Abstract
Actin-binding LIM protein 1 (ABLIM1), a member of the LIM-domain protein family, mediates interactions between actin filaments and cytoplasmic targets. However, the role of ABLIM1 in osteoclast and bone metabolism has not been reported. In the present study, we investigated the role of ABLIM1 in the receptor activator of NF-κB ligand (RANKL)-mediated osteoclastogenesis. ABLIM1 expression was induced by RANKL treatment and knockdown of ABLIM1 by retrovirus infection containing Ablim1-specific short hairpin RNA (shAblim1) decreased mature osteoclast formation and bone resorption activity in a RANKL-dose dependent manner. Coincident with the downregulated expression of osteoclast differentiation marker genes, the expression levels of c-Fos and the nuclear factor of activated T-cells cytoplasmic 1 (NFATc1), critical transcription factors of osteoclastogenesis, were also decreased in shAblim1-infected osteoclasts during RANKL-mediated osteoclast differentiation. In addition, the motility of preosteoclast was reduced by ABLIM1 knockdown via modulation of the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/Akt/Rac1 signaling pathway, suggesting another regulatory mechanism of ABLIM1 in osteoclast formation. These data demonstrated that ABLIM1 is a positive regulator of RANKL-mediated osteoclast formation via the modulation of the differentiation and PI3K/Akt/Rac1-dependent motility.
Keywords: ABLIM1, LIM domain, Motility, Osteoclast, RANKL
INTRODUCTION
Osteoclasts are the primary bone resorbing cells and are critical for bone remodeling and bone metabolism. Mature multinucleated osteoclasts differentiate from monocyte/macrophage lineage hematopoietic cells through multiple processes of osteoclastogenesis, including cell proliferation, adhesion, migration, podosome formation, and cell–cell fusion (1). These differentiation processes are mainly controlled by two essential osteoclastogenic cytokines: the receptor activator of the nuclear factor κB ligand (RANKL) and the macrophage colony-stimulating factor (M-CSF) (1). The interaction between RANKL and RANK increases c-Fos expression, the early transcription factor for osteoclast differentiation, which leads sequentially to the upregulation of the nuclear factor of activated T-cells cytoplasmic 1 (NFATc1) expression (2). NFATc1 is known as a master transcription factor that regulates all of the osteoclastogenesis steps by modulating osteoclast-specific genes such as acid phosphatase 5-tartrate resistant (Acp5), osteoclast-associated receptor (Oscar), and ATPase H+ transporting V0 subunit d2 (Atp6v0d2) (3, 4). For bone remodeling, mononucleated osteoclast precursors are first recruited on bone turnover sites. The preosteoclasts then fuse to form multinucleated mature osteoclasts, which are capable of resorbing the bone matrix as they actively migrate to the bone surface (5, 6). Osteoclasts undergo immense structural changes, such as cytoskeleton modification, while migrating on the bone matrix. Previously, the PI3K/Akt pathway was shown to be involved in the extracellular calcium-mediated migration of osteoclast precursor Raw 264.7 cells (7). The migration of osteoclast precursors or mature osteoclasts depends on the characteristics and distinguishable pathways of PI3K/PKCα-PKCδ/RhoA-Rac1 signaling (8). In addition, Rac1 is selectively activated by RANKL and is more critical than Rac2 in regulating the actin cytoskeleton, as shown by in vivo analysis with Rac1 or Rac2-null mice (9, 10).
The LIM domain is a cysteine-rich sequence motif and unique protein-protein interacting module, having a consensus sequence defined as CX2CX16–23HX2CX2CX2CX16–21CX2(C/H/D) (11). This domain is found in a variety of proteins with diverse functions, including regulation of cell growth, motility, and division (11, 12). In humans, the LIM superclass has been divided into 14 classes based on sequence, function, characteristic domain architectures, and cellular localization (13). Among the LIM classes, the actin-binding LIM (ABLIM) protein family consist of four amino-terminal LIM domains and a carboxyl-terminal dematin like domain, including a villin headpiece (VHP) domain that binds to actin filaments (13). To date, three subtypes, ABLIM1, ABLIM2, and ABLIM3, have been reported in mammals (14–16). ABLIM1 was initially cloned and identified from the inner segment and outer plexiform layer of the retina, including the outer limited membrane, brain, and muscle tissue (14). The ablation of a retina-specific isoform of ABLIM1 by gene targeting resulted in an absence of morphological and functional defects in retinal neurons (17). Therefore, the function of ABLIM1 remains unclear. The two other members of the ABLIM protein family, ABLIM-2 and ABLIM 3, display distinct tissue specific expression patterns, showing especially high expression in muscle and neuronal tissue (15). In addition, these ABLIM proteins bind F-actin intensively and synergistically stimulate the striated muscle activator of Rho signaling (STARS) dependent activation of the serum-response factor (SRF) (15). These studies indicated that ABLIM proteins are involved in the organization of the actin cytoskeleton, possibly in a small GTPase dependent manner. However, no reports have been found that demonstrate the function of ABLIM proteins in osteoclasts, which undergo massive cytoskeletal reorganization.
We performed transcriptomic analysis between bone marrow-derived macrophages (BMMs) and mature osteoclasts by using high throughput RNA sequencing to find new regulatory factors related to cytoskeleton reorganization which is an important process for osteoclast formation (data not shown). Among the cytoskeleton-related genes, Ablim1 was the most increased gene and selected to study its role on osteoclastogenesis. In the present study, we describe the identification and characterization of ABLIM1 as a positive regulator of RANKL-mediated osteoclastogenesis. We analyzed the expression of ABLIM1, and determined the effect of knockdown using Ablim1-specific shRNA (shAblim1) on osteoclast differentiation, bone-resorbing activity, and migration.
RESULTS
ABLIM1 expression and regulatory involvement of ABLIM1 in RANKL-induced osteoclast differentiation and activation
First, we assessed the expression levels of ABLIM1 during osteoclast formation. BMMs were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 d. The mRNA and protein expression levels of ABLIM1 were analyzed using reverse transcription-polymerase chain reaction (RT-PCR) and immunoblotting, respectively. The ABLIM1 mRNA and protein expression levels gradually increased with time and reached approximately 10- to 12-fold and 4- to 5-fold, respectively, at day 4 compared with the non-induced control (Fig. 1A and 1B). We then examined whether ABLIM1 participates in osteoclast differentiation and activation. The role of ABLIM1 in osteoclatogenesis was investigated by using retroviral vector expressing Ablim1-specific RNAi (shAblim1) which could efficiently downregulated ABLIM1 expression during osteoclastogenesis (Supplementary Fig. 1). BMMs infected with shGFP or shAblim1 were cultured with various concentrations of RANKL in the presence of M-CSF. Although the formation of the tartrate-resistant acid phosphatase (TRAP) positive multinuclear mature osteoclasts (MNCs) increased along with increasing RANKL concentrations in both shGFP- and shAblim1-infected osteoclasts, the formation of TRAP+-MNCs from BMMs was considerably decreased in the shAblim1-infected cells compared with the results in the shGFP-infected cells at all tested concentrations of RANKL (Fig. 2A and 2B). Moreover, the total TRAP activity was also inhibited in the shAblim1-infected osteoclasts compared with that in the control cells (Fig. 2C). To assess osteoclast activity, the formation of the F-actin ring and resorption activity were measured. ShAblim1-infected osteoclasts exhibited a dramatic reduction in the resorption pit formation on a bone resorption assay plate, as well as decreased numbers of osteoclasts having an F-actin ring (Fig. 2D). Taken together, these data implied that ABLIM1 induced by RANKL has a regulatory role in RANKL-mediated osteoclast differentiation, formation, and activation.
Fig. 1.
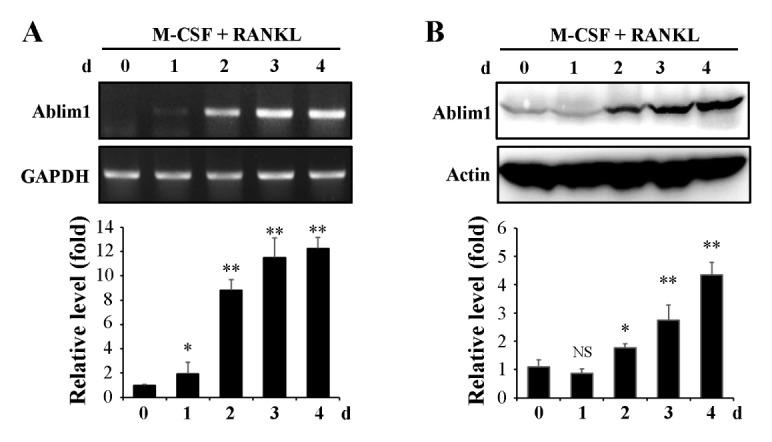
Induction of Ablim1 expression during RANKL-mediated osteoclastogenesis. BMMs were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for the indicated times. The mRNA (A) and protein (B) expression level of ABLIM1 was determined by RT PCR and Western blot analysis, respectively. The relative expression level was measured using Gapdh and Actin as loading controls (lower panels). Data are expressed as mean ± SD from triplicate experiments. NS, not significant. *P < 0.05, **P < 0.01 versus control (Day 0).
Fig. 2.
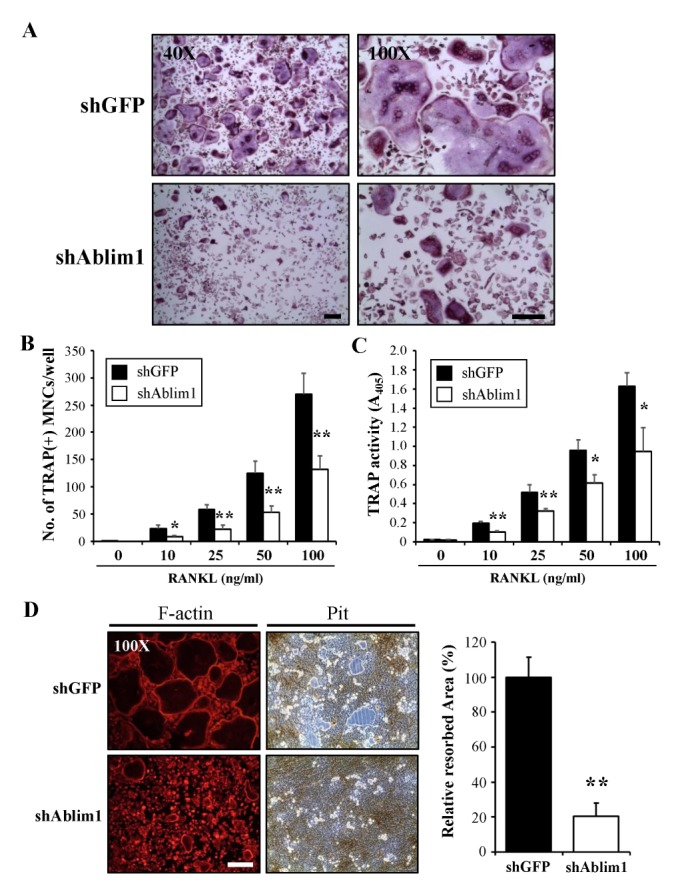
Effects of ABLIM1-specific knockdown on RANKL-induced osteoclast differentiation and activation. shGFP (control vector)- or shAblim1-infected BMMs were cultured with various concentrations of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 d. (A) TRAP of shGFP- or shAblim1-infected osteoclasts was stained. (B) TRAP+-multinuclear mature cells (MNCs) with ≥ three nuclei were counted as mature osteoclasts. (C) Total TRAP activity was measured by absorbance at 405 nm. (D) F-actin ring formation was determined by rhodamine-phalloidin staining at 4 d. Pit formation by shGFP- or shAblim1-infected osteoclasts cultured on hydroxyapatite-coated plates was measured at 7 d. The relative resorbed area is presented (right panel). Data represent the mean ± SD and are representative of at least three experiments. *P < 0.05, **P < 0.01 versus control (shGFP). Scale bar = 200 μm.
Effect of ABLIM1 on the expression of osteoclast differentiation marker genes and transcription factors
To verify the regulatory effect of ABLIM1 in osteoclastogenesis, alterations in the expression levels of osteoclast differentiation marker genes such as Acp5, Oscar, CtsK, Atp6v0d2, Dc-stamp, and Nfatc1, were examined using quantitative real-time PCR. As expected, the expression of these genes was significantly increased by RANKL treatment. However, the expression levels of the RANKL-induced genes were considerably inhibited by ABLIM1 knockdown compared with those of the control (Fig. 3A). We then assessed whether ABLIM1 downregulation affected the expression of pivotal transcription factors for osteoclastogenesis using immunoblotting analysis. The levels of c-Fos and NFATc1 were significantly suppressed, by approximately 50% in the shAblim1-infected osteoclasts, compared with those of the control cells, during RANKL-induced osteoclast differentiation (Fig. 3B). Consistent with previous data concerning osteoclast formation and activity, these results demonstrated that ABLIM1 regulates osteoclastogenesis via modulation of the expression of the osteoclast differentiation genes and transcription factors.
Fig. 3.
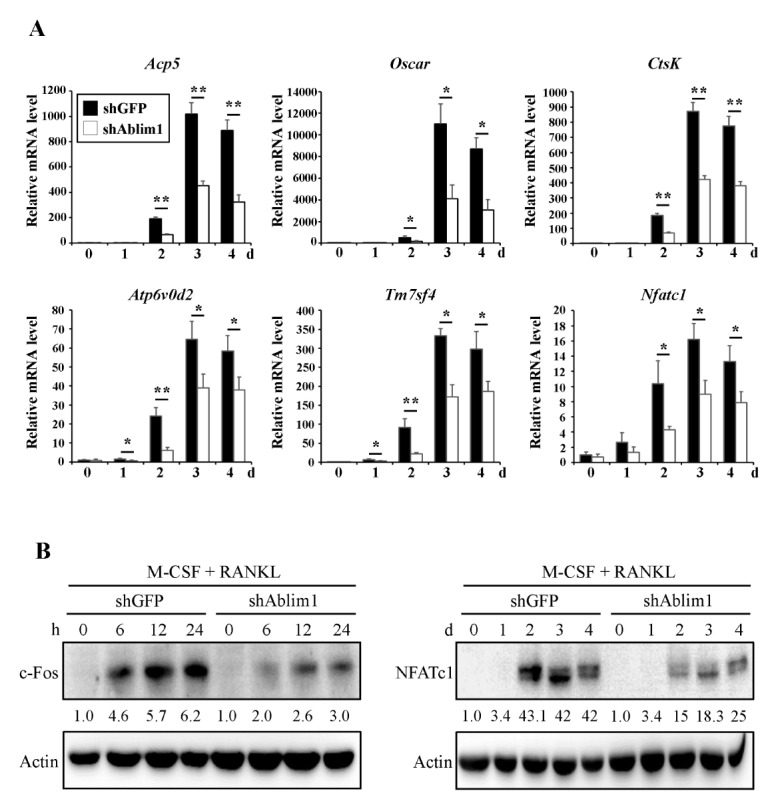
Effects of ABLIM1-specific knockdown on expression of osteoclast differentiation marker genes and transcriptional factors. Control vector (shGFP)- or shAblim1-infected BMMs were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for indicated times. (A) The relative mRNA expression levels of osteoclast differentiation marker genes were measured by real-time PCR and normalized to the expression of Gapdh. (B) c-Fos and NFATc1 expression levels were detected by anti-c-Fos and anti-NFATc1 antibodies, respectively, and normalized to the actin level. Data are expressed as the representative mean ± SD from at least three independent experiments. *P < 0.05, **P < 0.01 versus control (shGFP).
Inhibitory effect of ABLIM1 knockdown on preosteoclast motility
Based on its actin binding capacity, ABLIM1 was suggested as a potential mediator between actin filaments and cytoplasmic targets. Therefore, we examined whether ABLIM1 was involved in another regulatory mechanism of RANKL-induced osteoclast differentiation and formation. The cell migration of shGFP- or shAblim1-infected preosteoclasts was measured using a wound-healing assay. Comparison of cell motility between shGFP- or shAblim1-infected preosteoclasts showed a marked inhibition (by approximately 40%) in shAblim1 infected cells (Fig. 4A). To confirm the inhibitory mechanism of ABLIM1 knockdown on migration, the downstream signaling of RANK/RANKL was examined in both shGFP- and shAblim1-infected preosteoclasts. Knockdown of ABLIM1 led to the inhibition of the RANKL-mediated activation of PI3K and Akt (Fig. 4B, left panel). We then examined whether an association with Rac1, an osteoclast migration-related protein, is required for the downstream signaling of PI3K/Akt during RANKL-induced osteoclast differentiation. The induction of Rac1 expression was downregulated in the shAblim1-infected osteoclast compared with that in the control cells (Fig. 4B, right panel). These results suggest that the ABLIM1 is associated with the regulation of preosteoclast motility via the PI3K/Akt/Rac1 pathway during RANKL-mediated osteoclastogenesis.
Fig. 4.
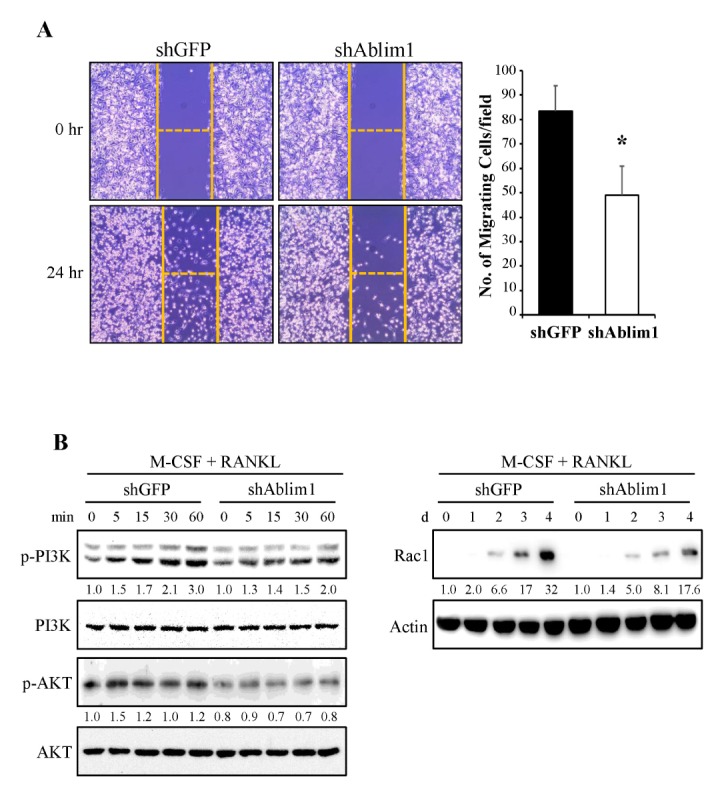
Effects of ABLIM1-specific knockdown on RANKL-mediated osteoclast mobility. (A) Cell migration of shGFP- or shAblim1-infected osteoclasts was measured as described in the Materials and Methods section. Data are expressed as the representative mean ± SD of the number of migrating cells from at least three independent experiments. *P < 0.05 versus control (shGFP). (B) To detect PI3K and AKT activation, shGFP- or shAblim1-infected BMMs cultured with M-CSF (30 ng/ml) overnight were stimulated by RANKL (100 ng/ml). To analyze Rac1 expression, BMMs infected with shGFP-or shAblim1 were cultured under treatment of M-CSF (30 ng/ml) and RANKL (100 ng/ml) for 4 d. Cell lysate (30 μg) was subjected to SDS-PAGE and analyzed by immunoblotting with their respective antibodies. The fold change was normalized to non-phosphorylated proteins (PI3K or AKT) or actin, respectively. Data are representative of three independent experiments.
DISCUSSION
The LIM domain has been identified in proteins from a wide variety of eukaryotic organisms. In the case of the human genome, 135 LIM-encoding sequences are located within 58 genes (18). The LIM domain is abundant in protein-interaction domains, such as the Src-homology-2 (SH2) and SH3 domains (19). Therefore, LIM domain-containing proteins participate in many cellular events, including cytoskeletal organization and focal adhesion to regulate cell growth, motility, and division (12, 20). It has been identified that several LIM proteins, including members of the zyxin, four-and-a-half LIM (FHL), and cysteine-rich protein (CRP) families, tend to shuttle between the nucleus and cytoplasm, which suggests that they influence gene expression related to cell development and fate at the transcriptional level (20–22). Nonetheless, most LIM proteins, including ABLIM proteins, are cytoskeleton-associated proteins that can directly or indirectly interact with the actin cytoskeleton (13). Previously, Feng et al. reported that a LIM-domain containing protein of the Ajuba/Zyxin family, LIMD1, affected RANKL-mediated osteoclast development. LIMD1 was upregulated during osteoclast differentiation and could bind to Traf6, leading to the activation of AP-1 (23).
In the present study, we investigated ABLIM1 as a positive regulator on RANKL-mediated osteoclastogenesis. ABLIM1 was markedly induced by RANKL treatment (Fig. 1). Osteoclast differentiation and bone resorptive activity, as well as the expression of osteoclast differentiation marker genes, were suppressed after the downregulation of ABLIM1 using shAblim1 (Figs. 2 and 3A). Moreover, the activation of JNK and the expression of c-Fos and NFATc1, critical transcriptional factors for osteoclastogenesis, were also decreased in the shAblim1-infected osteoclasts (Fig. 3B and Supplmentary Fig. 2).
UNC-115, the Caenorhabditis elegans homolog of ABLIM1, has been identified as a mediator of axon guidance from the results of a genetic experiment in which the mutant exhibited neuronal development defects leading to guidance errors (24). In addition, UNC-115 acts as a downstream cytoskeleton effector of the Rac Rho-family GTPases signaling during axon guidance (24). Experiments using mice lacking the retina-specific variant of ABLIM suggested the essential role of ABLIM-M in the regulation of actin dynamics during axon guidance (17). In the present study, we examined whether shAblim1-infected osteoclasts form a F-actin ring, which is an important construct for bone resorbing activity. Although some osteoclasts formed a F-actin ring, knockdown of ABLIM1 significantly reduced the number of mature osteoclasts having an F-actin ring (Fig. 2D). Consistent with its regulatory role in actin dynamics, this result might be caused by the inhibition of the actin-binding capacity of ABLIM1.
The Rho subfamily of small GTPases including Rho, Rac, and Cdc42 regulate cellular differentiation, and the organization of actin and adhesion structures in various cell types, leading to their migration (25–27). Among the Rac subfamily of small GTPases, including Rac1, Rac2, and Rac3, the Rac1 isoform is essential for regulating osteoclast differentiation and function through the modulation of lamellipodia formation and cell spreading (9, 10, 28). Recently, a more definite regulatory pathway for osteoclast migration in specialized stages during osteoclastogenesis was reported. The migration of osteoclast precursors depends on PI3K/PKCα-PKCδ signaling without RhoA and Rac1 activation, whereas preosteoclast and mature osteoclast migration depend on signaling of the PI3K/PKCα-PKCδ/RhoA-Rac1 axis (8). To assess the role of ABLIM1 on osteoclast migration, we measured cell motility using a wound healing assay and investigated molecular intracellular signaling in shAblim1- or control-infected osteoclasts. Preosteoclasts infected with Ablim1-specific shRNAs exhibited a markedly reduced number of migrating cells compared with the control cells (Fig. 4A). In addition, the activation of PI3K/Akt and Rac1 expression was dramatically decreased by ABLIM1 knockdown (Fig. 4B).
The results of the present study indicated that ABLIM1 directly regulates RANKL mediated osteoclastogenesis through the modulation of preosteoclast motility via the PI3K/Akt/Rac1 signal axis. Although further extensive in vitro and in vivo studies are necessary to identify the precise role of ABLIM1 in bone metabolism and its clinical application, ABLIM1 might be a candidate therapeutic target for bone diseases accompanied by bone destruction, such as osteoporosis, rheumatoid arthritis, and periodontitis.
MATERIALS AND METHODS
Experimental animals and reagents
C57BL/6J mice were purchased from Orient Bio Inc. (Seongnam, Korea) and all mouse protocols were approved by the Animal Care and Use Committee of Wonkwang University (WKU17-14). All cell culture media and supplements, including fetal bovine serum (FBS), were purchased from Hyclone (Rockford, IL, USA). Fast Red Violet LB Salt and Naphthol As-MX phosphate for TRAP staining were purchased from Sigma (St. Louis, MO, USA). TRIzol for RNA isolation was obtained from Invitrogen (Carlsbad, CA, USA). Recombinant human M-CSF and soluble recombinant mouse RANKL were donated by Dr. T Kim (KIOM, Daejeon, Korea). Antibodies against p-ERK/ERK, p-JNK/JNK, p-p38/p38, p-IκBα/IκBα, p-PI3K/PI3K, and p-Akt/Akt were purchased from Cell signaling Technology (Danvers, MA, USA). Antibodies recognizing NFATc1, c-Fos, Rac1, and actin were purchased from Santa Cruz Biotechnology (Dallas, Texas, USA).
Osteoclast culture, retroviral infection, F-actin staining, and resorption assay
Bone marrow cells isolated from femurs and tibiae of 6 to 8-week-old C57BL/6N mice were cultured in minimal essential medium alpha (MEM-α) containing 10% FBS and 1% antibiotics (penicillin/streptomycin) supplemented with 30 ng/ml M-CSF for 3 d. Adherent BMMs were used as osteoclast precursor cells. To produce multinucleated osteoclasts, BMMs were cultured with RANKL (100 ng/ml) and M-CSF (30 ng/ml) for 4 d, with replacement of fresh medium at 3 d. For the knockdown experiment, retroviral vector containing Ablim1-specific shRNAs (shAblim1) and green fluorescent protein (gfp)-specific shRNAs (shGFP) as a negative control, were constructed and infected as presented in the Supplementary Information section. Retroviral-infected BMMs were incubated with RANKL (100 ng/ml) and M-CSF (30 ng/ml) for 4 d to generate osteoclasts. Cells were then fixed with 10% formalin and stained with rhodamine phalloidin (Invitrogen) in order to label the F-actin ring. TRAP solution assay and TRAP staining were then performed. TRAP-positive cells with three or more nuclei were counted as mature osteoclasts. Total TRAP activity, as assessed by the TRAP solution assay, was measured as the absorbance at 405 nm after substrate (p-nitrophenyl phosphate) treatment. To measure the bone resorption activity, BMMs were cultured on a bone resorption assay plate 24 (COSMO BIO, Tokyo, Japan) in the presence of RANKL (100 ng/ml) and M-CSF (30 ng/ml) for 5 d. The cells were then cleared using bleach, and the resorption pit area was analyzed using Image J (NIH, version 1.62).
Reverse transcription polymerase chain reaction (RT-PCR) and quantitative real-time PCR
Total RNA (1 μg), prepared from BMMs using the TRIzol reagent, was reverse transcribed to cDNA with random primers using the SuperScript II RT-PCR System (Invitrogen) according to the manufacturer’s instructions, followed by PCR for Ablim1. Real-time PCR for osteoclast marker genes was performed using the VeriQuest SYBR Green qPCR Master Mix (Thermo Scientific, Waltham, MA, USA) and the StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) as previously described (29, 30). The relative expression levels were normalized to the level of Gapdh mRNA. The primer sequences used in this study are listed in Table S1.
Western blot analysis
BMMs infected with viral vectors were cultured with M-CSF (30 ng/ml) and RANKL (100 ng/ml) for the indicated times. In some experiments, BMMs cultured with M-CSF (30 ng/ml) for 12 h were induced by RANKL (100 ng/ml) treatment. The cells were lysed, and proteins were extracted using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific), supplemented with protease inhibitor cocktail (Roche, Mannheim, Germany) and phosphatase inhibitor (Thermo Scientific). Extracted proteins (30–50 μg) were subjected to SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes (Hybond-P, GE-Healthcare Life Science, Pittsburgh, PA, USA) as previously described (29, 30). After the application of primary and secondary antibodies, signals were visualized using the Enhanced Chemiluminescence detection system (Thermo Scientific) according to the manufacturer’s instructions. The immunoreactive bands were then quantified using ImageJ 1.62.
Cell motility assay
The cell motility assay was performed as described previously (31), with some modifications. Briefly, retroviral-infected BMMs (70 μl) were seeded at a density of 5 × 105 cells/ml onto a Culture-Insert 2 Well in a 35-mm μ-Dish (IBIDI GmbH, Planegg, Germany) and cultured with M-CSF (30 ng/ml) and RANKL (50 ng/ml) overnight. The next day, culture inserts were removed to make ‘wounds’ and the cells were fed with fresh medium containing M-CSF and RANKL for 24 h. Wound healing was monitored and images were captured at the indicated time points under a Leica DMIL microscope system (Leica, Wetzlar, Germany).
Statistical analysis
Data are presented as the mean ± SD from at least three independent experiments for each condition. Unless stated otherwise, the statistical analysis was performed using Student’s t-test to analyze the differences between the groups. A P-value < 0.05 was considered statistically significant.
Supplementary Information
ACKNOWLEDGEMENTS
This work was supported by the National Research Foundation of Korea (NRF) grants which were funded by the Korean goverment (NRF-2011-0030130, NRF-2016R1D1A1B03932 658 to S.H.L.), and partly by NIH grants (AR067726, AR0 69546 to Y.C.).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Walsh MC, Kim N, Kadono Y, et al. Osteoimmunology: interplay between the immune system and bone metabolism. Annu Rev Immunol. 2006;24:33–63. doi: 10.1146/annurev.immunol.24.021605.090646. [DOI] [PubMed] [Google Scholar]
- 2.Takayanagi H. The role of NFAT in osteoclast formation. Ann N Y Acad Sci. 2007;1116:227–237. doi: 10.1196/annals.1402.071. [DOI] [PubMed] [Google Scholar]
- 3.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7:292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 4.Kim K, Kim JH, Lee J, et al. Nuclear factor of activated T cells c1 induces osteoclast-associated receptor gene expression during tumor necrosis factor-related activation-induced cytokine-mediated osteoclastogenesis. J Biol Chem. 2005;280:35209–35216. doi: 10.1074/jbc.M505815200. [DOI] [PubMed] [Google Scholar]
- 5.Baroukh B, Cherruau M, Dobigny C, Guez D, Saffar JL. Osteoclasts differentiate from resident precursors in an in vivo model of synchronized resorption: a temporal and spatial study in rats. Bone. 2000;27:627–634. doi: 10.1016/S8756-3282(00)00372-0. [DOI] [PubMed] [Google Scholar]
- 6.Roodman GD. Advances in bone biology: the osteoclast. Endocr Rev. 1996;17:308–332. doi: 10.1210/edrv-17-4-308. [DOI] [PubMed] [Google Scholar]
- 7.Boudot C, Saidak Z, Boulanouar AK, et al. Implication of the calcium sensing receptor and the Phosphoinositide 3-kinase/Akt pathway in the extracellular calcium-mediated migration of RAW 264.7 osteoclast precursor cells. Bone. 2010;46:1416–1423. doi: 10.1016/j.bone.2010.01.383. [DOI] [PubMed] [Google Scholar]
- 8.Kim JM, Kim MY, Lee K, Jeong D. Distinctive and selective route of PI3K/PKCalpha-PKCdelta/RhoA-Rac1 signaling in osteoclastic cell migration. Mol Cell Endocrinol. 2016;437:261–267. doi: 10.1016/j.mce.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 9.Lee NK, Choi HK, Kim DK, Lee SY. Rac1 GTPase regulates osteoclast differentiation through TRANCE-induced NF-kappa B activation. Mol Cell Biochem. 2006;281:55–61. doi: 10.1007/s11010-006-0333-y. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, Lebowitz D, Sun C, Thang H, Grynpas MD, Glogauer M. Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J Bone Miner Res. 2008;23:260–270. doi: 10.1359/jbmr.071013. [DOI] [PubMed] [Google Scholar]
- 11.Schmeichel KL, Beckerle MC. The LIM domain is a modular protein-binding interface. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 12.Bach I. The LIM domain: regulation by association. Mech Dev. 2000;91:5–17. doi: 10.1016/S0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 13.Koch BJ, Ryan JF, Baxevanis AD. The diversification of the LIM superclass at the base of the metazoa increased subcellular complexity and promoted multicellular specialization. PLoS One. 2012;7:e33261. doi: 10.1371/journal.pone.0033261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roof DJ, Hayes A, Adamian M, Chishti AH, Li T. Molecular characterization of abLIM, a novel actin-binding and double zinc finger protein. J Cell Biol. 1997;138:575–588. doi: 10.1083/jcb.138.3.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrientos T, Frank D, Kuwahara K, et al. Two novel members of the ABLIM protein family, ABLIM-2 and -3, associate with STARS and directly bind F-actin. J Biol Chem. 2007;282:8393–8403. doi: 10.1074/jbc.M607549200. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda M, Yamashita JK, Tsukita S, Furuse M. abLIM3 is a novel component of adherens junctions with actin-binding activity. Eur J Cell Biol. 2010;89:807–816. doi: 10.1016/j.ejcb.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Lu C, Huang X, Ma HF, et al. Normal retinal development and retinofugal projections in mice lacking the retina-specific variant of actin-binding LIM domain protein. Neuroscience. 2003;120:121–131. doi: 10.1016/S0306-4522(03)00263-X. [DOI] [PubMed] [Google Scholar]
- 18.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 19.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, Zhao Y. The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein-protein interaction. Biol Cell. 2007;99:489–502. doi: 10.1042/BC20060126. [DOI] [PubMed] [Google Scholar]
- 21.Muller JM, Metzger E, Greschik H, et al. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 2002;21:736–748. doi: 10.1093/emboj/21.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang DF, Belaguli NS, Iyer D, et al. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–118. doi: 10.1016/S1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Zhao H, Luderer HF, et al. The LIM protein, Limd1, regulates AP-1 activation through an interaction with Traf6 to influence osteoclast development. J Biol Chem. 2007;282:39–48. doi: 10.1074/jbc.M607399200. [DOI] [PubMed] [Google Scholar]
- 24.Struckhoff EC, Lundquist EA. The actin-binding protein UNC-115 is an effector of Rac signaling during axon pathfinding in C. elegans. Development. 2003;130:693–704. doi: 10.1242/dev.00300. [DOI] [PubMed] [Google Scholar]
- 25.Huang Q, Shen HM, Ong CN. Emodin inhibits tumor cell migration through suppression of the phosphatidylinositol 3-kinase-Cdc42/Rac1 pathway. Cell Mol Life Sci. 2005;62:1167–1175. doi: 10.1007/s00018-005-5050-2. [DOI] [PubMed] [Google Scholar]
- 26.Zhang LJ, Tao BB, Wang MJ, Jin HM, Zhu YC. PI3K p110alpha isoform-dependent Rho GTPase Rac1 activation mediates H2S-promoted endothelial cell migration via actin cytoskeleton reorganization. PLoS One. 2012;7:e44590. doi: 10.1371/journal.pone.0044590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itzstein C, Coxon FP, Rogers MJ. The regulation of osteoclast function and bone resorption by small GTPases. Small GTPases. 2011;2:117–130. doi: 10.4161/sgtp.2.3.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda A, Hikita A, Wakeyama H, et al. Regulation of osteoclast apoptosis and motility by small GTPase binding protein Rac1. J Bone Miner Res. 2005;20:2245–2253. doi: 10.1359/JBMR.050816. [DOI] [PubMed] [Google Scholar]
- 29.Oh SJ, Gu DR, Jin SH, Park KH, Lee SH. Cytosolic malate dehydrogenase regulates RANKL-mediated osteoclastogenesis via AMPK/c-Fos/NFATc1 signaling. Biochem Biophys Res Commun. 2016;475:125–132. doi: 10.1016/j.bbrc.2016.05.055. [DOI] [PubMed] [Google Scholar]
- 30.Kim I, Kim JH, Kim K, Seong S, Kim N. Tusc2/Fus1 regulates osteoclast differentiation through NF-κB and NFATc1. BMB Rep. 2017;50:454–459. doi: 10.5483/BMBRep.2017.50.9.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ford CE, Jary E, Ma SS, Nixdorf S, Heinzelmann-Schwarz VA, Ward RL. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS One. 2013;8:e54362. doi: 10.1371/journal.pone.0054362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


