Abstract
Objective:
Evidence suggests that the nature and magnitude of some genetic effects on alcohol use vary by age. We tested for moderation in the effect of an alcohol metabolizing polygenic score by time across the college years.
Method:
Participants (total n = 2,214) were drawn from three cohorts of undergraduate college students, who were assessed annually for up to 4 years starting in their freshman year. Polygenic risk scores (PRSs) were calculated from genes involved in the metabolism of alcohol, as many of these markers are among the best replicated in association studies examining alcohol use phenotypes. Linear mixed effects models were fit by maximum likelihood to test the main effects of time and the PRS on alcohol consumption, as well as moderation of the PRS effect on alcohol consumption by time.
Results:
In the main effects model, the fixed effects for time and the PRS were positively associated with alcohol consumption. The interaction term testing moderation of the PRS effect by time reached statistical significance and remained statistically significant after other relevant interaction effects were controlled for. The main effect of the PRS accounted for 0.2% of the variance in alcohol consumption, whereas the interaction of PRS effect and time accounted for 0.05%.
Conclusions:
Alcohol metabolizing genetic effects on alcohol use appear to be more influential in later years of college than in earlier years. Shifting environmental contexts, such as increased access to alcohol as individuals approach the legal age to purchase alcohol, may account for this association.
Emerging adulthood represents a critical period in the acquisition of maladaptive patterns of alcohol use. Rates of alcohol use typically peak in this developmental period, converging with a variety of social and biological factors to elevate the risk of alcohol-related problems (Godette et al., 2006). As a subset of emerging adults, college students experience further elevated risk of alcohol-related problems, drinking more frequently and reporting a greater prevalence of alcohol abuse, according to criteria from the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (American Psychiatric Association, 1994), than their non–college-attending peers (Slutske, 2005). A range of novel developmental tasks demarcates emerging adulthood as a period with unique salience to individuals’ progression into adulthood—for example, establishing financial independence, reducing high-risk behaviors common in adolescence, and developing close social bonds (Schulenberg et al., 2004). Elevated alcohol use within this developmental stage may stall progress on these tasks and, subsequently, produce difficulty with adjustment into adult life.
Furthermore, there is considerable heterogeneity in rates of drinking over time, including by demographic and social characteristics. Most research indicates that alcohol use increases steadily over the course of students’ enrollment in college and declines after graduation (Auerbach & Collins, 2006; Lanza & Collins, 2006; White et al., 2005), although these broader trends may vary by gender (Klein, 1994) and ethnicity (Godette et al., 2006). Genetic influences on alcohol use may also offer some insight into heterogeneity in alcohol use and problems among emerging adult college students. Although meta-analytic results suggest that alcohol use disorder is approximately 50% heritable in adult samples (Verhulst et al., 2015), the nature and magnitude of genetic influences on drinking vary across development. For example, Dick et al. (2014a) found that a variant in GABRA2 is more strongly associated with the frequency of drunkenness in individuals age 19 or older, compared with younger individuals. Elsewhere, evidence suggests that the magnitude of the effect of the ALDH2*2 allele on drinking in East Asian populations is moderated by age, with its protective effect absent in adolescence and emerging in early adulthood (Irons et al., 2012). Changing developmental conditions could provide the context for genetic variation to become more influential, shaping both the phenotypes that manifest and the magnitude of association. Potential mechanisms for this process include increasing the availability of alcohol and declining social control over alcohol use as individuals reach the legal drinking age, allowing more opportunity for predispositions toward alcohol use to present.
The influence of alcohol metabolizing genes on drinking behaviors is well substantiated in the literature, with select markers reaching genome-wide significance in association with alcohol dependence (Gelernter et al., 2014). Functionally, the ADH and ALDH gene cluster influences drinking behaviors through interference with the metabolism of ethanol into acetaldehyde or acetaldehyde into acetate, respectively (Hurley & Edenberg, 2012). Both faster conversion of ethanol to acetaldehyde and slower conversion of acetaldehyde to acetate result in more unpleasant side effects associated with drinking and, subsequently, diminish chances of developing alcohol dependence (Hurley & Edenberg, 2012). Alcohol dehydrogenase (ALDH1B1, ALDH1A1, ALDH2) and aldehyde dehydrogenase (ADH1B, ADH1C) genes are among the best replicated for their association with alcohol use behaviors, with ethnic variation in allele frequency playing a crucial role in which genes exert measureable influence. Past work has identified an association between the ADH1B*2 (rs1229984) allele and alcohol dependence in European American populations (Bierut et al., 2012; Gelernter et al., 2014; Sherva et al., 2009). Other genes involved in alcohol metabolism also show association with alcohol use phenotypes in individuals of European ancestry, including ADH4 (Edenberg et al., 2006; Guindalini et al., 2005; Luo et al., 2005), AHD5 (Luo et al., 2006) and CYP2E1 (Webb et al., 2011). Wall et al. (2016) indicate that the alcohol dehydrogenase and aldehyde dehydrogenase systems act in tandem with one another to influence the risk of alcohol dependence. A more comprehensive assessment of the polygenic influence of the alcohol metabolizing cluster in this population may describe its relationship with alcohol use more completely.
The current analyses examined the effect of a polygenic risk score (PRS), generated using single nucleotide polymorphisms (SNPs) that map to gene clusters associated with the process of alcohol metabolism, on alcohol consumption in a population of emerging adult college students and also tested for differences in the effect of this score over time. Although there is no direct evidence that all SNPs in the score are involved in this process, for the purpose of this article we refer to this score as an alcohol metabolizing PRS. Riskenhancing SNPs contribute to a higher PRS, whereas protective SNPs contribute to a lower PRS. We hypothesized that this alcohol metabolizing score would demonstrate a positive association with alcohol consumption. In concordance with previous research, we also hypothesized that the PRS would demonstrate a stronger effect in later years, relative to students’ earlier years in college. Supplementary analyses include an assessment of the singular effect of ADH1B*2 on drinking, as a means to distinguish the well-replicated effect of this marker in European Americans from the polygenic effect of the broader alcohol metabolizing cluster.
Method
Data source and sample
Data are from a longitudinal survey of behavioral and emotional health in a sample of undergraduate college students attending an urban university in the mid-Atlantic United States (Dick et al., 2014b). The project was launched in 2011 and currently includes four cohorts of undergraduate students. One cohort of freshman subjects was enrolled each year from 2011 to 2014. Initial self-report data were collected in the fall semester of freshman year, with follow-up assessments at every subsequent spring semester. All participants were age 18 or older at induction. The sample is representative of the university’s student population in terms of gender and ethnicity. Self-report data were collected using an electronic survey programmed in the Research Electronic Data Capture (REDcap) software (Harris et al., 2009). This study was approved by the university’s Institutional Review Board. Participants were presented with consent documentation and indicated that they understood the potential risks and benefits of participating.
Genetic data were collected from consenting participants. Details regarding DNA collection and extraction (Dick et al., 2014b) and genotyping, quality control, and imputation (Webb et al., 2017) are available elsewhere. In brief, saliva samples were collected from each participant in Oragene collection tubes, and DNA was isolated according to the manufacturer’s instructions. Samples with DNA concentrations of at least 20 ng/ul in 1000 ml were retained for analysis. Genotyping was performed at Rutgers University Cell and DNA Repository (RUCDR) using the Affymetrix BioBank array. Pre-imputation quality control removed Off Target Variants identified in SNPolisher, SNPs missing more than 5% of genotypes, samples missing more than 2% of genotypes, and SNPs missing more than 2% of genotypes after sample filtering. Imputation was conducted using the 1000 Genomes phase 3 reference panel (Sudmant et al., 2015; The 1000 Genomes Project Consortium, 2015).
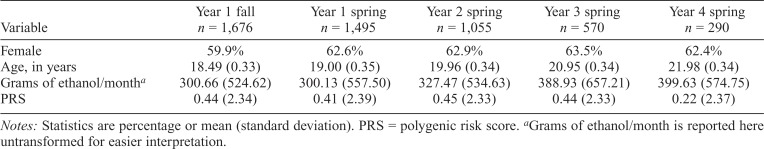
The current analyses focus on a subset (n = 2,214) of the broader, multiethnic study sample (N = 9,892). This smaller subset was selected based on age, availability of phenotypic and genotypic data, endorsement of lifetime alcohol use, and European American ancestry. These participants started the study in the fall of their freshman year when they were 18 to 20.48 years of age. Ancestry was determined by a genetic ancestry analysis detailed in Webb et al. (2017), which involved using SmartPCA (Eigenstrat) to match each DNA sample to the best fitting 1000 Genomes reference population (The 1000 Genomes Project Consortium, 2015) using Mahalanobis distance. The current analyses focus exclusively on subjects of European ancestry for compatibility with an independent European ancestry sample that was used to derive weights for PRS calculation (Edwards et al., 2015). For full explication of the current sample composition, see Table 1. Women accounted for more than half of the sample at all five time points. Participants also tended to consume more alcohol in later time points than in earlier time points. Because of the broader study’s cohort and longitudinal structure, data were available for three cohorts at Times 1, 2 and 3, two cohorts at Time 4, and one cohort at Time 5, reflected by varying sample size across time points. Subjects enrolled in Cohort 4 have not been genotyped and were excluded from analyses. Participation rates across time points are described here to supplement the table. First, participation was higher in Cohort 1 at each time point. Of the 1,676 participants present at Time 1, 73.7%–61.1% were present at Time 2, 46.2%–43.0% at Time 3, 40.3%–33.3% at Time 4, and 34.0% (Cohort 1 only) at Time 5.
Table 1.
Study sample composition and assessment time point
| Year 1 fall | Year 1 spring | Year 2 spring | Year 3 spring | Year 4 spring | |
| Variable | n = 1,676 | n = 1,495 | n = 1,055 | n = 570 | n = 290 |
| Female | 59.9% | 62.6% | 62.9% | 63.5% | 62.4% |
| Age, in years | 18.49 (0.33) | 19.00 (0.35) | 19.96 (0.34) | 20.95 (0.34) | 21.98 (0.34) |
| Grams of ethanol/montha | 300.66 (524.62) | 300.13 (557.50) | 327.47 (534.63) | 388.93 (657.21) | 399.63 (574.75) |
| PRS | 0.44 (2.34) | 0.41 (2.39) | 0.45 (2.33) | 0.44 (2.33) | 0.22 (2.37) |
Notes: Statistics are percentage or mean (standard deviation). PRS = polygenic risk score.
Grams of ethanol/month is reported here untransformed for easier interpretation.
Measures
Gender was measured with a single item, with response options 1 (male) and 0 (female).
Time was measured as a discrete variable, corresponding to the five study assessment time points. We adopted the following coding scheme: Year 1 Fall = 0; Year 1 Spring = 1; Year 2 Spring = 2; Year 3 Spring = 3; and Year 4 Spring = 4.
The polygenic risk score (PRS) included SNPs from 11 genes in the alcohol metabolizing systems identified in Hodgkinson et al. (2008) as a cohesive alcohol-metabolizing cluster: ALDH1, ALDH2, ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, ADH7, CAT, and CYP2E1. The PRS was calculated using dosage data for the current sample. Only markers with INFO > 0.5 were included. Gene base-pair boundaries were drawn from the UCSC Genome Browser, February 2009, GRCh37/hg19 assembly (The Genome Sequencing Consortium, 2001). Base pairs 50kb upstream and downstream of the transcriptional unit identified in the UCSC Genome Browser were selected, to include a potential 1,110,803 non-overlapping base pairs. Past work examining candidate gene polygenic scores have typically used basepair boundaries that stretch upstream and downstream of the transcriptional unit to capture effect markers that may lie in regulatory regions of the genes of interest (Cosgrove et al., 2017; Walter et al., 2013).
SNP weights were derived from a genome-wide association study (GWAS) of alcohol problems conducted in 4,304 participants from the Avon Longitudinal Study of Parents and Children (ALSPAC; Edwards et al., 2015). Initially, 3,093 SNPs were found in common between the current study and ALSPAC GWAS after indels and complementary SNPs were removed. Application of an inclusion threshold of p < .50 and pruning for linkage disequilibrium at r2 ≥ 0.20 with a sliding 500kb window identified 171 independent SNPs. PLINK’s clump procedure was used to preferentially select SNPs with a stronger association in the ALSPAC GWAS. Wray et al. (2014) recommend using a higher p-value threshold (e.g., p < .50) when weights are derived from a small GWAS (i.e., those that identify relatively few genome-wide significant markers) in order to maximize the scores’ predictive ability. See Supplemental Table A for a full explication of SNPs retained by these criteria. Using these remaining SNPs, the PRS was calculated in PLINK as a linear function of the number of effect alleles that an individual possessed, weighted by the product of the sign of the SNP effect and the negative logarithm (base 10) of the associated GWAS p-value—that is, -1 or 1*((-1)*LOG(p-value)). The distribution of the resulting PRS was approximately normal.
Grams of ethanol consumed per month is our outcome variable. It was calculated from separate measures of frequency and quantity of alcohol consumption. Frequency was measured with one item, “[During the past 30 days] how often do you have a drink containing alcohol?”, with response options never, monthly or less, 2 to 4 times a month, 2 to 3 times a week, and 4 or more times a week. Quantity was measured with one item, “[During the past 30 days] how many drinks containing alcohol do you have on a typical day when you are drinking?” with response options 1 or 2; 3 or 4; 5 or 6; 7, 8, or 9; and 10 or more. Together, these ranges were converted to a measure of grams of ethanol consumed per month using a procedure described in past works (Dawson, 2000; Salvatore et al., 2016). Briefly here, frequency categories were converted to the median of the described range to reflect drinking occasions per month—that is, never = 0, monthly or less = 0.5, 2–4 times a month = 3, 2–3 times a week = 10.7. The 4 or more times a week was set equal to 23.54, based on the average count of 4.28 weeks per month in all months of the year, for a range of 17.12–29.96 drinking days per month. Drinking quantity was quantified using a similar procedure—that is, 1 or 2 = 1.5, 3 or 4 = 3.5, 5 or 6 = 5.5, and 7, 8, or 9 = 8. The 10 or more category was set equal to 15 based on specification of 21 as the upper limit of drinks per drinking occasion. The product of recoded frequency and quantity was then multiplied by 14, which represents the grams of ethanol contained in a single, standard drink.
Data analysis
All data were structured in a one-row-per-measurement (“long”) format, and analyses were conducted in R (R Development Core Team, 2008). The “lme” function from the “nlme” package (Pinheiro et al., 2017) was used for linear mixed-effects modeling. All models were fit by maximum likelihood estimation and controlled for gender and cohort. Mixed-effects modeling allowed for the use of subjects with incomplete longitudinal data, provided that data were available for all relevant measures within a given time point. The total analytic sample size is n = 2,214, which includes subjects who had phenotypic data at later time points after endorsing lifetime alcohol use or who were missing relevant measures at some points. Three principal component measures (PCs) were used to control for within-ancestry group population stratification. We retained the first PC because it accounted for the largest proportion of within-ancestry genetic variability. We also regressed our outcome variable onto the set of 10 PCs, and those demonstrated to be significant (p < .05) were retained for the analyses. To allow the five time points to be modeled simultaneously, random intercepts were calculated for individuals, and random slopes were calculated for the effect of time within individuals. Grams of ethanol consumed per month was natural log transformed, ln (grams of ethanol + 1), to better satisfy model assumptions.To test our hypotheses about the relationships between time, the PRS, and alcohol consumption, we tested three iterations of our model. We first estimated the relationship between our main effects (time and PRS) and alcohol consumption. Next, an interaction between time and the PRS was added to the model. Finally, a series of control interactions were added to the model (Gender × PRS and Gender × Time), in line with principles discussed in previous reviews (Dick et al., 2015; Keller, 2014). Interactions were tested, as products of the two included variables, for departure from additivity. Coefficients are presented as unstandardized estimates and can be interpreted approximately as the percentage change in grams of ethanol consumed per month, per unit increase in the corresponding predictor measure. A plot of the Time × PRS interaction was constructed using the “effects” (Fox, 2003) and “ggplot2” (Wickham, 2009) packages in R. Pseudo-R2 values defined in Nakagawa and Schielzeth (2013) were calculated using the piecewiseSEM (Lefcheck, 2016) package in R. As a means to differentiate the wellreplicated effect of ADH1B*2 in European Americans from the polygenic effect of the broader alcohol metabolizing cluster, supplemental analyses with this single variant as a predictor—coded as (0) no minor alleles, (1) 1 or 2 minor alleles—were conducted to mirror the above three models. Last, additional analyses were conducted to test whether our findings were robust to the removal of the last two time points of data (i.e., Year 3 Spring and Year 4 Spring), which had relatively small sample sizes.
Results
Main effects of time and polygenic risk scores
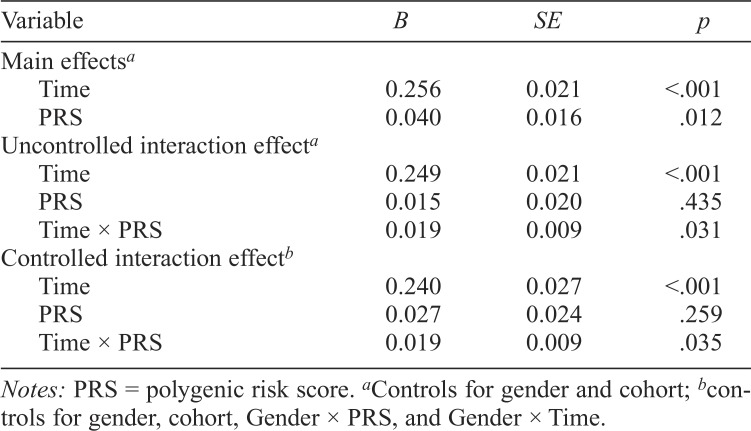
Table 2 presents coefficient estimates for fixed effects from each model. The fixed effect for time demonstrated a positive association with alcohol consumption, B = 0.256, SE = 0.021, t(2871) = 12.088, p < .001, showing that students drank more at later time points than at earlier time points. The fixed effect for the PRS was also positively associated with alcohol consumption, B = 0.040, SE = 0.016, t(2206) = 2.515, p = .012. Students with higher scores (i.e., a greater number of effect alleles, weighted by past GWAS p value) consumed more alcohol each month. The PRS accounted for a small proportion of the variance in monthly alcohol consumption (pseudo-R2 = .002), which conforms to past findings that suggest PRSs account for limited variability in complex phenotypes (Wray et al., 2014), particularly when in this analysis the PRS focused on a smaller subset of markers involved in alcohol metabolism.
Table 2.
Time and PRS main effects and interaction effect models
| Variable | B | SE | p |
| Main effectsa | |||
| Time | 0.256 | 0.021 | <.001 |
| PRS | 0.040 | 0.016 | .012 |
| Uncontrolled interaction effecta | |||
| Time | 0.249 | 0.021 | <.001 |
| PRS | 0.015 | 0.020 | .435 |
| Time × PRS | 0.019 | 0.009 | .031 |
| Controlled interaction effectb | |||
| Time | 0.240 | 0.027 | <.001 |
| PRS | 0.027 | 0.024 | .259 |
| Time × PRS | 0.019 | 0.009 | .035 |
Notes: PRS = polygenic risk score.
Controls for gender and cohort
controls for gender, cohort, Gender × PRS, and Gender × Time.
Interaction effect between time and polygenic risk scores
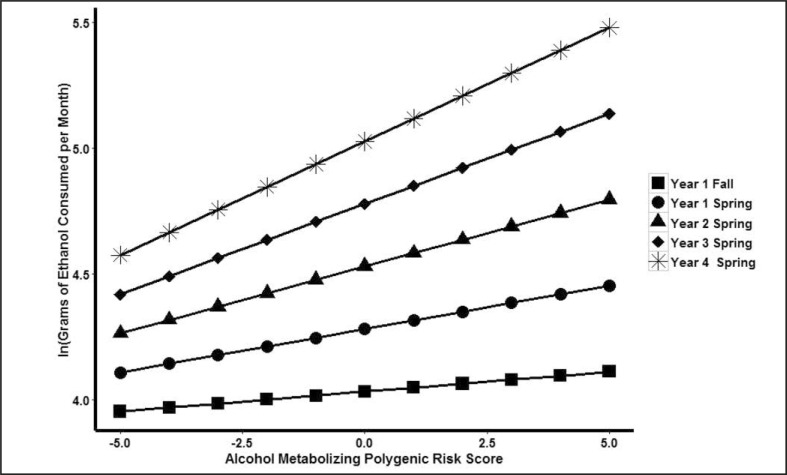
The interaction term testing moderation of the effect of the PRS by time reached statistical significance in both the uncontrolled, B = 0.019, SE = 0.009, t(2870) = 2.162, p = .031, and controlled, B = 0.019, SE = 0.009, t(2869) = 2.110, p = .035, interaction models. Further investigation of the interaction showed that the PRS exerted greater influence on drinking behaviors in later time points than in earlier time points (Figure 1). Similar to the PRS main effect, this interaction term accounted for only a small proportion of the variance in alcohol consumption in both the uncontrolled interaction (pseudo-R2 = .0006) and the controlled interaction models (pseudo-R2 = .0005). The Gender × PRS, B = -0.028, SE = 0.033, t(2205) = -0.865, p = .387, and Gender × Time, B = 0.023, SE = 0.043, t(2869) = 0.548, p = .584, control interactions were not statistically significant.
Figure 1.
Plot of time moderating the effect of an alcohol metabolizing polygenic risk score (PRS) on monthly alcohol consumption
Supplemental analyses
The fixed effect for ADH1B*2 was not significantly associated with alcohol consumption in the main effects model, B = 0.097, SE = 0.133, t(2206) = 0.731, p = .465. The Time × ADH1B*2 interaction term similarly did not reach statistical significance in either the uncontrolled, B = -0.044, SE = 0.072, t(2870) = -0.608, p = .543, or the controlled interaction models, B = -0.044, SE = 0.072, t(2869) = -0.610, p = .542. This finding may relate to relatively low minor allele frequency for ADH1B*2 in the current sample (minor allele frequency = 0.1196), which may reduce power to detect these effects (Hong & Park, 2012).
Results from models with the last two time points removed from analysis mirrored results from previous models using all available times. The fixed effect for time, B = 0.206, SE = 0.031, t(2059) = 6.702, p < .001, and for the PRS, B = 0.036, SE = 0.017, t(2158) = 2.154, p = .031, demonstrated positive associations with alcohol consumption in the main effects model. The interaction term for PRS × Time reached statistical significance in both the uncontrolled, B = 0.030, SE = 0.013, t(2058) = 2.295, p = .022, and the controlled, B = 0.030, SE = 0.013, t(2057) = 2.274, p = .023, interaction models.
Discussion
In this study we created a PRS encompassing genetic variation across genes involved in alcohol metabolism, and we tested for the effect of this score on alcohol consumption among college students and whether it varied across time. In support of our hypotheses, we found that the alcohol metabolizing PRS demonstrated a positive association with alcohol consumption, and the association of the PRS with drinking was stronger in later years relative to students’ earlier college years. Consistent with past work (Auerbach & Collins, 2006; Lanza & Collins, 2006; White et al., 2005), we also observed increased alcohol use in later years of college. Our findings were similar to those of previous studies showing that the nature and magnitude of genetic influences vary across development (Dick et al., 2014a; Irons et al., 2012).
Changing environmental contexts during students’ 4 years of college may help to explain the increased association of the alcohol metabolizing system on alcohol consumption over time. Access to alcohol is a prerequisite for genetic predispositions toward alcohol consumption related to alcohol metabolism to manifest. Therefore, declining social controls over alcohol use during college may enable genetic effects to become more salient in predicting alcohol use. For example, as individuals reach the legal drinking age in the later years of college and are able to obtain alcohol without legal restriction, it may allow greater opportunity to express genetic predispositions. Evidence suggests that trends in emerging adult drinking have varied historically by the legal drinking age; in times when emerging adults had legal access to alcohol before age 21, alcohol use was more common among these individuals (Gruenewald, 2011). It may stand to reason that the effectiveness of the minimum legal drinking age in controlling emerging adult alcohol use is parallel to its potential capacity to limit the influence of genetic effects on alcohol use.
Although legal access to alcohol is a clear and intuitive benchmark in the loosening of formal social control, other environmental factors may also influence the availability of alcohol. Access to alcohol through informal channels may begin to explain this stable increase in alcohol use. As individuals age, the age of their typical peer groups is likely to increase as well. It may follow that, when individuals reach the legal drinking age, their own formal access to alcohol translates to increased informal access to alcohol within their peer group, exposing more underage students to consistent sources of alcohol. Similar effects have been observed in sibling pairs, where older siblings of legal drinking age may often facilitate alcohol use in younger siblings (Samek et al., 2015). A range of other environmental factors may play a complementary role in expanding access to alcohol as emerging adults age, including greater independence and increasingly limited parental influence (Bountress et al., 2017).
The current analysis has several methodological strengths. The first is this study’s focus on the alcohol metabolizing system of genes. The well-substantiated role of the alcohol metabolizing system in alcohol use outcomes marks these genes as a reasonable candidate for an exploration of variability in effect over time. Supplemental analyses suggest that the effect of alcohol metabolizing genes on alcohol use is not solely driven by the ADH1B*2 variant. Second, the Spit for Science project, which enrolls cohorts of incoming freshmen and follows them across the entirety of their college years, provides an opportunity to test for longitudinal gene–environment interaction effects among college students. This may be crucial to accurately assessing the nature of genetic effects on alcohol use during the college years.
Results should be interpreted with respect to a number of methodological limitations. Foremost is that the available sample size declines at later time points because of the cohort structure and longitudinal study design. However, the association between the PRS and alcohol consumption and the increase in the PRS effect on drinking over time was replicated in models that excluded the last two time points, suggesting that our findings are robust to this potential sample size issue. In addition, we acknowledge that our grams of ethanol consumed variable, which was calculated from two ordinal measures of monthly quantity and frequency, represents an approximation of subjects’ monthly alcohol consumption.
Further examination of the polygenic nature of the alcohol metabolizing system by time in ancestry population groups other than individuals of European ancestry is warranted. Known population stratification in allele frequencies of alcohol metabolizing genes may produce substantially different results for scores calculated for other population groups. For example, the most pronounced effects of alcohol metabolizing genes on alcohol consumption are typically observed in association with ALDH2* and in East Asian populations (Goedde et al., 1992). Evidence also suggests that European American college students tend to drink more alcohol overall (Cacciola & Nevid, 2014) and demonstrate starker increases in drinking during this developmental period (Chen & Jacobson, 2013), suggesting that variability in drinking patterns over time by ethnicity may be an important consideration in generalizing these findings.
Despite reaching statistical significance, the main effect for the PRS and the PRS by time interaction term accounted for only a limited proportion of the variability in alcohol consumption (0.2% and 0.05%, respectively). Twin studies suggest that genetic effects specific to alcohol risk account for only a small portion of the heritability of alcohol dependence (Kendler et al., 2007), with much of the predisposition to alcohol consumption and problems coming through a broader predisposition to externalizing behavior. It should also be noted that this PRS was calculated to tag regions upstream and downstream of the transcriptional unit of the targeted genes and also used a relatively high p value threshold (p < .5) for inclusion. Some SNPs included in the PRS may not be directly involved in the metabolism of alcohol and may not contribute substantially to alcohol use.
In conclusion, we find that a PRS indexing risk for alcohol problems and generated from variants within genes involved in alcohol metabolism is associated with alcohol consumption in a large sample of college students of European American ancestry. The association of the PRS with alcohol use becomes stronger across time. This finding parallels previous studies (Dick et al., 2014a; Irons et al., 2012), indicating that genetic effects on consumption, across a variety of biological systems, become more strongly associated with consumption and problems across emerging adulthood. We hypothesize that this increase in the importance of genetic factors is attributable to released social control and increased access to alcohol, which allows greater opportunity for genetic predispositions to affect patterns of use. Future analyses could examine other environmental conditions that may moderate the effect of polygenic risk on consumption in emerging adulthood and whether these vary among population groups.
Acknowledgments
The authors thank the Virginia Commonwealth University students for making this study a success, as well as the many faculty members, students, and staff members who contributed to the design and implementation of the project.
Footnotes
This work is supported by National Institute on Alcohol Abuse and Alcoholism (NIAAA) Grant K01AA021145. Spit for Science: The Virginia Commonwealth University Student Survey has been supported by Virginia Commonwealth University; NIAAA Grants P20AA107828, R37AA011408, K02AA018755, K01AA021399, and P50AA022537; and National Center for Research Resources and National Institutes of Health Roadmap for Medical Research Grant UL1RR031990.
References
- American Psychiatric Association. 4th ed. Washington, DC: Author; 1994. Diagnostic and statistical manual of mental disorders. [Google Scholar]
- Auerbach K. J., Collins L. M. A multidimensional developmental model of alcohol use during emerging adulthood. Journal of Studies on Alcohol. 2006;67:917–925. doi: 10.15288/jsa.2006.67.917. doi:10.15288/jsa.2006.67.917. [DOI] [PubMed] [Google Scholar]
- Bierut L. J., Goate A. M., Breslau N., Johnson E. O., Bertelsen S., Fox L., Edenberg H. J. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Molecular Psychiatry. 2012;17:445–450. doi: 10.1038/mp.2011.124. doi:10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bountress K., Chassin L., Lemery-Chalfant K. Parent and peer influences on emerging adult substance use disorder: A genetically informed study. Development and Psychopathology. 2017;29:121–142. doi: 10.1017/S095457941500125X. doi:10.1017/S095457941500125X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacciola E. E. T., Nevid J. S. Alcohol consumption in relation to residence status and ethnicity in college students. Psychology of Addictive Behaviors. 2014;28:1278–1283. doi: 10.1037/a0038362. doi:10.1037/a0038362. [DOI] [PubMed] [Google Scholar]
- Chen P., Jacobson K. C. Longitudinal relationships between college education and patterns of heavy drinking: A comparison between Caucasians and African-Americans. Journal of Adolescent Health. 2013;53:356–362. doi: 10.1016/j.jadohealth.2013.04.003. doi:10.1016/j.jadohealth.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove D., Harold D., Mothersill O., Anney R., Hill M. J., Bray N. J., Donohoe G. the Wellcome Trust Case Control Consortium. MiR-137-derived polygenic risk: Effects on cognitive performance in patients with schizophrenia and controls. Translational Psychiatry. 2017;7:e1012. doi: 10.1038/tp.2016.286. doi:10.1038/tp.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson D. A. US low-risk drinking guidelines: An examination of four alternatives. Alcoholism: Clinical and Experimental Research. 2000;24:1820–1829. doi:10.1111/j.1530-0277.2000.tb01986.x. [PubMed] [Google Scholar]
- Dick D. M., Agrawal A., Keller M. C., Adkins A., Aliev F., Monroe S., Sher K. J. Candidate gene–environment interaction research: Reflections and recommendations. Perspectives on Psychological Science. 2015;10:37–59. doi: 10.1177/1745691614556682. doi:10.1177/1745691614556682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick D. M., Cho S. B., Latendresse S. J., Aliev F., Nurnberger J. I., Jr., Edenberg H. J., Kuperman S. Genetic influences on alcohol use across stages of development: GABRA2 and longitudinal trajectories of drunkenness from adolescence to young adulthood. Addiction Biology. 2014a;19:1055–1064. doi: 10.1111/adb.12066. doi:10.1111/adb.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick D. M., Nasim A., Edwards A. C., Salvatore J. E., Cho S. B., Adkins A., Kendler K. S. Spit for Science: Launching a longitudinal study of genetic and environmental influences on substance use and emotional health at a large US university. Frontiers in Genetics. 2014b;5:47. doi: 10.3389/fgene.2014.00047. doi:10.3389/fgene.2014.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg H. J., Xuei X., Chen H.-J., Tian H., Wetherill L. F., Dick D. M., Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: A comprehensive analysis. Human Molecular Genetics. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. doi:10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edwards A. C., Aliev F., Wolen A. R., Salvatore J. E., Gardner C. O., McMahon G., Kendler K. S. Genomic influences on alcohol problems in a population-based sample of young adults. Addiction. 2015;110:461–470. doi: 10.1111/add.12822. doi:10.1111/add.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J. Effect displays in R for generalised linear models. Journal of Statistical Software. 2003;8:1–27. doi:10.18637/jss.v008.i15. [Google Scholar]
- Gelernter J., Kranzler H. R., Sherva R., Almasy L., Koesterer R., Smith A. H., Farrer L. A. Genome-wide association study of alcohol dependence: Significant findings in African- and EuropeanAmericans including novel risk loci. Molecular Psychiatry. 2014;19:41–49. doi: 10.1038/mp.2013.145. doi:10.1038/mp.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godette D. C., Headen S., Ford C. L. Windows of opportunity: Fundamental concepts for understanding alcohol-related disparities experienced by young Blacks in the United States. Prevention Science. 2006;7:377–387. doi: 10.1007/s11121-006-0044-3. doi:10.1007/s11121-006-0044-3. [DOI] [PubMed] [Google Scholar]
- Goedde H. W., Agarwal D. P., Fritze G., Meier-Tackmann D., Singh S., Beckmann G., Czeizel C. Distribution of ADH2 and ALDH2 genotypes in different populations. Human Genetics. 1992;88:344–346. doi: 10.1007/BF00197271. doi:10.1007/BF00197271. [DOI] [PubMed] [Google Scholar]
- Gruenewald P. J. Regulating availability: How access to alcohol affects drinking and problems in youth and adults. Alcohol Research & Health. 2011;34:248–256. [PMC free article] [PubMed] [Google Scholar]
- Guindalini C., Scivoletto S., Ferreira R. G., Breen G., Zilberman M., Peluso M. A., Zatz M. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. American Journal of Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. doi:10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Harris P. A., Taylor R., Thielke R., Payne J., Gonzalez N., Conde J. G. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. doi:10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson C. A., Yuan Q., Xu K., Shen P. H., Heinz E., Lobos E. A., Goldman D. Addictions biology: Haplotype-based analysis for 130 candidate genes on a single array. Alcohol and Alcoholism. 2008;43:505–515. doi: 10.1093/alcalc/agn032. doi:10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. P., Park J. W. Sample size and statistical power calculation in genetic association studies. Genomics & Informatics. 2012;10:117–122. doi: 10.5808/GI.2012.10.2.117. doi:10.5808/GI.2012.10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley T. D., Edenberg H. J. Genes encoding enzymes involved in ethanol metabolism. Alcohol Research: Current Reviews. 2012;34:339–344. doi: 10.35946/arcr.v34.3.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons D. E., Iacono W. G., Oetting W. S., McGue M. Developmental trajectory and environmental moderation of the effect of ALDH2 polymorphism on alcohol use. Alcoholism: Clinical and Experimental Research. 2012;36:1882–1891. doi: 10.1111/j.1530-0277.2012.01809.x. doi:10.1111/j.1530-0277.2012.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M. C. Gene × environment interaction studies have not properly controlled for potential confounders: The problem and the (simple) solution. Biological Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. doi:10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K. S., Myers J., Prescott C. A. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Archives of General Psychiatry. 2007;64:1313–1320. doi: 10.1001/archpsyc.64.11.1313. doi:10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- Klein H. Changes in college students’ use and abuse of alcohol, and in their attitudes toward drinking over the course of their college years. Journal of Youth and Adolescence. 1994;23:251–269. doi:10.1007/BF01537448. [Google Scholar]
- Lanza S. T., Collins L. M. A mixture model of discontinuous development in heavy drinking from ages 18 to 30: The role of college enrollment. Journal of Studies on Alcohol. 2006;67:552–561. doi: 10.15288/jsa.2006.67.552. doi:10.15288/jsa.2006.67.552. [DOI] [PubMed] [Google Scholar]
- Lefcheck J. S. piecewiseSEM: Piecewise structural equation modelling in R for ecology, evolution, and systematics. Methods in Ecology and Evolution. 2016;7:573–579. doi:10.1111/2041-210X.12512. [Google Scholar]
- Luo X., Kranzler H. R., Zuo L., Wang S., Schork N. J., Gelernter J. Diplotype trend regression analysis of the ADH gene cluster and the ALDH2 gene: Multiple significant associations with alcohol dependence. American Journal of Human Genetics. 2006;78:973–987. doi: 10.1086/504113. doi:10.1086/504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Kranzler H. R., Zuo L., Yang B. Z., Lappalainen J., Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: Results from family controlled and population-structured association studies. Pharmacogenetics and Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. doi:10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods in Ecology and Evolution. 2013;4:133–142. doi:10.1111/j.2041-210x.2012.00261.x. [Google Scholar]
- Pinheiro J., Bates D., DebRoy S., Sarkar D. R Core Team. nlme: Linear and nonlinear mixed effects models. 2017 Retrieved from https://CRAN.R-project.org/package=nlme.
- R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. 2008 Retrieved from http://www.r-project.org.
- Salvatore J. E., Thomas N. S., Cho S. B., Adkins A., Kendler K. S., Dick D. M. The role of romantic relationship status in pathways of risk for emerging adult alcohol use. Psychology of Addictive Behaviors. 2016;30:335–344. doi: 10.1037/adb0000145. doi:10.1037/adb0000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samek D. R., McGue M., Keyes M., Iacono W. G. Sibling facilitation mediates the association between older and younger sibling alcohol use in late adolescence. Journal of Adolescent Research. 2015;25:638–651. doi: 10.1111/jora.12154. doi:10.1111/jora.12154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulenberg J. E., Bryant A. L., O’Malley P. M. Taking hold of some kind of life: How developmental tasks relate to trajectories of well-being during the transition to adulthood. Development and Psychopathology. 2004;16:1119–1140. doi: 10.1017/s0954579404040167. doi:10.1017/S0954579404040167. [DOI] [PubMed] [Google Scholar]
- Sherva R., Rice J. P., Neuman R. J., Rochberg N., Saccone N. L., Bierut L. J. Associations and interactions between SNPs in the alcohol metabolizing genes and alcoholism phenotypes in European Americans. Alcoholism: Clinical and Experimental Research. 2009;33:848–857. doi: 10.1111/j.1530-0277.2009.00904.x. doi:10.1111/j.1530-0277.2009.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske W. S. Alcohol use disorders among US college students and their non-college-attending peers. Archives of General Psychiatry. 2005;62:321–327. doi: 10.1001/archpsyc.62.3.321. doi:10.1001/archpsyc.62.3.321. [DOI] [PubMed] [Google Scholar]
- Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A., Huddleston J., Korbel J. O. the 1000 Genomes Project Consortium. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. doi:10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. doi:10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst B., Neale M. C., Kendler K. S. The heritability of alcohol use disorders: A meta-analysis of twin and adoption studies. Psychological Medicine. 2015;45:1061–1072. doi: 10.1017/S0033291714002165. doi:10.1017/S0033291714002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall T. L., Luczak S. E., Hiller-Sturmhöfel S. Biology, genetics, and environment: Underlying factors influencing alcohol metabolism. Alcohol Research: Current Reviews. 2016;38:59–68. doi: 10.35946/arcr.v38.1.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S., Glymour M. M., Koenen K., Liang L., Tchetgen Tchetgen E. J., Cornelis M., Kubzansky L. D. Performance of polygenic scores for predicting phobic anxiety. PLoS ONE. 2013;8(11):e80326. doi: 10.1371/journal.pone.0080326. doi:10.1371/journal.pone.0080326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A., Lind P. A., Kalmijn J., Feiler H. S., Smith T. L., Schuckit M. A., Wilhelmsen K. The investigation into CYP2E1 in relation to the level of response to alcohol through a combination of linkage and association analysis. Alcoholism: Clinical and Experimental Research. 2011;35:10–18. doi: 10.1111/j.1530-0277.2010.01317.x. doi:10.1111/j.1530-0277.2010.01317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B. T., Edwards A. C., Wolen A. R., Salvatore J. E., Aliev F., Riley B. P., Kendler K. S. Molecular genetic influences on normative and problematic alcohol use in a population-based sample of college students. Frontiers in Genetics. 2017;8:30. doi: 10.3389/fgene.2017.00030. doi:10.3389/fgene.2017.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White H. R., Labouvie E. W., Papadaratsakis V. Changes in substance use during the transition to adulthood: A comparison of college students and their noncollege age peers. Journal of Drug Issues. 2005;35:281–306. doi:10.1177/002204260503500204. [Google Scholar]
- Wickham H. New York, NY: Springer-Verlag; 2009. ggplot2: Elegant graphics for data analysis. [Google Scholar]
- Wray N. R., Lee S. H., Mehta D., Vinkhuyzen A. A., Dudbridge F., Middeldorp C. M. Research review: Polygenic methods and their application to psychiatric traits. Journal of Child Psychology and Psychiatry, and Allied Disciplines. 2014;55:1068–1087. doi: 10.1111/jcpp.12295. doi:10.1111/jcpp.12295. [DOI] [PubMed] [Google Scholar]