Abstract
AIM
To reveal the insight mechanism of liver metastasis in uveal melanoma, we investigated cell functions of microRNA-21 in three different uveal melanoma cell lines and analyze the relationship of target gene p53 and its downstream targets.
METHODS
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to detect microRNA-21 expression in normal uveal tissue and uveal melanoma cell lines. Lenti-virus expression system was used to construct OCM-1, MuM-2B and M619 cell line with stable overexpression and inhibition of microRNA-21. In vitro cell function tests such as cell proliferation, cell apoptosis, cell circle and abilities of migration and invasion were examined by MTT, BrdU assay, flow cytometry, transwell assay and Matrigel invasion assay respectively. The target gene was predicted by bioinformatics and confirmed by using a dual luciferase reporter assay. The expression of p53 and its suspected downstream targets LIM and SH3 protein 1 (LASP1) and glutathione S transferase pi (GST-Pi) were determined by qRT-PCR in mRNA level and Western blotting analysis in protein level. Finally, the effect of microRNA-21 in a xenograft tumor model was assessed in four-week-old BALB/c nude mice.
RESULTS
Compared to normal uveal melanoma, expressions of microRNA-21 were significantly higher in uveal melanoma cell lines. Overexpression of microRNA-21 promoted proliferation, migration, and invasion of OCM-1, M619 and MuM-2B cells, while inhibition of microRNA-21 reveal opposite effects. Wild type p53 was identified as a target gene of microRNA-21-3p, and proved by dual luciferase reporter assay. Up-regulated microRNA-21 inhibited the expression of wild type p53 gene, and the increased expression of LASP1 in mRNA level and protein level, while down-regulated microRNA-21 presented opposite way. However, GST-pi showed the potential pattern as expected, but relative mRNA level showed no statistically significant difference in OCM-1 cells. Furthermore, the mRNA expression of GST-pi was decreased in microRNA-21 overexpressing MuM-2B, and increased in M619 cells with inhibition of microRNA-21. In vivo, inhibition of microRNA-21 reduced tumor growth with statistically significant difference.
CONCLUSION
These findings provide novel insight into molecular etiology of microRNA-21 in uveal melanoma cell lines, and suggest that microRNA-21 might be a potential candidate for the diagnosis and prognostic factor of human uveal melanoma.
Keywords: uveal melanoma, microRNA-21, p53, LIM and SH3 protein 1, Glutathione S Transferase pi
INTRODUCTION
Uveal melanoma, the most common primary intraocular malignancies in adults, is usually found in the people who are in their 40's to 50's. The morbidity of the uveal melanoma is around seven millionths and most people are diagnosed as unilateral disease[1]. The uveal melanoma which accounts for 5%-6% of primary melanoma ranks at the second place next to skin melanoma[2]. There are miscellaneous treatments for uveal melanoma, however, these therapies are mainly focused on the local treatment of the eyeball, which does not improve the status of metastatic patients. The uveal melanoma has one prominent feature that no matter what successful local treatment of the tumor in the eye, about 50% of the patients still die from the distant metastasis of the tumor. After the metastasis of the patient, most of the patients die within a few months. The survival period is only 2 to 7mo. Only 13% of the patients survive for more than 1y, and the average survival time is less than 6mo[3].
Uveal melanoma not only seriously threatens the patient's vision, but also seriously intimidates the patient's life. Although in the past nearly 30y, the diagnosis and treatment of uveal melanoma has had significant progress, unfortunately, it not fundamentally solved the key problems of tumor metastasis, the 5-year survival rate of patients with no obvious improvement. The crux of the problem is that for uveal melanoma spreading to the liver, has yet to fully understand, cannot be effective intervention; as early as 1978, Zimmerman et al[4] put forward “The metastasis of uveal melanoma may be an early event, and the metastasis may occur before the tumor is ready to be removed.” Therefore, metastasis has become a bottleneck restricting the therapeutic effect of uveal melanoma. The underlying cause of the disease is that there is no understanding of the cause and mechanism of metastasis, therefore early intervention and treatment of uveal melanoma cannot be performed from pathogenesis.
In our previous study[5], the microRNA profiling of uveal melanoma performed by using Agilent microRNA array demonstrated that expressions of many microRNAs were up-regulated and down-regulated significantly compared with normal uveal tissue. MicroRNA-21, a well-known oncogene, found overexpress in multiple cytology types of uveal melanoma tissue. MicroRNA-21, first found highly expressing in malignant glioma[6], blocks expression of apoptosis-associating key genes. MicroRNA-21 regulates and participates in multiple steps of tumor metastasis[7], involving in the tumorigenesis of liver cancer, gastrointestinal malignancy, breast cancer, cutaneous melanoma, and other tumors[8]–[13], while mechanisms of microRNA-21 in uveal melanoma were haven't been researched.
In this report, the effect of overexpression and inhibition of microRNA-21 on uveal melanoma cell line behavior was evaluated. The relationship of p53, a target of microRNA-21, and its downstream proteins was investigated. Finally, in vivo study revealed that a miR-21 inhibition could restrain melanoma tumor growth.
MATERIALS AND METHODS
Cell Culture Condition, Uveal Tissues, and RNA Extraction
Human uveal melanoma cell lines OCM-1, M619 and MuM-2B were obtained commercially, and were cultured in Roswell Park Memorial Institute 1640 (RPMI 1640) medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were maintained at 37°C and 5% CO2 in an incubator with 95% humidity. The cell culture medium was replaced every second day, and cells were passaged at 85%-90% confluence. Normal uveal samples, obtained from the Beijing Tongren Eye Bank (Beijing, China), were separated from the eye within 18h after death. Total RNA containing small RNA was extracted from cultured OCM-1, M619, MuM-2B cells and normal uveal tissue using Trizol reagent (Invitrogen, USA). MicroRNA was obtained, according to the manufacturer's instructions, using the mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA). The purity and concentration of RNA were determined from OD260/280 readings using spectrophotometer (NanoDrop 2000/2000c, ThermoFisher).
Nude Mice
BALB/c nude mice weighing 20 to 22 g, age between 4 to 6wk were obtained from the Beijing Huafukang Bioscience Co. Inc. The animals were maintained in a specific pathogen-free environment. The animals were fed with an autoclaved laboratory rodent diet. The mice were maintained on a daily 12-h light-12-h dark cycle. Our study was approved by the ethics committee of Beijing Tongren Hospital of capital medical University. Animal studies were conducted in accordance with the University's Institutional Animal Care and Use Committee Guidelines.
Quantitative Reverse Transcription Polymerase Chain Reaction for microRNA-21 Validation
A cDNA synthesis was carried out using miScript Reverse Transcription Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. A quantitative PCR was performed using a miScript SYBR-Green PCR Kit (Qiagen). Expression analysis was performed in triplicate for each sample. The small nuclear RNA U6 was used as the normalization control. The miRNAs expression level was quantified using the ABI 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). In each sample, we calculated a Delta Ct (target-reference, ΔCt), which is equal to the difference between threshold cycles for microRNA-21 (target) and the threshold cycle for U6 RNA (reference). The fold-change between cell samples and a normal control for microRNA-21 was calculated with the 2−ΔΔCt method. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was repeated in triplicate for each sample, an average 2−ΔΔCt value was calculated for each sample relative to the normal control for expression of microRNA-21.
Lentiviral Vector Construction and Infection
The human hsa-miR-21 precursor sequences and a sequence-specific hsa-miR-21-3p-inhibition synthesized chemically via technical support from Genechem (Shanghai) were cloned and inserted into the EcoR I/EcoR I sites and Age I/EcoR I sites in GV217 and GV280 vector (Genechem, Shanghai), respectively, and identified by restriction endonuclease digestion and nucleotide sequencing. Lentivirus packaging and infection were performed according to standard protocols as recommended by the manufacturer. Uveal melanoma cell lines were infected with 1×107 lentivirus transducing units in the presence of 5 µg/mL polybrene (Sigma Aldrich, St. Louis, Missouri, USA). For stably infected cells, puromycin (2 µg/mL, Invitrogen) were added to select for 48h after 72h been transfected. The expression level of miR-21-3p was identified by quantitative qRT-PCR. Experiments were divided into four groups for each uveal melanoma cell line as empty vector infected cell line as negative control group (NC-up) and lenti-virus infected uveal melanoma cells as hsa-miR-21 group (micro up) as well as hsa-miR-21-3p-inhibition group (micro down) and its negative control group (NC-down).
Cell Proliferation Assay
Total of 2000 stable infected uveal melanoma cells per well were plated in 96-well plates cultured in normal conditions. Cell proliferation was assessed every 24h using MTT assay (Sigma, USA) to determine relative cell growth. Twenty microliters of 5 mg/mL MTT was added to the media for 4h incubation at 37°C and 5% CO2. Following removal of the culture medium, the remaining crystals were dissolved in 150 µL DMSO (Sigma) and the absorbance at 490 nm was measured. BrdU assay (Roche, Sigma-Aldrich, China) was used to measure the DNA synthesis of every groups on the first day and the fourth day according to test principle as recommended by the manufacturer. The assay is based on BrdU incorporation into the DNA of proliferating cells.
Cell Apoptosis and Cell Cycle Assay
Cell apoptosis was detected using AnnexinV-APC apoptosis Detection Kit (eBioscience, USA) on BD FACSCalibur Flow Cytometer System. Results were calculated by the percentage of apoptotic cells in all cells counted. Cells were washed with phosphate buffer solution (PBS) and fixed with 70% ethanol at 4°C. After overnight fixation, the cells were washed with PBS again and stained with proper amount of propidium iodide (PI) (Sigma, USA) and RNase A (MBI Fermentas), in which 40×PI (2 mg/mL):100×RNase (10 mg/mL):1×PBS=25:10:1000 for 30min. Cell cycle features were analyzed using BD FACSCalibur Flow Cytometer System (BD BioSciences). Data analysis was performed using CellQuest Pro software (BD BioSciences). The experiments were repeated at least thrice.
Transwell Migration and Invasion Assay
Each uveal melanoma cells were washed twice with serum-free RPMI-1640 medium and re-suspended in the same medium. OCM-1 (1×105), M619 (80 000) and MuM-2B (1×105) cells were seeded each well into the upper chambers of transwell culture plates, each with an 8-µm pore membrane insert (Corning, Shanghai, China). RPMI-1640 medium supplemented with 10% FBS was placed in the lower chambers as a chemo-attractant. After incubation for 24h, cells that had penetrated through to the lower surface of the membrane were fixed with 4% paraformaldehyde for 20min, stained with crystal violet for 20min at ambient temperature, photographed, and counted under inverted microscope (ShangHai Caikon Optical Instrument Co., Ltd., China) at ×200 and magnification in nine randomly chosen fields.
The cell invasion assay was similar to the migration assay except that the transwell chambers were coated with matrigel solution (40 µL per chamber; matrigel:serum-free medium ratio 1:10). OCM-1 (1×105), M619 (80 000) and MuM-2B (80 000) cells were seeded into the upper chambers of the transwells and RPMI-1640 medium with 20% FBS (600 µL) was added to the lower chambers. After 20h incubation, the cells that had penetrated the matrigel and moved to the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with crystal violet. Cells adhering to the upper-surface of the membrane were removed with a cotton swab. The cells attached to the lower surface were counted and photographed under an inverted microscope (ShangHai Caikon Optical Instrument Co., Ltd., China) at ×200 and magnification in nine randomly chosen fields.
Vector Construction and Dual Luciferase Reporter Assay
Constructing the microRNA-21 expression vector (pre-hsa-miR-21), two fragments encompassing the mature microRNA-21 sequence and its 5′- and 3′-flanking regions were synthesized chemically and then cloned into the XhoI and EcoRI sites in GV262 (Genechem, Shanghai). The full-length wild-type 3′ untranslated region (UTR) of p53 and mutant 3′ UTR of p53 containing predicted miR-21-3p target sites were amplified by PCR from human monocyte total RNA and then cloned into the XhoI and SacI sites of GV126 vector (Genechem, Shanghai). All plasmids were confirmed by DNA sequencing. These plasmid vectors were cotransfected using Lipofectamine 2000 (Invitrogen) respectively into 293T cells. Luciferase was measured 48h after transfection using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer's instructions. The firefly luciferase activity was then normalized to β-galactosidase activity. Experiments were repeated three times independently.
Quantitative Reverse Transcription Polymerase Chain Reaction and Western Blotting
A cDNA synthesis was carried out using a PrimeScript II 1st strand cDNA Synthesis Kit (TaKaRa) according to the manufacturer's instructions. A quantitative RT-PCR was performed on the Mx3005P ™ Detection System (Stratagene, California, USA) in a volume of 20 µL containing 10 µL GoTaqR qPCR Master Mix (Promega) and 0.2 µm each primer of the tested target genes. The reaction profile consisted of 40 cycles of 95°C for 2min, 55°C for 30s, and 72°C for 30s. The primers used in RT-PCR are as follows: p53, 5′-CCT CCT CAG CAT CTT ATC C-3′ and 5′-ACA AAC ACG CAC CTC AAA-3′; LIM and SH3 protein 1 (LASP1), 5′-AAA CCT TCG CCT CAA GCA AC-3′ and 5′-CTG CCA CTA CGC TGA AAC CTT-3′; glutathione S transferase pi (GST-pi), 5′-GTC CAA TAC CAT CCT GCG TCA C-3′ and 5′-CAT CCT TGC CCG CCT CAT AG-3′; GAPDH, 5′-TGA CTT CAA CAG CGA CAC CCA-3′ and 5′-CAC CCT GTT GCT GTA GCC AAA-3′. Real-time PCR was performed according to the manufacturer's instructions. The relative gene expression was calculated via a 2−ΔΔCt method. The housekeeping GAPDH was used as the normalization control.
The spectrophotometer determined the total protein extracted from each groups of uveal melanoma cells by homogenizing and extracting in RIPA lysis buffer and protease inhibitor cocktail (Roche), then 20 µg total protein sample was electrophoresed on polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membranes. The transferred nitrocellulose blot was blocked with 5% fat-free milk powder in TBS-T (pH 7.6; 20 mmol/L Tris-HCl, 100 mmol/L NaCl, and 0.01% Tween 20) at room temperature for 2h. After being washed with TBS-T solution three times, the membrane was incubated with 1:1000 anti-beta actin monoclonal antibody for human β-actin (Origene, Beijing, China), 1:500 rabbit anti-p53 monoclonal antibody for human target protein (Abcam, England), 1:100 rabbit GST-pi antibody - C-terminal polyclonal antibody for human target protein (Abcam), or 1:400 rabbit anti-LASP1 polyclonal antibody for human target protein (Abcam) in TBS-T at 4°C overnight. Then, the membrane was washed three times and hybridized with horseradish peroxidase-conjugated secondary antibody (dilution factor 1:2000) (Santa Cruz Biotechnology, Texas, USA) at room temperature for 2h. At last, detection was performed using the reagents provided in the enhanced chemiluminescence plus kit. The experiment was performed in triplicate.
Xenograft Model in Nude Mice to Exam Antitumor Effects of microRNA-21-3p-inhibition
Four-week-old BALB/c nude mice were subcutaneously injected in the right side of axillary with 4×106 OCM-1 cells stably transfected by microRNA-21-3p inhibition vector (micro down) or its negative control vector (NC-down). The mice were also injected with an empty vector in the left flank. The tumor volumes and body weights of the animals were measured and recorded every 3-5d for every week. The tumor volume was evaluated by in vivo bioluminescence imaging on the last day. All mice were sacrificed according to a standard procedure after 25d, and the tumors were harvested and weighed. The volume of each tumor was calculated according to the formula V=π/6×L×W×W where L and W are the length and width of the tumor, respectively, measured by a sliding caliper.
Statistical Analysis
All analyses were carried out with SPSS statistical software (version 19.0 for Windows), with P<0.05 being considered significant for a two-tailed test. All experiments data were represented as means±standard deviation. Statistical significance was tested using one-way analysis of variance. P<0.05 was considered statistically significant.
RESULTS
Original Expression Level of microRNA-21 in Human Uveal Melanoma Cell Lines
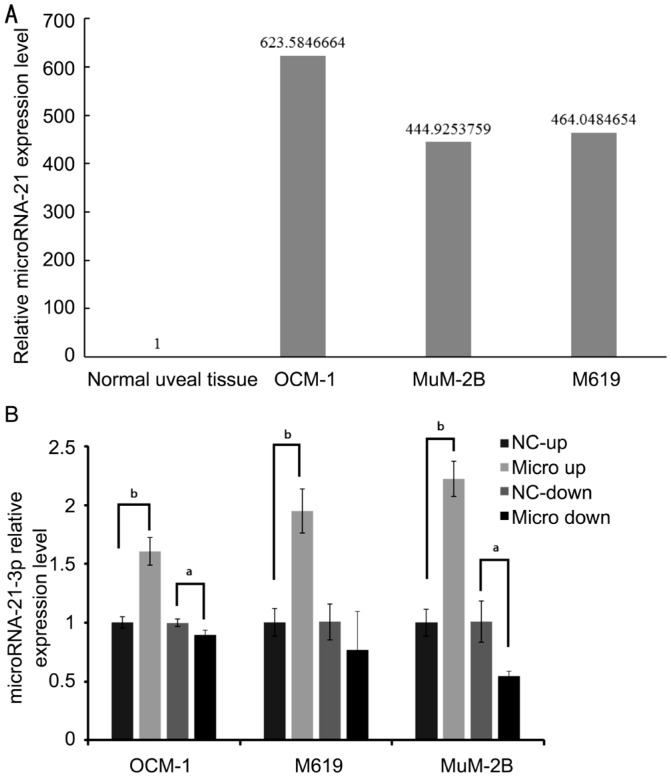
To investigate whether hsa-miR-21 was up-regulated in human uveal melanoma cell lines, we performed the qRT-PCR analysis. As shown in Figure 1A, our data confirmed that hsa-microRNA-21 was strongly up-regulated in human uveal melanoma cell lines compared with normal uveal tissues. Uveal melanoma cell lines showed remarkable expression of hsa-miR-21, while the relative expression level of hsa-miR-21 was 1 in human normal uveal tissues.
Figure 1. microRNA-21 is significantly up-regulated in uveal melanoma cell lines and the expression level can be regulated after being infected by lenti-virus infection system.
A: qRT-PCR assay of microRNA-21 expression in uveal melanoma cell lines; B: Uveal melanoma cell lines with stable microRNA-21 overexpression and inhibition. Compared with negative control of each group, microRNA-21-3p express level were significantly different. aP<0.05, bP<0.01 compared with NC.
Establishing Uveal Melanoma Cell Lines with Stable microRNA-21 Overexpression and Inhibition by Lenti-virus Infection System
Lenti-virus expression system was used to establish cell lines with stable overexpression and inhibition of miR-21. Three commercially available uveal melanoma cell lines, OCM-1, M619 and MuM-2B, were transfected with lenti-virus system including P-miR-21 vector (as micro up group), hsa-miR-21-3p-inhibition vector (as micro down group), and their empty vectors as negative control (as NC-up and NC-down group), to test the potential function of miR-21 in each uveal melanoma cells. The expression levels of miR-21 in transfected OCM-1, M619 and MuM-2B cells were determined by the qRT-PCR assay (Figure 1B).
Cell Proliferation, Invasion and Metastasis of Infected Uveal Melanoma Cell Lines in Vitro
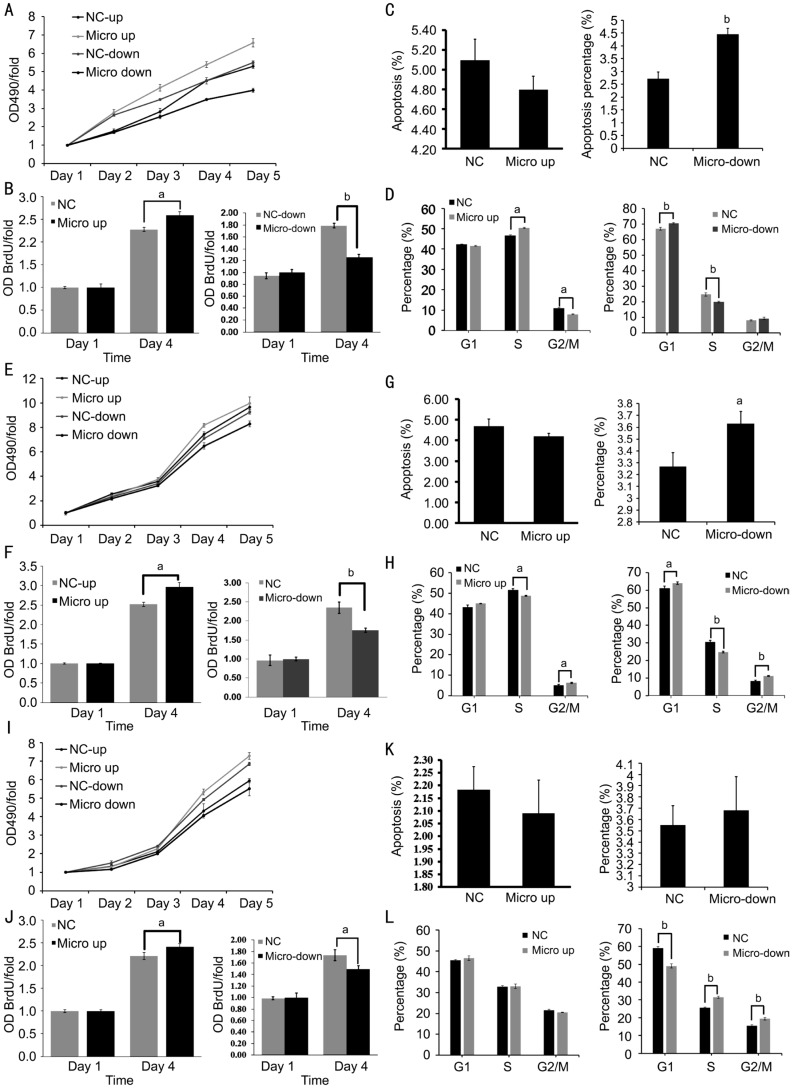
Expression level of microRNA-21 significantly (P<0.01) effected the cell proliferation in every cell lines on the fourth and fifth day in MTT assay result (Figure 2A, 2E, 2I). According to MTT test results above, the BrdU test was further conducted on the first day and the fourth day, and the results showed that the proliferation values of overexpression and inhibition of miR-21 in each uveal melanoma cell lines on the fourth day were statistically different from their negative control groups (Figure 2B, 2F, 2J). Annexin V detection and APC staining showed that miR-21 overexpression decreased OCM-1, M619 and MUM-2B cell apoptosis, but didn't show significantly different compared with NC-down groups. Inhibition of miR-21 increased OCM-1, M619 and MuM-2B cell apoptosis, and show significantly different compared with NC-down groups in OCM-1 (P=0.001) and M619 (P=0.028; Figure 2C, 2G and 2K), probably by blocking OCM-1 cell at G1 phase, MuM-2B cell at S and G2 phase and M619 cell at G2 phase (Figure 2D, 2H, 2L).
Figure 2. microRNA-21 overexpression and inhibition influence uveal melanoma cell function.
A: MTT assays of infected OCM-1 cell line; B: BrdU assay confirmed the result of MTT assay and showed significantly different; C: Apoptosis assay of lenti-virus system infected OCM-1 cell; D: PI staining showed that OCM-1 cells were blocked at G1 phase by inhibition of microRNA-21; E: MTT assays of infected M619 cell; F: BrdU assay confirmed the result of MTT assay; G: Apoptosis assay of infected M619 cell; H: PI staining showed that M619 cells were blocked at G2 phase by inhibition of microRNA-21; I: MTT assays of infected MuM-2B cell; J: BrdU assay confirmed the result of MTT assay; K: Apoptosis assay of infected MuM-2B cell; L: PI staining showed that MuM-2B cells were blocked at S and G2 phase by inhibition of microRNA-21. aP<0.05, bP<0.01 versus NC group.
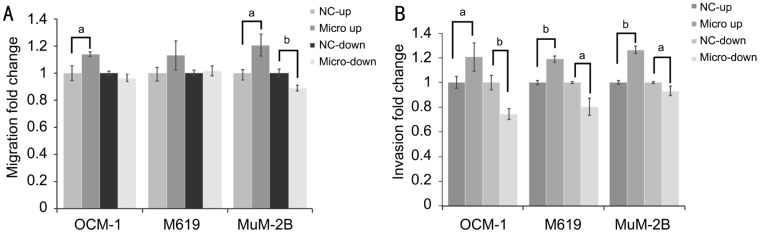
In cell migration assay (Figure 3A), we observed that cells' ability to metastasize was significantly strengthened (P<0.05) by stable overexpression of microRNA-21 in MuM-2B (P=0.037) and OCM-1 (P=0.035) cell lines, and weaken by stable inhibition of microRNA-21. However, metastatic rate of M619 cell was improved but not showing statistical significant different. In Matrigel invasion assay (Figure 3B), results showed that cell invasive abilities of OCM-1, M619 and MuM-2B, these three different uveal melanoma cell lines with different cytological properties, were all significantly strengthened and weaken by stable overexpression and inhibition of microRNA-21. These results suggested that microRNA-21 may be important in the progression of uveal melanoma through enhancing cell invasion and migration that can make highly invasive tumor cells as MuM-2B still been improved by microRNA-21 overexpression.
Figure 3. microRNA-21 overexpression evoked cell migrant and invasive abilities in uveal melanoma cells.
A: Metastatic rate quantification of uveal melanoma cells that cross through transwell chamber in OCM-1, MuM-2B and M619; B: Invasive rate quantification of uveal melanoma cells that invaded through matrigel-coated membrane in OCM-1, MuM-2B and M619. aP<0.05, bP<0.01 versus NC group.
Wild p53, the Direct Target of hsa-microRNA-21-3p
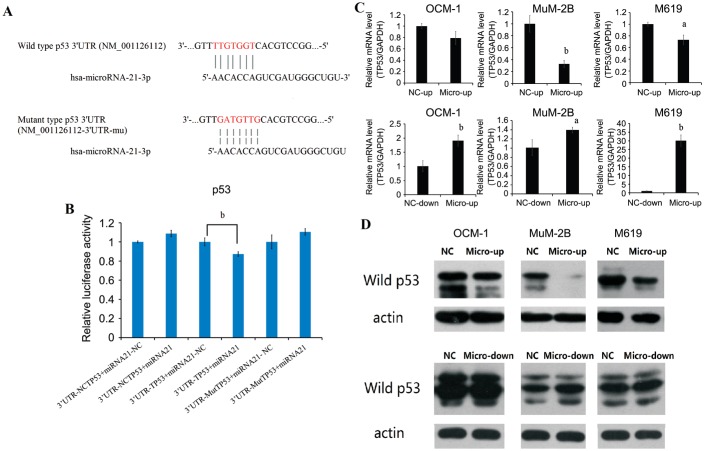
To explore the mechanisms by which miR-21 promote tumor cell proliferation and invasion, we used several computational methods to help identify miR-21 targets in humans. We found that miR-21 has binded in the 3′UTR of p53 (Figure 4A). In order to determine whether p53 were a direct target of miRNA-21, we carried out luciferase reporter assays. Dual luciferase assay system was used to test the interaction between miRNA-21 and wild type p53 mRNAs. The luciferase activities of wild type p53 3′UTR are decreased compared with its mutant type by cotransfected with P-microRNA-21 (Figure 4B). As expected, miR-21 inhibited luciferase activity downstream of the wild-type 3′UTR of p53. These data demonstrated that microRNA-21 might inhibit wild type p53 expression by targeting 3′UTR of p53. The effect of miR-21 overexpression and inhibition on mRNA and protein levels of p53 in uveal melanoma cell lines were been evaluated. MicroRNA-21 overexpression did cause degradation of wild p53 mRNA. In contrast, inhibition of microRNA-21 up regulated wild p53 mRNA with significantly different (Figure 4C). Protein levels of wild P53 were determined by Western blot in lenti-virus infected uveal melanoma cell lines. The result showed that a clear reduction in the level of endogenous wild p53 protein was observed in micro up group of OCM-1, MuM2B and M619 cell lines, especially in MuM-2B. In contrast, micro down groups reveal darker strips compared with NC-down groups (Figure 4D).
Figure 4. Dual luciferase reporter assay confirms that wild p53 is a target of hsa-microRNA-21-3p.
The affect presents on mRNA level and protein level. A: The putative target genes and mutant complementary sequences of the p53 mRNA 3′UTR were predicted by microRNA-21 sequences; B: Results of dual luciferase reporter assay; C: Real-time reverse transcription PCR (RT-PCR) data for p53 mRNA in human uveal melanoma cell line transfected with overexpression and inhibition of miR-21. Data were normalized to GAPDH mRNA; D: Western blot of P53 in human uveal melanoma cell line transfected with overexpression and inhibition of miR-21. aP<0.05, bP<0.01 versus NC group.
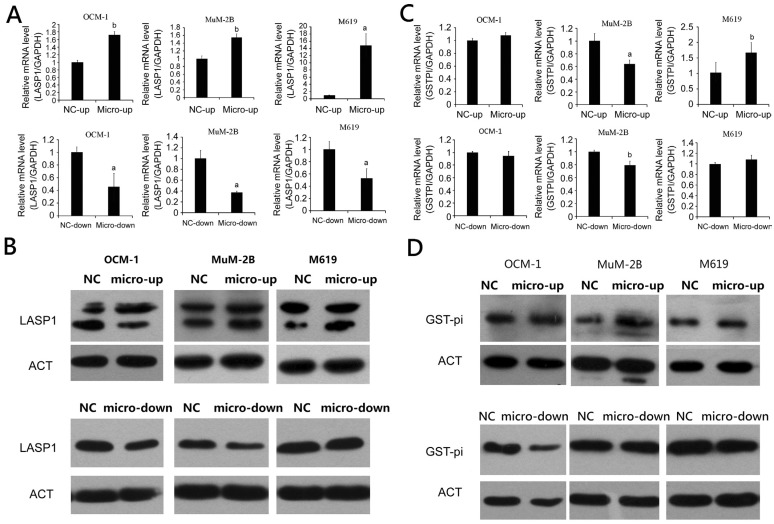
Downstream Effective Proteins of p53
In our previous study[14], protein level of LASP1 and GST-pi had been found overexpression in uveal melanoma tissue sample there for the expectation about LASP1 and GST-pi as downstream proteins of wild p53 to influence the metastasis of uveal melanoma were formed. To determine this idea, effects of microRNA-21 expression on mRNA and protein levels of LASP1 and GST-pi in uveal melanoma cell lines OCM-1, M619 and MuM-2B were been evaluated using qRT-PCR and Western blot on each group of uveal melanoma. Results of RT-PCR showed that LASP1 were up-regulated in micro up groups and down-regulated in micro down groups (Figure 5A) as expecting, and showed significantly statistical dif-ference. Results of Western blot (Figure 5B) supported the mRNA regulating as well. However, GST-pi didn't show the same pattern as we expected. The mRNA express level of GST-pi in OCM-1 cells with overexpression and inhibition of microRNA-21 were up-regulated and down-regulated respectively, but showed no significant difference. GST-pi was decreased in microRNA-21 overexpressing MuM-2B, and increased in M619 cells with inhibition of microRNA-21. This result did not match the Western blot results (Figure 5C, 5D).
Figure 5. mRNA and protein expression of LASP1 and GST-pi in human uveal melanoma cell line transfected with overexpression and inhibition of miR-21.
aP<0.05, bP<0.01 versus NC group.
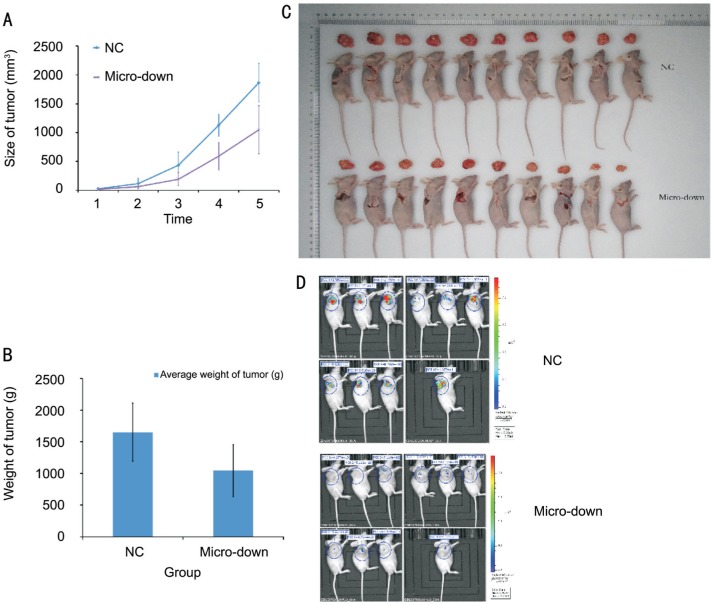
microRNA-21 Inhibition in BALB/c Nude Mice
Tumor mass of micro down group of OCM-1 cell line were grow slower than NC-down group, and the weight of tumors were lighter as well (Figure 6).
Figure 6. miRNA-21-3p inhibition decreased OCM-1 tumor growth in BALB/c nude mice.
A: OCM-1 tumor mass grow slower in micro-down group than in negative control (NC-down); B: Tumor weight of group micro-down was significantly lighted than NC-down group (P=0.00583); C: Tumor-dissected mice and the dissected tumor; D: The tumor volume evaluated by in vivo bioluminescence imaging showed that total radiant efficiency of group micro-down were significantly less than NC-down group (P=0.02861) on the last day.
DISCUSSION
Choosing OCM-1, a spindle cell line with low invasiveness, M619, an in situ epithelial cell line with high invasiveness, and MuM-2B, an invasive epithelial cell lines with high liver metastasis in the experiment, we studied the variations of the cellular functions in the uveal melanoma cell line under a situation with overexpression and inhibition of microRNA-21. Emerging evidences suggest that miRNA-21 promoted proliferation, migration, and invasion of OCM-1, M619, and MuM-2B cells in vitro by targeting p53. Expression of p53 gene would be inhibited when the expression of microRNA-21 increased, and the downstream protein which were LASP-1 and GST-pi would be highly expression. On the other hand, down-regulation of microRNA-21 results in opposite consequence. Furthermore, inhibition of microRNA-21-3p decreased tumor growth in vivo.
MicroRNA-21 were found over-expressing in various solid tumor diseases such as lung cancers, breast cancers, glioblastoma, stomach cancers, pancreatic cancers, pancreatic neuroendocrine tumors, liver cancers, bile duct cancers, colon cancers and prostate cancers[15]. Through this study, we learned that microRNA-21 promoted the proliferation and metastases of uveal melanoma cell lines, and functions of the cells were consistent with that in the scientific literatures. In glioma cells, microRNA-21 can play its oncogenic role by acting on sophisticated tumor suppressing signal pathways. This theory was confirmed by various studies indicated that tropomyosin 1 (TPM1)[16], programmed cell death factor 4 (PDCD4)[17], phosphatase and tensin homolog deleted on chromosome ten (PTEN) and p53 signal path[18] could be all participating. Using online target gene predicting softwares to find out that p53 in the target gene of microRNA-21-3p, we verified the binding site of p53 and microRNA-21-3p by the dual-luciferase reporter gene detection method. qRT-PCR result shows that mRNA level of p53 in the uveal melanoma cell lines with up-regulated microRNA-21 was down-regulated. This is consistent with the report[19] that microRNA-21 can negatively regulate p53 in tumor cells, demonstrating that microRNA-21 can also be used to inhibit tumor apoptosis and promote proliferation and metastasis by down-regulating p53 expression in uveal melanoma cell lines. The modulating mechanisms might be that microRNA-21 performed directly on the 3′UTR of p53 managing the stabilities of p53 to achieve the relevant regulations; furthermore, microRNA-21 regulated p53 by adjusting the upstream genes of p53. p53 is involved in constituting its numerous downstream genes as a promoter which can all be found a conservative binding site on p53[20]. Through these promoters, p53 regulates transcription of a large amount of downstream genes, including microRNAs. Furthermore, P53 can also be used as a binding protein of RNA, which can indirectly regulate microRNAs by influencing the recruitment and binding process of microRNAs for mRNA.
In our previous study[14], comparative proteomics were designed to exam the pre-stage of metastases in progress of uveal melanoma. Comparing surgical excised uveal melanoma tissues with normal uveal tissue, for the first time we discovered that GST-pi overexpressed in uveal melanoma tissues, and LASP1 only expressed in uveal melanoma tissues not normal uveal tissues. Both proteins are targets of wild-type p53[21]–[22]. GST-pi and LASP1 were associated with tumor invasions and metastases; furthermore, they might be connected with liver metastases. Simultaneously, the expressions of GST-pi were up-regulated in proteomic analyses, which were both found in the uveal melanoma metastatic cell lines and primary cell lines[23].
LASP1 as an adhesion protein, was found up-regulated in miscellaneous malignancies reported in many studies. It was initially identified from a cDNA library of breast cancer metastases[24]. LASP1, as a dynamic adhesion spot protein[25], combined directly with the carboxy terminal domain of chemokine receptor CXCR and became a pre-migration modulator that managed malignant tumor cell migration[26]. According to transwell assays and scratching experiments, LASP1 could enhance cellular invasive and migratory potential. Immunofluorescence technique revealed that LASP1 might interfere cytoskeleton remodeling processes such as actin to alter cell motile potential leading to tumor metastases[27]. Comparing LASP1 expressions in the cytoplasm and nucleus of the hepatocellular carcinoma with normal liver cells, results showed that the express level of LASP1 in hepatocellular carcinoma was higher than adjacent non-neoplastic tissues. Furthermore, LASP1 expression related to hepatitis B surface antigen (HBsAg) and serum alpha-fetoprotein (AFP) in patients with hepatocellular carcinoma. Highly expressions of LASP1 in cytoplasm should take respond to deteriorating clinical prognoses. Multivariable analysis demonstrated that the intracellular LASP1 express level was an independent prognostic factor to patients' survival rate. LASP1 could encourage cell proliferation and migration, which made cancer cells develop into an aggressive phenotype[28]. Wang et al[29] discovered LASP1 as a transcriptional target of wild type p53, which restrained LASP1 expression. Missense of mutant p53-r175h[29]–[30] in the key region of the DNA binding domain destroying the p53-mediated LASP1 expression inhibition revealed that p53 might affect tumor metastases by interfering the mechanism of LASP1.
GST-pi is a phase II metabolic enzyme that is important in the detoxification of a wide range of electrophiles through oxidative metabolism to protect tumor cells from cytotoxic drugs[31]–[32]. Wang et al[33] found that GST-pi is up-regulated in the metastatic and invasive potential of gastric cancer cells by transforming growth factor-β1 (TGF-β1), which may be involved in TGF-β1-induced enhanced gastric cancer transfer potential. Investigating glutathione S-transferase isoforms within colorectal cancer tissue, normal liver tissues and liver metastasis tumor tissue from the patients with colorectal cancer, Mulder et al[21] learned GST-pi was found to be higher expressed in all liver metastasis tumor tissue than normal liver tissues. GST-pi reveals only in tumor cells and, except for the biliary epithelium and hepatocytes. It indicated colorectal cancer liver metastasis is related to GST-pi expression and activation. GST-pi expression is regulated by the activator protein 1 (AP-1) transcription factor activated by the stress-activated JNK pathway and is involved in the signaling pathway of apoptosis[34]. However, GST-pi interacts with activated JNK1 or JNK2 activates transcription factor 2 (ATF2), which directly inhibits the phosphorylation of ATF2 by activating JNK by binding to ATF2 or JNK and ATF2 complexes, thereby inhibiting apoptosis and causing tumor growth and metastasis[35]. Studies had revealed that GST-pi closely related to the tumor invasion and metastases. Zhang et al[36] reported that GST-pi was found to be up-regulated in colorectal adenocarcinoma with positive expression of tumor suppressor gene p53 mutant protein, indicating that GST-pi expression may be related to p53. Jardim et al[37] also reported the same discoveries in breast cancer tissues and explained that the higher expressions of GST-pi the better resistance to chemotherapy drugs, which would be a judgmental marker of prognoses. Under the condition of cell damage stress, antioncoprotein P53[38] would play central role in maintaining genomic stability by inhibiting the expression of GST-pi which were closely associated with antitumor drug resistance and metastasis.
In this study, western blotting was used to detect microRNA-21 overexpression and inhibition in uveal melanoma cell lines, OCM-1, M619 and MuM2B. The results showed that the protein level of LASP1 and GST-pi were up-regulated in micro up groups and down-regulated in micro down groups. According to studies which were mentioned before, the up-regulated LASP1 and GST-pi could be the reason for proliferation and metastasis of tumor. This idea was verified by the results of detection of LASP1 and GST-pi mRNA with qRT-PCR in levius infected uveal melanoma cell lines. LASP1 in microRNA-21 overexpression of uveal melanoma cell lines all showed a statistically significant increase, especially in the highly invasive M619 cells while down-regulation of microRNA-21 statistically significant decrease. However, the mRNA express level of GST-pi in OCM-1 cells with overexpression and inhibition of microRNA-21 were up-regulated and down-regulated respectively, but showed no significant difference. GST-pi was decreased in microRNA-21 overexpressing MuM-2B, and increased in M619 cells with inhibition of microRNA-21. This result did not match the Western blot results, which might indicate that partial molecular mechanism may be influenced by characteristics of cytotype differentiation which might be different form driving point as a target.
Recent studies show that due to the high level of bio-technology research and bioinformatics commonality, microRNAs in tumor differential expression research is full of possibilities. MicroRNA-21 has been extensively studied in colorectal cancer, breast cancer, glioblastoma of the human brain, and digestive system tumors. Because of its population, region and tumor staging, the clinical manifestations of microRNA-21 are clear. It has been reported that microRNA-21 can be identified as a specific biomarker in a given tumor. In a variety of brain tumors, breast cancer and various gastrointestinal tumors in patients with circulating blood microRNA-21 concentrations can be used as predictors of poor prognosis biomarkers[39]–[42]; in colorectal tumors, circulating microRNA-21 is a biomarker with moderate sensitivity and specificity[43]. Other studies point out that microRNA-21 can be used as a diagnostic tool to screen early cancers[44] and can be used as a translational medicine.
The present study demonstrated that in uveal melanoma the up-regulated microRNA-21 may inhibit the expression of wild type p53 gene, which could inactivate its inhibition on GST-pi and LASP1, and the increased expression of LASP1 might promote the metastasis and invasion of uveal melanoma cells subsequently in vitro. In vivo, inhibition of microRNA-21 reduced tumor growth. These findings provide novel insight into molecular etiology of microRNA-21 in uveal melanoma cell lines, and suggest that microRNA-21 might be a potential candidate for the diagnosis and prognostic factor of human uveal melanoma in future.
Acknowledgments
Foundations: Supported by the National Natural Science Foundation of China (No.81570891; No.81272981); Beijing Natural Science Foundation (No.7151003); Advanced Health Care Professionals Development Project of Beijing Municipal Health Bureau (No.2014-2-003); the Capital Health Research and Development of Special (No.2016-1-2051); Hospitals' Ascent Plan (No.DFL20150201).
Conflicts of Interest: Wang YC, None; Yang X, None; Wei WB, None; Xu XL, None.
REFERENCES
- 1.Sagus M, Bedikian AY. Uveal melanoma in the first 4 decades of life. South Med J. 2018;108(3):158–163. doi: 10.14423/SMJ.0000000000000248. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay C, Kim DW, Gombos DS, Oba J, Qin Y, Williams MD, Esmaeli B, Grimm EA, Wargo JA, Woodman SE, Patel SP. Uveal melanoma: from diagnosis to treatment and the science in between. Cancer. 2016;122(15):2299–2312. doi: 10.1002/cncr.29727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaborative Ocular Melanoma Study Group. Ten-year follow-up of fellow eyes of patients enrolled in Collaborative Ocular Melanoma Study randomized trials: COMS report no. 22. Ophthalmology. 2004;111(5):966–976. doi: 10.1016/j.ophtha.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman LE, McLean IW, Foster WD. Does enucleation of the eye containing a malignant melanoma prevent or accelerate the dissemination of tumour cells? Br J Ophthalmol. 1978;62(6):420–425. doi: 10.1136/bjo.62.6.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang CH, Wei WB. The miRNA expression profile of the uveal melanoma. Sci China Life Sci. 2011;54(4):351–358. doi: 10.1007/s11427-011-4149-y. [DOI] [PubMed] [Google Scholar]
- 6.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 7.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs-the micro steering wheel of tumor metastases. Nat Rev Cancer. 2009;9(4):293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 8.Lin PL, Wu DW, Huang CC, He TY, Chou MC, Sheu GT, Lee H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis. 2014;35(10):2175–2182. doi: 10.1093/carcin/bgu110. [DOI] [PubMed] [Google Scholar]
- 9.Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu S, Hamada T, Fukuyama T, Nakano R, Uchiyama A, Kawamoto M, Yamaguchi K, Hashimoto H. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol. 2012;25(1):112–121. doi: 10.1038/modpathol.2011.142. [DOI] [PubMed] [Google Scholar]
- 10.Eto K, Iwatsuki M, Watanabe M, Ida S, Ishimoto T, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, Yoshida N, Baba H. The microRNA-21/PTEN pathway regulates the sensitivity of HER2-positive gastric cancer cells to trastuzumab. Ann Surg Oncol. 2014;21(1):343–350. doi: 10.1245/s10434-013-3325-7. [DOI] [PubMed] [Google Scholar]
- 11.Zhao D, Tu Y, Wan L, Bu L, Huang T, Sun X, Wang K, Shen B. In vivo monitoring of angiogenesis inhibition via down-regulation of mir-21 in a VEGFR2-luc murine breast cancer model using bioluminescent imaging. PLoS One. 2013;8(8):e71472. doi: 10.1371/journal.pone.0071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, Sun Q, Yan F, Yan C, Li H, Ren X. Diagnostic and prognostic value of circulating miR-21 for cancer: a systematic review and meta-analysis. Gene. 2014;533(1):389–397. doi: 10.1016/j.gene.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Martin del Campo SE, Nicholas Latchana, Levine KM, Grignol VP, Fairchild ET, Jaime-Ramirez AC, Dao TV, Karpa VI, Carson M, Ganju A, Chan AN, Carson WE., 3rd MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PLoS One. 2015;10(1):e0115919. doi: 10.1371/journal.pone.0115919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang MX, Wei WB, Shi XH, Cui L. Cancer-related protein discovery of uveal melanoma with comparative proteomic analysis. Ophthalmology in China. 2008;17(4):230–235. [Google Scholar]
- 15.Fu X, Han Y, Wu Y, Zhu X, Lu X, Mao F, Wang X, He X, Zhao Y, Zhao Y. Prognostic role of microRNA-21 in various carcinomas: a systematic review and meta-analysis. Eur J Clin Invest. 2011;41(11):1245–1253. doi: 10.1111/j.1365-2362.2011.02535.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPMl) J Biol Chem. 2007;282(19):14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Xin S, He Z, Che X, Wang J, Xiao X, Chen J, Song X. MicroRNA-21(miR-21) post-transcriptionally downregulates tumor suppressor PDCD4 and promotes cell transformation, proliferation, and metastasis in renal cell carcinoma. Cell Physiol Biochem. 2014;33(6):1631–1642. doi: 10.1159/000362946. [DOI] [PubMed] [Google Scholar]
- 18.Costa PM, Cardoso AL, Nóbrega C, Pereira de Almeida LF, Bruce JN, Canoll P, Pedroso de Lima MC. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum Mol Genet. 2013;22(5):904–918. doi: 10.1093/hmg/dds496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin S, Cha HJ, Lee EM, Jung JH, Lee SJ, Park IC, Jin YW, An S. MicroRNAs are significantly influenced by p53 and radiation in HCT116 human colon carcinoma cells. Int J Oncol. 2009;34(6):1645–1652. doi: 10.3892/ijo_00000295. [DOI] [PubMed] [Google Scholar]
- 20.Xi Y, Shalgi R, Fodstad O, Pilpel Y, Ju J. Differentially regulated micro-RNAs and actively translated messenger RNA transcripts by tumor suppressor p53 in colon cancer. Clin Cancer Res. 2006;12(7 Pt 1):2014–2024. doi: 10.1158/1078-0432.CCR-05-1853. [DOI] [PubMed] [Google Scholar]
- 21.Mulder TP, Roelofs HM, Peters WH, Wagenmans MJ, Sier CF, Verspaget HW. Glutathione S-transferases in liver metastases of colorectal cancer. A comparison with normal liver and primary carcinomas. Carcinogenesis. 1994;15(10):2149–2153. doi: 10.1093/carcin/15.10.2149. [DOI] [PubMed] [Google Scholar]
- 22.Zhao L, Wang H, Liu C, Wang X, Wang S, Sun X, Li J, Deng Y, Jiang Y, Ding Y. Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 2010;59(9):1226–1235. doi: 10.1136/gut.2009.202739. [DOI] [PubMed] [Google Scholar]
- 23.Ramasamy P, Murphy CC, Clynes M, Horgan N, Moriarty P, Tiernan D, Beatty S, Kennedy S, Meleady P. Proteomics in uveal melanoma. Exp Eye Res. 2014;118:1–12. doi: 10.1016/j.exer.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Schreiber V, Masson R, Linares JL, Mattei MG, Tomasetto C, Rio MC. Chromosomal assignment and expression pattern of the murine Lasp-1 gene. Gene. 1998;207(2):171–175. doi: 10.1016/s0378-1119(97)00622-7. [DOI] [PubMed] [Google Scholar]
- 25.Lin YH, Park ZY, Lin D, Brahmbhatt AA, Rio MC, Yates JR, 3rd, Klemke RL. Regulation of cell migration and survival by focal adhesion targeting of Lasp-1. J Cell Biol. 2004;165(3):421–432. doi: 10.1083/jcb.200311045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raman D, Sai J, Neel NF, Chew CS, Richmond A. LIM and SH3 protein-1 modulates CXCR2-mediated cell migration. PLoS One. 2010;5(4):e10050. doi: 10.1371/journal.pone.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber V, Moog-Lutz C, Régnier CH, Chenard MP, Boeuf H, Vonesch JL, Tomasetto C, Rio MC. Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol Med. 1998;4(10):675–687. [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Li W, Jin X, Cui S, Zhao L. LIM and SH3 protein 1, a promoter of cell proliferation and migration, is a novel independent prognostic indicator in hepatocellular carcinoma. Eur J Cancer. 2013;49(4):974–983. doi: 10.1016/j.ejca.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 29.Wang B, Feng P, Xiao Z, Ren EC. LIM and SH3 protein 1 (LASP1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol. 2009;50(3):528–537. doi: 10.1016/j.jhep.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 30.Wang B, Xiao ZW, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci U S A. 2009;106(34):14373–14378. doi: 10.1073/pnas.0903284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awasthi S, Bajpai KK, Piper JT, Singhal SS, Ballatore A, Seifert WE, Jr, Awasthi YC, Ansari GA. Interactions of melphalan with glutathione and the role of glutathione S-transferase. Drug Metab Dispos. 1996;24(3):371–374. [PubMed] [Google Scholar]
- 32.Kulaksiz-Erkmen G, Dalmizrak O, Dincsoy-Tuna G, Dogan A, Ogus IH, Ozer N. Amitriptyline may have a supportive role in cancer treatment by inhibiting glutathione S-transferase pi (GST-π) and alpha (GST-α) J Enzyme Inhib Med Chem. 2013;28(1):131–136. doi: 10.3109/14756366.2011.639017. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Li J, Zhen C, Zhou J, Xiao D, Liu J, Liu Y, Jiang H, Chen C, Wen J. Enhanced invasive and metastatic potential induced by transforming growth factor-beta1 might be correlated with glutathione-S-transferase-pi, cofilin and heat shock protein 27 in SGC-7901 gastric cancer cells. Acta Biochim Biophy Sin (Shanghai) 2007;39(7):520–526. doi: 10.1111/j.1745-7270.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- 34.Tsai CW, Chen HW, Yang JJ, Sheen LY, Lii CK. Diallyl disulfide and diallyl trisulfide up-regulate the expression of the pi class of glutathione S-transferase via an AP-1-dependent pathway. J Agric Food Chem. 2007;55(3):1019–1126. doi: 10.1021/jf061874t. [DOI] [PubMed] [Google Scholar]
- 35.Thévenin AF, Zony CL, Bahnson BJ, Colman RF. GST pi modulates JNK activity through a direct interaction with JNK substrate, ATF2. Protein Sci. 2011;20(5):834–848. doi: 10.1002/pro.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Z, Deng X, Ren X, Zhang B, Chen X, Yang J, Ding H, Sui J, Song X. Expression of mutant p53 and of the multidrug resistant proteins P-glycoprotein and glutathione S-transferase-pi correlated in colorectal adenocarcinoma. Scand J Gastroenterol. 2010;45(7-8):925–934. doi: 10.3109/00365521003734117. [DOI] [PubMed] [Google Scholar]
- 37.Jardim BV, Moschetta MG, Gelaleti GB, Leonel C, Regiani VR, de Santi Neto D, Bordin-Junior NA, Perea SA, Zuccari DA. Glutathione transferase pi (GSTpi) expression in breast cancer: an immunohistochemical and molecular study. Acta Histochem. 2012;114(5):510–517. doi: 10.1016/j.acthis.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Song L, Gao M, Dong W, Hu M, Li J, Shi X, Hao Y, Li Y, Huang C. p85α mediates p53 K370 acetylation by p300 and regulates its promoter-specific transactivity in the cellular UVB response. Oncogene. 2011;30(11):1360–1371. doi: 10.1038/onc.2010.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He XY, Liao YD, Guo XQ, Wang R, Xiao ZY, Wang YG. Prognostic role of microRNA-21 expression in brain tumors: a Meta-analysis. Mol Neurobiol. 2016;53(3):1856–1861. doi: 10.1007/s12035-015-9140-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Zhang Y, Pan C, Ma F, Zhang S. Prediction of poor prognosis in breast cancer patients based on microRNA-21 expression: a meta-analysis. PLoS One. 2015;10(2):e0118647. doi: 10.1371/journal.pone.0118647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of MicroRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668–1674. doi: 10.12659/MSM.892096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye TT, Yang YL, Liu XY, Ji QQ, Pan YF, Xiang YQ. Prognostic value of circulating microRNA-21 in digestive system cancers: a meta-analysis. Int J Clin Exp Med. 2014;7(4):873–878. [PMC free article] [PubMed] [Google Scholar]
- 43.Xu F, Xu L, Wang M, An G, Feng G. The accuracy of circulating microRNA-21 in the diagnosis of colorectal cancer: a systematic review and meta-analysis. Colorectal Dis. 2015;17(5):O100–107. doi: 10.1111/codi.12917. [DOI] [PubMed] [Google Scholar]
- 44.Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol. 2013;19(34):5615–5621. doi: 10.3748/wjg.v19.i34.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]