was to evaluate the impact of the mentioned UGT and transporter polymorphisms on the steady‐state disposition of lamotrigine and the valproate–lamotrigine interaction in adult and adolescent Southern‐Central European patients. In this respect, the present study has several limitations: first, it was based on morning trough concentrations as a part of routine TDM and not on a detailed pharmacokinetic evaluation. This, however, does not diminish its potential clinical relevance, since morning troughs are recommended indicators in TDM of AEDs 2; next, certain treatment‐by‐variant allele carriage subsets were rather small – there were only six (2.9%) variant allele carriers at UGT1A4*2 70C>A, precluding any meaningful analysis; finally, some other potentially interesting polymorphisms such as UGT2B7 372A>G 12, and UGT1A4 –219C>T / 163G>A 35 were not considered. The present results should be viewed within the scope of these limitations. By contrast, the study included a reasonable sample of patients and inclusion/exclusion criteria, the timing of TDM sampling and analysis of measured (with adjustment for dose) and dose‐adjusted troughs (multivariate models simultaneously accounting for all polymorphisms and relevant demographics) ascertained a fair level of control for potential confounders. Therefore, the observed effects should be considered fairly accurate.
The main present finding is an interaction between variant allele at ABCG2 421C>A and valproate in their effects on steady‐state lamotrigine disposition. While in the entire cohort (lamotrigine monotreatment and valproate cotreatment approximately 2:1) variant allele had no effect, in the subset of patients on monotreatment, it was associated with around 20–25% lower troughs vs. wild type homozygosity (accounting for body mass index and delivered dose). This is a mild effect unlikely to affect efficacy of lamotrigine since the range of recommended lamotrigine TDM troughs expected to result in seizure control is rather wide 2, 3. In the subset of valproate cotreated patients, by contrast, variant allele was associated with 60–70% higher troughs (vs. wild type homozygosity). While again this could be judged as a moderate effect with an uncertain clinical relevance, it does convey an increased risk of lamotrigine adverse effects which is known to increase considerably at concentrations >58 μmol l–1 2, 3. However, this change from 20–25% lower to 60–75% higher lamotrigine disposition conditional on presence of valproate actually results in a 2.0–2.3‐fold difference {exp[–1x (ln 0.75 or 0.80) + (ln 1.70 or 1.60)]}. Its practical meaning is more intuitive for interpretation when the reasoning is reversed. Valproate increases exposure to lamotrigine around two‐fold, hence dosing guidelines for lamotrigine anticipate twice lower (by combination of frequency and dosage) doses in patients cotreated with valproate 1. These estimates were probably obtained in patient samples with a small number of ABCG2 421C>A variant allele carriers since its frequency is estimated at around 30% in Asians, 10–15% in Caucasians and 2% in African‐Americans 36, 37. Hence, in the present study, in line with expectations, cotreatment with valproate in wild type ABCG2 421C>A homozygotes resulted in around 2.2‐times higher steady‐state lamotrigine troughs compared to lamotrigine monotreatment. However, in the variant allele carriers, valproate effect was 2.0–2.3‐times greater (quantitative interaction) – cotreatment with valproate resulted in 4.3–5.3‐times higher lamotrigine troughs than in lamotrigine‐only patients. Therefore, ABCG2 421C>A variant allele carriers needing lamotrigine‐valproate cotreatment might actually require even lower than recommended lamotrigine doses. The ATP‐binding cassette subfamily G member 2, ABCG2 (breast cancer resistance protein, BCRP) is an efflux transporter expressed in the small intestine, liver, blood–brain barrier, testis, placenta and mammary glands 38. It functions primarily as an apical efflux pump in enterocytes, as a canalicular efflux pump transporting substrates from hepatocytes into the bile, as a transporter in the brain microvascular endothelial cells, renal proximal tubular cells and placental syncytiotrophoblasts contributing to the absorption, distribution and elimination of compounds as well as to tissue protection against xenobiotic exposure 39. This transporter has now been recognized as one of the key drug transporters involved in clinically relevant drug disposition 40, 41. A large number of ABCG2 inhibitors have been identified 42 and there are some convincing data that point to ABCG2 as an important mediator of drug–drug interactions in humans 43. Single nucleotide polymorphism of ABCG2 421C>A (p.Q141K) results in its reduced activity 44 – mRNA expression is maintained, but protein expression and function is reduced by 50–70% due to enhanced susceptibility to proteasomal degradation 45, 46. Variant allele carriage is associated with a higher systemic exposure to several statins and sulfasalazine 44, 47 and with an increased incidence of adverse effects of statins 48, 49 and gefitinib 50. The present data suggest that it enhances the effect of valproate on exposure to lamotrigine. In vitro, valproate is not a substrate for human ABCG2 21, hence an interaction at the transporter level does not seem likely. A possible mechanism could include increased absorption of lamotrigine due to a reduced enterocyte apical efflux. Combined with the reported stimulation of ABCG2 expression by valproate in vitro [26] the observed trend of increasing variant allele effect with increasing exposure to valproate makes it tempting to speculate about other, more complex mechanisms. Lamotrigine elimination is only scarcely by renal excretion of the unchanged drug (≤10%) 1 and the mechanism has not been disclosed – should it involve ABCG2‐mediated transport, kidneys could be an additional site of the lamotrigine–valproate interaction. The observation that in the lamotrigine‐only treated patients variant allele carriage was associated with mildly (20–26%) lower morning troughs appears confusing and raises a question about potentially opposing effects of ABCG2 421C>A polymorphism on lamotrigine transport that is conditional on valproate.
In agreement with observations in a sample of patients from a similar population 12, we found no effect of MDR1/ABCB1 1236C>T (rs1128503) polymorphism on lamotrigine disposition. This is in a contrast with previously reported mildly lower lamotrigine troughs in variant allele carriers 22; however, it should be noted that the previous sample (n = 222) included around 75% of patients cotreated with UGT inducers, valproic acid or a combination of more than two AEDs, and no effect of this polymorphism was observed in a subset of lamotrigine‐only treated patients. Therefore, a potential role of this polymorphism in lamotrigine disposition should be more thoroughly investigated. We were also unable to detect any relevant impact of UGT1A4*3 or UGT2B7 –161C>T on lamotrigine troughs, while UGT1A4*2 variant allele carriers were too few for any meaningful analysis. Discrepancies vs. reported effects of these polymorphisms 7, 8, 9, 10, 11, 12, 19 might be due to several factors, including rather limited/small sample sizes, differences in minor allele frequency or (not) accounting for confounding (genetic or other).
As by‐findings, the present study also provides observations about the effects of several UGT/transporter and CYP polymorphisms on valproate steady‐state disposition. However, it was not conceived specifically for this purpose, hence there were no patients treated only with valproic acid and several potentially interesting polymorphisms were not addressed. Under these circumstances, the present data do not indicate any relevant impact of CYP2C9 or CYP2C19 polymorphisms, ABCG2 421C>A, MDR1/ABCB1 1236C>T, or UGT1A4*3 and UGT2B7 –161C>T polymorphisms on valproate disposition. This is in a disagreement with some of the published data indicating a significant influence of UGT2B7 –161C>T 51, 52 or CYP2C9*3 53 variants on VPA concentrations but predominantly in Asian populations. Besides, some data indicate that other genes/variants such as UGT2B7 A268G 11, UGT1A3*5 54, UGT1A4 –219C>T / 163G>A 35 or UGT1A6 552A>G 55 may affect valproate serum concentrations.
Conclusion
To our knowledge, the present study is the first to indicate a potentially relevant interaction between ABCG2 421C>A polymorphism and valproate in their effects on lamotrigine disposition in patients with epilepsy. The present findings contribute to a still mostly unexplored field of the practical relevance of ABCG2 and its polymorphisms in drug–drug interactions.
Competing Interests
I.K.D. is an employee of the Croatian Agency for Medicinal Products and Medical Devices. The other authors have no competing interests to declare.
Financially supported the Croatian Agency for Medicinal Products and Medical Devices (project: Pharmacogenomics and Pharmacovigilance ‐ Prevention of Side Effects by Individualization of Therapy). We would like to thank Zrinka Mirkovic and Maja Mezak Herceg for technical support.
Contributors
All authors were fully involved in manuscript development and assume responsibility for the direction and content. I.K.D., M.L., V.T., Ž.P.G. and N.B., conceived the study. M.L., L.G. and N.B. performed and reviewed the analyses. V.T. performed and wrote the statistical analysis report. I.K.D., M.L., V.T., I.Č. and N.B., assisted in the preparation of the manuscript. All authors reviewed the manuscript and provided their approval for submission.
Supporting information
Table S1 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state lamotrigine troughs or dose‐adjusted troughs: main analysis, unadjusted models
Table S2 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state lamotrigine troughs or dose‐adjusted troughs: main analysis, adjusted models
Table S3 Effects of variant allele at ABCG2 421C>A on steady‐state lamotrigine troughs or dose‐adjusted troughs: supportive analysis with valproate cotreatment represented by valproate troughs
Table S4 Effects of variant allele at ABCG2 421C>A on steady‐state lamotrigine troughs or dose‐adjusted troughs: supportive analysis separately in patients treated only with lamotrigine and patients treated with lamotrigine+valproate
Figure S1 Effects of CYP2C9 and CYP2C19 polymorphisms on steady‐state valproate troughs or dose‐adjusted troughs
Figure S2 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state valproate troughs or dose‐adjusted troughs
Klarica Domjanović, I. , Lovrić, M. , Trkulja, V. , Petelin‐Gadže, Ž. , Ganoci, L. , Čajić, I. , and Božina, N. (2018) Interaction between ABCG2 421C>A polymorphism and valproate in their effects on steady‐state disposition of lamotrigine in adults with epilepsy. Br J Clin Pharmacol, 84: 2106–2119. 10.1111/bcp.13646.
References
- 1. Medicines.org.uk Lamictal . Summary of product characteristics 2017. Available at http://www.medicines.org.uk/emc/medicine/4228 (last accessed 15 August 2017).
- 2. Patsalos PN, Berry DJ, Bourgeois BFD, Cloyd JC, Glauser TA, Johannessen SI, et al Antiepileptic drugs – best practice guidelines for therapeutic drug monitoring: a position paper by the subcommission on therapeutic drug monitoring, ILAE commission on therapeutic strategies. Epilepsia 2008; 49: 1239–1276. [DOI] [PubMed] [Google Scholar]
- 3. Patsalos PN. Lamotrigine In: Antiepileptic drug interactions, 3rd edn. Springer: Cham, 2016; 55–60. [Google Scholar]
- 4. Rowland A, Elliot DJ, Williams JA, Mackenzie PI, Dickinson RG, Miners JO. In vitro characterization of lamotrigine N2‐glucuronidation and the lamotrigine‐valproic acid interaction. Durg Metab Dispos 2006; 34: 1055–1062. [DOI] [PubMed] [Google Scholar]
- 5. May TW, Rambeck B, Jürgens U. Influence of oxcarbazepine and methsuximide on lamotrigine concentrations in epileptic patients with and without valproic acid comedication: results of a retrospective study. Ther Drug Monit 1999; 21: 175–181. [DOI] [PubMed] [Google Scholar]
- 6. Patsalos PN. Drug interactions with the newer antiepileptic drugs (AEDs) – part 1: pharmacokinetic and pharmacodynamics interactions between AEDs. Clin Pharmacokinet 2013; 52: 927–966. [DOI] [PubMed] [Google Scholar]
- 7. Chang Y, Yang LY, Zhang MC, Liu SY. Correlation of the UGT1A4 gene polymorphism with serum concentration and therapeutic efficacy of lamotrigine in Han Chinese of northern China. Eur J Clin Pharmacol 2014; 70: 941–946. [DOI] [PubMed] [Google Scholar]
- 8. Reimers A, Sjursen W, Helde G, Brodtkorb E. Frequencies of UGT1A4*2 (P24T) and *3 (L48V) and their effects on serum concentrations of lamotrigine. Eur J Drug Metab Pharmacokinet 2016; 41: 149–155. [DOI] [PubMed] [Google Scholar]
- 9. Gulcebi MI, Ozkaynakcı A, Goren MZ, Aker RG, Ozkara C, Onat FY. The relationship between UGT1A4 polymorphism and serum concentration of lamotrigine in patients with epilepsy. Epilepsy Res 2011; 95: 1–8. [DOI] [PubMed] [Google Scholar]
- 10. Liu L, Zhao L, Wang Q, Qiu F, Wu X, Ma Y. Influence of valproic acid concentration and polymorphism of UGT1A4*3, UGT2B7–161C>T and UGT2B7*2 on serum concentration of lamotrigine in Chinese epileptic children. Eur J Clin Pharmacol 2015; 71: 1341–1347. [DOI] [PubMed] [Google Scholar]
- 11. Du Z, Jiao Y, Shi L. Association of UGT2B7 and UGT1A4 polymorphisms with serum concentration of antiepileptic drugs in children. Med Sci Monit 2016; 22: 4107–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Milosheska D, Lorber B, Vovk T, Kastelic M, Dolžan V, Grabnar I. Pharmacokinetics of lamotrigine and its metabolite N‐2‐glucuronide: influence of polymorphism of UDP‐glucuronosyltransferases and drug transporters. Br J Clin Pharmacol 2016; 82: 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sawyer MB, Innocenti F, Das S, Cheng C, Ramirez J, Pantle‐Fisher FH, et al A pharmacogenetics study of uridine diphosphate‐glucuronosyltransferase 2B7 in patients receiving morphine. Clin Pharmacol Ther 2003; 73: 566–574. [DOI] [PubMed] [Google Scholar]
- 14. Hu DG, Meech R, Lu L, McKinnon RA, Mackenzie PI. Polymorphisms and haplotypes of the UDP‐glucuronosyltransferase 2B7 gene promoter. Drug Metab Dispos 2014; 42: 854–862. [DOI] [PubMed] [Google Scholar]
- 15. Daly AK. Pharmacogenetics of drug metabolizing enzymes in the United Kingdom population: review of current knowledge and comparison with selected European populations. Drug Metab Pers Ther 2015; 30: 165–174. [DOI] [PubMed] [Google Scholar]
- 16. Innocenti F, Liu W, Fackenthal D, Ramírez J, Chen P, Ye X, et al Single nucleotide polymorphism discovery and functional assessment of variation in the UDP464 glucuronosyltransferase 2B7 gene. Pharmacogenet Genomics 2008; 18: 683–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang P, Lin XQ, Cai WK, Xu GL, Zhou MD, Yang M, et al Effect of UGT2B7 genotypes on plasma concentration of valproic acid: a meta‐analysis. Eur J Clin Pharmacol 2018; 74: 433–442. [DOI] [PubMed] [Google Scholar]
- 18. Blanca Sánchez M, Herranz JL, Leno C, Arteaga R, Oterino A, Valdizán EM, et al UGT2B7_‐161C>T polymorphism is associated with lamotrigine concentration‐to‐dose ratio in a multivariate study. Ther Drug Monit 2010; 32: 177–184. [DOI] [PubMed] [Google Scholar]
- 19. Singkham N, Towanabut S, Lertkachatarn S, Punyawudho B. Influence of the UGT2B7–161C>T polymorphism on the population pharmacokinetics of lamotrigine in Thai patients. Eur J Clin Pharmacol 2013; 69: 1285–1291. [DOI] [PubMed] [Google Scholar]
- 20. Luna‐Tortós C, Fedrowitz M, Löscher W. Several major antiepileptic drugs are substrates for human P‐glycoprotein. Neuropharmacology 2008; 55: 1364–1375. [DOI] [PubMed] [Google Scholar]
- 21. Römermann K, Helmer R, Löscher W. The antiepileptic drug lamotrigine is a substrate of mouse and human breast cancer resistance protein (ABCG2). Neuropharmacology 2015; 93: 7–14. [DOI] [PubMed] [Google Scholar]
- 22. Lovrić M, Božina N, Hajnšek S, Kuzman MR, Sporiš D, Lalić Z, et al Association between lamotrigine concentrations and ABCB1 polymorphisms in patients with epilepsy. Ther Drug Monit 2012; 34: 518–525. [DOI] [PubMed] [Google Scholar]
- 23. Cerveny L, Pavek P, Malakova J, Staud F, Fendrich Z. Lack of interactions between breast cancer resistance protein (bcrp/abcg2) and selected antiepileptic agents. Epilepsia 2006; 47: 461–468. [DOI] [PubMed] [Google Scholar]
- 24. Zhou Y, Wang X, Li H, Zhang J, Chen Z, Xie W, et al Polymorphisms of ABCG2, ABCB1 and HNF4α are associated with lamotrigine trough concentrations in epilepsy patients. Drug Metab Pharmacokinet 2015; 30: 282–287. [DOI] [PubMed] [Google Scholar]
- 25. Shen CH, Zhang YX, Lu RY, Jin B, Wang S, Liu ZR, et al Specific OCT1 and ABCG2 polymorphisms are associated with lamotrigine concentrations in Chinese patients with epilepsy. Epilepsy Res 2016; 127: 186–190. [DOI] [PubMed] [Google Scholar]
- 26. Rubinchik‐Stern M, Shmuel M, Eyal S. Antiepileptic drugs alter the expression of placental carriers: an in vitro study in a human placental cell line. Epilepsia 2015; 56: 1023–1032. [DOI] [PubMed] [Google Scholar]
- 27. Ghodke‐Puranik Y, Thorn CF, Lamba JK, Leeder JS, Song W, Birnbaum AK. Valproic acid pathway: pharmacokinetics and pharmacodynamics. Altm Pharmacogenet Genomics 2013; 23: 236–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hussein Z, Posner J. Population pharmacokinetics of lamotrigine monotherapy in patients with epilepsy: retrospective analysis of routing monitoring data. Br J Clin Pharmacol 1997; 43: 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haslemo T, Loryan I, Ueda N, Mannheimer B, Bertilsson L, Ingelman‐Sundberg M, et al UGT1A4*3 encodes significantly increased glucuronidation of olanzapine in patients on maintenance treatment and in recombinant systems. Clin Pharmacol Ther 2012; 92: 221–227. [DOI] [PubMed] [Google Scholar]
- 30. Ganoci L, Božina T, Mirošević Skvrce N, Lovrić M, Mas P, Božina N. Genetic polymorphisms of cytochrome P450 enzymes: CYP2C9, CYP2C19, CYP2D6, CYP3A4 and CYP3A5 in the Croatian population. Drug Metab Pers Ther 2017; 32: 11–21. [DOI] [PubMed] [Google Scholar]
- 31. Gaunt TR, Rodriguez S, Day NMI. Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool “CubeX”. BMC Bioinformatics 2007; 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harding SD, Sharman JL, Faccenda E, Southan C, Pawson AJ, Ireland S, et al The IUPHAR/BPS guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY. Nucl Acids Res 2018; 46: D1091–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alexander SPH, Kelly E, Marrion NV, Peters JA, Faccenda E, Harding SD, et al The Concise Guide to PHARMACOLOGY 2017/18: Transporters. Br J Pharmacol 2017; 174: S360–S446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alexander SPH, Fabbro D, Kelly E, Marrion NV, Peters JA, Faccenda E, et al The Concise Guide to PHARMACOLOGY 2017/18: Enzymes. Br J Pharmacol 2017; 174: S272–S359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Q, Liang M, Dong Y, Yun W, Qiu F, Zhao L, et al Effects of UGT1A4 genetic polymorphisms on serum lamotrigine concentrations in Chinese children with epilepsy. Drug Metab Pharmacokinet 2015; 30: 209–213. [DOI] [PubMed] [Google Scholar]
- 36. Sakiyama M, Matsuo H, Takada Y, Nakamura T, Nakayama A, Takada T, et al Ethnic differences in ATP‐binding cassette transporter, sub‐family G, member 2 (ABCG2/BCRP): genotype combinations and estimated functions. Drug Metab Pharmacokinet 2014; 29: 490–492. [DOI] [PubMed] [Google Scholar]
- 37. Keskitalo JE, Pasanen MK, Neuvonen PJ, Niemi M. Different effects of the ABCG2 c.421C>a SNP on the pharmacokinetics of fluvastatin, pravastatin and simvastatin. Pharmacogenomics 2009; 10: 1617–1624. [DOI] [PubMed] [Google Scholar]
- 38. Endres CJ, Hsiao P, Chung FS, Unadkat JD. The role of transporters in drug interactions. Eur J Pharm Sci 2006; 27: 501–517. [DOI] [PubMed] [Google Scholar]
- 39. Poirier A, Portmann R, Cascais AC, Bader U, Walter I, Ullah M, et al The need for human breast cancer resistance protein substrate and inhibition evaluation in drug discovery and development: why, when, and how? Drug Metab Dispos 2014; 42: 1466–1477. [DOI] [PubMed] [Google Scholar]
- 40. Fda.gov . US Food and Drug Administration: guidance for industry: drug interaction studies—study design, data analysis, implications for dosing, and labeling recommendations (2017). Available at http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM292362.pdf (last accessed 10 November 2017).
- 41. Ema.Europa.Eu European medicines agency, London, UK: guideline on the investigation of drug interactions CPMP/EWP/560/95/rev. 1 Corr. 2** (2012). Available at http://www.ema.europa.eu/docs/en_GB/document_library/Scientific_guideline/2012/07/WC500129606.pdf (last accessed 15 August 2017).
- 42. Mao Q, Unadkat JD. Role of the breast cancer resistance protein (ABCG2) in drug transport. AAPS J 2005; 7: E118–E133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Giacomini KM, Huang SM, Tweedie DJ, Benet LZ, Brouwer KL, Chu X, et al International transporter consortium membrane transporters in drug development. Nat Rev Drug Discov 2010; 9: 215–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giacomini KM, Balimane PV, Cho SK, Eadon M, Edeki T, Hillgren KM, et al International transporter consortium commentary on clinically important transporter polymorphisms. Clin Pharmacol Ther 2013; 94: 23–26. [DOI] [PubMed] [Google Scholar]
- 45. Kondo C, Suzuki H, Itoda M, Ozawa S, Sawada J, Kobayashi D, et al Functional analysis of SNPs variants of BCRP/ABCG2. Pharm Res 2004; 21: 1895–1903. [DOI] [PubMed] [Google Scholar]
- 46. Furukawa T, Wakabayashi K, Tamura A, Nakagawa H, Morishima Y, Osawa Y, et al Major SNP (Q141K) variant of human ABC transporter ABCG2 undergoes lysosomal and proteasomal degradations. Pharm Res 2009; 26: 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Elsby R, Martin P, Surry D, Sharma P, Fenner K. Solitary inhibition of the breast cancer protein efflux transporter results in a clinically significant drug‐drug interaction with Rosuvastatin by causing up to a 2‐fold increase in statin exposure. Drug Metab Dispos 2016; 44: 398–408. [DOI] [PubMed] [Google Scholar]
- 48. Miroševic Skvrce N, Božina N, Zibar L, Barišic I, Pejnovic L, Macolic Šarinic V. CYP2C9 and ABCG2 polymorphisms as risk factors for developing adverse drug reactions in renal transplant patients taking fluvastatin: a case‐control study. Pharmacogenomics 2013; 14: 1419–1431. [DOI] [PubMed] [Google Scholar]
- 49. Mirošević Skvrce N, Macolić Šarinić V, Šimić I, Ganoci L, Muačević Katanec D, Božina N. ABCG2 gene polymorphisms as risk factors for atorvastatin adverse reactions: a case‐control study. Pharmacogenomics 2015; 16: 803–815. [DOI] [PubMed] [Google Scholar]
- 50. Cusatis G, Gregorc V, Li J, Spreafico A, Ingersoll RG, Verweij J, et al Pharmacogenetics of ABCG2 and adverse reactions to gefitinib. J Natl Cancer Inst 2006; 98: 1739–1742. [DOI] [PubMed] [Google Scholar]
- 51. Wang Q, Zhao L, Liang M, Dong Y, Yun W, Qiu F, et al Effects of UGT2B7 genetic polymorphisms on serum concentrations of valproic acid in Chinese children with epilepsy comedicated with lamotrigine. Ther Drug Monit 2016; 38: 343–349. [DOI] [PubMed] [Google Scholar]
- 52. Inoue K, Suzuki E, Yazawa R, Yamamoto Y, Takahashi T, Takahashi Y, et al Influence of uridine diphosphate glucuronosyltransferase 2B7–161 C>T polymorphism on the concentration of valproic acid in pediatric epilepsy patients. Ther Drug Monit 2014; 36: 406–409. [DOI] [PubMed] [Google Scholar]
- 53. Tan L, Yu JT, Sun YP, Ou JR, Song JH, Yu Y. The influence of cytochrome oxidase CYP2A6, CYP2B6, and CYP2C9 polymorphisms on the plasma concentrations of valproic acid in epileptic patients. Clin Neurol Neurosurg 2010; 112: 320–323. [DOI] [PubMed] [Google Scholar]
- 54. Chu XM, Zhang LF, Wang GJ, Zhang SN, Zhou JH, Hao HP. Influence of UDP‐glucuronosyltransferase polymorphisms on valproic acid pharmacokinetics in Chinese epilepsy patients. Eur J Clin Pharmacol 2012; 68: 1395–1401. [DOI] [PubMed] [Google Scholar]
- 55. Munisamy M, Tripathi M, Behari M, Raghavan S, Jain DC, Ramanujam B, et al The effect of uridine diphosphate glucuronosyltransferase (UGT) 1A6 genetic polymorphism on valproic acid pharmacokinetics in Indian patients with epilepsy: a pharmacogenetic approach. Mol Diagn Ther 2013; 17: 319–326. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state lamotrigine troughs or dose‐adjusted troughs: main analysis, unadjusted models
Table S2 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state lamotrigine troughs or dose‐adjusted troughs: main analysis, adjusted models
Table S3 Effects of variant allele at ABCG2 421C>A on steady‐state lamotrigine troughs or dose‐adjusted troughs: supportive analysis with valproate cotreatment represented by valproate troughs
Table S4 Effects of variant allele at ABCG2 421C>A on steady‐state lamotrigine troughs or dose‐adjusted troughs: supportive analysis separately in patients treated only with lamotrigine and patients treated with lamotrigine+valproate
Figure S1 Effects of CYP2C9 and CYP2C19 polymorphisms on steady‐state valproate troughs or dose‐adjusted troughs
Figure S2 Effects of variant alleles at genotyped UGT and ABC transporter loci on steady‐state valproate troughs or dose‐adjusted troughs


