Abstract
Four new limonoid-type nortriterpenoids, 1-detigloyl-1-O-methacryloylsalannin (1), 28-deoxo-2,3-dihydronimbolide (2), 12-acetoxy-3-O-acetyl-7-O-tigloylvilasinin (3) and 12-acetoxy-3-O-acetyl-7-O-methacryloylvilasinin (4), along with five known ones, were isolated from seeds of Azadirachta indica A. Juss. Their structures were elucidated by various spectroscopic methods, including UV, IR, MS, NMR, X-ray crystallography, quantum chemical calculation, as well as by comparison of their spectroscopic data with those reported. In the in vitro cytotoxic assay, 2 showed inhibitory activity against human breast cancer MDA-MB-231 cell line with IC50 value of 7.68±1.74 μmol/L, and 5 inhibited growth of human cervical cancer Hela cell line, melanoma A375 cell line and promyelocytic leukemia HL-60 cell line, with IC50 12.00±2.08, 17.44±2.11, and 13.95±5.74 μmol/L, respectively.
KEY WORDS: Azadirachta indica, Nortriterpenoid, Limonoid, Spectroscopy, Cytotoxic activity
Graphical abstract
Four new limonoids, together with five known ones, were isolated from dry seeds of neem (Azadirachta indica A. Juss). Their structures were elucidated by spectroscopic methods and quantum chemical calculation. Compounds 2 and 5 selectively inhibited growth of human cancer cells.
1. Introduction
Neem tree (Azadirachta indica A. Juss.) belonging to the meliaceae family is indigenous to India and Burma1, 2. Neem has been used as a folk medicinal plant in India for over thousands of years. Almost all parts of this magical tree—leaves, stems, barks, roots, seeds, fruits, flowers, etc.—could be natural drugs in treatment of cough, wounds, leprosy, skin ulcers, intestinal worms, diabetes, cholera, diarrheal, etc.1, 3, 4. Therefore, neem received a bulk of attention of phytochemists and their efforts led to discovery of more than 300 compounds from this plant. Over 130 of these compounds belong to limonoid-type triterpenoids5.
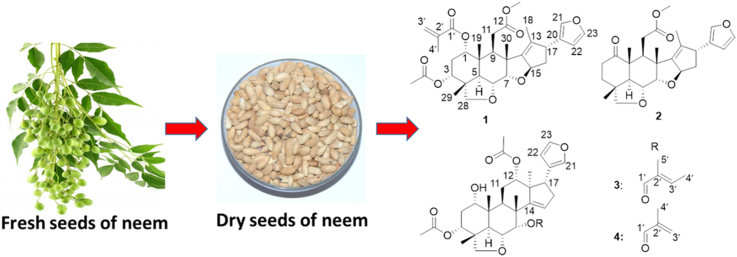
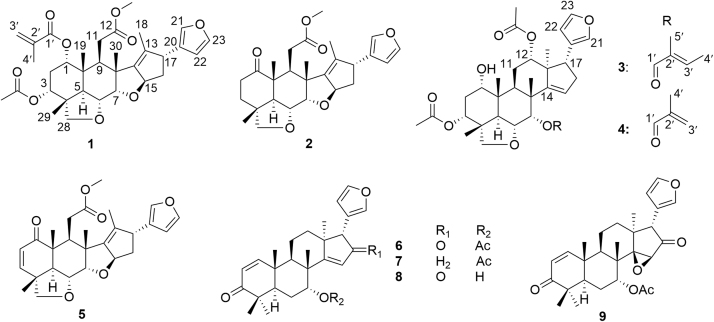
Limonoids are derived from apotirucallane- or apoeuphane-type triterpenoids, the side chains of which lose four terminal carbons and in the most cases form a furan ring. Over a thousand of limonoids have been found in nature. Due to their structural features, limonoids are classified into many subgroups, including ring intact limonoids, ring-seco limonoids, rearranged limonoids and limonoid derivatives5. Limonoids have shown a variety of activities, including anticancer5, anti-inflammation6, 7, antimalaria8, antibacteria9, antiprotozoal5, etc. Especially in the studies on anticancer activity of limonoids, many of them exhibited inhibitory activity against various human cancer cell lines. For examples, both 1-O-deacetylohchinolide B10 and 15-O-deacetylnimbolindin B11 can remarkably inhibit growth of HeLa S3 human cervical adenocarcinoma cells with IC50 of 0.10 μmol/L. Notably, cytotoxic mechanisms of nimbolide and azadirone, two main limonoid constituents in neem, were extensively studied. Nimbolide can inhibit cancer cell growth through inducing ROS-mediated apoptosis12, inhibiting P13K/Akt signaling13, 14, and upregulating reversion-inducing cysteine-rich protein with Kazal motifs (RECK)15. Azadirone can enhance the efficacy of tumor necrosis factor-related apoptosis-inducing ligand against cancer cells16. Therefore, limonoids are a group of promising compounds for discovery of new anticancer agents. Herein, we report four new limonoids, together with five known ones (Fig. 1), from neem seeds and their cytotoxic activity.
Figure 1.
The structures of compounds 1—9.
2. Result and discussion
Compound 1 (Fig. 1) was obtained as a white amorphous powder with [α]25D +74 (c 0.10, MeOH). Its molecular formula of C33H42O9 was determined by the quasi-molecular ion peak at m/z 605.2719 [M + Na]+ (Calcd. 605.2721) observed in the HR-ESI-MS. The 1H and 13C NMR spectra of 1 showed a methacrylyl group [δH 6.21 (s, 1H, H-3′a), 5.60 (s, 1H, H-3′b) and 2.05 (s, 3H, H-4′); δC 166.1 (C-1′), 136.8 (C-2′), 125.6 (C-3′) and 18.1 (C-4′)], an acetyl group [δH 1.96 (s, 3H, 3-OCOMe); δC 170.4 (3-OCOMe) and 20.9 (3-OCOMe)] and an oxygen-bearing methyl group [δH 3.25 (s, 3H, 12-OMe) and δC 51.5 (12-OMe)], which were confirmed by the HMBC correlations from H-4′ to C-1′, C-2′ and C-3′, and from H-3-OCOMe to C-3-OCOMe. The rest NMR data, including a group of characteristic signals assigned to a β-substituted furan ring [δH 7.24 (brs, 1H, H-21), 6.28 (brs, 1H, H-22) and 7.31 (brs, 1H, H-23); δC 127.0 (C-20), 138.8 (C-21), 110.6 (C-22) and 142.9 (C-23)], are very similar to those of the limonoid skeleton of salannin17, 18.
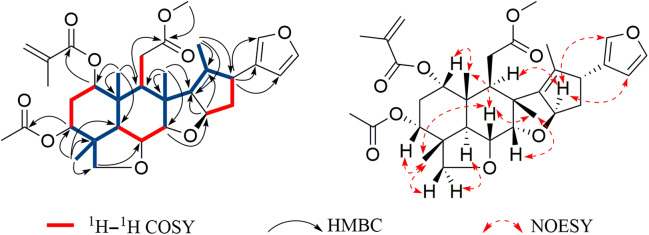
The structure of 1 was finally constructed by its 2D NMR spectra. The proton and protonated carbon resonances in the NMR spectra of 1 were firstly assigned by the HSQC experiment. The 1H—1H COSY correlations H-1/H2-2/H-3, H-5/H-6/H-7, H-9/H2-11 and H-15/H2-16/H-17 indicated four structural fragments labeled in red in Fig. 2. In the HMBC spectrum, the correlations from H3-29 to C-3, C-4, C-5 and C-28, from H3-19 to C-1, C-5, C-9 and C-10, from H3-30 to C-7, C-8, C-9 and C-14, and from H3-18 to C-13, C-14 and C-17 tethered four red fragments together with the blue ones shown in Fig. 2. The HMBC correlation of H-9/C-12, together with the chemical shift of C-12 indicated that C-12 was linked to C-11 and oxidized into a carboxyl group which was methylated due to the HMBC correlation between H-12-OMe and C-12. The methacrylyl group, acetyl group and furan ring located at C-1, C-3 and C-17, respectively, based on the HMBC correlations of H-1/C-1′, H-3/C-3-OCOMe, H-17/C-20, H-17/C-21 and H-17/C-22. Finally, two tetrahydrofuran rings were constructed by the correlations between H-28 and C-6, and between H-7 and C-15 in the HMBC spectrum. Therefore, the planar structure of 1 was elucidated.
Figure 2.
Selective 2D NMR correlations of compound 1.
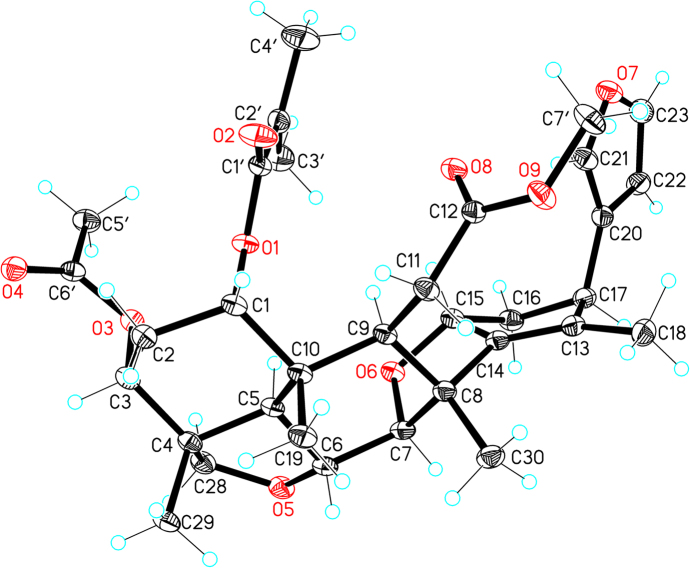
The relative configuration of 1 was determined by its NOESY spectrum. The NOE correlations of H-6/H3-19/H-1, H-6/H3-29/H-1/H-3, H-6/H3-30/H-7, H3-19/H2-11/H3-30 and H3-29/H-28β indicated these protons were oriented on the same side of the ring system and arbitrarily assigned as β. H-5 was assigned as α due to the correlation between it and H-28α. H-9 strongly correlated with H-15 in the NOESY experiment, whereas H-11 correlated with both H3-19 and H3-30, which indicated that H-9 and H-15 have α configurations. Finally, the NOE correlations from H-15 to H-22 indicated that the furan ring has an α orientation as well. The absolute configuration of 1 was finally determined by X-ray crystallography as shown in Fig. 3.
Figure 3.
ORTEP drawing of the compound 1.
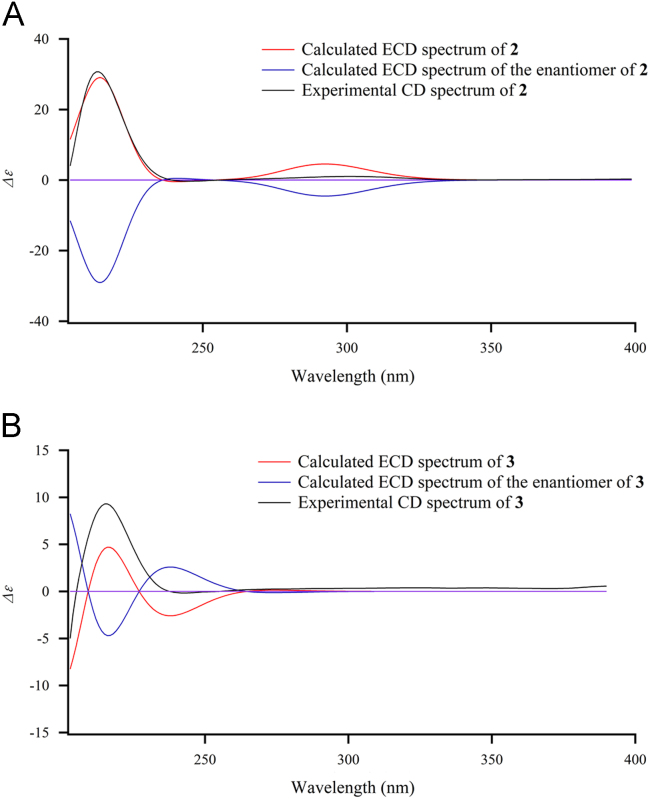
Compound 2 was obtained as a white amorphous powder with [α]25D +102 (c 0.20, MeOH). Its quasi-molecular ion peak at m/z 477.2240 [M + Na]+ (Calcd. 477.2248) was observed in the HR-ESI-MS, which indicated its molecular formula was C27H34O6. Except for a methoxyl group, there were 26 carbon signals in the 13C NMR spectrum, including four sp2 carbons [δC 126.9 (C-20), 138.9 (C-21), 110.5 (C-22) and 142.9 (C-23)] belonging to a furan ring, which implied that 2 was a limonoid-type nortriterpenoid as well. In the HMBC spectrum of 2, H3-19 (δH 1.22) correlated to a keto group with a chemical shift at δC 213.0, indicating C-1 was oxidized into a keto group. Based on this clue, it was found that the NMR data of 2 were very similar to those of 2,3-dihydronimbolide19 except that C-28 (a carbonyl carbon) of 2,3-dihydronimbolide was replaced by an isolated oxygen-bearing methylene [δH 3.53 (d, J=7.5 Hz, H-28α) and 3.75 (d, J=7.5 Hz, H-28β), and δC 82.8 (C-28)], implying C-28 was reduced into a methylene in 2. This deduction was confirmed by the HMBC correlations from H3-29 to C-3, C-4, C-5 and C-28, and from H-28α and 28β to C-5 and C-6. In the NOESY spectrum of 2, the NOE correlations of H-11a/H3-19/H3-29/H-6/H3-30/H-7/H-16β/H-17, as well as the correlations of H-5/H-9/H-15, indicated that 2 possessed the same relative configuration profile as that of 2,3-dihydronimbolide19. And the optical rotation of 2 is positive, which is also same as those of 2,3-dihydronimbolide19, 28-deoxonimbolide20 and nimbolide20. All these data implied that the absolute stereochemistry of 2 was consistent with those of the known nimbolide-type limonoids. Finally, the absolute stereochemistry of 2 was confirmed by comparison of its experimental CD spectrum with the calculated ones, as shown in Fig. 4. Therefore, the structure of 2 was determined as 28-deoxo-2,3-dihydronimbolide.
Figure 4.
Experimental CD spectra of compounds 2 and 3 (black) overlaid with the calculated ECD spectra of 2 and 3 (red) and their enantiomers (blue).
Compound 3 was obtained as a white amorphous powder with [α]25D —25 (c 0.10, MeOH). Its molecular formula, C35H46O9, was also determined by the quasi-molecular ion peak at m/z 633.3032 shown in its HR-ESI-MS spectrum. The 1H and 13C NMR data of 3, in combination with its HMBC spectrum, indicated that there were two acetyl [δH 2.05 (s, 3H), δC 169.3 and 21.1; δH 1.91, δC 171.2 and 21.6] and one tigloyl [δH 6.88 (q, J=7.0 Hz, H-3′), 1.79 (d, J=7.0 Hz, H3-4′) and 1.86 (s, H3-5′); δC 166.8 (C-1′), 128.9 (C-2′), 137.1 (C-3′), 14.7 (C-4′) and 12.4 (C-5′)] groups in 3. The rest NMR data showed 26 carbons and 33 protons, including a set of the characteristic signals attributed to a furan ring, indicating a limonoid-type nortriterpenoid backbone, too. Comparison of NMR data between 3 and 1,3-diacetyl-12-hydroxy-7-tigloylvilasinin21 showed the chemical shifts of C-1 and C-11 were shifted 1.0 and 5.7 ppm upfield in 3, while those of C-2 and C-12 were shifted 2.8 and 1.9 ppm downfield, which indicated that 12-OH was acetylated in 3 instead of acetylation of 1-OH in 1,3-diacetyl-12-hydroxy-7-tigloylvilasinin. This deduction was confirmed by the HMBC correlation from H-12 to C-12-OCOMe. The relative configuration of 3 was approved the same as that of 1,3-diacetyl-12-hydroxy-7-tigloylvilasinin by the NOESY experiment in which the NOE correlations of H-3/H3-29/H-6/H3-19/H3-30, H-1/H3-19/H-11a and H-7/H3-30/H-12 assigned them β orientation, while the correlations of H-5/H-9/H3-18/H-22 indicated these protons and the furan ring possessed α orientation. Finally, the absolute stereochemistry of 3 was confirmed by comparison of its experimental CD spectrum with the calculated ones, as shown in Fig. 4.
Compound 4 was obtained as a white amorphous powder with [α]25D —87 (c 0.08, MeOH). Based on its HR-ESI-MS spectrum, the molecular formula of 4 was C34H44O9 which was CH2 less than that of 3. And the NMR data of 3 and 4 were very similar, except for the presence of a methylacryloyl [δH 6.13 (brs, H-3a′), 5.54 (brs, H-3b′) and 1.97 (s, H3-4′); δC 165.9 (C-1′), 136.7 (C-2′), 124.9 (C-3′) and 18.5 (C-4′)] in 4 instead of a tigloyl group in 3, which was confirmed by the HMBC correlations from H-4′ to C-1′, C-2′ and C-3′. Therefore, 4 was determined as 12-acetoxy-3-O-acetyl-7-O-methacryloylvilasinin.
By comparison of the NMR data of five known compounds with those reported in the literature, they were identified as 28-deoxonimbolide (5)20 azadiradione (6)22, azadirone (7)23, nimbocinol (8)22 and epoxyazadiradione (9)24, respectively.
In the in vitro cytotoxic assay, 2 showed inhibitory activity against MDA-MB-231 cell line with IC50 value of 7.68±1.74 μmol/L; and 5 inhibited growth of Hela cell line, A375 cell line and HL-60 cell line, with IC50 12.00±2.08, 17.44±2.11 and 13.95±5.74 µmol/L, respectively. Cisplatin was used as the positive control. It inhibited growth of A375, Hela, HL-60, MDA-MB-231, HepG2, K562 and MCF-7 cells with IC50 of 2.50±0.50, 8.96±1.59, 1.54±0.38, 1.70±0.43, 2.46±0.14, 8.17±2.31 and 11.43±2.79 μmol/L, respectively.
3. Experimental section
3.1. General experimental procedures
Optical rotations were measured with a JASCO P-2000 polarimeter (Jasco, Tokyo, Japan). UV spectra were recorded on a JASCO V-550 spectrophotometer (Jasco, Tokyo, Japan). IR spectra were performed on a FT/IR-6600 spectrometer (Jasco, Tokyo, Japan). NMR spectra were acquired on a Bruker Avance III-600 and a Bruker Avance III-300 instruments (Bruker, Bremerhaven, Germany). High-resolution mass spectra were obtained on a LCQ Advantage MAX (Finnign, USA). CD spectrum was measured on a Chirascan spectropolarimeter (Applied Photophysics, Ltd.). X-ray Crystallography was collected at 100 K on a Rigaku Oxford Diffraction Supernova Dual Source, Cu at Zero equipped with an AtlasS2 CCD using Cu Kα radiation. Silica gel (80−100 and 200−300 mesh, Qingdao Haiyang, Qingdao, China), Sephadex LH-20 (Pharmadex), and RP-C18 (AA12S50, YMC) were used for column chromatography. Preparative HPLC was carried out using an Ultimate 3000 instrument (Thermo Scientific, USA) with a Waters XBridge RP-C18 column (250 mm×10 mm). Analytical HPLC was run on using an Agilent 1260 instrument (Agilent, USA) with a Phenomenex Synergi RP-C18 column (250 mm×4.6 mm).
3.2. Botanical material
The seeds of Azadirachta indica A. Juss. were collected from Yunnan province and were authenticated by Prof. Hanhong Xu, College of Resource and Environmental Engineering, South China Agricultural University. A voucher specimen was deposited in the College of Pharmacy, Jinan University.
3.3. Extraction and isolation
The air-dried, powdered seeds of A. indica (30 kg) were macerated for 12 h with 150 L of 95% EtOH and refluxed for 2 h in twice. Then the combined EtOH extracts were concentrated under reduced pressure to give a crude residue (2.5 kg). The crude extract was suspended in 8 L distilled H2O and partitioned with petroleum ether (PE), EtOAc and n-BuOH (3 × 8 L for each solvent). After removal of the solvent under vacuum, the EtOAc (431 g) fraction was subjected to a silica gel column chromatography eluted with a gradient of increasing acetone (0–50%) in PE, followed by a gradient of increasing MeOH (5%–100%) in CHCl3, to afford 18 fractions (Fr.1—18). Fr.10 (40 g) was chromatographed on a silica gel column with increasing amounts of EtOAc:PE (1:10–1:0) to provide 26 sub-fractions (Fr.10a—10z). Fr.10t (2.6 g) was further fractionated via an ODS column (RP-18, AA12S50, 200 g) by eluting with a gradient of MeOH (55%–65%–75%–85%–100%) in H2O to obtain 30 sub-sub-fractions (Fr.10t1—Fr.10t30). Fr.10t10 (276 mg) was purified by reversed-phase semi-preparative HPLC (4 mL/min, 53% MeOH) to obtain compounds 5 (28 mg, tR 23.35 min), 2 (10 mg, tR 30.16 min) and 1 (19 mg, tR 46.56 min). Fr.10t14 (53 mg) was purified by semi-preparative HPLC (4 mL/min, 59% MeOH in H2O) to yield compounds 3 (3 mg, tR 55.58 min) and 4 (2 mg, tR 46.30 min). Fr.10w (1.1 g) was separated by Sephadex LH-20 to afford 6 sub-sub-fractions (Fr.10w1—Fr.10w6), using PE:CH2Cl2:MeOH (5:4:1) as mobile phase. Fr.10w1 and Fr.10w5 were two pure compounds, i.e. compounds 8 (16 mg) and 6 (24 mg), respectively; Fr.10w2 (108 mg) was purified by semi-preparative HPLC (4 mL/min, 50% MeOH in H2O) to yield compounds 9 (13 mg, tR 21.37 min) and 7 (4 mg, tR 41.54 min).
1-O-Detigloyl-1-O-methacryloylsalannin (1): white amorphous powder; [α]25D +74 (c 0.10, MeOH); UV (MeOH) λmax (logε) 206.5 (4.12) nm; IR (KBr) νmax 2981, 2954, 2873, 1727, 1633, 1434, 1369, 1291, 1242, 1167, 1050, 949, 874 cm−1; 1H and 13C NMR data, see Table 1; HR-ESI-MS m/z 605.2719 [M + Na]+ (Calcd. for C33H42O9Na, 605.2721).
Table 1.
NMR data of compounds 1–4 in CDCl3 (δ in ppm, J in Hz).
| Position |
1a |
2a |
3a |
4b |
||||
|---|---|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | δH | δC | |
| 1 | 4.79 (t, 2.7) | 71.6 | 213.0 | 3.50 (t, 2.7) | 72.1 | 3.50 (t, 2.7) | 71.8 | |
| 2α | 2.21 (m, 2H) | 27.6 | 2.24 (m) | 34.7 | 2.00 (dt, 16.3, 2.8) | 30.3 | 2.00 (dt, 16.3, 2.8) | 30.0 |
| 2β | 2.85 (td, 14.5, 6.1) | 2.31 (dt, 16.2, 3.2) | 2.31 (dt, 16.2, 3.2) | |||||
| 3α | 4.96 (t, 2.7) | 71.2 | 1.83 (td, 13.0, 4.4) | 35.8 | 5.09 (t, 2.7) | 73.9 | 5.09 (t, 3.2) | 73.6 |
| 3β | 1.94 (dd, 13.0, 6.1) | |||||||
| 4 | 42.7 | 39.3 | 42.5 | 42.3 | ||||
| 5 | 2.78 (d, 12.6) | 39.9 | 2.36 (d, 12.0) | 52.2 | 2.40 (d, 12.4) | 40.6 | 2.40 (d, 12.4) | 40.3 |
| 6 | 3.98 (dd, 12.6, 3.2) | 72.6 | 4.09 (dd, 12.0, 2.7) | 72.3 | 4.20 (dd, 12.4, 2.9) | 72.9 | 4.22 (d, 2.9) | 72.7 |
| 7 | 4.16 (d, 3.2) | 85.5 | 4.13 (d, 2.7) | 85.3 | 5.67 (d, 2.9) | 73.7 | 5.65 (d, 2.9) | 73.9 |
| 8 | 49.1 | 49.9 | 44.4 | 44.1 | ||||
| 9 | 2.73 (dd, 8.4, 4.0) | 39.4 | 2.59 (t, 5.1) | 41.1 | 2.98 (dd, 8.4, 4.0) | 36.4 | 2.98 (dd, 8.4, 4.0) | 36.1 |
| 10 | 40.5 | 50.7 | 40.4 | 40.1 | ||||
| 11a | 2.30 (m) | 30.6 | 2.79 (dd, 15.5, 5.1) | 33.1 | 2.21 (m) | 24.5 | 2.21 (m) | 24.2 |
| 11b | 2.19 (m) | 2.29 (dd, 15.5, 5.1) | 1.46 (m) | 1.46 (m) | ||||
| 12 | 172.8 | 173.4 | 5.05 (dd, 9.1, 7.3) | 78.14 | 5.05 (dd, 9.1, 7.3) | 77.9 | ||
| 13 | 135.0 | 134.8 | 51.9 | 51.6 | ||||
| 14 | 146.4 | 146.1 | 155.9 | 155.6 | ||||
| 15 | 5.42 (t, 7.2) | 87.9 | 5.49 (t, 6.7) | 87.8 | 5.62 (t, 2.4) | 122.9 | 5.63 (t, 2.6) | 122.7 |
| 16α | 2.22 (m) | 41.3 | 2.19 (dd, 12.1, 6.6) | 41.4 | 2.36 (m, 2H) | 36.8 | 2.38 (m, 2H) | 36.6 |
| 16β | 2.11 (m) | 2.11 (m) | ||||||
| 17 | 3.62 (d, 8.8) | 49.4 | 3.63 (d, 8.7) | 49.4 | 2.96 (d, 8.8) | 50.6 | 2.96 (d, 8.8) | 50.3 |
| 18 | 1.65 (s, 3H) | 13.0 | 1.67 (s, 3H) | 12.9 | 1.01 (s, 3H) | 15.9 | 1.02 (s, 3H) | 15.7 |
| 19 | 0.98 (s, 3H) | 15.1 | 1.22 (s, 3H) | 14.5 | 0.96 (s, 3H) | 15.6 | 0.96 (s, 3H) | 15.3 |
| 20 | 127.0 | 126.9 | 124.9 | 124.7 | ||||
| 21 | 7.24 (br s) | 138.8 | 7.24 (br s) | 138.9 | 7.19 (br s) | 140.5 | 7.19 (br s) | 140.2 |
| 22 | 6.28 (br s) | 110.6 | 6.33 (br s) | 110.5 | 6.23 (br s) | 112.1 | 6.23 (br s) | 111.8 |
| 23 | 7.31 (br s) | 142.9 | 7.31 (br s) | 142.9 | 7.31 (br s) | 142.1 | 7.31 (br s) | 141.9 |
| 28α | 3.66 (d, 7.5) | 77.6 | 3.53 (d, 7.5) | 82.8 | 3.27 (d, 7.6) | 78.11 | 3.27 (d, 7.7) | 77.8 |
| 28β | 3.57 (d, 7.5) | 3.75 (d, 7.5) | 3.50 (d, 7.6) | 3.50 (m) | ||||
| 29 | 1.21 (s, 3H) | 19.5 | 1.38 (s, 3H) | 18.9 | 1.16 (s, 3H) | 19.1 | 1.17 (s, 3H) | 18.8 |
| 30 | 1.29 (s, 3H) | 16.9 | 1.25 (s, 3H) | 17.3 | 1.18 (s, 3H) | 27.3 | 1.19 (s, 3H) | 27.0 |
| 1′ | 166.1 | 166.8 | 165.9 | |||||
| 2′ | 136.8 | 128.9 | 136.7 | |||||
| 3′a | 6.21 (s) | 125.6 | 6.88 (q, 7.0) | 137.1 | 6.13 (br s) | 124.9 | ||
| 3′b | 5.60 (s) | 5.54 (br s) | ||||||
| 4′ | 2.05 (s, 3H) | 18.1 | 1.79 (d, 7.0, 3H) | 14.7 | 1.97 (s, 3H) | 18.5 | ||
| 5′ | 1.86 (s, 3H) | 12.4 | ||||||
| 3-OCOMe | 170.4 | 169.3 | 169.1 | |||||
| 3-OCOMe | 1.96 (s, 3H) | 20.9 | 2.05 (s, 3H) | 21.1 | 2.05 (s, 3H) | 20.9 | ||
| 12-OCOMe | 171.2 | 171.0 | ||||||
| 12-OCOMe | 1.91 (s, 3H) | 21.6 | 1.91 (s, 3H) | 21.3 | ||||
| 12-OMe | 3.25 (s, 3H) | 51.5 | 3.55 (s, 3H) | 51.6 | ||||
600 MHz for 1H NMR and 150 MHz for 13C NMR;
300 MHz for 1H NMR and 75 MHz for 13C NMR.
28-Deoxo-2,3-dihydronimbolide (2): white amorphous powder; [α]25D +102 (c 0.20, MeOH); UV (MeOH) λmax (logε) 205 (4.36) nm; IR (KBr) νmax 2947, 2875, 1736, 1709, 1458, 1434, 1209, 1162, 1054, 1033, 950, 754 cm−1; CD (c 0.22 mmol/L, MeOH) λ (∆ε) 208 (+37.52), 198 (—28.07), 192 (—15.00) nm; 1H and 13C NMR data, see Table 1; HR-ESI-MS m/z 477.2240 [M+Na]+ (Calcd. for C27H34O6Na, 477.2248).
12-Acetoxy-3-O-acetyl-7-O-tigloylvilasinin (3): white amorphous powder; [α]25D —25 (c 0.10, MeOH); UV (MeOH) λmax (logε) 204 (4.91), 221 (4.54) nm; IR (KBr) ν max 3582, 2972, 2924, 2851, 1730, 1709, 1648, 1376, 1248, 1137, 1081, 1036, 876 cm−1; CD (c 0.08 mmol/L, MeOH) λ (∆ε) 214 (+10.33), 196 (−49.14), 192 (−23.43) nm; 1H and 13C NMR data, see Table 1; HR-ESI-MS m/z 633.3032 [M + Na]+ (Calcd. for C35H46O9Na 633.3034).
12-Acetoxy-3-O-acetyl-7-O-methacryloylvilasinin (4): white amorphous powder; [α]25D —87 (c 0.08, MeOH); UV (MeOH) λmax (logε) 204 (4.30) nm; IR (KBr) νmax 3568, 2973, 2928, 2888, 2842, 1727, 1373, 1245, 1161, 1079, 1027 cm−1; CD (c 0.08 mmol/L, MeOH) λ (∆ε) 214 (+6.08), 199 (−47.52) nm; 1H and 13C NMR data, see Table 1; HR-ESI-MS m/z 619.2880 [M + Na]+ (Calcd. for C34H44O9Na 619.2878).
3.4. Quantum chemical calculation
Conformational analysis was carried out by using the MMFF94 molecular mechanics force field via the MOE software package as previously reported25. All quantum computations were performed using Gaussian 09 program package, on an IBM cluster machine. Conductor-like polarizable continuum model (CPCM) was adopted to consider solvent effects using the dielectric constant of MeOH (ε=32.6).
3.5. X-ray crystallography
The data of 1 was collected at 100 K on a Rigaku Oxford Diffraction Supernova Dual Source, Cu at Zero equipped with an AtlasS2 CCD using Cu Kα radiation. Data reduction was carried out with the diffractometer's software26. The structures were solved by direct methods using Olex2 software27, and the non-hydrogen atoms were located from the trial structure and then refined anisotropically with SHELXL-201428 using a full-matrix least squares procedure based on F2. The weighted R factor, wR and goodness-of-fit S values were obtained based on F2. The hydrogen atom positions were fixed geometrically at the calculated distances and allowed to ride on their parent atoms. Crystallographic data for the structure reported in this paper have been deposited at the Cambridge Crystallographic Data Center and allocated with the deposition numbers: CCDC 1589663.
3.6. Cytotoxic assay
Human breast cancer cell line (MCF-7), human malignant melanoma cell line (A375), human liver carcinoma cell line (HepG2), human Caucasian chronic myelogenous leukaemia cell line (K562), human promyelocytic leukemia cell line (HL-60), human breast cancer MDA-MB-231 cell line and human cervix epithelioid carcinoma cell line (HeLa) were provided by Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, China. As previously reported29, all cells were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics (Penicillin 100 IU/mL, Streptomycin 100 μg/mL), and incubated at 37 °C in a humidified atmosphere containing 5% CO2. Cisplatin was used as the positive control.
Acknowledgments
This work was supported by grants from Thousand Young Talents Program of China and National Natural Science Foundation of China (No. 81673530).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Supplementary data associated with this article can be found in the online version at 10.1016/j.apsb.2017.12.009.
Contributor Information
Rongmin Yu, Email: tyrm@jnu.edu.cn.
Hanhong Xu, Email: hhxu@scau.edu.cn.
Jiachen Zi, Email: jiachen_zi@163.com.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Kumar V.S., Navaratnam V. Neem (Azadirachta indica): prehistory to contemporary medicinal uses to humankind. Asian Pac J Trop Biomed. 2013;3:505–514. doi: 10.1016/S2221-1691(13)60105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senthil-Nathan S. Physiological and biochemical effect of neem and other Meliaceae plants secondary metabo lites against Lepidopteran insects. Front Physiol. 2013:4. doi: 10.3389/fphys.2013.00359. [article 359]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thakurta P., Bhowmik P., Mukherjee S., Hajra T.K., Patra A., Bag P.K. Antibacterial, antisecretory and antihemorrhagic activity of Azadirachta indica used to treat cholera and diarrhea in India. J Ethnopharmacol. 2007;111:607–612. doi: 10.1016/j.jep.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Brahmachari G. Neem–an omnipotent plant: a retrospection. ChemBioChem. 2004;5:408–421. doi: 10.1002/cbic.200300749. [DOI] [PubMed] [Google Scholar]
- 5.Tan Q.G., Lou X.D. Meliaceous limonoids: chemistry and biological activities. Chem Rev. 2011;111:7437–7522. doi: 10.1021/cr9004023. [DOI] [PubMed] [Google Scholar]
- 6.Akihisa T., Noto T., Takahashi A., Fujita Y., Banno N., Tokuda H. Melanogenesis inhibitory, anti-inflammatory, and chemopreventive effects of limonoids from the seeds of Azadirachta indicia A. Juss. (Neem) J Oleo Sci. 2009;58:581–594. doi: 10.5650/jos.58.581. [DOI] [PubMed] [Google Scholar]
- 7.Alam A., Haldar S., Thulasiram H.V., Kumar R., Goyal M., Iqbal M.S. Novel anti-inflammatory activity of epoxyazadiradione against macrophage migration inhibitory factor. Inhibition of tautomerase and proinflammatory activities of macrophage migration inhibitory factor. J Biol Chem. 2012;287:24844–24861. doi: 10.1074/jbc.M112.341321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khalid S.A., Duddeck H., Gonzalez-Sierra M. Isolation and characterization of an antimalarial agent of the neem tree Azadirachta indica. J Nat Prod. 1989;52:922–927. doi: 10.1021/np50065a002. [DOI] [PubMed] [Google Scholar]
- 9.Maneerat W., Laphookhieo S., Koysomboon S., Chantrapromma K. Antimalarial, antimycobacterial and cytotoxic limonoids from Chisocheton siamensis. Phytomedicine. 2008;15:1130–1134. doi: 10.1016/j.phymed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Zhou H.L., Hamazaki A., Fontana J.D., Takahashi H., Esumi T., Wandscheer C.B. New ring C-seco limonoids from Brazilian Melia azedarach and their cytotoxic activity. J Nat Prod. 2004;67:1544–1547. doi: 10.1021/np040077r. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H.L., Hamazaki A., Fontana J.D., Takahashi H., Wandscheer C.B., Fukuyama Y. Cytotoxic limonoids from Brazilian Melia azedarach. Chem Pharm Bull. 2005;53:1362–1365. doi: 10.1248/cpb.53.1362. [DOI] [PubMed] [Google Scholar]
- 12.Subramani R., Gonzalez E., Arumugam A., Nandy S., Gonzalez V., Medel J. Nimbolide inhibits pancreatic cancer growth and metastasis through ROS-mediated apoptosis and inhibition of epithelial-to-mesenchymal transition. Sci Rep. 2016;6:19819. doi: 10.1038/srep19819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sophia J., Kiran Kishore T.K., Kowshik J., Mishra R., Nagini S. Nimbolide, a neem limonoid inhibits phosphatidyl inositol-3 kinase to activate glycogen synthase kinase-3β in a hamster model of oral oncogenesis. Sci Rep. 2016;6:22192. doi: 10.1038/srep22192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raja Singh P., Sugantha Priya E., Balakrishnan S., Arunkumar R., Sharmila G., Rajalakshmi M., Arunakaran J. Inhibition of cell survival and proliferation by nimbolide in human androgen-independent prostate cancer (PC-3) cells: involvement of the PI3K/Akt pathway. Mol Cell Biochem. 2017;427:69–79. doi: 10.1007/s11010-016-2898-4. [DOI] [PubMed] [Google Scholar]
- 15.Kowshik J., Mishra R., Sophia J., Rautray S., Anbarasu K., Reddy G.D. Nimbolide upregulates RECK by targeting miR-21 and HIF-1α in cell lines and in a hamster oral carcinogenesis model. Sci Rep. 2017;7:2045. doi: 10.1038/s41598-017-01960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta S.C., Francis S.K., Nair M.S., Mo Y.Y., Aggarwal B.B. Azadirone, a limonoid tetranortriterpene, induces death receptors and sensitizes human cancer cells to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) through a p53 protein-independent mechanism. Evidence for the role of the ROS-ERK-CHOP-death receptor pathway. J Biol Chem. 2013;288:32343–32356. doi: 10.1074/jbc.M113.455188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg H.S., Bhakuni D.S. Salannolide, a meliacin from Azadirachta indica. Phytochemistry. 1984;23:2383–2385. [Google Scholar]
- 18.Xie F., Zhang C.F., Zhang M., Wang Z.T., Yu B.Y. Two new limonoids from Melia toosendan. Chin Chem Lett. 2008;19:183–186. [Google Scholar]
- 19.Cui B., Chai H., Constant H.L., Santisuk T., Reutrakul V., Beecher C.W. Limonoids from Azadirachta excelsa. Phytochemistry. 1998;47:1283–1287. doi: 10.1016/s0031-9422(97)00711-5. [DOI] [PubMed] [Google Scholar]
- 20.Kigodi P.G.K., Blaskó G., Thebtaranonth Y., Pezzuto J.M., Cordell G.A. Spectroscopic and biological investigation of nimbolide and 28-deoxonimbolide from Azadirachta indica. J Nat Prod. 1989;52:1246–1251. doi: 10.1021/np50066a008. [DOI] [PubMed] [Google Scholar]
- 21.Kumar Ch.S.S.R., Srinivas M., Yakkundi S. Limonoids from the seeds of Azadirachta indica. Phytochemistry. 1996;43:451–455. [Google Scholar]
- 22.Siddiqui S., Faizi S., Siddiqui B.S. Studies on the chemical constituents of Azadirachta indica A. Juss. (Meliaceae), Part VII. Z Naturforsch B. 1986;41:922–924. [Google Scholar]
- 23.Rodríguez B. Complete assignments of the 1H and 13C NMR spectra of 15 limonoids. Magn Reson Chem. 2003;41:206–212. [Google Scholar]
- 24.Mulholland D.A., Osborne R., Roberts S.L., Taylor D.A.H. Limonoids and triterpenoid acids from the bark of Entandrophragma delevoyi. Phytochemistry. 1994;37:1417–1420. [Google Scholar]
- 25.Meng L.J., Guo Q.L., Liu Y.F., Shi J.G. 8,4′-Oxyneolignane glucosides from anaqueous extract of “ban lan gen” (Isatis indigotica root) and their absolute configurations. Acta Pharm Sin B. 2017;7:638–646. doi: 10.1016/j.apsb.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agilent Technologies, CrysAlisPRO, Version 1.171.36.28, 2013.
- 27.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J Appl Cryst. 2009;42:339–341. [Google Scholar]
- 28.Kratzert D., Holstein J.J., Krossing I.J. DSR: enhanced modelling and refinement of disordered structures with SHELXL. J Appl Cryst. 2015;48:933–938. doi: 10.1107/S1600576715005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Z.F., Bao H., Zhou F.Y., Liu J.X., Meng F.C., Feng L. Cytotoxic cassane diterpenoids from the seeds of Caesalpinia sappan. Chin Chem Lett. 2017;28:1711–1715. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material