Abstract
Phage PHB02 specifically infects Pasteurella multocida capsular serogroup A strains. In this study, we found that capsule deletion mutants were not lysed by PHB02, suggesting that the capsule of P. multocida serogroup A strains might be the primary receptor. Based on sequence analysis, a gene encoding a phage-associated putative depolymerase was identified. The corresponding recombinant depolymerase demonstrated specific activity against capsular serogroup A strains but did not strip capsule deletion mutants. In vivo experiments showed that PHB02 was retained at detectable levels in the liver, spleen, kidneys, lung, and blood, at 24 h post-administration in mice. Depolymerase plus serum significantly reduced the number of viable wild-type P. multocida strain HB03 cells (3.5–4.5 log decrease in colony-forming units). Moreover, treatment with phage or purified depolymerase resulted in significantly increased survival of mice infected with P. multocida HB03, and an absence of increase of eosinophils and basophils or other pathological changes when compared with the control group. These results show that phage PHB02 and its putative depolymerase represent a novel strategy for controlling P. multocida serogroup A strains.
Keywords: Bacteriophage, Pasteurella multocida, mutants, receptor, putative capsule depolymerase
Introduction
Gram-negative bacterium Pasteurella multocida is part of the normal respiratory microbiota of many animals. However, it is potentially pathogenic in domestic and agricultural animals (e.g., cats, dogs, cattle, pigs, and rabbits) (Ewers et al., 2006; García-Alvarez et al., 2015) and in some bird species (Wilkie et al., 2012). Importantly, human P. multocida infections are not common and are usually associated with the older adult or immunocompromised individuals (Baillot et al., 2011; Abreu et al., 2018). P. multocida is generally classified into five capsular serogroups (A, B, D, E, and F) (Carter, 1955) and 16 somatic lipopolysaccharide (LPS) serotypes (1–16) (Heddleston et al., 1972). Capsular serogroup A strains are most often associated with bovine hemorrhagic septicemia and avian cholera (Chung et al., 2001; Borrathybay et al., 2003). It is estimated that the cost associated with treating bovine hemorrhagic septicemia is greater than $500 million per year in North America (Miles, 2009). In addition, some birds, such as turkeys and waterfowl, are more susceptible to serious diseases caused by capsular serogroup A strains (Samuel et al., 2005; Wilkie et al., 2012). Serogroups B and E cause hemorrhagic septicemia in cattle and buffalo (Boyce and Adler, 2000), while serogroup D is responsible for atrophic rhinitis in pigs (Davies et al., 2003), and serogroup F is usually associated with poultry cholera (Jonas et al., 2001; Jaglic et al., 2011).
Polysaccharide capsules are produced by a wide range of bacteria, and they provide protection against the host immune system (Jann and Jann, 1987; Petruzzi et al., 2017). Moreover, the capsules of many pathogenic bacteria impair phagocytosis and reduce or inhibit complement-mediated killing (Boyce and Adler, 2000; Scholl and Merril, 2005). Some reports show that loss of the capsule in P. multocida is associated with reduction or loss of virulence (Sthitmatee et al., 2011). For example, mutant P. multocida strain P-1059 (serovar A:3) was completely attenuated in chickens (Sthitmatee et al., 2011).
In recent years, the emergence of multidrug-resistant bacteria has gained much attention. Previous studies have isolated P. multocida strains, showing resistance to chloramphenicol, enrofloxacin, lincomycin, norfloxacin, and doxycycline-HCl, from various animals, including chicken, ducks, turkeys, quails, and geese (Shivachandra et al., 2004; Sarangi et al., 2015; Maynou et al., 2017). Therefore, there is a need to identify and develop new therapeutic strategies against these multidrug-resistant P. multocida strains.
Phages have been isolated from all environments in which bacteria exist. Phages demonstrate high specificity and effectiveness in killing bacterial pathogens, especially multidrug-resistant bacteria (Bhetwal et al., 2017; Reindel and Fiore, 2017; Cha et al., 2018). There are a few reports on P. multocida phages, with only a few temperate phage genomes studied in depth (Campoy et al., 2006). However, our previous studies have shown that phage PHB02, isolated from wastewater, is a lytic phage specific for P. multocida serogroup A strains (Chen et al., 2018).
Recently, there have been several reports on the successful application of lytic phages in the treatment of clinical multidrug-resistant bacterial strains (Bhetwal et al., 2017; Reindel and Fiore, 2017; Cha et al., 2018). These phages inhibited or killed the bacterial pathogens but were harmless to the animal or human host (Brunel and Guery, 2017; Cheng et al., 2018). Furthermore, phage-derived proteins, such as lysate endolysin and lyase, have been used independently for pathogen control (Drulis-Kawa et al., 2015; Benešík et al., 2018; Ha et al., 2018). There is also increasing interest in using phage depolymerases as biocontrol agents (Majkowska-Skrobek et al., 2016; Guo et al., 2017; Hsieh et al., 2017). Phage depolymerases can specifically recognize and degrade capsular polysaccharides, extracellular polysaccharides, and O-antigen (Corbett and Roberts, 2008). Importantly, Rakhuba et al. (2010) reported that the bacterial capsule can act as a primary receptor for phage, which often possesses tail fibers or tail spikes with capsule depolymerization activity. Previously, Scholl and Merril (2005) reported that phage K1F encodes a depolymerase that allows the phage to recognize and degrade the polysaccharide capsule of Escherichia coli. In addition, phages may encode several different capsule depolymerases to degrade the capsules of multiple bacterial species (Hsieh et al., 2017). Moreover, bacterial capsules can function as primary receptors for different types of phages (Hsieh et al., 2017). As such, unencapsulated strains are not infected by phages and are not degraded by phage-derived depolymerases.
Previously, we described the biological characteristics and genomic properties of phage PHB02 (Chen et al., 2018). PHB02 is specific for P. multocida serogroup A strains and can form plaque-surrounding halos on agar plates. This typical halo ring is associated with phage-derived depolymerases (Latka et al., 2017). Depolymerases (or phages) have proven to be effective for the prevention or eradication of biofilms (Gutiérrez et al., 2016; Guo et al., 2017), and treatment with recombinant depolymerase derived from phage NTUH-K2044-K1-1 resulted in significantly increased survival of mice infected with capsular type K1 Klebsiella pneumoniae (Lin et al., 2014). Based on these results, we hypothesized that depolymerases may have considerable potential as biocontrol agents.
In this study, we isolated putative depolymerase Dep-ORF8 from P. multocida phage PHB02 and demonstrated that neither the phage nor the isolated putative depolymerase had activity against a capsule-minus P. multocida HB03 deletion mutant strain. We also investigated the effects of phage PHB02 or putative depolymerase Dep-ORF8 treatment in mice infected with P. multocida.
Materials and Methods
Bacterial Strains and Animals
Pasteurella multocida strains were grown at 37°C in tryptic soy broth (TSB; Becton, Dickinson and Company, Glencoe, MD, United States) or tryptic soy agar (TSA; Becton, Dickinson and Company, Glencoe, MD, United States) medium supplemented with 10% (vol/vol) sterile, defibrinated sheep blood (Jiulongbio, Zhengzhou, China). Soft agar was prepared by the addition of 0.75% (wt/vol) agar to TSB medium. All P. multocida strains used in this study are listed in Supplementary Table S1. Escherichia coli BL21 (DE3) was cultured in Luria-Bertani broth (LB; 10 g tryptone, 5 g yeast extract, 10 g NaCl per liter) at 37°C. If required, antibiotics were added to the medium (kanamycin, 50 μg/ml; ampicillin 100 μg/ml).
A capsule-deficient hexA-knockout mutant was generated from wild-type P. multocida strain HB03 by the method described by Tabatabaei et al. (2002). Briefly, forward and reverse primers were designed from the hexA sequence of P. multocida HB03 (GenBank accession number CP003328; Supplementary Table S2). The hexA fragments were removed as SalI/EcoRI- and EcoRI/SacI-digested fragments and cloned into the suicide vector pBC-SK that was cut with the same enzymes. The kanamycin resistance cassette amplified from plasmid pUC-4k digested with EcoRI was inserted into pBC-SK to generate hexA-Km-pBC-SK. Plasmid hexA-Km-pBC-SK was inserted into P. multocida HB03 by electroporation (Zhao et al., 2016).
Five-week-old female BALB/c mice (20–25 g in weight) were purchased from the Experimental Animal Centre of Huazhong Agricultural University, Wuhan, China. All animal procedures were performed in strict accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals, approved through the State Council of the People’s Republic of China (1988.11.1), and with the approval of the Animal Welfare and Research Ethics Committee at Huazhong Agricultural University.
Identification and Analysis of Putative Depolymerase-Encoding Genes
The complete genome sequence of phage PHB02 (GenBank accession number MF034659) has been described earlier (Chen et al., 2018). The Position-Specific Iterative Basic Local Alignment Search Tool1 was used to identify putative depolymerase-encoding genes in the PHB02 genome, and Phyre22 was used to predict the structure of the depolymerase (Kelley et al., 2015).
Expression and Purification of the Phage-Derived Putative Depolymerase
Phage PHB02 open reading frame 8 (orf 8) was amplified by polymerase chain reaction using primers A2-KpnI (5′-CGGGGTACCGTGGAGTTTCTGATTCCCTT-3′) and A2-BamHI (5′-CGGGATCCTCATGCTAGCTTCTTTGTCTT-3′). The purified amplicon was cloned into plasmid pCold TF, generating pCold-ORF8, which was then transformed into E. coli BL21(DE3). The recombinant His-tagged PHB02-ORF8 protein was expressed by induction with 0.1 mM isopropyl β-D-1-thiogalactopyranosid for 18 h at 16°C and was then purified as described by Guo et al. (2017). Briefly, PHB02-ORF8 was purified from the soluble fraction using a Ni-nitrilotriacetic acid column (Genscript, Wuhan, China) before being eluted and dialyzed overnight at 4°C against a 1000-fold volume of phosphate-buffered saline (PBS) buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH = 7.4). The hexa-histidine tag was then removed from the purified protein by digestion with thrombin (Solarbio, Shanghai, China). The purified protein solution was again passed through the column and then concentrated to 0.36 μg/μl by centrifugation over a 30-kDa ultrafiltration tube (Solarbio, Shanghai, China). The resulting purified PHB02-ORF8 protein was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis as described by Guo et al. (2017). Different doses were obtained by dilution of the stock depolymerase, which was stored at −80°C.
Spot Test
Depolymerase Dep-ORF8 activity against host strains was determined by spot test, as described by Guo et al. (2017). Briefly, molten soft TSA (0.75% agar) supplemented with 10% (vol/vol) sterile defibrinated sheep blood was inoculated with 300 μl of the host bacterial strain grown to exponential phase. This mixture was then poured onto the surface of a TSA (1.5% agar) plate supplemented with 10% (vol/vol) sterile defibrinated sheep blood and allowed to set. After drying, aliquots of a serial dilution of purified PHB02-ORF8 protein (360 to 1.4 ng) were spotted onto the surface of the double-layer agar plates. Elution buffer diluted in PBS was used as a control. To determine the specificity of the Dep-ORF8 depolymerase, capsular type D or F P. multocida strains were used. Briefly, 5-μl aliquots of PHB02-ORF8 protein (360 ng) were spotted onto double-layer agar plates as described above. The plates were observed for the formation of semi-clear spots for 6 h at 37°C. This experiment was repeated three times.
Phage Killing Assay
In vitro phage killing assays against the P. multocida HB03 strains were conducted as described by Dalmasso et al. (2016). Briefly, phage PHB02 was added to the P. multocida HB03 strains or capsule mutant strains (105 colony-forming units (CFU)/ml) at a multiplicity of infection of 10 or 100, and then cultured at 37°C for 30 or 60 min, respectively. Bacterial counts were determined by plating serial dilutions of the cultures. This experiment was repeated three times. The experimental data were analyzed by using a two-way analysis of variance. The data obtained are expressed as the mean ± standard deviation (SD).
Serum Sensitivity Assay
Serum sensitivity was determined as previously described (Lin et al., 2017). Briefly, exponentially growing bacteria [1–3 × 107 colony-forming units (CFU)/ml] mixed with depolymerase (100 μg/ml) were incubated with different sera, including 75% mouse serum (Hengyuan, Shanghai, China), 75% human serum (Hengyuan, Shanghai, China), heat-inactivated mouse or human serum (56°C, 30 min), 75% mouse whole blood (Hengyuan, Shanghai, China), and PBS (as a control) at 37°C for 3 h. Bacterial counts were determined by plating serial dilutions of the cultures. Experimental data were analyzed by one-way analysis of variance. The data obtained are expressed as the mean ± SD.
Acute Toxicity Assay
To study the toxicity of the phage, nine 5-week-old female BALB/c mice were randomly divided into three groups. The groups were inoculated intraperitoneally with 100 μl of phage [109 plaque-forming units (PFU)/ml], 100 μl of depolymerase Dep-ORF8 (36 μg), or an equal volume of PBS buffer. The mice were then observed daily for 7 days.
Health scores were determined as previously described (Zhang et al., 2016). Briefly, the health status of each group of mice was given a score between zero and five, as follows: five: normal health, condition unremarkable; four: decreased physical activity and ruffled fur; three: lethargy and hunched back; two: exudative accumulation around partially closed eyes; one: near death; zero: dead. The total score for each group was recorded at least three times per day. The mice were euthanized by CO2 at 7 days post-inoculation and subjected to histopathological examination. The liver, spleen, kidney, and lungs were removed and immediately placed in 4% formalin, then dehydrated with different concentrations of alcohol, and treated with wintergreen oil overnight. Paraffin-embedded tissue was cut into slices (∼5 μm thick) with a microtome (ZENDA, United States). Samples were dewaxed and then stained with hematoxylin and eosin or toluidine blue, as described previously (Zhang et al., 2016). The data describing the health status of each group of mice are expressed as the mean ± SD.
Phage in vivo Distribution Test
To examine phage distribution patterns in mice, 15 5-week-old female BALB/c mice were randomly divided into three groups. The groups were inoculated intraperitoneally with 100 μl of phage PHB02 (109 pfu/ml). Individual mice were then euthanized at 6, 12, 24, 48, and 72 h post-inoculation. The amount of phage present in the liver, lungs, kidney, and spleen was measured and reported as PFU/g of tissue, while blood counts were measured and reported as PFU/ml. The titration of phage particles was conducted via the double-layer agar method. The data are expressed as the mean ± SD.
Mouse Infection and Treatment
To examine the protective effects of phage and capsule depolymerase therapy, the minimum lethal dose (MLD) of wild-type P. multocida HB03 was first determined as described previously (Xia et al., 2015). Briefly, 15 5-week-old female BALB/c mice were randomly divided into five groups and inoculated intraperitoneally with 100 μl of different doses of P. multocida (10 CFU, 20 CFU, 40 CFU, 80 CFU, and 160 CFU, respectively). Once the MLD was determined, 2× the MLD was used as the challenge dose. Forty-eight 5-week-old female BALB/c mice were then randomly divided into eight groups (a–h). Groups a–g were inoculated intraperitoneally with 100-μl doses of 80 CFU of wild-type P. multocida HB03, while the control group h animals were inoculated intraperitoneally with an equivalent volume of PBS. Groups b and c were then inoculated intraperitoneally with 100 μl of phage PHB02 (109 pfu/ml) at 6 h and 12 h post-bacterial inoculation, while group d mice were inoculated intraperitoneally with 100 μl of phage PHB02 (109 pfu/ml) at 12 h following bacterial inoculation, and then once daily for 5 days. Similarly, groups e and f were inoculated intraperitoneally with 100 μl of depolymerase Dep-ORF8 (36 μg) at 6 h and 12 h post-bacterial inoculation, whereas group g mice were inoculated intraperitoneally with 100 μl of depolymerase Dep-ORF8 (36 μg) at 12 h after bacterial inoculation, and then once daily for 5 days. The mice were then observed daily for 21 days. The data pertaining to the health status of each group of mice are expressed as the mean ± SD. Survival was analyzed by Kaplan–Meier analysis with a log-rank test (statistically significant at P < 0.05).
Results
Analysis of Putative Depolymerase-Encoding Genes
In a previous study, we observed that phage PHB02 produced halos around the plaques, which we suspected was related to a putative phage-derived depolymerase. As phage depolymerases are usually part of the tail spike or tail fiber, we hypothesized that the tail fiber of phage PHB02, encoded by orf8, might be related to phage depolymerase. We therefore used Phyre2 to predict the secondary structure of Dep-ORF8, which consisted of 24 α-helices and 47 β-structures, as well as disordered regions. In addition, the model of Dep-ORF8 exhibited limited similarity to other known proteins but showed a confidence value of 97.8% (13% coverage) with homologous protein Clp9Ha (Supplementary Figure S1).
Expression and Purification of the Phage Depolymerase
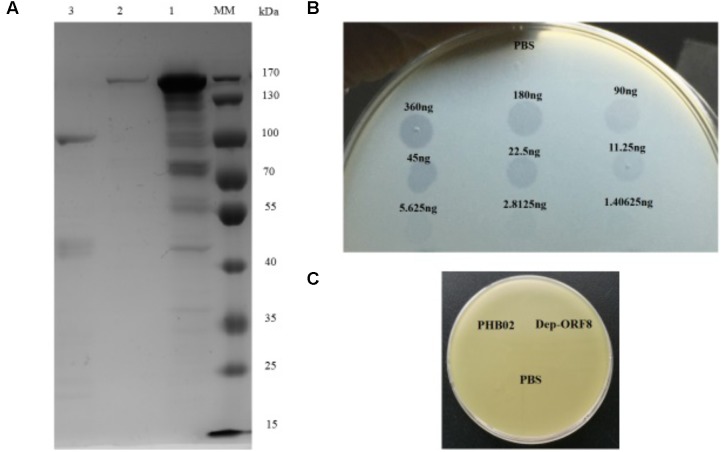
Dep-ORF8 was predicted to be 109.6 kDa in size and, following purification, the concentration of the recombinant protein was 0.36 μg/μl (Figure 1A). Spot tests showed that at amounts ≥ 2.8125 ng, purified recombinant Dep-ORF8 produced a translucent spot in the lawn of host bacteria (Figure 1B). The assay indicated that all 31 P. multocida capsular type A strains were sensitive to the recombinant Dep-ORF8 protein (Supplementary Table S1). Neither the phage nor the purified Dep-ORF8 protein hydrolyzed the capsule-deficient mutant strains (Figure 1C).
FIGURE 1.
Dep-ORF8 overexpression and activity. (A) Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of purified Dep-ORF8. Lane MM, protein marker; lane 1, 20 μg of supernatant from the induced E. coli BL21(DE3) cells; lane 2, 200 ng of purified Dep-ORF8 (containing the his-tag); lane 3, 200 ng of purified Dep-ORF8 (minus the his-tag). (B) Activity of depolymerase Dep-ORF8 against wild-type P. multocida strain HB03. Serial dilutions of Dep-ORF8 were spotted onto the plate containing P. multocida HB03. Elution buffer diluted in PBS was used as a control. (C) Spot tests examining the ability of phage PHB02 and depolymerase Dep-ORF8 to lyse the capsule-deficient mutant strain. Aliquots (5 μl) of phage PHB02 at a concentration of 108 pfu/ml and depolymerase Dep-ORF8 (360 ng) were spotted onto a plate containing the mutant strains. The plates were observed for 6 h at 37°C.
Phage Killing Assay
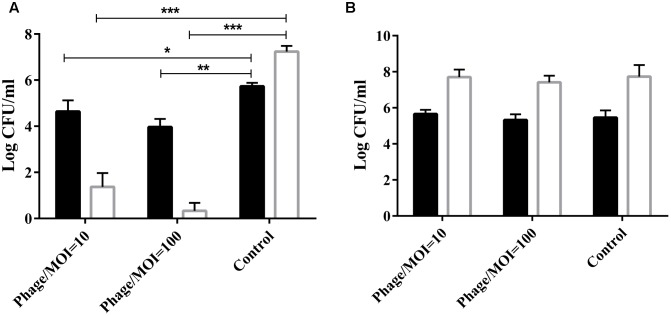
The phage was co-cultured with P. multocida HB03 for 1 h at a multiplicity of infection (MOI) of 10 or 100. Results showed that at a MOI of 100, phage PHB02 significantly reduced the number of viable wild-type HB03 cells when compared with the control treatment after 60 min of incubation (P < 0.01) (Figure 2A). The phage had no effect on the survival of the capsule-mutant strains (Figure 2B).
FIGURE 2.
In vitro activity of the phage against the wild-type and capsule-deficient mutant strains. (A)
In vitro activity of the phage against wild-type P. multocida strain HB03. The phage was mixed with the bacteria at a MOI of 10 or 100 and then cultured at 37°C for 30 min ( ) or 60 min (
) or 60 min ( ). Results show the number of viable bacteria following incubation. Experimental data were analyzed by two-way analysis of variance and are expressed as the mean ± SD. The viable cell counts in each of the treatment groups were compared with that of the control, with significant differences indicated by ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001. (B)
In vitro activity of the phage against the capsule-deficient P. multocida strain. Conditions and data interpretation were the same as described in (A).
). Results show the number of viable bacteria following incubation. Experimental data were analyzed by two-way analysis of variance and are expressed as the mean ± SD. The viable cell counts in each of the treatment groups were compared with that of the control, with significant differences indicated by ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001. (B)
In vitro activity of the phage against the capsule-deficient P. multocida strain. Conditions and data interpretation were the same as described in (A).
Serum Sensitivity Assay
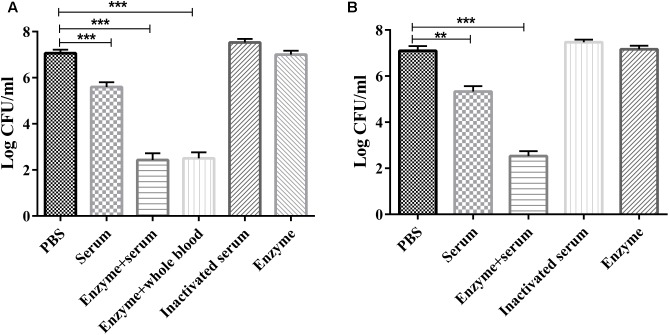
The serum sensitivity assay showed that purified Dep-ORF8 alone had no significant effect on bacterial survival (Figures 3A,B). Mouse serum, mouse whole-blood, and human serum alone had a small bactericidal effect, with a 1.2–1.7 log CFU decrease in viable cell counts. Enzyme plus serum significantly reduced the number of viable wild-type P. multocida strain HB03 cells (3.5–4.5 log CFU decrease). There was no significant difference in viable cell counts between the enzyme plus mouse serum group and the enzyme plus mouse whole-blood group (Figures 3A,B). Meanwhile, heat-inactivated serum slightly increased bacterial survival (Figures 3A,B). Depolymerase alone had no effect on capsule-mutant HB03 strains, while serum alone or mouse whole-blood alone killed more than 95% of capsule-mutant bacterial cells within 2 h (data not shown).
FIGURE 3.
Serum sensitivity assay. (A) P. multocida strain HB03 mixed with depolymerase was incubated with mouse serum or whole-blood. (B) P. multocida strain HB03 with depolymerase was incubated with human serum. The mixtures were incubated at 37°C for 2 h. The number of viable bacteria was determined by plating and enumeration of serial dilutions of the suspension. Assays were repeated at least three times. Experimental data were analyzed by one-way analysis of variance and are expressed as the mean ± SD. The viable cell counts in each of the treatment groups were compared with that of the PBS control, with significant differences indicated by ∗P < 0.05; ∗∗P < 0.01; and ∗∗∗P < 0.001.
Acute Toxicity
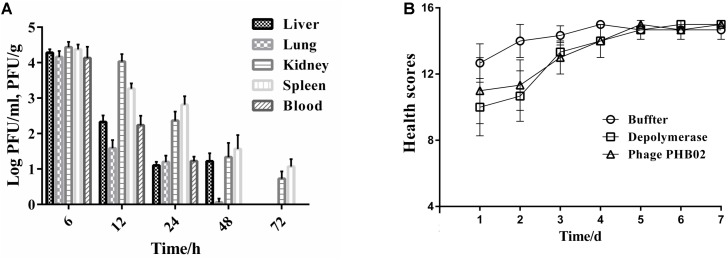
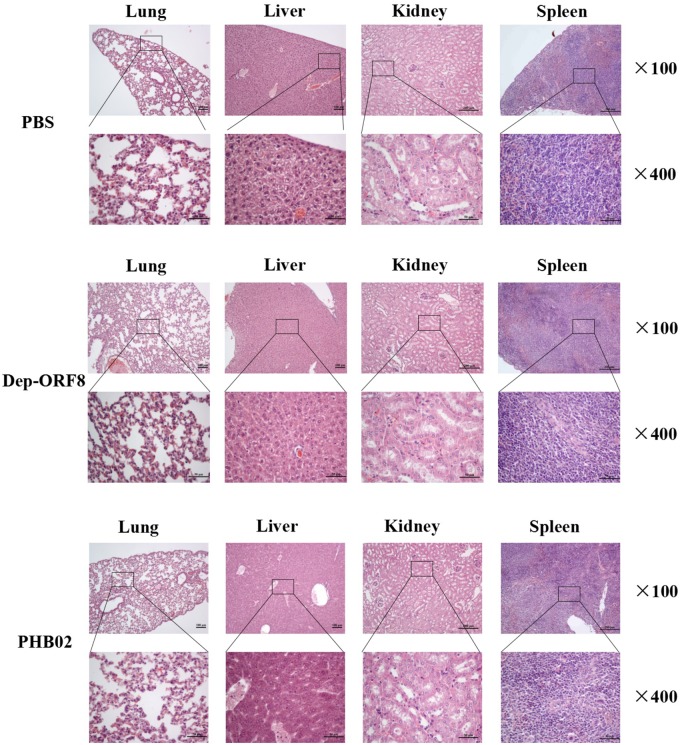
The distribution of phage in mice was studied following intraperitoneal injection of PHB02 at a dose of 1.0 × 108 PFU. Phage was detected in the liver, spleen, kidney, and lungs, but was absent from blood at 48 h following phage inoculation. At 72 h post-inoculation, the phage was only detected in the spleen and kidney (Figure 4A). The appearance and behavior of mice was examined daily for 7 days following PHB02 inoculation. We did not observe any behavioral changes in the mice following inoculation (Figure 4B). More importantly, none of the tissues isolated from mice immunized with phage PHB02 or Dep-ORF8 showed an increase in eosinophils or basophils or other pathological changes when compared with the control group (Figure 5).
FIGURE 4.
In vivo assays. (A) Distribution of phage PHB02 in mice. The experiment was repeated three times and results are expressed as the mean ± SD. (B) Scores of clinical symptoms post-challenge. The health status of each group of mice was given a score between zero and five, as follows: five: normal health, condition unremarkable; four: decreased physical activity and ruffled fur; three: lethargy and hunched back; two: exudative accumulation around partially closed eyes; one: near death; zero: dead. The total score for each group was recorded at least three times per day. The data are expressed as the mean ± SD.
FIGURE 5.
Pathological changes and histopathology of the mice challenged with phage and depolymerase. The liver, spleen, kidney, and lungs from the mice challenged with phage and depolymerase were removed and immediately placed in 4% formalin. Tissue sections were stained with hematoxylin and eosin or toluidine blue and observed under 100× and 400× magnification.
Phage and Capsule Depolymerase Therapy
Analysis of the MLD of wild-type P. multocida showed that more than 66% of mice died when inoculated with a bacterial dose of 40–160 CFU (Supplementary Table S3). Therefore, we concluded that the MLD of P. multocida HB03 in mice was 40 CFU, and accordingly used 80 CFU (2× MLD) as the challenge dose. The efficacy of depolymerase and phage treatment was then evaluated in mice infected with P. multocida HB03. This depolymerase therapy study showed that the survival rate of group e mice (treated with Dep-ORF8 at 6 h post-bacterial infection) was significantly increased when compared with that of control group a (P = 0.048), whereas the survival rate of group f (treated with Dep-ORF8 at 12 h post-bacterial infection) was not significantly increased (P = 0.18) (Figure 6A). However, mice treated with purified depolymerase at 12 h post-bacterial infection, and then daily for 5 days (group g), showed a significantly increased survival rate when compared with control group a (P = 0.0029). No deaths were recorded for PBS control group h (Figure 6A). The study also showed that none of the mice in treatment groups b or d died, while two mice from treatment group c died during the course of the experiment (on days 6 and 7, respectively) (Figure 6B). Treatment groups b and d had significantly greater survival than treatment group a (mice infected with HB03 alone) (P = 0.0006) (Figure 6B). No changes in behavior were observed in the 12 days following infection, after which point no further deaths occurred (Figures 6C,D).
FIGURE 6.
Protective effects of depolymerase and phage in mice challenged with wild-type P. multocida strain HB03. (A) Treatment efficacy of purified recombinant depolymerase. (B) Treatment efficacy of phage PHB02. (C) Clinical symptom scores post-challenge with purified recombinant depolymerase. (D) Clinical symptom scores post-challenge with phage PHB02. Treatment groups were as follows: a, infected with P. multocida strain HB03; b, phage treatment 6 h post-infection; c, phage treatment 12 h post-infection; d, multiple-dose phage treatment 12 h post-infection; e, Dep-ORF8 treatment 6 h post-infection; f, Dep-ORF8 treatment 12 h post-infection; g, multiple-dose Dep-ORF8 treatment 12 h post-infection; h, inoculated with PBS only. The total score for the health status of each group was recorded at least three times per day. The data are expressed as the mean ± SD.
Discussion
Pasteurella multocida phage were first reported by Kirchner and Eisenstark (1956); however, there have since been very few reports regarding lytic phages in P. multocida. To our knowledge, this is the first study to demonstrate that capsular material in P. multocida serogroup A strains acts as a receptor for P. multocida phage, and that the putative depolymerase from phage PHB02 can specifically degrade capsular serogroup A strains.
The interaction of the phage with the bacterial surface is the first step in the infection process. When infecting a bacterium, the phage first needs to interact with the bacterial surface (Leiman et al., 2007; Leiman and Molineux, 2008). Polysaccharides on the cell surface normally act as a physical barrier to prevent the phage from entering the bacterial cell. However, the polysaccharides also act as primary receptors for phages, along with other protruding cell wall structures such as pili and flagella (Leiman et al., 2007; Leiman and Molineux, 2008; Li et al., 2016; Latka et al., 2017). For example, Caudovirales phages can recognize and attach to the bacterial surface by specifically digesting the polysaccharides on the surface via the tail-associated protein (Yan et al., 2014). Therefore, the tail apparatus of Caudovirales phages plays an important role in the infection of host cells. Previous research shows that the polysaccharide-degrading activity of a phage depolymerase is related to the tail structure (Scholl and Merril, 2005; Lai et al., 2016; Hsieh et al., 2017). To further verify the “primary receptor” function of polysaccharides on the bacterial cell surface, phage PHB02 was used in the current study to infect capsule-mutant strain P. multocida HB03. The results showed that the phage is ineffective against the capsule-mutant strain, indicating that polysaccharides are required for phage infection.
Phage PHB02 appears to be specific for capsular serotype A P. multocida strains, lysing 30 out of 31 tested isolates in the current study. In addition, PHB02 produces a halo around the plaque but has a narrow host range. Similar phage specificity has been reported in K. pneumoniae (Lin et al., 2014; Volozhantsev et al., 2016; Hsieh et al., 2017) and E. coli (Scholl and Merril, 2005; Leiman et al., 2007; Guo et al., 2017). Phages specifically recognize the polysaccharides of their host bacteria, and this specificity can be used for bacterial typing in epidemiological studies. In addition, phage typing can quickly and reliably identify capsular antigens. In the current study, putative depolymerase Dep-ORF8 showed activity against all 31 strains tested, suggesting that using Dep-ORF8 for capsular typing may provide a more consistent result than phage alone. In addition, depolymerase can be used as a research tool to help determine polysaccharide structure and can even be used for industrial production of complex carbohydrates.
After the bacterial capsule is stripped by depolymerase, cells are more susceptible to attack by host immune defenses (Mushtaq et al., 2004; Finlay and McFadden, 2006; Lin et al., 2014). In addition, research shows that serum can kill bacteria (Mushtaq et al., 2004). Therefore, combining depolymerase and serum may kill more bacteria than each treatment individually. To investigate this, we compared the bactericidal effects of depolymerase plus mouse serum and depolymerase plus mouse whole-blood. The results showed that there was no significant difference between the two groups. Although four of the six mice in group g survived in the in vivo assays (mice were treated with 100 μl of Dep-ORF8 (36 μg) at 12 h after bacterial inoculation and then once daily for 5 days), optimizing the depolymerase concentration could further increase the survival rate. Normally, bacterial infection with simultaneous treatment is important for the rescue of mice. However, to mimic a clinical treatment situation, we administered phage or depolymerase at least 6 h after bacterial infection. The results show that simultaneous treatment with phage or depolymerase is not necessary for survival following P. multocida infection. Therefore, it is feasible to administer phage or depolymerase at some time after initial infection.
In addition to the well-known antibacterial actions, phages are also able to mediate some anti-inflammatory activities (Jończyk-Matysiak et al., 2017; Miȩdzybrodzki et al., 2017). Therefore, the levels of inflammatory cytokines can be used as markers for monitoring treatment efficacy. Zimecki et al. (2009) demonstrated that the application of phage resulted in a down-regulation in the levels of proinflammatory cytokines (particularly TNF-α) in mice infected with Staphylococcus aureus. Chadha et al. (2017) showed that treatment with a phage cocktail resulted in a significant reduction of inflammatory cytokine levels, including IL-1β and TNF-α, when compared with the infection control group in a model of K. pneumoniae-mediated burn wound infection. Therefore, future work should be conducted to examine host inflammatory responses following phage inoculation, such as measuring the induction of IL-6 and TNF-α. Moreover, although phage specifically kill bacteria, biosafety issues cannot be ignored. Temperate phage genomes usually contain many genes with unknown functions, some of which may be virulence or resistance genes, which are difficult to predict using the limited number of sequences in the databases. In contrast, purified recombinant depolymerase can only degrade bacterial capsule, thus making it a much safer option for use in vivo. Application of the purified protein would strip the natural protective layer from the bacterial cells, making them much more susceptible to killing by conventional therapies both in vitro and in vivo.
Author Contributions
YC drafted the main manuscript and performed the data analysis. YC, ES, LY, and JS planned and performed experiments. YC and BW were responsible for experimental design.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yigang Tong for plasmid pBC-SK and pUC-4k, which was used for constructing P. multocida ΔhexA, and Can Zhang for plasmid pCold TF, which was used for constructing plasmid pCold-ORF8.
Funding. This study was funded by Research projects of agricultural public welfare industry of China (201403054), and National Science supported planning projects of Hubei Province of China (2014BBB010).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01678/full#supplementary-material
References
- Abreu F., Rodríguez-Lucas C., Rodicio M. R., Vela A. I., Fernández-Garayzábal J. F., Leiva P. S., et al. (2018). Human Pasteurella multocida infection with likely zoonotic transmission from a pet dog, Spain. Emerg. Infect. Dis. 24 1145–1146. 10.3201/eid2406.171998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillot R., Voisine P., Côté L., Longtin Y. (2011). Deep sternal wound infection due to Pasteurella multocida: the first case report and review of literature. Infection 39 575–578. 10.1007/s15010-011-0141-5 [DOI] [PubMed] [Google Scholar]
- Benešík M., Nováèek J., Janda L., Dopitová R., Pernisová M., Melková K., et al. (2018). Role of SH3b binding domain in a natural deletion mutant of Kayvirus endolysin LysF1 with a broad range of lytic activity. Virus Genes 54 130–139. 10.1007/s11262-017-1507-2 [DOI] [PubMed] [Google Scholar]
- Bhetwal A., Maharjan A., Shakya S., Satyal D., Ghimire S., Khanal P. R., et al. (2017). Isolation of potential phages against multidrug-resistant bacterial isolates: promising agents in the rivers of Kathmandu, Nepal. Biomed Res. Int. 2017:3723254. 10.1155/2017/3723254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrathybay E., Sawada T., Kataoka Y., Okiyama E., Kawamoto E., Amao H. (2003). Capsule thickness and amounts of a 39 kDa capsular protein of avian Pasteurella multocida type A strains correlate with their pathogenicity for chickens. Vet. Microbiol. 97 215–227. 10.1016/j.vetmic.2003.09.011 [DOI] [PubMed] [Google Scholar]
- Boyce J. D., Adler B. (2000). The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68 3463–3468. 10.1128/IAI.68.6.3463-3468.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunel A. S., Guery B. (2017). Multidrug resistant (or antimicrobial-resistant) pathogens - alternatives to new antibiotics? Swiss Med. Wkly. 147:w14553. [DOI] [PubMed] [Google Scholar]
- Campoy S., Aranda J., Àlvarez G., Barbé J., Llagostera M. (2006). Isolation and sequencing of a temperate transducing phage for Pasteurella multocida. Appl. Environ. Microbiol. 72 3154–3160. 10.1128/AEM.72.5.3154-3160.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter G. R. (1955). Serotyping of Pasteurella multocida. I. A hemagglutination test for the identification of serological type. Am. J. Vet. Res. 6 481–484. [PubMed] [Google Scholar]
- Cha K., Oh H. K., Jang J. Y., Jo Y., Kim W. K., Ha G. U., et al. (2018). Characterization of two novel bacteriophages infecting multidrug-resistant (MDR) Acinetobacter baumannii and evaluation of their therapeutic efficacy in vivo. Front. Microbiol. 9:696. 10.3389/fmicb.2018.00696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadha P., Katare O. P., Chhibber S. (2017). Liposome loaded phage cocktail: enhanced therapeutic potential in resolving Klebsiella pneumoniae mediated burn wound infections. Burns 43 1532–1543. 10.1016/j.burns.2017.03.029 [DOI] [PubMed] [Google Scholar]
- Chen Y. B., Sun E. C., Song J. Y., Yang L., Wu B. (2018). Complete genome sequence of a novel T7-like bacteriophage from a Pasteurella multocida capsular type A isolate. Curr. Microbiol. 75 574–579. 10.1007/s00284-017-1419-3 [DOI] [PubMed] [Google Scholar]
- Cheng M., Zhang L., Zhang H., Li X., Wang Y., Xia F., et al. (2018). An ointment consisting of the phage lysin LysGH15 and apigenin for decolonization of methicillin-resistant Staphylococcus aureus from skin wounds. Viruses 10:E244. 10.3390/v10050244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J. Y., Wilkie I., Boyce J. D., Townsend K. M., Frost A. J., Ghoddusi M., et al. (2001). Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69 2487–2492. 10.1128/IAI.69.4.2487-2492.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett D., Roberts I. S. (2008). Capsular polysaccharides in Escherichia coli. Adv. Appl. Microbiol. 65 1–26. 10.1016/S0065-2164(08)00601-1 [DOI] [PubMed] [Google Scholar]
- Dalmasso M., Strain R., Neve H., Franz C. M., Cousin F. J., Ross R. P., et al. (2016). The new Escherichia coli phages from the human gut show promising potential for phage therapy. PLoS One 11:e0156773. 10.1371/journal.pone.0156773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies R. L., MacCorquodale R., Baillie S., Caffrey B. (2003). Characterization and comparison of Pasteurella multocida strains associated with porcine pneumonia and atrophic rhinitis. J. Med. Microbiol. 52 59–67. 10.1099/jmm.0.05019-0 [DOI] [PubMed] [Google Scholar]
- Drulis-Kawa Z., Majkowska-Skrobek G., Maciejewska B. (2015). Bacteriophages and phage-derived proteins-application approaches. Curr. Med. Chem. 22 1757–1773. 10.2174/0929867322666150209152851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewers C., Lübke-Becker A., Bethe A., Kiebling S., Filter M., Wieler L. H. (2006). Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet. Microbiol. 114 304–317. 10.1016/j.vetmic.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Finlay B. B., McFadden G. (2006). Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124 767–782. 10.1016/j.cell.2006.01.034 [DOI] [PubMed] [Google Scholar]
- García-Alvarez A., Chaves F., Fernández A., Sanz C., Borobia M., Cid D. (2015). An ST11 clone of Pasteurella multocida, widely spread among farmed rabbits in the Iberian Peninsula, demonstrates respiratory niche association. Infect. Genet. Evol. 34 81–87. 10.1016/j.meegid.2015.07.018 [DOI] [PubMed] [Google Scholar]
- Guo Z., Huang J., Yan G., Lei L., Wang S., Yu L., et al. (2017). Identification and characterization of Dpo 42, a novel depolymerase derived from the Escherichia coli phage vB_EcoM_ECOO78. Front. Microbiol. 8:1460 10.3389/fmicb.2017.01460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez D., Rodríguez-Rubio L., Martínez B., Rodríguez A., García P. (2016). Bacteriophages as weapons against bacterial biofilms in the food industry. Front. Microbiol. 7:825. 10.3389/fmicb.2016.00825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha E., Son B., Ryu S. (2018). Clostridium perfringens virulent bacteriophage CPS2 and its thermostable endolysin LysCPS2. Viruses 10:E251. 10.3390/v10050251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heddleston K. L., Gallagher J. E., Rebers P. A. (1972). Fowl cholera: gel diffusion precipitin test for serotyping Pasteurella multocida from avian species. Avian Dis. 16 925–936. 10.2307/1588773 [DOI] [PubMed] [Google Scholar]
- Hsieh P. F., Lin H. H., Lin T. L., Chen Y. Y., Wang J. T. (2017). Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci. Rep. 7:4624. 10.1038/s41598-017-04644-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglic Z., Jeklova E., Christensen H., Leva L., Register K., Kummer V., et al. (2011). Host response in rabbits to infection with Pasteurella multocida serogroup F strains originating from fowl cholera. Can. J. Vet. Res. 5 200–208. [PMC free article] [PubMed] [Google Scholar]
- Jann K., Jann B. (1987). Polysaccharide antigens of Escherichia coli. Rev. Infect. Dis. Suppl. 5 S517–S526. 10.1093/clinids/9.Supplement_5.S517 [DOI] [PubMed] [Google Scholar]
- Jonas M., Morishita T. Y., Angrick E. J., Jahja J. (2001). Characterization of nine Pasteurella multocida isolates from avian cholera outbreaks in Indonesia. Avian Dis. 45 34–42. 10.2307/1593009 [DOI] [PubMed] [Google Scholar]
- Jończyk-Matysiak E., Weber-Dąbrowska B., Owczarek B., Miȩdzybrodzki R., Łusiak-Szelachowska M., Łodej N., et al. (2017). Phage-phagocyte interactions and their implications for phage application as therapeutics. Viruses 9:E150. 10.3390/v9060150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner C., Eisenstark A. (1956). Lysogeny in Pasteurella multocida. Am. J. Vet. Res. 17 547–548. [PubMed] [Google Scholar]
- Lai M. J., Chang K. C., Huang S. W., Luo C. H., Chiou P. Y., Wu C. C., et al. (2016). The tail associated protein of Acinetobacter baumannii phage ΦAB6 is the host specificity determinant possessing exopolysaccharide depolymerase activity. PLoS One 11:e0153361. 10.1371/journal.pone.0153361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latka A., Maciejewska B., Majkowska-Skrobek G., Briers Y., Drulis-Kawa Z. (2017). Bacteriophage-encoded virion associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 101 3103–3119. 10.1007/s00253-017-8224-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman P. G., Battisti A. J., Bowman V. D., Stummeyer K., Mühlenhoff M., Gerardy-Schahn R., et al. (2007). The structures of bacteriophages K1E and K1-5 explain processive degradation of polysaccharide capsules and evolution of new host specificities. J. Mol. Biol. 371 836–849. 10.1016/j.jmb.2007.05.083 [DOI] [PubMed] [Google Scholar]
- Leiman P. G., Molineux I. J. (2008). Evolution of a new enzyme activity from the same motif fold. Mol. Microbiol. 69 287–290. 10.1111/j.1365-2958.2008.06241.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Koç C., Kühner P., Stierhof Y. D., Krismer B., Enright M. C., et al. (2016). An essential role for the baseplate protein Gp45 in phage adsorption to Staphylococcus aureus. Sci. Rep. 6:26455. 10.1038/srep26455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Paff M. L., Molineux I. J., Bull J. J. (2017). Therapeutic application of phage capsule depolymerases against K 1, K 5, and K30 capsulated E. coli in mice. Front. Microbiol. 16:2257 10.3389/fmicb.2017.02257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T. L., Hsieh P. F., Huang Y. T., Lee W. C., Tsai Y. T., Su P. A., et al. (2014). Isolation of a bacteriophage and its depolymerase specific for K1 capsule of Klebsiella pneumoniae: implication in typing and treatment. J. Infect. Dis. 210 1734–1744. 10.1093/infdis/jiu332 [DOI] [PubMed] [Google Scholar]
- Majkowska-Skrobek G., Łątka A., Berisio R., Maciejewska B., Squeglia F., Romano M., et al. (2016). Capsule-targeting depolymerase, derived from Klebsiella KP36 phage, as a tool for the development of anti-virulent strategy. Viruses 8:E324. 10.3390/v8120324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynou G., Bach A., Terré M. (2017). Feeding of waste milk to Holstein calves affects antimicrobial resistance of Escherichia coli and Pasteurella multocida isolated from fecal and nasal swabs. J. Dairy Sci. 100 2682–2694. 10.3168/jds.2016-11891 [DOI] [PubMed] [Google Scholar]
- Miȩdzybrodzki R., Borysowski J., Kłak M., Jończyk-Matysiak E., Obmińska-Mrukowicz B., Suszko-Pawłowska A., et al. (2017). In vivo studies on the influence of bacteriophage preparations on the autoimmune inflammatory process. Biomed Res. Int. 2017:3612015. 10.1155/2017/3612015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles D. G. (2009). Overview of the North American beef cattle industry and the incidence of bovine respiratory disease (BRD). Anim. Health Res. Rev. 10 101–103. 10.1017/S1466252309990090 [DOI] [PubMed] [Google Scholar]
- Mushtaq N., Redpath M. B., Luzio J. P., Taylor P. W. (2004). Prevention and cure of systemic Escherichia coli K1 infection by modification of the bacterial phenotype. Antimicrob. Agents Chemother. 48 1503–1508. 10.1128/AAC.48.5.1503-1508.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruzzi B., Briggs R. E., Swords W. E., De Castro C., Molinaro A., Inzana T. J. (2017). Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. mBio 8:e1843-17. 10.1128/mBio.01843-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakhuba D. V., Kolomiets E. I., Dey E. S., Novik G. I. (2010). Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 59 145–155. [PubMed] [Google Scholar]
- Reindel R., Fiore C. R. (2017). Phage therapy: considerations and challenges for development. Clin. Infect. Dis. 64 1589–1590. 10.1093/cid/cix188 [DOI] [PubMed] [Google Scholar]
- Samuel M. D., Shadduck D. J., Goldberg D. R., Johnson W. P. (2005). Avian cholera in waterfowl: the role of lesser snow and Ross’s geese as disease carriers in the Playa Lakes region. J. Wildl. Dis. 41 48–57. 10.7589/0090-3558-41.1.48 [DOI] [PubMed] [Google Scholar]
- Sarangi L. N., Thomas P., Gupta S. K., Priyadarshini A., Kumar S., Nagaleekar V. K., et al. (2015). Virulence gene profiling and antibiotic resistance pattern of Indian isolates of Pasteurella multocida of small ruminant origin. Comp. Immunol. Microbiol. Infect. Dis. 38 33–39. 10.1016/j.cimid.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Scholl D., Merril C. (2005). The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 187 8499–8503. 10.1128/JB.187.24.8499-8503.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivachandra S. B., Kumar A. A., Biswas A., Ramakrishnan M. A., Singh V. P., Srivastava S. K. (2004). Antibiotic sensitivity patterns among Indian strains of avian Pasteurella multocida. Trop. Anim. Health Prod. 36 743–750. 10.1023/B:TROP.0000045950.35070.7f [DOI] [PubMed] [Google Scholar]
- Sthitmatee N., Kataoka Y., Sawada T. (2011). Inhibition of capsular protein synthesis of Pasteurella multocida strain P-1059. J. Vet. Med. Sci. 73 1445–1451. 10.1292/jvms.11-0282 [DOI] [PubMed] [Google Scholar]
- Tabatabaei M., Liu Z., Finucane A., Parton R., Coote J. (2002). Protective immunity conferred by attenuated aroA derivatives of Pasteurella multocida B:2 strains in a mouse model of hemorrhagic septicemia. Infect. Immun. 70 3355–3362. 10.1128/IAI.70.7.3355-3362.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volozhantsev N. V., Myakinina V. P., Popova A. V., Kislichkina A. A., Komisarova E. V., Knyazeva A. I., et al. (2016). Complete genome sequence of novel T7-like virus vB_KpnP_KpV289 with lytic activity against Klebsiella pneumoniae. Arch. Virol. 161 499–501. 10.1007/s00705-015-2680-z [DOI] [PubMed] [Google Scholar]
- Wilkie I. W., Harper M., Boyce J. D., Adler B. (2012). Pasteurella multocida: diseases and pathogenesis. Curr. Top. Microbiol. Immunol. 361 1–22. 10.1007/82_2012_216 [DOI] [PubMed] [Google Scholar]
- Xia F., Li X., Wang B., Gong P., Xiao F., Yang M., et al. (2015). Combination therapy of LysGH15 and apigenin as a new strategy for treating pneumonia caused by Staphylococcus aureus. Appl. Environ. Microbiol. 82 87–94. 10.1128/AEM.02581-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Mao J., Xie J. (2014). Bacteriophage polysaccharide depolymerases and biomedical applications. Biodrugs 28 265–274. 10.1007/s40259-013-0081-y [DOI] [PubMed] [Google Scholar]
- Zhang L., Li D., Li X., Hu L., Cheng M., Xia F., et al. (2016). LysGH15 kills Staphylococcus aureus without being affected by the humoral immune response or inducing inflammation. Sci. Rep. 6:29344. 10.1038/srep29344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Liu Q., Xiao K., Hu Y., Liu X., Li Y., et al. (2016). Identification of the crp gene in avian Pasteurella multocida and evaluation of the effects of crp deletion on its phenotype, virulence and immunogenicity. BMC Microbiol. 16:125. 10.1186/s12866-016-0739-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimecki M., Artym J., Kocieba M., Weber-Dabrowska B., Borysowski J., Górski A. (2009). Effects of prophylactic administration of bacteriophages to immunosuppressed mice infected with Staphylococcus aureus. BMC Microbiol. 9:169. 10.1186/1471-2180-9-169 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.