Abstract
HnRNPK is a multifunctional protein that participates in chromatin remodeling, transcription, RNA splicing, mRNA stability and translation. Here, we uncovered the function of hnRNPK in regulating the proliferation and differentiation of myoblasts. hnRNPK was mutated in the C2C12 myoblast cell line using the CRISPR/Cas9 system. A decreased proliferation rate was observed in hnRNPK-mutated cells, suggesting an impaired proliferation phenotype. Furthermore, increased G2/M phase, decreased S phase and increased sub-G1 phase cells were detected in the hnRNPK-mutated cell lines. The expression analysis of key cell cycle regulators indicated mRNA of Cyclin A2 was significantly increased in the mutant myoblasts compared to the control cells, while Cyclin B1, Cdc25b and Cdc25c were decreased sharply. In addition to the myoblast proliferation defect, the mutant cells exhibited defect in myotube formation. The myotube formation marker, myosin heavy chain (MHC), was decreased sharply in hnRNPK-mutated cells compared to control myoblasts during differentiation. The deficiency in hnRNPK also resulted in the repression of Myog expression, a key myogenic regulator during differentiation. Together, our data demonstrate that hnRNPK is required for myoblast proliferation and differentiation and may be an essential regulator of myoblast function.
Keywords: C2C12 myoblasts, CRISPR/Cas9, Differentiation, hnRNPK, Proliferation
INTRODUCTION
HnRNPK (Heterogeneous nuclear ribonucleoprotein k) is one of the most extensively studied hnRNP family members. It has been detected in the nucleus, cytoplasm, mitochondria and plasma membrane (1). It contains three K homology (KH) domains, one N-terminal bipartite nuclear localization signal (NLS), one K protein interactive region (KI) domain, and one nuclear-cytoplasmic shuttling domain (KNS) (1, 2). As an RNA recognition motif, the KH domain is responsible for DNA/RNA binding and participates in transcription, RNA splicing and mRNA translation regulation (3). The KI domain, which is evolutionarily conserved between Xenopus laevis and mammals, allows K proteins to gain new functions, such as protein-protein interactions (4). In addition, hnRNPK is subject to several post-translational modifications, such as phosphorylation, methylation, sumoylation, and acetylation, which can regulate its interactions with different molecules and influence its functions (2, 5). Therefore, hnRNPK has several integrated functions in many biological process including transcription, mRNA translation, RNA splicing, DNA repair, and translation (5, 6). HnRNPK plays key roles in hematopoiesis (7), the nervous system (8), organogenesis (9) and carcinogenesis (2, 6). Nevertheless, how hnRNPK works in skeletal myogenesis is still poorly understood.
Our previous proteomic research on porcine skeletal muscle development has shown that hnRNPK was exclusively and highly expressed in the 65 dpc porcine longissimus dorsi muscle, and was downregulated sharply from the 65 dpc to the 120 day (10). The proteomic analysis of early bovine myogenesis and differentiating human myoblasts also indicated that the expression of hnRNPK decreased significantly during myogenesis (11, 12). Additionally, in Xenopus laevis, the in situ hybridization analysis showed that hnRNPK was especially abundant in aligned nuclei of somitic muscle (13). These findings imply that hnRNPK may potentially play a role in myogenesis or skeletal muscle development, but how and why it has this effect remains unknown. To reveal possible roles for hnRNPK, we specifically targeted hnRNPK exon 12 by CRISPR/Cas9 mutagenesis in C2C12 cells. The proliferation and differentiation potential of the resulting clones was evaluated. We found that hnRNPK-mutated myoblasts exhibited a decreased proliferation rate, G2/M phase arrest, and sub-G1 phase accumulation. In addition to the myoblast proliferation defect, the decreased expression of Myog and MHC and abolished myotube formation were detected in the differentiating hnRNPK mutated cells. Our results suggest that hnRNPK may be an essential regulator for myoblast proliferation and differentiation.
RESULTS
HnRNPK is downregulated during myogenesis
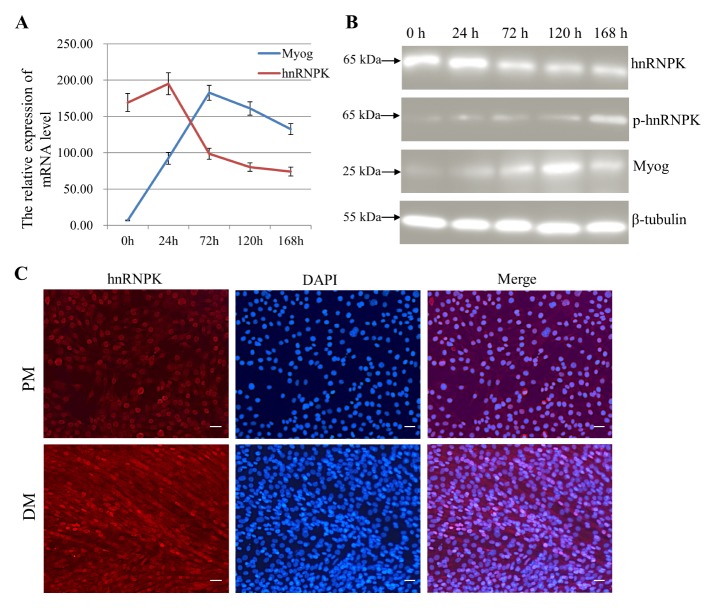
To investigate the expression profile of hnRNPK during myogenesis, we examined the expression pattern of hnRNPK during the C2C12 cell differentiation process. As shown by qRT-PCR and western blot, both the mRNA and protein level of hnRNPK had a relatively high expression, and reached a maximum at 24 h, then were sharply decreased from 24 h to 168 h (Fig. 1A, 1B). However, unlike the total hnRNPK protein level, phospho-hnRNPK (ser284) was upregulated after C2C12 cells were transferred into differentiation medium. Furthermore, immunofluorescence (IF) analysis showed that in proliferating C2C12 myoblasts, hnRNPK was predominantly distributed in the nucleus, while in differentiating C2C12 myoblasts, hnRNPK exhibited a homogeneous cytoplasmic and nuclear localization (Fig. 1C), suggesting a translocation of nuclear hnRNPK protein to the cytoplasm during C2C12 myoblast differentiation. These results demonstrate that the total hnRNPK protein and phospho- hnRNPK showed differential expression patterns during C2C12 myogenesis, indicating that this protein could play a role in both proliferation and differentiation of C2C12 myoblasts.
Fig. 1.
hnRNPK is upregulated during C2C12 myogenesis. (A) qRT-PCR analysis was performed on hnRNPK and Myog from C2C12 cells at different times post differentiation. Gapdh was used as an internal control. (B) Western blot analysis of hnRNPK was performed in C2C12 cells grown in growth medium, as well as after 24, 72, 120 and 168 h in DM. (C) hnRNPK localisation changes in C2C12 myoblasts during differentiation. Cells were proliferating (PM) or differentiating for 72 h (DM). Cellular hnRNPK (red) was detected by indirect IF. Cells were DAPI-stained to reveal DNA (blue) prior to confocal fluorescence microscopy. Scale bar = 100 μm.
Generation of hnRNPK-mutated myoblast cell line using CRISPR/Cas9
From a list of several candidates, we chose two target sequences (spanning bases 8646–8665 and 8754–8773 respectively) of the mouse hnRNPK genomic DNA (NC_000079.6). These nucleotides were positioned immediately 5′ to the trinucleotide PAM sequence “GGG” and “AGG”, respectively (Supplementary Fig. 1). Once identified, two oligo pairs containing the guide sequence were separately cloned into pCAG-T7-Cas9 plasmid.
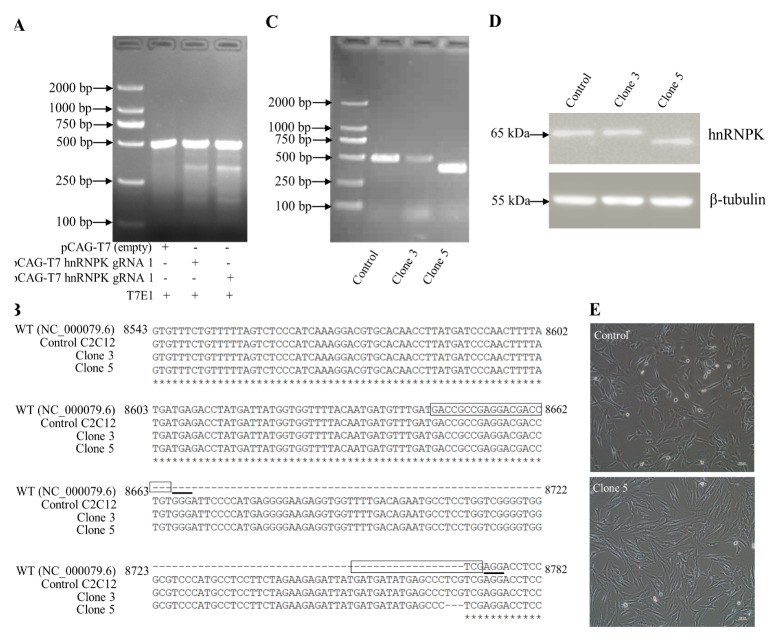
To test the effectiveness of the hnRNPK gRNA at triggering Cas9-mediated gene editing at the target site, C2C12 cells were transiently transfected with one of the pCAG-T7- Cas9-gRNA plasmids, resulting in the generation of a mixed population of edited and non-edited cells. The results showed that in genomic DNA extracts from hnRNPK gRNA-transfected cells, the T7E1 enzyme cleaved exon 12 of hnRNPK at a position targeted by the hnRNPK gRNA, resulting in the generation of a lower molecular weight DNA fragment of ~300 bp or ~200 bp (Fig. 2A).
Fig. 2.
Generation and validation of hnRNPK-mutated cell lines. (A) Mismatch-specific endonuclease assay. Genomic PCR products spanning exon 12 of hnRNPK were amplified from the C2C12 cells transfected with pCAG-T7-Cas9-hnRNPK guide. (B) After clone isolation, gPCR amplicons encompassing the modified locus of genomic DNA were run on a 1.5% agarose gel. (C) Cell lysates of control and clonal hnRNPK mutated isolates, probed with specific hnRNPK antibodies. (D) Sequence alignment of bases 8543–8782 of exon 12 of the mouse hnRNPK sequence (NC_000079.6), a WT allele from control C2C12 cells, and two mutant alleles identified from each of the two hnRNPK-mutated clones. The complementary plus-strand sequences corresponding to the 20-nt target and 3-nt PAM are highlighted in a box and underlined, respectively. (E) The morphological changes of WT control and clone 5 cells. Scale bar = 100 μm.
A new batch of C2C12 cells was cotransfected with the two pCAG-T7-Cas9-gRNA plasmids, and transfected cells were isolated by selectable and reporter markers (puromycin and GFP). Following transfection of C2C12 cells, a number of puromycin resistant colonies were selected. The sorted cells were then plated as individual clones in 96-well plates and expanded. A total of 20 clones were identified by PCR and DNA sequencing. From this screen, clones 3 and 5 were selected for further analysis. Clone 3 showed a 3 bp deletion (g.8768–8770) in the second target sequence, which caused a one-amino acid (AA) deficiency (p.Arg286) in hnRNPK protein (Fig. 2B). Clone 5 showed a 108 bp deletion (g.8663–8770) between the two targets compared with the control (Fig. 2B, 2C and Supplementary Fig. 2). However, the mutation didn’t cause frame shifts within hnRNPK protein, and a 36 AA deficiency was created (p.Val251-Arg286) within the KI domain. Compared with the control, a lower molecular-weight of the protein was directly detected in clone 5 (Fig. 2D). Despite the 36 AA deficiency of hnRNPK protein, microscopic observation indicated that the cellular morphology of the clone 5 was grossly indistinguishable from controls (Fig. 2E).
Myoblast proliferation is impeded by the 36 AA deficiency mutation of hnRNPK
During the routine culture of clone 5 cells, we observed a much slower growth rate of these cells than the control cells in the growth medium. To quantify this growth retardation, equal amounts of clone 5 and control cells were cultured in the growth medium to assess the cell growth rate difference by MTT colorimetric assay. This difference in cell vitality was augmented as cells started to grow exponentially 12 h after synchronization. Compared with the control, the obtained MTT assay data showed that clone 5 had a significant inhibitory effect on the growth of C2C12 cells (P < 0.05) (Supplementary Fig. 3A). Ki67 pulse-labeling at the 24 h time point also confirmed impeded cell proliferation in clone 5 in the form of fewer Ki67 positive clone 5 cells (20.5 ± 3.2%) compared to control cells (47.2 ± 5.1%) (Supplementary Fig. 3B, 3C).
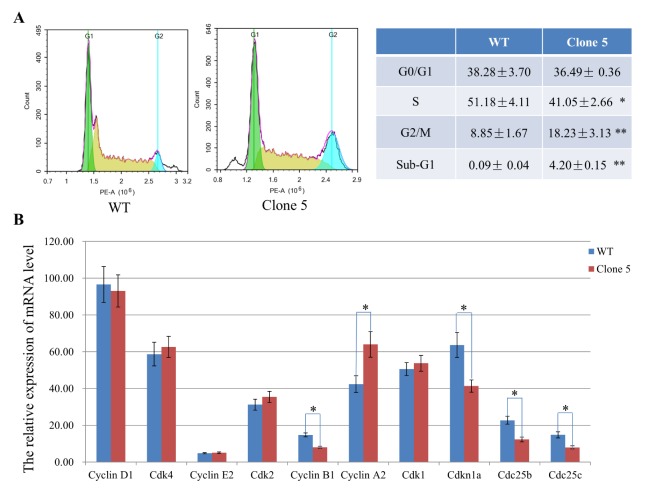
To examine the effects of hnRNPK 36 AA deficiency on cell cycle profile and aneuploidy, we further performed PI staining and flow cytometry (Fig. 3A). Compared with the control, the populations of clone 5 displayed a significant increase in G2/M phase (18.23 ± 3.13% vs 8.85 ± 1.67%), an obvious decrease in S phase (41.05 ± 2.66% vs 51.18 ± 4.11%), and the emergence of the sub-G1 phase. However, the ratio of G0/G1 phase had no significant difference between the two groups (P > 0.05). These data indicate that the 36 AA deficiency of hnRNPK blocked or delayed cell cycle progression from G2/M to G1 phases and induced cells toward apoptosis.
Fig. 3.
The 36 AA deficiency of hnRNPK protein results in G2/M phase arrest and accumulation sub-G1 phase myoblasts. (A) Flow cytometry analysis showed more cells in G2/M phase, fewer in S phase, and more in sub-G1 phase in the clone 5 group compared to the WT control. The percentage of clone 5 and WT myoblasts in each cell cycle phases is shown in the right table. (B) The mRNA expression of key cell cycle regulators in clone 5 and WT control cells were detected by qRT-PCR. Data are presented as the mean ± s.d., n = 3. *P < 0.05, **P < 0.01 compared to WT control.
To evaluate the mechanism by which the hnRNPK 36 AA deficiency enhanced G2/M and sub-G1 phase arrest, we examined the mRNA expression of key cell cycle regulators. As shown in Fig. 3B, the expression level of Cyclin A2 was significantly increased in the mutant myoblasts, while Cyclin B1, Cdc25b, Cdc25c and Cdkn1a were decreased sharply (P < 0.05). No statistically significant difference in Cyclin D1, Cyclin E2, Cdk1, Cdk2, or Cdk4 was observed between the clone 5 and WT control groups (P > 0.05). These results indicated that the hnRNPK 36 AA deficiency-induced G2/M phase cell cycle arrest in C2C12 cells was associated with a marked decline in the mRNA levels of cyclin B1, Cdc25b, and Cdc25c, but not Cdk1 and Cdkn1a.
Myoblast differentiation is impaired in clone 5
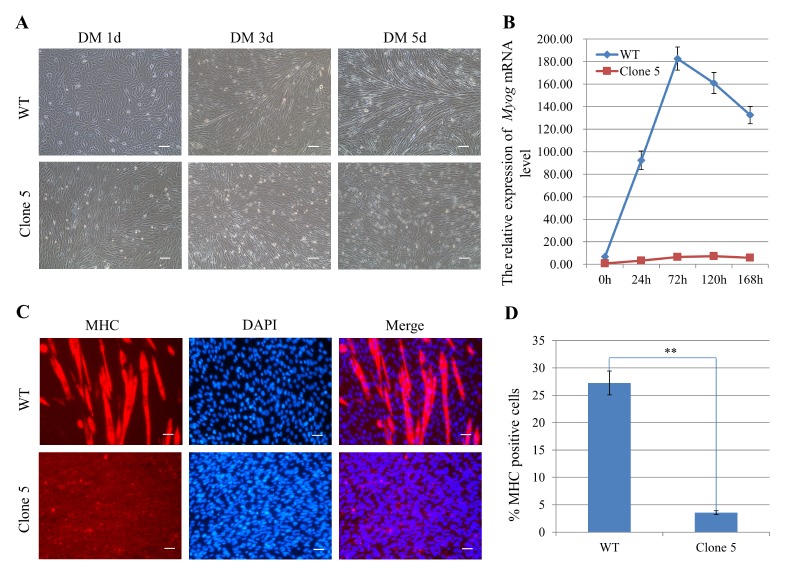
To further examine the phenotype of clone 5, we induced myotube formation by culturing the cells in medium containing 2% horse serum. After 5 days in DM, the hnRNPK mutated cells exhibited a deficiency in myoblast differentiation and had nearly no elongated myofibers, in contrast to control (Fig. 4A). Immunofluorescence staining for MHC and statistical analysis of the differentiation index confirmed the impaired differentiation of hnRNPK-mutated cells (Fig. 4C, D). qRT-PCR detection of Myog also suggested that the differentiation efficiency was lower in clone 5 than in control (Fig. 4B). These data demonstrate that hnRNPK is required for the differentiation of C2C12 myoblasts in vitro.
Fig. 4.
Myogenesis is impaired in clone 5 myoblasts. (A) Myotube formation was disrupted in clone 5 cells. Myogenesis in WT control and clone 5 was induced by culturing in medium containing 2% horse serum. Scale bar = 100 μm. (B) qRT-PCR analysis for myoblast differentiation showed lower Myog in clone 5 compared to the WT control. (C) The clone 5 and WT control were induced to differentiate and immunostained for MHC (red) and DAPI (blue) at 5 days after differentiation. Scale bar =100 μm. (D) Graph illustrating the percentage of MHC-positive cells per field of vision in Fig. 4C as the differentiation index. Data are presented as the mean ± s.d., n = 3. **P < 0.01
DISCUSSION
Cell cycle distribution analysis revealed an increased accumulation of G2/M phase and sub-G1 phase cells in clone 5, indicating a retardation of cell-cycle progression. We observed a decreased proliferation rate with cell cycle arrest at the G2/M phase and cell accumulation in the sub-G1 phase, as well as decreased expression levels of the major cell cycle checkpoint regulators, cyclin B1, Cdc25b and Cdc25c. In cycling cells, Cyclin B1/Cdk1 kinase activity dominates G2/M phase transition and the G2/M phase arrest is induced via inhibitory phosphorylation of Cdk1 at Thr14 and Tyr15 (14). The Cdc25 family comprises dual-specificity phosphatases that promote entry into mitosis by removing the inhibitory phosphates on Cdk1 (15, 16). Therefore, the downregulation of Cyclin B1, Cdc25b and Cdc25c is critical for the G2/M phase arrest in clone 5 and was achieved by the 36 AA deficiency within the KI domain in. Through the KI domain, hnRNPK interacts with more than 100 known protein partners, such as RNA-binding proteins, translation factors, transcription factors, DNA-binding proteins, splicing factors, and protein kinase (17). In this way, hnRNPK protein is involved in various cellular events including DNA transcription, RNA splicing, RNA stability, and translation. The KI domain mediated gene expression regulation could have a key role in controlling the expression of these cell cycle regulators. The partial dysfunction of hnRNPK was most likely attributed to the loss of protein-protein interaction.
P21, also known as cyclin-dependent kinase inhibitor Cdkn1a, is an inhibitor of the cell cycle and can arrest the cell cycle progression at the G1/S and G2/M transitions by inhibiting cyclin-Cdk2, -Cdk1, and -Cdk4/6 complexes (18). Our results showed that p21 was downregulated in clone 5. HnRNPK plays key roles in coordinating transcription as a cofactor of p53 through protein-protein interactions when it is transiently recruited to the promoters of p53-responsive genes such as p21 (9). The dysfunction of the hnRNPK KI domain could have impaired the transcriptional induction of p21 in C2C12 myoblasts. However, the decreased expression of p21 conflicted with the cell cycle G2/M arrest phenotype. This could be because p21 does not play a direct role in inhibiting cyclin-Cdk1 complexes and has low affinity toward Cdk1 (16). These findings suggest that p21 might play a significant role through different mechanisms to regulate cell proliferation in C2C12 myoblast. Moreover, p21 has an important function in the expression of genes involved in DNA repair and protecting cells from apoptosis (18). The induced accumulations of the sub-G1 population (apoptotic cell) in clone 5 might be due to the downregulation of p21.
In addition to the myoblast proliferation defect in clone 5, impaired cell differentiation also resulted from hnRNPK dysfunction. HnRNPK undergoes multiple covalent modifications which regulated its activity and interactions with molecular partners (1). The ERK-regulated phosphorylation at Ser284 is important for the cytoplasmic accumulation of hnRNPK protein (19). Phosphorylation of hnRNPK at Ser284 is also detected in differentiated C2C12 cells, which may be central to regulating C2C12 cellular differentiation (20). Cytoplasmic hnRNPK is part of a signaling complex that may be involved in growth stimulated post-transcriptional regulation (21). Moreover, Ser284 was located within the 36 AA deficiency of hnRNPK protein created here. We also detected an upregulation of Ser284 phospho-hnRNPK and the cytoplasmic accumulation of hnRNPK protein in myogenesis. In the cytoplasm, hnRNPK functions as a translational regulator of specific mRNAs, such as c-myc, c-Src, Ucp2, renin, Tak1 mRNA (2, 7, 22). Therefore, it was likely that a connection existed between the clone 5 and the phenotype of impaired cell differentiation. However, besides the function of Ser-284 phosphorylation, the gene expression process regulated by hnRNPK’s interactions with proteins could also play an important role in myoblast differentiation. HnRNPK interacts with the bifurcated SET domain-containing histone methyltransferase Setdb1, and function in H3K9me3-mediated transcriptional silencing (23). Setdb1 has been associated with transcriptional silencing of euchromatic genes through chromatin modification, and it is required for myogenic differentiation by maintaining the level of MyoD (24, 25). Thus, the hnRNPK-Setdb1 interaction might give hnRNPK its function in the regulation of myoblast differentiation. If so, future investigation of the hnRNPK-Setdb1 interaction and the downstream targets of Setd1 during myogenesis may help identify these pathways.
In conclusion, we reported a simple approach using programmable CRISPR/Cas9 nucleases to generate mutated cell lines. The new hnRNPK-mutated model generated via this method highlights a role for hnRNPK in myoblast proliferation and differentiation in vitro, providing new insights into the complex networks involved in myogenesis. Future investigations of the changes in global gene expression regulation and hnRNPK protein-protein interactions in hnRNPK-mutated myoblasts may provide valuable information about its role in muscle differentiation. In vivo models also need to be developed to fully test the role of hnRNPK in muscle maintenance or development.
MATERIALS AND METHODS
hnRNPK single-guide RNA expression vector construction
For the main functional elements of the hnRNPK gene, a synthetic guide RNA (sgRNA) sequence was designed to disrupt a region within the KI domain of the mouse hnRNPK protein, located in exon 12 of the coding sequence. The design tool ChopChop (https://chopchop.rc.fas.harvard.edu/) was used to scan the input sequence. Then, two 20-bp guide sequences (5′-GACCGCCGAGGACGACCTGT-3′ and 5′-GAT GATATGAGCCCTCGTCG-3′) targeting the predicted high-specificity protospacer adjacent motif (PAM) were selected from a published database. Then two complementary primer pairs (sgRNA 1 forward primer: 5′-AAACACCGGACCGCCG AGGACGACCTGT-3′, sgRNA 1 reverse primer: 5′-CTCTAAAA CACAGGTCGTCCTCGGCGGTC-3′, sgRNA 2 forward primer: 5′-AAACACCGGATGATATGAGCCCTCGTCG-3′ and sgRNA 2 reverse: 5′-CTCTAAAACCGACGAGGGCTCATATCATC-3′) and sticky end adapters were synthesized by GenScript (Nanjing, China). 1 μl of each oligo (10 μM) was annealed using T4 polynucleotide kinase (New England Biolabs) in a Bio-Rad C100 PCR amplifier. The cycling conditions were 95°C for 3 min, followed by a ramp to 25°C at 5°C/min. The annealed oligo was ligated into the digested pCAG-T7-Cas9 vector (Viewsolid Biotech) using Quick-Ligase (New England Biolabs). The ligation mixture was transformed into competent E. coli cells. After plasmid DNA extraction, the sequence of the construct was verified by automated DNA sequence analysis performed at GenScript (Nanjing, China).
T7E1 endonuclease cleavage assay
C2C12 cells in six-well dishes were cultured as described previously to 40–50% confluence. Cells were transfected with 2 μg of hnRNPK sgRNA plasmid and 8 μl of FuGENE® HD/well. At 72 h posttransfection, the cells were harvested, and genomic DNA was extracted. A region of exon 12 of the hnRNPK gene was amplified with genomic DNA-specific primers (Hnrnpk-g, Supplementary table 1). The hybrid PCR product was incubated with T7E1 nuclease (New England Biolabs) for 30 min at 37°C. The result was checked on a 1.5% agarose gel.
Establishment of hnRNPK-mutated cell lines
C2C12 cells were co-transfected with the two hnRNPK exon 12 CRISPR/Cas9 targeting vectors. At 48 h posttransfection, puromycin was added into the growth medium to select colonies for 7 days. Clonal selection by serial dilutions into 96-well plates was undertaken to generate colonies from single cells. To test for mutations generated by NHEJ, exon 12 of hnRNPK was amplified using the hnRNPK-specific PCR primers described above. The PCR products were analyzed by DNA sequencing.
Cell proliferation assay
For the cell proliferation assay, cells were seeded in 96-well plates with 2000 cells per well and measured at 7 different growth times by MTT assay. Briefly, we added 20 μl MTT (4 mg/ml) to each well and incubated them at 37°C for 4 h. Then, we discarded the supernatants and dissolved the remains with 150 μl DMSO per well. The absorbance was measured at 490 nm using a SpectraMax M5 unit (Molecular Devices).
Flow cytometry analysis
At least 106 synchronized C2C12 cells grown in GM were prepared for the cell cycle analyses by FACS as described previously. Briefly, cells were fixed in 70% ice-cold ethanol for 30 min at 4°C and washed in PBS. The cells were subsequently treated with RNase A and stained with PI. The intensity was measured by FACS using the ACEA NovoCyteTM system and the data were collected and processed using NovoExpressTM software to determine cell populations in the G0/G1, S, and G2/M phases.
Immunofluorescence assay
For immunofluorescence analysis of C2C12 myoblasts or myotubes, cells were grown or differentiated to the required time points, and then briefly fixed with 4% PFA. The cells were then treated with a blocking solution before incubations with antibodies were performed. The following primary antibodies were anti- hnRNPK (sc-28380, Santa Cruz Biotechnology), anti-MHC (sc-376157, Santa Cruz Biotechnology) and anti-Ki67 (550609, BD Pharmingen). The secondary antibody used was goat anti-mouse IgG antibody coupled with Alexa Fluor 555 (A0460, Beyotime). Nuclei were visualized by DAPI, and images were acquired by fluorescence microscopy.
Statistical analysis
All statistical analysis was performed using SPSS 18.0. Data are expressed as the means ± standard errors of the means. The non-paired t test was used and a value of P < 0.05 was considered statistically significant.
Detailed methods of cell culture, C2C12 myoblast differentiation, RNA isolation, quantitative RT-PCR, and western blotting are provided in the supplementary methods.
Supplementary Information
ACKNOWLEDGEMENTS
This work was financially supported by the National Natural Sciences Foundation of China (U1204326 and 31601167), Department of Science and Technology in Henan Province (182102310211 and 182300410027), the Nanhu Scholars Program of XYNU, and the Program of Youth Learning Backbone Teacher in Henan province (2015GGJS-139).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 2.Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer Lett. 2014;352:152–159. doi: 10.1016/j.canlet.2014.06.019. [DOI] [PubMed] [Google Scholar]
- 3.Siomi H, Matunis MJ, Michael WM, et al. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostareck-Lederer A, Ostareck DH, Rucknagel KP, et al. Asymmetric arginine dimethylation of heterogeneous nuclear ribonucleoprotein K by protein-arginine methyltransferase 1 inhibits its interaction with c-Src. J Biol Chem. 2006;281:11115–11125. doi: 10.1074/jbc.M513053200. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Zhou X, Liu N, et al. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett. 2008;582:1761–1765. doi: 10.1016/j.febslet.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 6.Gallardo M, Hornbaker MJ, Zhang X, et al. Aberrant hnRNP K expression: All roads lead to cancer. Cell Cycle. 2016;15:1552–1557. doi: 10.1080/15384101.2016.1164372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ostareck-Lederer A, Ostareck DH. Precision mechanics with multifunctional tools: how hnRNP K and hnRNPs E1/E2 contribute to post-transcriptional control of gene expression in hematopoiesis. Curr Protein Pept Sci. 2012;13:391–400. doi: 10.2174/138920312801619484. [DOI] [PubMed] [Google Scholar]
- 8.Hutchins EJ, Szaro BG. c-Jun N-terminal kinase phosphorylation of heterogeneous nuclear ribonucleoprotein K regulates vertebrate axon outgrowth via a posttranscriptional mechanism. J Neurosci. 2013;33:14666–14680. doi: 10.1523/JNEUROSCI.4821-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallardo M, Lee HJ, Zhang X, et al. hnRNP K Is a Haploinsufficient Tumor Suppressor that Regulates Proliferation and Differentiation Programs in Hematologic Malignancies. Cancer Cell. 2015;28:486–499. doi: 10.1016/j.ccell.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y, Qian H, Feng X, et al. Differential proteome and transcriptome analysis of porcine skeletal muscle during development. J Proteomics. 2012;75:2093–2108. doi: 10.1016/j.jprot.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Chaze T, Meunier B, Chambon C, et al. In vivo proteome dynamics during early bovine myogenesis. Proteomics. 2008;8:4236–4248. doi: 10.1002/pmic.200701101. [DOI] [PubMed] [Google Scholar]
- 12.Gonnet F, Bouazza B, Millot GA, et al. Proteome analysis of differentiating human myoblasts by dialysis-assisted two-dimensional gel electrophoresis (DAGE) Proteomics. 2008;8:264–278. doi: 10.1002/pmic.200700261. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Gervasi C, Szaro BG. A crucial role for hnRNP K in axon development in Xenopus laevis. Development. 2008;135:3125–3135. doi: 10.1242/dev.022236. [DOI] [PubMed] [Google Scholar]
- 14.Porter LA, Donoghue DJ. Cyclin B1 and CDK1: nuclear localization and upstream regulators. Prog Cell Cycle Res. 2003;5:335–347. [PubMed] [Google Scholar]
- 15.Giono LE, Resnick-Silverman L, Carvajal LA, St Clair S, Manfredi JJ. Mdm2 promotes Cdc25C protein degradation and delays cell cycle progression through the G2/M phase. Oncogene. 2017;36:6762–6773. doi: 10.1038/onc.2017.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charrier-Savournin FB, Chateau MT, Gire V, et al. p21-Mediated nuclear retention of cyclin B1-Cdk1 in response to genotoxic stress. Mol Biol Cell. 2004;15:3965–3976. doi: 10.1091/mbc.e03-12-0871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikula M, Dzwonek A, Karczmarski J, et al. Landscape of the hnRNP K protein-protein interactome. Proteomics. 2006;6:2395–2406. doi: 10.1002/pmic.200500632. [DOI] [PubMed] [Google Scholar]
- 18.Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Habelhah H, Shah K, Huang L, et al. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3:325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 20.Puente LG, Voisin S, Lee RE, et al. Reconstructing the regulatory kinase pathways of myogenesis from phosphopeptide data. Mol Cell Proteomics. 2006;5:2244–2251. doi: 10.1074/mcp.M600134-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Laury-Kleintop LD, Tresini M, Hammond O. Compartmentalization of hnRNP-K during cell cycle progression and its interaction with calponin in the cytoplasm. J Cell Biochem. 2005;95:1042–1056. doi: 10.1002/jcb.20486. [DOI] [PubMed] [Google Scholar]
- 22.Xu H, Xu Y, Liang X, et al. The Biological Function of Heterogeneous Nuclear Ribonucleoprotein K (hnRNP K) and Its Roles in Spermatogenisis. Journal of Agricultural Biotechnology. 2015;23:661–670. [Google Scholar]
- 23.Bao X, Wu H, Zhu X, et al. The p53-induced lincRNA-p21 derails somatic cell reprogramming by sustaining H3K9me3 and CpG methylation at pluripotency gene promoters. Cell Res. 2015;25:80–92. doi: 10.1038/cr.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song YJ, Choi JH, Lee H. Setdb1 is required for myogenic differentiation of C2C12 myoblast cells via maintenance of MyoD expression. Mol Cells. 2015;38:362–372. doi: 10.14348/molcells.2015.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyer S, Pontis J, Schirwis E, et al. Canonical Wnt signalling regulates nuclear export of Setdb1 during skeletal muscle terminal differentiation. Cell Discov. 2016;2:16037. doi: 10.1038/celldisc.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.