Abstract
MicroRNAs (miRNAs) are dysregulated in many tumors; however, miRNA regulation in parathyroid tumors remains poorly understood. To identify differentially expressed miRNAs between sporadic and hereditary parathyroid tumors and to analyze their correlation with clinicopathological features, a microarray containing 887 miRNAs was performed; then, the differentially expressed miRNAs were validated by qRT-PCR using 25 sporadic and 12 hereditary parathyroid tumors and 24 normal parathyroid tissue samples. A receiver operating characteristic curve (ROC) analysis was applied to evaluate the utility of the miRNAs for distinguishing parathyroid tumor types. Compared to the miRNAs in the normal parathyroid tissues, 10 miRNAs were differentially expressed between the sporadic and hereditary parathyroid tumors. Seven of these miRNAs (let-7i, miR-365, miR-125a-3p, miR-125a-5p, miR-142-3p, miR-193b, and miR-199b-5p) were validated in the parathyroid tumor samples. Among these miRNAs, only miR-199b-5p was differentially expressed (P < 0.001); miR-199b-5p was significantly downregulated and negatively associated with PTH levels (γ = −0.579, P = 0.002) in the sporadic tumors but was upregulated in the hereditary tumors. This miRNA showed 67% sensitivity and 100% specificity for distinguishing sporadic and hereditary parathyroid tumors. These results reveal altered expression of a miRNA between sporadic and hereditary parathyroid tumors and the potential role of miR-199b-5p as a novel biomarker for distinguishing these two types of parathyroid tumors.
Introduction
Primary hyperparathyroidism (PHPT) is a relatively common endocrine disease with a prevalence of three per one thousand in the general population1. PHPT occurs sporadically in up to 90% of cases but may also be a major component of familial syndromes, such as multiple endocrine neoplasia type 1 (MEN1)2. MEN1 is an autosomal dominant inherited disease characterized by the occurrence of several endocrine tumors, particularly in the parathyroid gland, endocrine pancreas and pituitary gland3. PHPT shows the highest penetrant expression in this syndrome; PHPT occurs in almost 100% of MEN1 patients by the age of 50 yrs, while the MEN1 frequency in PHPT patients is estimated to be 1–18%4.
Clinical features, such as age of onset, sex ratio, severity of bone involvement and recurrence rates after parathyroidectomy, are different between sporadic and hereditary parathyroid tumors5,6. The discrimination of these tumor types is important because the treatment and disease courses are quite different7,8. However, most of the current studies have analyzed the PHPT clinical data without distinguishing the different etiologies6,9. Theoretically, PHPT in MEN1 patients is present with multinodular hyperplasia of the parathyroid glands; in contrast, parathyroid adenoma is present in sporadic PHPT. However, the histopathological discrimination between sporadic and hereditary parathyroid tumors is difficult due to a lack of specific abnormalities10.
miRNAs are small non-coding RNAs with roles in a wide range of cellular processes in tumorigenesis11,12. Currently, many miRNAs have been demonstrated to be diagnostic and prognostic biomarkers for multiple cancer types13–16. The potential uses of miRNAs for the effective diagnosis and optimal treatment of parathyroid tumors have also been investigated17–20. However, there is little data on parathyroid tumor miRNAs that are differentially expressed between the sporadic and hereditary forms. The purpose of the present study was to identify and analyze differentially expressed miRNAs and to determine their correlation with the clinicopathological features of sporadic and hereditary parathyroid tumors. We determined whether miRNA profiling could serve as a potential biomarker for distinguishing these tumor types.
Results
Differences in clinical manifestations between sporadic and hereditary parathyroid tumors
We compared the clinical and biochemical parameters of sporadic and MEN1 parathyroid tumors. The laboratory results showed higher PTH and calcium levels in patients with parathyroid tumors than in normal controls. Patients with sporadic parathyroid tumors had larger tumor sizes and higher levels of PTH and calcium than patients with hereditary parathyroid tumors (Table 1). Consistent with a previous report on the correlation between PTH and tumor volume21,22, PTH levels were correlated significantly with tumor size in patients with sporadic parathyroid tumors (γ = 0.592, P = 0.002) but not significantly in patients with hereditary parathyroid tumors.
Table 1.
Clinical and biochemical characteristics.
| Normal parathyroid tissues | Sporadic parathyroid tumors | Hereditary parathyroid tumors | |
|---|---|---|---|
| Patient No. | 24 | 25 | 12 |
| Age | 51.2 ± 10.1 | 54.8 ± 12.4 | 48.1 ± 12.1 |
| Sex (M:F) | 6:18 | 10:15 | 4:8 |
| PTH (pg/mL) | 31.5 ± 11.0 | 238.3 ± 178.9*† | 131.3 ± 73.1 |
| Ca (mg/dL) | 9.2 ± 0.5 | 11.6 ± 0.9*† | 10.7 ± 1.0* |
| P (mg/dL) | 3.7 ± 0.6 | 2.6 ± 0.3* | 2.7 ± 0.4* |
| Cr (mg/dL) | 0.7 ± 0.2 | 0.8 ± 0.2 | 0.8 ± 0.1 |
| Tumor size (cm) | — | 2.0 ± 1.0 | 1.7 ± 1.0 |
The data are presented as the means ± SD. One-way between-groups ANOVA with Tukey’s post hoc test. *P < 0.05 vs. normal; †P < 0.05 sporadic vs. hereditary parathyroid tumors.
Differential expression of miRNA between sporadic and MEN1 parathyroid tumors
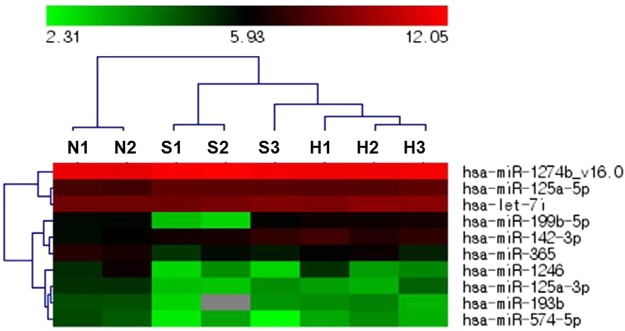
A microarray-based supervised cluster analysis for 10 differentially expressed miRNAs in sporadic and hereditary parathyroid tumors versus normal parathyroid tissues is shown (FDR < 0.05) (Fig. 1). Four miRNAs, including miR-365, miR-125a-3p, miR-574-5p, and miR-1246, were significantly downregulated in sporadic parathyroid tumors, whereas miR-142-3p, let-7i, miR-125a-5p, miR-199b-5p, and miR-1274b_v16.0 were significantly upregulated; miR-193b was downregulated in MEN1 parathyroid tumors.
Figure 1.
Supervised cluster analysis of miRNA levels in parathyroid tumors. N, normal parathyroid tissue; S, sporadic parathyroid tumor; H, hereditary parathyroid tumor. The data normalized to RNU6 were hierarchically clustered. Red indicates an increase relative to all data in this set, and green indicates a decrease relative to all data in this set.
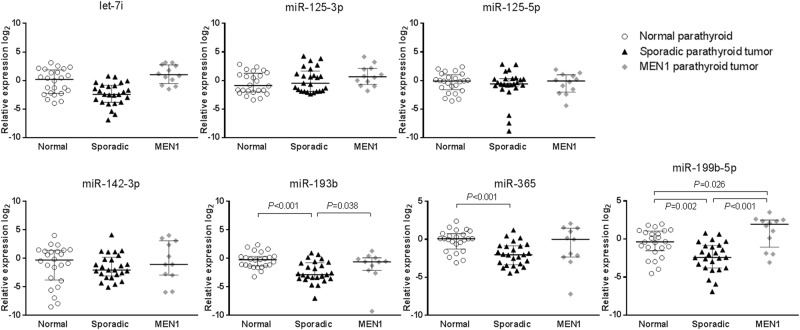
Next, seven commercially available miRNAs were used to validate the results in sporadic and MEN1 parathyroid tumors compared with normal parathyroid tissues using quantitative real-time PCR (qRT-PCR) (Fig. 2). The expression levels of miR-193b and miR-365 were lower in sporadic parathyroid tumors than in normal parathyroid tissues, and miR-193b expression was higher in MEN1 parathyroid tumors than in sporadic tumors. Interestingly, only miR-199b-5p had significantly different expression between the two parathyroid tumor types; compared with that in normal parathyroid tissue, miR-199b-5p was downregulated in the sporadic form (median fold change of 0.2) and upregulated in the hereditary form (median fold change of 3.9).
Figure 2.
Validation of most relevant miRNAs by qRT-PCR in parathyroid tumors. Scatterplots show relative expression levels of let-7i, miR-365, miR-125a-3p, miR-125a-5p, miR-142-3p, miR-193b, and miR-199b-5p in 24 normal parathyroid tissues, 25 sporadic, and 12 MEN1 parathyroid tumor samples. Horizontal bars represent the median and interquartile range. P values were calculated using the Mann-Whitney U-test.
Discriminating value of miR-199b-5p in parathyroid tumors
The diagnostic relevance of miR-199b-5p was analyzed using a ROC curve analysis (Fig. 3). miR-199b-5p had a large area under the concentration-time curve (AUC = 0.863, P < 0.001) with a sensitivity of 67% and a specificity of 100% for discriminating sporadic and hereditary parathyroid tumors.
Figure 3.
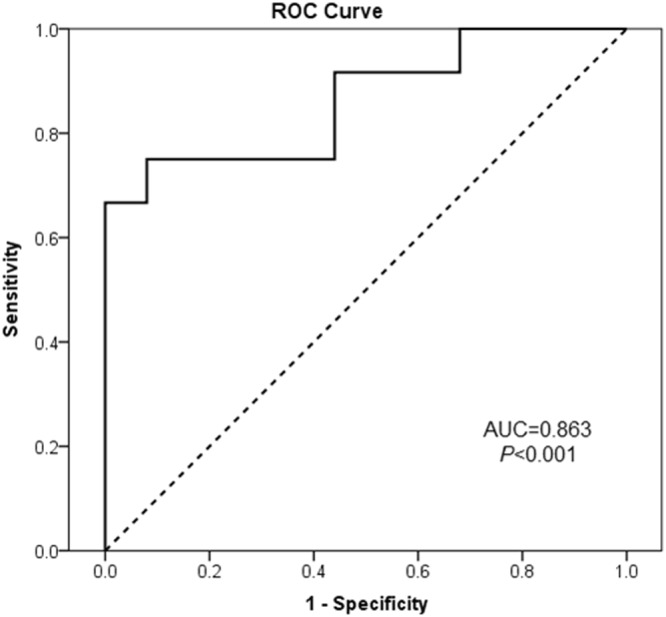
Receiver operator characteristic (ROC) curves of miR-199b-5p showing the discrimination between sporadic and hereditary parathyroid tumors.
Clinical implication of miR-199b-5p in parathyroid tumors
Given that miR-199b-5p was differentially expressed according to the parathyroid tumor type, we further analyzed the correlation between miR-199b-5p and PTH levels. Interestingly, different correlations between miR-199b-5p and PTH levels in sporadic and hereditary parathyroid tumors were identified: there was a negative association in the sporadic form (γ = −0.579, P = 0.002) and no significant correlation in the hereditary form (Fig. 4).
Figure 4.
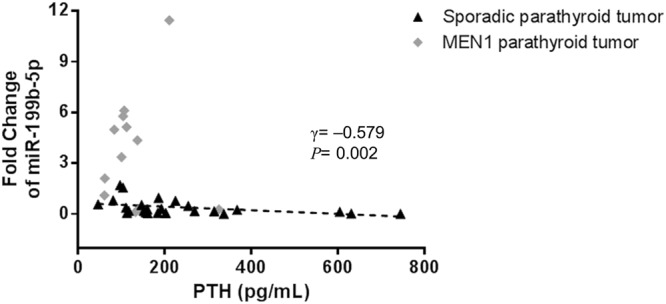
Different correlations between the relative expression of miR-199b-5p and serum PTH levels in parathyroid tumors. A negative association of miR-199b-5p and PTH levels was found in sporadic parathyroid tumors (γ = −0.579, P = 0.002), but there was no significant correlation for the hereditary parathyroid tumors.
miRNA target genes and biological function analysis
To inspect the function of miR-199b-5p in parathyroid tumorigenesis, we collected 113 validated miR-199b-5p targets based on miRWalk and miRTarBase (data not shown). Gene ontology (GO) analysis of the candidate target genes showed that DNA-templated regulation of transcription, regulation of cell growth, angiogenesis, regulation of transcription from RNA polymerase II promoter, regulation of cell proliferation, and response to drug are the most significantly enriched GO terms (Table 2). Moreover, KEGG pathway enrichment analysis revealed that the miRNA-targets were significantly associated with pathways in cancer, focal adhesion, and central carbon metabolism in cancer (Table 3).
Table 2.
GO functional annotation for validated targets of miR-199b-5p according to biological process.
| GO Term | Count | % | P-value | Genes |
|---|---|---|---|---|
| GO:0006351 Transcription, DNA-templated | 35 | 31.2 | 3.0E-8 | DDX3X, E2F3, NAA15, NAB2, POLR2F, ZFP1, CSNK2A1, CCNL1, ERBB2, HES1, HIF1A, MAP3K9, NLK, PAX8, SIRT1, TFDP2, ZBTB37, ZNF117, ZNF195, ZNF215, ZNF286B, ZNF394, ZNF415, ZNF440, ZNF468, ZNF525, ZNF544, ZNF584, ZNF611, ZNF625, ZNF669, ZNF772, ZNF791, ZNF844, ZNF846 |
| GO:0006355 Regulation of transcription, DNA-templated | 29 | 25.9 | 1.8E-7 | AKAP17A, SETD2, TSC22D1, ZFP1, HES1, HIF1A, MAP3K9, NLK PAX8, ZBTB37, ZNF117, ZNF195, ZNF215, ZNF286B, ZNF394, ZNF415, ZNF440, ZNF468, ZNF525, ZNF544, ZNF584, ZNF611, ZNF625, ZNF669, ZNF772, ZNF791, ZNF844, ZNF846 |
| GO:0030307 Positive regulation of cell growth | 5 | 4.5 | 2.1E-3 | DDX3X TAF9B, CSNK2A1, ERBB2, EXTL3 |
| GO:0001525 Angiogenesis | 7 | 6.2 | 3.2E-3 | NAA15, SETD2, HIF1A, JAG1, PLXND1, SIRT1, VAV3 |
| GO:0045944 Positive regulation of transcription from RNA polymerase II promoter | 14 | 12.5 | 1.0E-2 | DDX3X, GATA6, JUNB, TAF9B, CCNL1, CDK9, HES1, HIF1A, JAG1, LIF, PAX8, PIN1, SIRT1, TFDP2 |
| GO:0008284 Positive regulation of cell proliferation | 9 | 8.0 | 1.0E-2 | CHRFAM7A, E2F3, KIT, CSNK2A1, DYNAP, HES1, KAMC2, LIF, SIRT1 |
| GO:0042493 Response to drug | 7 | 6.2 | 1.4E-2 | ABCC1, GATA6, JUNB, CDK9, ITGA3, SLC8A1, VAV3 |
Table 3.
KEGG pathway for most significantly associated targets of miR-199b-5p.
| KEGG pathway | Count | % | P-value | Genes |
|---|---|---|---|---|
| Pathways in cancer | 7 | 6.5 | 0.019 | E2F3, KIT, ERBB2, HIF1A, ITGA3, KAMC2, PAX8 |
| Focal adhesion | 5 | 4.5 | 0.026 | ERBB2, ITGA3, LAMC2, VASP, VAV3 |
| Central carbon metabolism in cancer | 3 | 2.7 | 0.048 | KIT, ERBB2, HIF1A |
Discussion
The differential diagnosis of sporadic and hereditary parathyroid tumors remains uncertain. To establish a precise differential diagnosis method for these parathyroid tumors, numerous parameters have been investigated from different clinicopathological conditions23–25, but the discrimination remains difficult. Even gene array data showed that hereditary parathyroid tumors are clustered with the sporadic form, indicating that these tumors may share a similar genetic pathway of tumorigenesis26. Consequently, new biomarkers are needed for the effective discrimination of these tumor types.
In the present study, miR-199b-5p showed good accuracy for distinguishing sporadic and hereditary parathyroid tumors. The differentiation of these tumor types is associated with improved therapeutic outcomes and decreased recurrence rates of hyperparathyroidism, which requires the reevaluation of uncertain prognoses. These results indicate the need for a more in-depth evaluation of miRNAs in parathyroid tumors. We also observed significantly lower expression of miR-199b-5p in sporadic parathyroid tumors, and miR-199b-5p expression was negatively associated with PTH levels, indicating a specific role for this miRNA in parathyroid tumorigenesis.
Little is known about the different molecular mechanisms between sporadic and hereditary parathyroid tumors that could explain the different disease progression profiles and phenotypes. Several responsible genetic germline changes associated with parathyroid tumors in familial syndromes, such as MEN1 in MEN 1, RET in MEN 2 A, and CDC73/HRPT2 in HPT-JS (hyperparathyroidism-jaw tumor syndrome), have been identified27,28. However, these genetic alterations have also been implicated in a subset of sporadic parathyroid tumors. Genetic alterations in the MEN1 gene have been reported in 20 to 30% of sporadic parathyroid tumors29. Therefore, the presence of constitutively mutated MEN1 alleles is not sufficient to explain the different tumor profiles between sporadic and hereditary parathyroid tumors.
Substantial advances in the study of miRNA involvement in parathyroid tumorigenesis have been achieved in recent years. Differentially expressed miRNAs between parathyroid carcinoma and adenoma were identified, including miR-139, miR-296, miR-222, miR-503 miR-26b, miR-30b, miR-126*, miR-517c, and miR-37217–19. This subset of miRNAs was further verified by Hu et al.20. Emerging evidence has shown that even the partial inactivation of tumor suppressors can importantly contribute to tumorigenesis11. In this context, miRNAs can be good candidates for the subtle regulation of gene expression based on a continuum model of tumor suppressor gene function. Interestingly, this presumption was confirmed, in part, by Luzi et al., showing that miR-24-1 could bind to the MEN1 mRNA and inhibit menin expression, closing a feedback loop30. Grolmusz et al. also reported miR-24 and miR-28 were differentially expressed between sporadic and MEN1 parathyroid tumors31. Subsequently, miR-4258, miR-664, and miR-1301 were demonstrated to involve in the MEN1 associated parathyroid tumors32. Unfortunately, however, miR-199b-5p was not identified in previous studies. This inconsistent result might be due to small sample size because of rarity of MEN1 related samples and genetic heterogenesis of parathyroid tumors20.
Several studies related to miRNAs have shown that miR-199b-5p is a putative tumor suppressor that targets several signaling pathways: Hes1 involved in both Notch and Hedgehog pathways in medulloblastoma33 and osteosarcoma34, PODXL and DDR1 in acute myeloid leukemia35, HIF-1α in hepatocellular carcinoma36, and HER2 and its downstream signaling ERK1/2 and AKT pathway in breast cancer37. Taken together, the overexpression of miR-199b-5p could significantly inhibit cell proliferation, migration, and clonogenicity. Interestingly, miR-199b-5p could be a fine tuner of target gene expression, suggesting its epigenetic control function during tumor development33. Although its exact functions in parathyroid tumors are unknown, miR-199b-5p was found to be negatively correlated with serum PTH which associated with tumor size in sporadic parathyroid tumors in our study. Moreover, GO analysis suggested that miR-199b-5p targeted genes may play roles in transcription regulation, regulation of cell growth and proliferation, and angiogenesis. The most significant pathway in KEGG analysis was pathways in cancer. Altogether suggests that miR-199b-5p could play a possible role in parathyroid tumorigenesis.
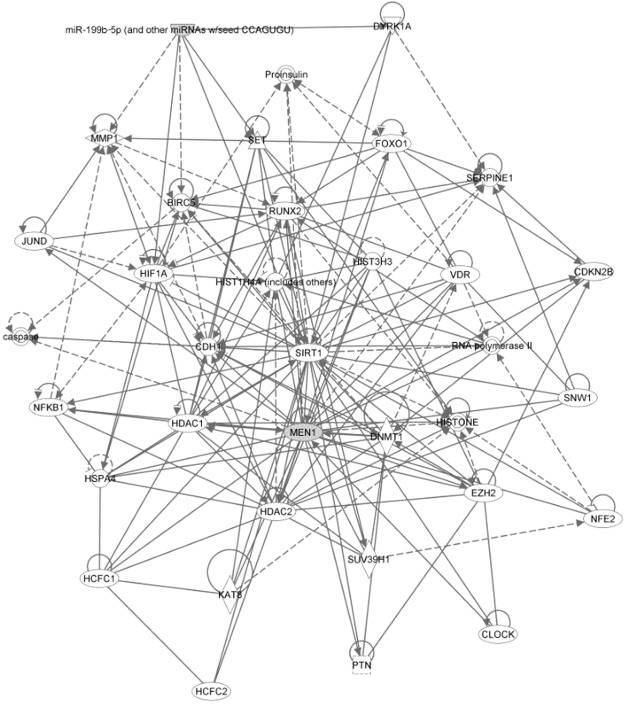
A significant direct association between miR-199b-5p and PTH levels in MEN1 parathyroid tumors was not observed in the present study, which may be due to the complex interactions between genetic backgrounds and other susceptibility factors affected by the MEN1 gene. To investigate the potential effects of miR-199b-5p on parathyroid tumorigenesis in different genetic backgrounds, we used bioinformatics to predict a network between miR-199b-5p and the MEN1 gene with Ingenuity Pathway Analysis (IPA) software (Ingenuity® Systems version 8.0, www.ingenuity.com) (Fig. 5). Interestingly, one pathway was identified: gene expression, cellular development, cellular growth and proliferation. According to the IPA results, miR-199b-5p directly targets the transcription regulators HIF-1α and SIRT1, which play a key role in promoting cell proliferation38,39 and have known interactions with MEN140. However, it is unclear how altered miR-199b-5p expression occurs within the context of dysregulated MEN1 in parathyroid tumors. The ability of miR-199b-5p to mediate the expression of these two genes may also explain the relationship between miR-199b-5p and MEN1. These complex relations must be confirmed in further investigations. There are some limitations to this study. First, the sample size of MEN1-related parathyroid tumors is small due to the rarity of this condition. Second, the lack of experimental validation for functional studies of miR-199b-5p and its predicted target genes is a further limitation.
Figure 5.
A network predicted to be regulated by miRNA-199b-5p and the MEN1 gene. One predicted network regulated by miRNA-199b-5p and the MEN1 gene was “Gene expression, cellular development, cellular growth and proliferation”.
In conclusion, we identified that miR-199b-5p is differentially expressed between sporadic and MEN1 parathyroid tumors and could be a potential diagnostic marker for distinguishing these tumors in different genetic backgrounds. Considering these data on the association between PTH and miR-199b-5p in parathyroid tumors, it will be important to focus future studies on the role of miR-199b-5p in parathyroid tumorigenesis.
Materials and Methods
Parathyroid tissue samples
We obtained a total of 61 parathyroid tissue samples and the associated clinical and histopathological data. Thirty-seven parathyroid tumor tissues were obtained from parathyroidectomy procedures. Twelve samples were obtained from MEN1 patients and confirmed to have germline mutations in the MEN1 gene. Twenty-four normal parathyroid tissues were used as controls; these tissues were incidentally removed during thyroidectomy in hyperthyroidism patients who had no evidence of PHPT. The present study was approved by the Institutional Review Board of Severance Hospital (4-2011-0613), and written informed consent was obtained from all patients. All experiments were performed in accordance with the relevant guidelines and regulations.
RNA isolation
Total RNA was extracted from the formalin-fixed and paraffin-embedded (FFPE) samples using TRIzol reagent (GIBCO, BRL, Gaithersburg, MD, USA) according to the manufacturer’s protocol. Following extraction, total RNA was quantified by an ND-1000 spectrophotometer (NanoDrop Technologies, Rockland, DE, USA).
MicroRNA microarray
For the miRNA microarray study, 3 sporadic and 3 MEN1-related parathyroid tumors and 2 normal parathyroid tissue samples were used. The quantitation of mature miRNA expression levels in parathyroid tissues was performed using a human miRNA Microarray Release 14.0, 8 × 15 K (Agilent, Waldbronn, Germany), which contains 887 human miRNAs with four duplicate probes per miRNA. The hybridization signals were detected by an Agilent SureScan microarray scanner. The scanner images were analyzed by Agilent feature extraction software. Data normalization was performed with Genowiz 4.0.5.6. An adjusted P-value controlling for a false discovery rate (FDR) of < 0.05 was used to identify miRNAs that were differentially expressed between parathyroid tumors and normal parathyroid tissues.
Quantitative real-time PCR
miRNAs were validated in 25 sporadic and 12 hereditary parathyroid tumors and 24 normal parathyroid tissues by qRT-PCR. The levels of mature hsa-let 7i, miR-125a-3p, miR-125a-5p, miR-142-3p, miR-193b, miR-199b-5p, and miR-365 were measured using individual TaqMan microRNA assays (Applied Biosystems) according to the manufacturer’s instructions. Total RNA (15 ng/15 µL of reaction) was converted into complementary DNA (cDNA) using a TaqMan miRNA reverse-transcription kit (Applied Biosystems, Carlsbad, CA, USA); then, the cDNA was subjected to amplification with TaqMan Universal PCR Master Mix and an ABI 7500 quantitative PCR machine (Applied Biosystems, Carlsbad, CA, USA). The following commercially available and prevalidated TaqMan® primers/probes for stem-loop miRNA were used: let-7i (002172), hsa-miR-125a-3p (002199), hsa-miR-125a-5p (002198), hsa- miR-142-3p (000464), hsa-miR-193b (002367), 199b-5p (000500), and hsa-miR-365 (001020). The expression miRNA levels in the samples were normalized to RNU6. miRNA expression levels were analyzed for relative fold-changes from the threshold cycle (Ct) values using the 2−ΔΔCt method38.
In silico miRNA target prediction and functional analysis
The validated target genes of miRNA were predicted via miRWalk41 and miRTarBase42. GO term and KEGG pathway analysis were performed for the candidate genes using the DAVID gene annotation tool (http://david.abcc.ncifcrf.gov). The enrichment P values of both GO and KEGG pathway enrichment analysis were set as significant when P < 0.05.
Statistical analysis
Statistical analyses were performed using SPSS 18.0 software (SPSS, Inc. Chicago, IL, USA). Differences in continuous variables between the three groups were tested by one-way ANOVA with Tukey’s post hoc test, and the differences between two groups were determined by the independent sample t-test or Mann-Whitney U-test. Spearman’s rank correlation coefficients were used to assess the associations between the relative miRNA expression and PTH levels. Receiver operating characteristic curve (ROC) analysis was applied to obtain the utility of the miRNA for distinguishing between sporadic and hereditary parathyroid tumors. A value of P < 0.05 was considered statistically significant.
Acknowledgements
The present study was supported by the Korean Association of Internal Medicine Research Fund (2017-151).
Author Contributions
S.H. and Y.R. designed the research; Y.J.C., S.Y.K. and Y.J.L. performed the experiments; S.H., J.J.J., S.H.K., Y.J.C., S.Y.K. and Y.R. analyzed the data; S.H., J.J.J., S.H.K., Y.J.C. and Y.R. wrote the manuscript. All authors reviewed the manuscript.
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Melton LJ., III Epidemiology of primary hyperparathyroidism. Journal of Bone and Mineral Research. 1991;6:S25–S30. doi: 10.1002/jbmr.5650061409. [DOI] [PubMed] [Google Scholar]
- 2.DeLellis RA, Mazzaglia P, Mangray S. Primary hyperparathyroidism: a current perspective. Arch Pathol Lab Med. 2008;132:1251–1262. doi: 10.5858/2008-132-1251-PHACP. [DOI] [PubMed] [Google Scholar]
- 3.Brandi ML, et al. Guidelines for diagnosis and therapy of MEN type 1 and type 2. J Clin Endocrinol Metab. 2001;86:5658–5671. doi: 10.1210/jcem.86.12.8070. [DOI] [PubMed] [Google Scholar]
- 4.Piecha G, Chudek J, Wiecek A. Primary hyperparathyroidism in patients with multiple endocrine neoplasia type 1. Int J Endocrinol. 2010;2010:928383. doi: 10.1155/2010/928383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lourenco DM, Jr, et al. Early-onset, progressive, frequent, extensive, and severe bone mineral and renal complications in multiple endocrine neoplasia type 1-associated primary hyperparathyroidism. J Bone Miner Res. 2010;25:2382–2391. doi: 10.1002/jbmr.125. [DOI] [PubMed] [Google Scholar]
- 6.Eller-Vainicher C, et al. Sporadic and MEN1-related primary hyperparathyroidism: differences in clinical expression and severity. J Bone Miner Res. 2009;24:1404–1410. doi: 10.1359/jbmr.090304. [DOI] [PubMed] [Google Scholar]
- 7.Norton JA, et al. Prospective study of surgery for primary Hyperparathyroidism (HPT) in multiple endocrine neoplasia-type 1 (MEN1), and Zollinger-Ellison syndrome (ZES): long-term outcome of a more virulent form of HPT. Annals of surgery. 2008;247:501. doi: 10.1097/SLA.0b013e31815efda5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giusti F, Tonelli F, Brandi ML. Primary hyperparathyroidism in multiple endocrine neoplasia type 1: when to perform surgery? Clinics. 2012;67:141–144. doi: 10.6061/clinics/2012(Sup01)23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katai M, Sakurai A, Ikeo Y, Hashizume K. Primary hyperparathyroidism in patients with multiple endocrine neoplasia type 1: comparison with sporadic parathyroid adenomas. Horm Metab Res. 2001;33:499–503. doi: 10.1055/s-2001-16944. [DOI] [PubMed] [Google Scholar]
- 10.Berger AC, et al. Heterogeneous gland size in sporadic multiple gland parathyroid hyperplasia. Journal of the American College of Surgeons. 1999;188:382–389. doi: 10.1016/S1072-7515(98)00317-2. [DOI] [PubMed] [Google Scholar]
- 11.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 14.Qi J, Wang J, Katayama H, Sen S, Liu S-m. Circulating microRNAs (cmiRNAs) as novel potential biomarkers for hepatocellular carcinoma. Neoplasma. 2013;60:135. doi: 10.4149/neo_2013_018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Sen S. MicroRNA functional network in pancreatic cancer: from biology to biomarkers of disease. Journal of biosciences. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y, et al. Expression profile and clinical significance of microRNAs in papillary thyroid carcinoma. Molecules. 2014;19:11586–11599. doi: 10.3390/molecules190811586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbetta S, et al. Differential expression of microRNAs in human parathyroid carcinomas compared with normal parathyroid tissue. Endocr Relat Cancer. 2010;17:135–146. doi: 10.1677/ERC-09-0134. [DOI] [PubMed] [Google Scholar]
- 18.Rahbari R, et al. Identification of differentially expressed microRNA in parathyroid tumors. Ann Surg Oncol. 2011;18:1158–1165. doi: 10.1245/s10434-010-1359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaira V, et al. The microRNA cluster C19MC is deregulated in parathyroid tumours. Journal of molecular endocrinology. 2012;49:115–124. doi: 10.1530/JME-11-0189. [DOI] [PubMed] [Google Scholar]
- 20.Hu Y, et al. Verification of candidate microRNA markers for parathyroid carcinoma. Endocrine. 2018;60:246–254. doi: 10.1007/s12020-018-1551-2. [DOI] [PubMed] [Google Scholar]
- 21.Zamboni WA, Folse R. Adenoma weight: a predictor of transient hypocalcemia after parathyroidectomy. Am J Surg. 1986;152:611–615. doi: 10.1016/0002-9610(86)90436-8. [DOI] [PubMed] [Google Scholar]
- 22.Rutledge R, Stiegel M, Thomas CG, Wild RE., Jr. The relation of serum calcium and immunoparathormone levels to parathyroid size and weight in primary hyperparathyroidism. Surgery. 1985;98:1107–1112. [PubMed] [Google Scholar]
- 23.Twigt BA, Scholten A, Valk GD, Rinkes IHB, Vriens MR. Differences between sporadic and MEN related primary hyperparathyroidism; clinical expression, preoperative workup, operative strategy and follow-up. Orphanet J Rare Dis. 2013;8:50. doi: 10.1186/1750-1172-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Laat JM, et al. Predicting the risk of multiple endocrine neoplasia type 1 for patients with commonly occurring endocrine tumors. European Journal of Endocrinology. 2012;167:181–187. doi: 10.1530/EJE-12-0210. [DOI] [PubMed] [Google Scholar]
- 25.Friedman E, et al. Clonality of parathyroid tumors in familial multiple endocrine neoplasia type 1. N Engl J Med. 1989;321:213–218. doi: 10.1056/NEJM198907273210402. [DOI] [PubMed] [Google Scholar]
- 26.Haven CJ, et al. Gene expression of parathyroid tumors: molecular subclassification and identification of the potential malignant phenotype. Cancer Res. 2004;64:7405–7411. doi: 10.1158/0008-5472.CAN-04-2063. [DOI] [PubMed] [Google Scholar]
- 27.Westin G, Bjorklund P, Akerstrom G. Molecular genetics of parathyroid disease. World J Surg. 2009;33:2224–2233. doi: 10.1007/s00268-009-0022-6. [DOI] [PubMed] [Google Scholar]
- 28.Alvelos MI, Mendes M, Soares P. Molecular alterations in sporadic primary hyperparathyroidism. Genet Res Int. 2011;2011:275802. doi: 10.4061/2011/275802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchino S, et al. Screening of the Men1 gene and discovery of germ-line and somatic mutations in apparently sporadic parathyroid tumors. Cancer Res. 2000;60:5553–5557. [PubMed] [Google Scholar]
- 30.Luzi E, et al. The negative feedback-loop between the oncomir Mir-24-1 and menin modulates the Men1 tumorigenesis by mimicking the “Knudson’s second hit”. PLoS One. 2012;7:e39767. doi: 10.1371/journal.pone.0039767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grolmusz VK, et al. MEN1 mutations and potentially MEN1-targeting miRNAs are responsible for menin deficiency in sporadic and MEN1 syndrome-associated primary hyperparathyroidism. Virchows Archiv. 2017;471:401–411. doi: 10.1007/s00428-017-2158-3. [DOI] [PubMed] [Google Scholar]
- 32.Luzi E, et al. Analysis of differentially expressed microRNAs in MEN1 parathyroid adenomas. American journal of translational research. 2017;9:1743. [PMC free article] [PubMed] [Google Scholar]
- 33.Andolfo I, et al. The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14:596–612. doi: 10.1093/neuonc/nos002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Won KY, et al. MicroRNA-199b-5p is involved in the Notch signaling pathway in osteosarcoma. Hum Pathol. 2013;44:1648–1655. doi: 10.1016/j.humpath.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 35.Favreau AJ, Cross EL, Sathyanarayana P. miR-199b-5p directly targets PODXL and DDR1 and decreased levels of miR-199b-5p correlate with elevated expressions of PODXL and DDR1 in acute myeloid leukemia. Am J Hematol. 2012;87:442–446. doi: 10.1002/ajh.23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C, et al. Underexpressed microRNA-199b-5p targets hypoxia-inducible factor-1alpha in hepatocellular carcinoma and predicts prognosis of hepatocellular carcinoma patients. J Gastroenterol Hepatol. 2011;26:1630–1637. doi: 10.1111/j.1440-1746.2011.06758.x. [DOI] [PubMed] [Google Scholar]
- 37.Jiang, G. et al. Expression levels of microRNA-199 and hypoxia-inducible factor-1 alpha in brain tissue of patients with intractable epilepsy. Int J Neurosci 1–29, 10.3109/00207454.2014.994209 (2014). [DOI] [PubMed]
- 38.Goda N, Dozier SJ, Johnson RS. HIF-1 in cell cycle regulation, apoptosis, and tumor progression. Antioxid Redox Signal. 2003;5:467–473. doi: 10.1089/152308603768295212. [DOI] [PubMed] [Google Scholar]
- 39.Cha YI, Kim HS. Emerging role of sirtuins on tumorigenesis: possible link between aging and cancer. BMB Rep. 2013;46:429–438. doi: 10.5483/BMBRep.2013.46.9.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fontaniere S, et al. Gene expression profiling in insulinomas of Men1 β-cell mutant mice reveals early genetic and epigenetic events involved in pancreatic β-cell tumorigenesis. Endocrine-Related Cancer. 2006;13:1223–1236. doi: 10.1677/erc.1.01294. [DOI] [PubMed] [Google Scholar]
- 41.Dweep, H., Gretz, N. & Sticht, C. miRWalk database for miRNA–target interactions in RNA Mapping 289–305 (Springer, 2014). [DOI] [PubMed]
- 42.Chou C-H, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic acids research. 2017;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]