Abstract
In plants, salicylic acid (SA) plays important roles in regulating immunity and programed cell death. Early studies revealed that increased SA accumulation is associated with the onset of hypersensitive reaction during resistance gene-mediated defense responses. SA was also found to accumulate to high levels in lesion-mimic mutants and in some cases the accumulation of SA is required for the spontaneous cell death phenotype. Meanwhile, high levels of SA have been shown to negatively regulate plant cell death during effector-triggered immunity, suggesting that SA has dual functions in cell death control. The molecular mechanisms of how SA regulates cell death in plants are discussed.
Keywords: salicylic acid, hypersensitive reaction, programed cell death, effector-triggered immunity, plant immunity
Salicylic acid (SA) is a plant hormone that plays key roles in defense signaling (Vlot et al., 2009). Pathogen infection induces SA biosynthesis and accumulation. Two groups of Arabidopsis mutants, salicylic acid induction deficient2 (sid2) and enhanced disease susceptibility5 (eds5), are deficient in pathogen-induced SA accumulation and exhibit increased susceptibility to biotrophic pathogens (Nawrath and Metraux, 1999; Dewdney et al., 2000). sid2 mutants carry mutations in the isochorismate synthase ICS1, suggesting that SA is synthesized from chorismate following pathogen infection via ICS1 (Wildermuth et al., 2001). EDS5 encodes a multi-antimicrobial extrusion protein (MATE) transporter (Nawrath et al., 2002). The exact role of EDS5 in SA metabolism is unclear. It is likely to be involved in exporting SA or a precursor of SA out of plastids (Serrano et al., 2013).
SA is perceived by two groups of receptors, NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) and NPR3/NPR4, all of which display high affinity with SA (Fu et al., 2012; Wu et al., 2012; Manohar et al., 2015; Ding et al., 2018). However, they have opposite roles in transcriptional regulation of defense gene expression (Ding et al., 2018). NPR1 functions as a transcriptional activator that promotes SA-induced defense gene expression and pathogen resistance (Fan and Dong, 2002). Loss of NPR1 results in reduced SA-induced PR gene expression and increased susceptibility to pathogens (Cao et al., 1994; Delaney et al., 1995). On the other hand, NPR3 and NPR4 serve as redundant transcriptional co-repressors that prevent activation of defense gene expression when the SA level is low (Ding et al., 2018). When SA levels are high, SA inhibits the transcriptional repression activity of NPR3/NPR4 to activate the expression of SA-responsive genes. The NPR4-4D mutant protein that is unable to bind SA constitutively represses defense gene expression and blocks SA-induced immunity, rendering the mutant plants with enhanced disease susceptibility (Ding et al., 2018). Regulation of defense genes by NPR1 and NPR3/NPR4 is directly facilitated by a group of redundant bZIP transcription factors, including TGA2, TGA5, and TGA6, which interact with both NPR1 and NPR3/NPR4 (Zhang et al., 1999, 2003, 2006; Despres et al., 2000; Zhou et al., 2000).
Increased SA accumulation is associated with hypersensitive response (HR), a form of programed cell death often induced by effector-triggered immunity (ETI), as well as spontaneous cell death in lesion-mimic mutants. Early studies showed that activation of N gene-mediated defense responses by tobacco mosaic virus led to about 20-fold increase in endogenous SA levels in the infected tobacco leaves (Malamy et al., 1990). Activation of ETI by Pseudomonas effectors AvrRpm1 and AvrRpt2 in Arabidopsis also results in dramatic increases in local SA levels in a SID2 and EDS5-dependent manner (Nawrath and Metraux, 1999). Meanwhile, in mutants with spontaneous cell death, SA accumulates at much higher levels than in wild type (Bruggeman et al., 2015). However, in autoimmune mutants with no spontaneous lesion formation, such as suppressor of npr1-1, constitutive1 (snc1) and defense, no death1 (dnd1), SA levels are still dramatically increased (Yu et al., 1998; Li et al., 2001), suggesting that cell death is not required for the activation of SA biosynthesis and high levels of SA alone are not sufficient to activate cell death.
Salicylic acid has been shown to be required for spontaneous cell death in several lesion-mimic mutants (Table 1). Treatment with low levels of SA activates runaway cell death in lesion simulating disease 1 (lsd1) (Dietrich et al., 1994). Blocking SA accumulation by expressing the SA hydroxylase encoded by the bacterial NahG gene suppresses lesion formation in lsd6, lsd7, accelerated cell death 6 (acd6), and acd11 mutants (Weymann et al., 1995; Rate et al., 1999; Brodersen et al., 2005). In the syntaxin of plants 121 (syp121) syp122 double mutant, spontaneous cell death is also attenuated when SA biosynthesis or SA perception is blocked (Zhang et al., 2007). However, not all lesion-mimic mutants require SA accumulation for activation of spontaneous cell death. For example, expression of NahG does not affect lesion formation in lsd2 and lsd4 mutants (Dietrich et al., 1994; Hunt et al., 1997).
Table 1.
SA levels and cell death phenotypes of Arabidopsis thaliana mutants.
| Mutant | SA levels | Cell death phenotype | Reference |
|---|---|---|---|
| lsd1 | High | Spontaneous cell death | Dietrich et al., 1994 |
| lsd2 | ND∗ | Spontaneous cell death | Dietrich et al., 1994 |
| lsd2 nahG | Low | Spontaneous cell death | Dietrich et al., 1994; Hunt et al., 1997 |
| lsd4 | ND∗ | Spontaneous cell death | Dietrich et al., 1994 |
| lsd4 nahG | Low | Spontaneous cell death | Dietrich et al., 1994; Hunt et al., 1997 |
| lsd6 | High | Spontaneous cell death | Weymann et al., 1995 |
| lsd6 nahG | Low | No spontaneous cell death | Weymann et al., 1995 |
| lsd7 | High | Spontaneous cell death | Weymann et al., 1995 |
| lsd7 nahG | Low | No spontaneous cell death | Weymann et al., 1995 |
| acd6 | High | Spontaneous cell death | Rate et al., 1999 |
| acd6 nahG | Low | No spontaneous cell death | Rate et al., 1999 |
| acd11 | High | Spontaneous cell death | Brodersen et al., 2005 |
| acd11 nahG | Low | No spontaneous cell death | Brodersen et al., 2005 |
| syp121 syp122 | High | Spontaneous cell death | Zhang et al., 2007 |
| syp121 syp122 nahG | Low | Reduced spontaneous cell death | Zhang et al., 2007 |
| syp121 syp122 sid2 | Low | Reduced spontaneous cell death | Zhang et al., 2007 |
| snc1 | High | No spontaneous cell death | Li et al., 2001 |
| dnd1 | High | No spontaneous cell death; reduced AvrRpt2-induced cell death | Yu et al., 1998 |
| dnd2 | High | No spontaneous cell death; reduced AvrRpt2-induced cell death | Jurkowski et al., 2004 |
| agd2 | High | Spontaneous cell death; reduced AvrRpt2- and AvrRpm1-induced cell death | Rate and Greenberg, 2001 |
| agd2 nahG | Low | Spontaneous cell death; restored AvrRpm1-induced cell death | Rate and Greenberg, 2001 |
| agd2 npr1 | ND∗ | Reduced spontaneous cell death; restored AvrRpt2-induced and AvrRpm1-induced cell death | Rate and Greenberg, 2001 |
| hrl1 | High | Spontaneous cell death; reduced AvrRpm1-induced cell death | Devadas and Raina, 2002 |
| hrl1 nahG | Low | Delayed spontaneous cell death; restored AvrRpm1-induced cell death | Devadas and Raina, 2002 |
| hrl1 npr1 | High | Delayed spontaneous cell death; restored AvrRpm1-induced cell death | Devadas and Raina, 2002 |
| npr3 npr4 | WT-like | No spontaneous cell death; reduced AvrRpt2-induced cell death | Zhang et al., 2006; Fu et al., 2012 |
∗ND, not determined; WT, wild type.
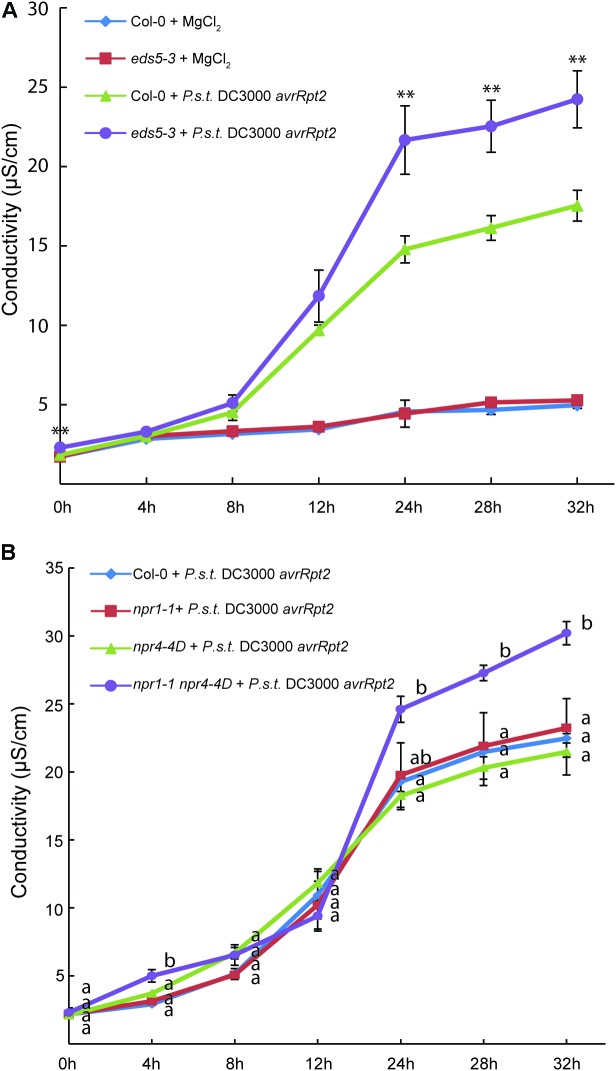
Interestingly, pre-treatment of Arabidopsis Col-0 plants with SA blocks HR activated by Pseudomonas syringae pv maculicola (P.s.m.) ES4326 carrying avrRpm1 (Devadas and Raina, 2002). In transgenic plants overexpressing NPR1, activation of cell death by the bacteria is also attenuated (Rate and Greenberg, 2001). In addition, increased ion leakage was observed in eds5-3 compared to wild type following treatment with Pseudomonas syringae pv tomato (P.s.t.) DC3000 with avrRpt2 (Figure 1A), indicating that AvrRpt2-induced cell death is enhanced in eds5-3. These findings suggest that activation of SA signaling plays an important role in negative regulation of cell death during ETI.
FIGURE 1.
Analysis of ion leakage in eds5-3, npr1-1, npr4-4D, and npr1-1 npr4-4D plants after treatment with P.s.t. DC3000 avrRpt2. Leaves of 4-week-old plants of the indicated genotypes grown under 12 h/12 h light/dark photoperiod at 23°C were infiltrated with mock (10 mM MgCl2) or P.s.t. DC3000 avrRpt2 (OD600 = 0.02). For each plant, two leaves were infiltrated and one leaf disk was cut from each leaf immediately after infiltration. The leaf disks were subsequently washed twice in distilled water. Six leaf disks from three plants, representing one biological replicate, were transferred into a 50-ml plastic tube containing 20 ml of distilled water and electrical conductivity was measured at different time points after infiltration using a VWR EC meter (Model 2052). Each data point on the graph represents the mean ± SD of three biological replicates. In (A), Two-tailed t-test was performed for each time point between wild type (Col-0) and eds5-3 plants treated with P.s.t. DC3000 avrRpt2 (∗∗p < 0.01). In (B), one way ANOVA with post hoc Tukey HSD test was performed for each time point among the different genotypes. Different letters (a,b) indicate statistically significant differences between the samples (p < 0.01).
Consistent with the role of pathogen-induced SA in negative regulation of cell death in ETI, enhanced cell death was observed in the npr1-1 mutant compared to wild type following treatment with P.s.m. ES4326 carrying avrRpm1 (Rate and Greenberg, 2001), suggesting that perception of SA by NPR1 is critical for the attenuation of AvrRpm1-induced cell death. When npr1-1, npr4-4D, and the npr1-1 npr4-4D double mutant plants were challenged with P.s.t. DC3000 carrying avrRpt2, cell death in the npr1-1 and npr4-4D single mutants was similar to that in wild type, whereas npr1-1 npr4-4D exhibited enhanced cell death (Figure 1B), suggesting that npr1-1 and npr4-4D have additive effect on AvrRpt2-induced cell death. These data also suggest that SA signaling mediated by both NPR1 and NPR3/NPR4 plays critical roles in dampening cell death during ETI.
Consistent with the effects of pathogen-induced SA accumulation on inhibition of HR, avirulent pathogen-induced cell death in several autoimmune mutants with high SA levels was found to be greatly reduced. For example, cell death induced by P.s.m. ES4326 strains carrying avrRpt2 or avrRpm1 is dramatically reduced in aberrant growth and death2 (agd2) plants (Rate and Greenberg, 2001). The reduced cell death can be restored back to wild type level by introducing NahG or npr1-1 into agd2, suggesting that the high SA level in agd2 is responsible for the suppression of cell death activated during ETI. In the hypersensitive response like lesions1 (hrl1) mutant, cell death induced by AvrRpt2 and AvrRpm1 is also greatly reduced (Devadas and Raina, 2002). Similarly, introducing NahG or npr1-1 into hrl1 leads to restoration of RPM1-mediated cell death. In another class of autoimmune mutants, including dnd1 and dnd2, gene-for-gene resistance is normal, but there is almost no HR following infection by avirulent bacterial pathogens (Yu et al., 1998; Jurkowski et al., 2004). Both dnd1 and dnd2 accumulate high levels of SA in the absence of pathogen infection, which is likely responsible for the lack of ETI-induced HR in these mutants.
Arabidopsis NPR3 and NPR4 function redundantly in negative regulation of defense gene expression. npr3 npr4 double mutants accumulate similar levels of SA as wild type plants, but constitutively express PR genes and exhibit enhanced resistance to virulent pathogens (Zhang et al., 2006). Interestingly, HR activated by AvrRpt2 is almost completely blocked in npr3 npr4 double mutant plants (Fu et al., 2012). AvrRpt2-induced HR is restored in the npr3 np4 npr1 triple mutant [9], suggesting that constitutive activation of SA response in npr3 npr4 mutants is responsible for the suppression of cell death activated by AvrRpt2. This is consistent with reduced ETI-induced cell death in autoimmune mutants with high SA levels.
In conclusion, SA plays dual roles in the regulation of programed cell death in plants. The exact mechanism of how SA regulates cell death is currently still unclear. Analysis of early SA-responsive genes by RNA-sequencing revealed that a large number of positive regulators of defense signaling are strongly up-regulated 1 h after SA treatment (Ding et al., 2018). Induction of these defense regulators may play critical roles in potentiating defense signaling leading to activation of cell death. Meanwhile, many known negative regulators of plant immunity are also rapidly induced after SA treatment. Induction of such negative immune regulators could lead to negative feedback regulation of defense responses and cell death, which is critical in controlling the magnitude of cell death and preventing the spread of cell death beyond the infection site. The key regulatory components downstream of the SA receptors that are involved in SA-mediated inhibition of ETI-induced cell death remain to be determined in the future.
Author Contributions
YZ designed the experiments. AR performed the experiments. All authors wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yuli Ding, Tongjun Sun, and Di Wu (The University of British Columbia) for assistance with the ion leakage analysis and Natural Sciences and Engineering Research Council (NSERC) of Canada and Canada Foundation for Innovation (CFI) for the financial support.
References
- Brodersen P., Malinovsky F. G., Hématy K., Newman M.-A., Mundy J. (2005). The role of salicylic acid in the induction of cell death in Arabidopsis acd11. Plant Physiol. 138 1037–1045. 10.1104/pp.105.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman Q., Raynaud C., Benhamed M., Delarue M. (2015). To die or not to die? Lessons from lesion mimic mutants. Front. Plant Sci. 6:24. 10.3389/fpls.2015.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Bowling S. A., Gordon A. S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592. 10.1105/tpc.6.11.1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney T. P., Friedrich L., Ryals J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92 6602–6606. 10.1073/pnas.92.14.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres C., DeLong C., Glaze S., Liu E., Fobert P. R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12 279–290. 10.1105/tpc.12.2.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devadas S. K., Raina R. (2002). Preexisting systemic acquired resistance suppresses hypersensitive response-associated cell death in Arabidopsis hrl1 mutant. Plant Physiol. 128 1234–1244. 10.1104/pp.010941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewdney J., Reuber T. L., Wildermuth M. C., Devoto A., Cui J., Stutius L. M., et al. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24 205–218. 10.1046/j.1365-313x.2000.00870.x [DOI] [PubMed] [Google Scholar]
- Dietrich R. A., Delaney T. P., Uknes S. J., Ward E. R., Ryals J. A., Dangl J. L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77 565–577. 10.1016/0092-8674(94)90218-6 [DOI] [PubMed] [Google Scholar]
- Ding Y., Sun T., Ao K., Peng Y., Zhang Y., Li X., et al. (2018). Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173 1454.e15–1467.e15. 10.1016/j.cell.2018.03.044 [DOI] [PubMed] [Google Scholar]
- Fan W., Dong X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14 1377–1389. 10.1105/tpc.001628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z. Q., Yan S., Saleh A., Wang W., Ruble J., Oka N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486 228–232. 10.1038/nature11162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M. D., Delaney T. P., Dietrich R. A., Weymann K. B., Dangl J. L., Ryals J. A. (1997). Salicylate-independent lesion formation in Arabidopsis lsd mutants. Mol. Plant Microbe Interact. 10 531–536. 10.1094/MPMI.1997.10.5.531 [DOI] [PubMed] [Google Scholar]
- Jurkowski G. I., Smith R. K., Jr., Yu I. C., Ham J. H., Sharma S. B., Klessig D. F., et al. (2004). Arabidopsis DND2, a second cyclic nucleotide-gated ion channel gene for which mutation causes the “defense, no death” phenotype. Mol. Plant Microbe Interact. 17 511–520. 10.1094/MPMI.2004.17.5.511 [DOI] [PubMed] [Google Scholar]
- Li X., Clarke J. D., Zhang Y., Dong X. (2001). Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant Microbe Interact. 14 1131–1139. 10.1094/MPMI.2001.14.10.1131 [DOI] [PubMed] [Google Scholar]
- Malamy J., Carr J. P., Klessig D. F., Raskin I. (1990). Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250 1002–1004. 10.1126/science.250.4983.1002 [DOI] [PubMed] [Google Scholar]
- Manohar M., Tian M., Moreau M., Park S.-W., Choi H. W., Fei Z., et al. (2015). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. 10.3389/fpls.2014.00777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Heck S., Parinthawong N., Metraux J. P. (2002). EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14 275–286. 10.1105/tpc.010376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C., Metraux J. P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 1393–1404. 10.2307/3870970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D. N., Cuenca J. V., Bowman G. R., Guttman D. S., Greenberg J. T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell 11 1695–1708. 10.1105/tpc.11.9.1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D. N., Greenberg J. T. (2001). The Arabidopsis aberrant growth and death2 mutant shows resistance to Pseudomonas syringae and reveals a role for NPR1 in suppressing hypersensitive cell death. Plant J. 27 203–211. 10.1046/j.0960-7412.2001.1075umedoc.x [DOI] [PubMed] [Google Scholar]
- Serrano M., Wang B., Aryal B., Garcion C., Abou-Mansour E., Heck S., et al. (2013). Export of salicylic acid from the chloroplast requires the multidrug and toxin extrusion-like transporter EDS5. Plant Physiol. 162 1815–1821. 10.1104/pp.113.218156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A. C., Dempsey D. A., Klessig D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47 177–206. 10.1146/annurev.phyto.050908.135202 [DOI] [PubMed] [Google Scholar]
- Weymann K., Hunt M., Uknes S., Neuenschwander U., Lawton K., Steiner H.-Y., et al. (1995). Suppression and restoration of lesion formation in Arabidopsis lsd mutants. Plant Cell 7 2013–2022. 10.1105/tpc.7.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M. C., Dewdney J., Wu G., Ausubel F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414 562–565. 10.1038/35107108 [DOI] [PubMed] [Google Scholar]
- Wu Y., Zhang D., Chu J. Y., Boyle P., Wang Y., Brindle I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1 639–647. 10.1016/j.celrep.2012.05.008 [DOI] [PubMed] [Google Scholar]
- Yu I. C., Parker J., Bent A. F. (1998). Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc. Natl. Acad. Sci. U.S.A. 95 7819–7824. 10.1073/pnas.95.13.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Cheng Y. T., Qu N., Zhao Q., Bi D., Li X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48 647–656. 10.1111/j.1365-313X.2006.02903.x [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fan W., Kinkema M., Li X., Dong X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96 6523–6528. 10.1073/pnas.96.11.6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tessaro M. J., Lassner M., Li X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15 2647–2653. 10.1105/tpc.014894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Feechan A., Pedersen C., Newman M. A., Qiu J. L., Olesen K. L., et al. (2007). A SNARE-protein has opposing functions in penetration resistance and defence signalling pathways. Plant J. 49 302–312. 10.1111/j.1365-313X.2006.02961.x [DOI] [PubMed] [Google Scholar]
- Zhou J. M., Trifa Y., Silva H., Pontier D., Lam E., Shah J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13 191–202. 10.1094/MPMI.2000.13.2.191 [DOI] [PubMed] [Google Scholar]