Abstract
The molecular mechanisms underlying osteoarthritis (OA) and Kashin-Beck disease (KBD) remain poorly understood. Hypertrophic chondrocytes serve an important role in the development of both OA and KBD, whereas oxidative stress can contribute to the pathological progression of cartilage damage. Therefore, the aim of the present study was to detect altered expression of osteogenesis-related genes in hypertrophic chondrocytes, following treatment with 3-morpholinosydnonimine (SIN-1). ATDC5 cells were induced to develop into hypertrophic chondrocytes via Insulin-Transferrin-Selenium. The appropriate concentration and time of SIN-1 treatment was determined via MTT assay. Following hypertrophic chondrocyte stimulation with SIN-1, a liquid chip was analyzed using a polymerase chain reaction (PCR) array. Reverse transcription-quantitative PCR was conducted on individual genes to validate the array-based data. Analyses of protein-protein interactions, gene ontology functions and Kyoto Encyclopedia of Genes and Genomes pathway enrichment of the differentially expressed genes were also performed. A total of 6 upregulated and 34 downregulated genes were identified, including the mothers against decapentaplegic homolog (Smad) family (Smad1-4), bone morphogenetic proteins and their receptors (Bmp2, Bmp3, Bmpr1α and Bmpr1β), and matrix metalloproteinases (MMP2,−9 and−10). These genes are associated with collagen biology, transcriptional control, skeletal development, bone mineral metabolism, and cell adhesion. SIN-1 induced death of hypertrophic chondrocytes likely through TGF-β/Smad or BMP/Smad pathways. Oxidative-stress-dependent induction of abnormal gene expression may be associated with chondronecrosis in the cartilage of patients with OA or KBD.
Keywords: gene expression, hypertrophic chondrocytes, 3-morpholinosydnonimine, Kashin-Beck disease, osteoarthritis
Introduction
Osteoarthritis (OA) is a common form of arthritis, a disease of the joints, considered the leading cause of disability worldwide (1). Kashin-Beck disease (KBD) is an endemic OA, prevailing in China, Eastern Siberia, and North Korea (2). OA and KBD not only affect physical function, but also cause emotional stress to patients (3). Although OA and KBD have similar clinical features, such as common articular cartilage lesions and chronic pain, the pathogenesis is quite different. KBD is characterized by degeneration and necrosis in the deep zone of articular cartilage and epiphyseal plate cartilage, whereas in OA, progressive articular cartilage degeneration and synovial inflammation are the major pathological processes (4). OA is more common in the elderly, whereas KBD typically affects children and adolescents (3). A clear understanding of the molecular mechanisms underlying both OA and KBD remains elusive, and prevents the development of effective therapeutic strategies.
Cartilage is composed of chondrocytes and the extracellular matrix (ECM). Chondrocytes have a strong biological activity and can differentiate and hypertrophy (5). The ECM has an important role in cartilage regeneration and degradation (5). The family of matrix metalloproteinases (MMPs) has been reported as the primary factor in arthritis (6). Integrin, as a transmembrane receptor, mediates the connection between the cell and its external environment (such as the ECM) (5). Therefore, when MMP activity is abnormal, the function of chondrocytes will inevitably be affected through integrins (7,8). In OA, cartilage may contain excessive numbers of hypertrophy-like chondrocytes (9); however, chondrocytes can exhibit dedifferentiation in KBD. Although the two diseases have certain differences, there is no doubt that chondrocyte hypertrophy is abnormal in both OA and KBD. Furthermore, OA and KBD are oxidative-stress-associated diseases. In affected joints, oxidative DNA damage occurring in chondrocytes accumulates with OA progression (10).
In a previous study, 4-hydroxy-2-nonenal and 8-hydroxydeoxyguanisine were demonstrated to accumulate in the articular cartilage of KBD patients (11). A generator of peroxynitrite (ONOO−) in an aqueous solution, 3-morpholinosydnonimine (SIN-1), has been widely used in studies on oxidative or nitrosative stress (12,13). Considering the instability of authentic ONOO− at physiological pH, SIN-1 was chosen as an NO donor in the present study. Although numerous studies have been performed on deregulated hypertrophic differentiation and oxidative stress in chondrocytes of joint cartilages, studies that explore the exact molecular mechanism of oxidative stress in hypertrophic chondrocytes have not been published in recent years. In the present study, expression of osteogenesis genes was detected in hypertrophic chondrocytes treated with SIN-1 using chip array, and the interactions between these deregulated genes and how signaling factors change in OA was discussed. These data will provide new insights into the pathogenesis of OA and KBD.
Materials and methods
Cell culture and establishment of the hypertrophic chondrocyte model
The murine chondrogenitor cell line ATDC5 is internationally used to study cartilage differentiation in vitro, and has previously been applied to set up a hypertrophic chondrocyte model (14). For the present study, the ATDC5 cell line was purchased from the European Collection of Cell Cultures (Salisbury, UK). ATDC5 cells were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium (DMEM/F-12 medium; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), supplemented with 5% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences), penicillin (100 U/ml; Hyclone; GE Healthcare Life Sciences), and streptomycin (100 µg/ml; Hyclone; GE Healthcare Life Sciences). The cells were maintained at 37°C. ATDC5 cells were driven to hypertrophy using Insulin-Transferrin-Selenium (ITS) differentiation medium [DMEM/F-12 containing 5% FBS, penicillin (100 U/ml), streptomycin (100 µg/ml) and 1% ITS (cat. no. 354352; BD Biosciences, Franklin Lakes, NJ, USA)]. The ITS differentiation medium was changed every other day.
Identification of the hypertrophic chondrocyte model via reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
ATDC5 cells were seeded at a density of 4×104 cells/well in a six-well plate. Total RNA was isolated at 7, 14 and 21 days following ITS addition, using an RNeasy Plus Mini Kit (cat. no. 74104; Qiagen GmbH, Hilden, Germany). The concentration and purity of total RNA were determined by spectrophotometric measurement on a NanoDrop ND-2000 (NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, Delaware, USA). A total of 1 µg of total RNA from each sample was reverse transcribed using a Revert Aid™ First cDNA Synthesis Kit (cat. no. K1622; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and the resulting cDNA was diluted five times in nuclease-free water. Type X collagen (Col X) and runt-related transcription factor 2 (Runx2), as marker genes of hypertrophy, were assessed during the induction of hypertrophy in ATDC5 cells using the QuantiFast SYBR Green PCR kit (cat. no. 204054; Qiagen GmbH, Hilden, Germany). The thermocycling conditions were as follows: One cycle at 95°C for 10 min; 40 cycles at 95°C for 5 sec, 60°C for 10 sec and 72°C for 10 sec. GAPDH mRNA served as an endogenous control. The synthetic oligonucleotide primers (Shanghai Boya Biotechnology Co., Ltd., Shanghai, China) for RT-qPCR are presented in Table I. The relative mRNA expression of the target genes were calculated using relative quantification (2−ΔΔCq) (15).
Table I.
Primers used in reverse transcription-quantitative polymerase chain reaction experiments.
| Genes | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| GAPDH | GGGCTCATGACCACAGTCCATGC | CCTTGCCCACAGCCTTGGCA |
| Col X | ACGCATCTCCCAGCACCAGAATC | GGGGCTAGCAAGTGGGCCCT |
| Runx2 | GGTTGTAGCCCTCGGAGAGG | GCCATGACGGTAACCACAGTC |
| Smad1 | AAAGACCTGTGGCTTCCGTCT | TTATCGTGGCTCCTTCGTCAG |
| Smad2 | ATGTCGTCCATCTTGCCATTC | AACCGTCCTGTTTTCTTTAGCTT |
| Smad3 | GCTGCCCTCCTAGCTCAG | GGTGCTGGTCACTGTCTGTC |
| Smad4 | GAGAACATTGGATGGACGACT | CACAGACGGGCATAGATCAC |
| Col2a1 | CCAGCTGACCTCGCCACTGC | GGGTCCAGGCGCACCCTTTT |
| MMP10 | GCAGCCCATGAACTTGGCCACT | AGGGACCGGCTCCATACAGGG |
| Vcam1 | GATAGACAGCCCACTAAACGCG | GAATCTCTGGATCCTTGGGG |
Col X, type X collagen; Runx2, runt-related transcription factor 2; Smad, mothers against decapentaplegic homolog; Col 2a, collagen, type II, α; MMP10, matrix metalloproteinase 10; Vcam1, vascular cell adhesion molecule 1.
Identification of the hypertrophic chondrocyte model by a western blotting assay
Proteins were harvested at 7, 14 and 21 days following ITS addition using radioimmunoprecipitation assay reagent (Beyotime Institute of Biotechnology, Haimen, China). Proteins were measured using a BCA kit (Beyotime Institute of Biotechnology). The protein levels of Col X and Runx2 were measured by western blotting. Equal amounts (20 µg) of total protein were separated by 10% SDS-PAGE, and were then transferred to an Immobilon polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). Following blocking in 5% skimmed milk for 30 min at room temperature, the membranes were incubated with primary antibodies against Col X (1:200; cat. no. ab58632; Abcam, Cambridge, UK), Runx2 (1:1,000; cat. no. 12556; Cell Signaling Technology, Inc., Danvers, MA, USA) and GAPDH (1:500; Boster Biological Technology, Pleasanton, CA, USA) for 40 min at 37°C and then overnight at 4°C. After washing, the secondary antibody [horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (1:10,000; cat. no. 120745; Jackson ImmunoResearch Laboratories Inc, West Grove, PA, USA)] was incubated for 30 min at 37°C. Following washing, the membrane was reacted with the Immobilon Chemiluminescent HRP substrate (EMD Millipore) in accordance with the manufacturer's protocol. Images were captured and analyzed by means of the SuperSignal Ultra Western blot chemiluminescence system (Gene Co., Ltd., Hong Kong, China).
MTT assay to determine suitable SIN-1 concentration and incubation time
ATDC5 cells were seeded in a 96-well plate (1×103 cells/well) and the ITS differentiation medium was used from the next day-day 21. The medium was changed every other day. Cells were incubated with 0, 1, 5 and 10 mM SIN-1 for 4 h, or 0, 1, 2, 3, 4 or 5 mM SIN-1 for 24 h at 37°C, and then further incubated for another 4 h in medium containing 0.5 mg/ml MTT (MP Biomedicals, LLC, Santa Ana, CA, USA) at 37°C. The cell culture medium was discarded and the intracellular purple formazan in each well was dissolved in 150 µl dimethyl sulfoxide. The purple crystals were quantified by measuring the absorbance at a wavelength of 490 nm in a microplate reader (3550; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
RNA preparation for PCR array analysis
Total RNA from hypertrophic chondrocytes stimulated by 0 or 3 mM SIN-1 for 24 h was isolated using the RNeasy Plus Mini Kit (Qiagen GmbH). The concentration and purity of total RNA were measured, and RNA quality and integrity were evaluated by 2% agarose gel electrophoresis. cDNA was synthesized from 1 µg total RNA from each sample with the RT2 First Strand Kit (cat. no. 330401; Qiagen GmbH). Eliminating genomic DNA contamination with the RT2 Profiler PCR Array was essential for obtaining optimal real-time gene expression profiling results. The mixed sample was incubated at 42°C for 15 min and 95°C for 5 min. A total of 91 µl RNase-free water was added to each 20 µl cDNA synthesis reaction. The samples were mixed by pipetting up and down several times. The mixtures were kept on ice until the PCR procedure or stored at−20°C until processing.
Liquid chip analysis using RT2 Profiler PCR Arrayss®
The 102 µl cDNA synthesis reaction was diluted with 1,248 µl RNase-free water and then added to 1,350 µl RT2 qPCR SYBR Green Mastermix (cat. no. 330522; Qiagen GmbH). A total of 25 µl PCR master mix was dispensed into each well of the 96-well Mouse Osteogenesis RT2 Profiler PCR Array (cat. no. PAMM-026Z; Qiagen GmbH). qPCR was performed on a Thermal Cycler Dice Real Time System (TP-800; Takara Bio, Inc., Otsu, Japan) via SYBR Green detection, and the following thermal cycling program: 1 cycle at 95°C for 10 min; 40 cycles of 95°C for 15 sec, 55°C for 40 sec, and 72°C for 30 sec; and 1 cycle at 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec. All data from the real-time instrument were interpreted via the PCR Array Data web tool (https://www.qiagen.com/cn/shop/genes-and-pathways/data-analysis-center-overview-page/?akamai-feo=off; SABiosciences; Qiagen GmbH).
Statistical analysis of PCR array data
Each PCR array included 5 housekeeping genes (Actb, B2m, GAPDH, Gusb, and Hsp90ab1) for normalization of the sample data. According to the manusfacturer's instructions of the aforementioned Mouse Osteogenesis RT2 Profiler PCR Array, if the Cq value of Mouse Genomic DNA Contamination (MGDA) control was >30, then no genomic DNA contamination was detectable. Although the PCR array was performed only once, the fold changes and P-values were calculated by means of the PCR Array Data web tool.
RT-qPCR validation
RT-qPCR was conducted to validate PCR array data. A total of 8 genes were selected, including 7 downregulated genes (Smad1-4, ColX, Vcam1 and MMP10) and 1 upregulated gene (Col2a1). The samples were prepared in the same way as described above. Total RNA was extracted using the RNeasy Plus Mini Kit (Qiagen GmbH). A total of 1 µg total RNA from each sample was reverse-transcribed with the Revert Aid™ First cDNA Synthesis kit (cat. no. K1622; Thermo Fisher Scientific, Inc.), and the resulting cDNA was diluted 5 times. GAPDH expression served as an endogenous control to normalize all samples for potential variations in mRNA content. The synthetic oligonucleotide primers for qPCR are presented in Table I. Single-stranded cDNA was amplified with the QuantiFast SYBR Green PCR Kit (cat. no. 204054; Qiagen GmbH). The thermal cycling program was the same as that in the PCR array experiment. The relative mRNA expression levels of the target genes were calculated using relative quantification (2−ΔΔCq) (15).
Analysis of protein-protein interactions (PPIs), gene ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
In the present study, PPIs, GO functions and KEGG pathway enrichment of 40 differentially expressed genes were analyzed in the STRING 10.05 software (https://string-db.org/) to understand their biological processes, molecular function, cellular components, KEGG pathways as well as their interactions at the protein level. The parameters of the reliability and the additional nodes could be adjusted according to the concrete analysis results. The PPI networks were constructed. A gene-pathway network analysis was conducted using Cytoscape 3.4.0 (http://www.cytoscape.org/).
Statistical analysis
All data were expressed as the mean ± standard deviation and analyzed using SPSS version 16.0 (SPSS, Inc., Chicago, IL, USA). Differences between two sampled were analyzed using Student's t-test. Data with more than two comparisons were analyzed using one-way analysis of variance followed by post hoc Fisher's least significant difference test or Bonferroni's multiple-comparison test, as required. P<0.05 was considered to indicate a statistically significant difference.
Results
Establishment of the hypertrophic chondrocyte model
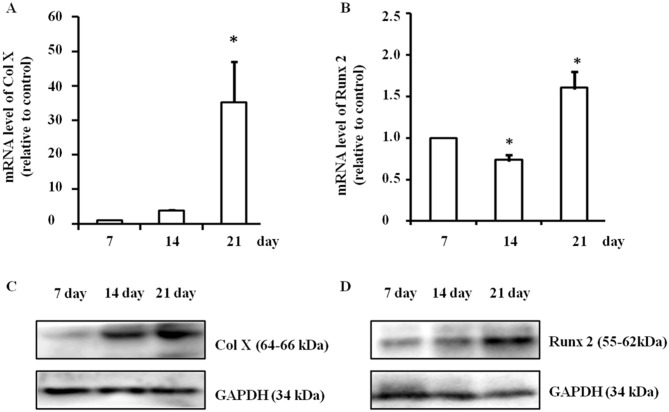
During the induction of hypertrophy, mRNA expression of Col X was significantly upregulated (P<0.05) and it peaked on day 21 (P<0.05; Fig. 1A). The mRNA expression of Runx2 was also significantly changed (P<0.05); it increased significantly on day 21 (P<0.05), but decreased on day 14 (P<0.05) compared with day 7 (P<0.05; Fig. 1B). Concurrently, protein expression levels of Col X and Runx2 also markedly increased on day 21, according to western blotting. Representative data from three independent experiments are presented (Fig. 1C and D). Thus, culturing ATDC5 cells for 21 days to set up the hypertrophic chondrocyte model was carried out for all subsequent experiments.
Figure 1.
Establishment of the hypertrophic chondrocyte model. The mRNA expression levels of (A) Col X and (B) Runx2 during the differentiation process were detected by reverse transcription-quantitative polymerase chain reaction. The group treated for 7 days served as a control. Simultaneously, protein expression levels of (C) Col X and (D) Runx2 were assessed via western blotting. Data are expressed as the mean ± standard deviation from three independent experiments. *P<0.05 vs. day 7. Col X, type X collagen; Runx2, runt-related transcription factor 2.
The effect of SIN-1 on vitality of hypertrophic chondrocytes
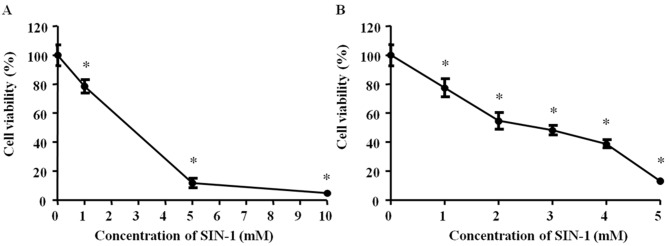
Subsequently, 0, 1, 5 or 10 mM SIN-1 was added to hypertrophic chondrocytes for 4 h incubation and cellular viability was measured via the MTT assay. All SIN-1 concentrations significantly decreased the cell viability (P<0.05). The results demonstrated a high degree of toxicity with 10 mM SIN-1 P<0.05; Fig. 2A). Subsequently, 0, 1, 2, 3, 4 or 5 mM SIN-1 was added to hypertrophic chondrocytes for 24 h incubation and cell viability was measured via the MTT assay. All SIN-1 concentrations significantly decreased the cell viability (P<0.05). There was a significant negative association between cellular viability and SIN-1 concentration, and 3 mM SIN-1 decreased viability by 50% (P<0.05; Fig. 2B). Therefore, 3 mM SIN-1 treatment was selected for 24 h as the standard procedure for the subsequent experiments.
Figure 2.
Viability of hypertrophic chondrocytes treated with SIN-1. (A) Cells were treated with 0, 1, 5 or 10 mM SIN-1 for 4 h. (B) Cells were treated with 0, 1, 2, 3, 4 or 5 mM SIN-1 for 24 h. Data are expressed as the mean ± standard deviation from three independent experiments. *P<0.05 vs. 0 mM. SIN-1, 3-morpholinosydnonimine.
Differentially expressed genes in the chip array
In the PCR array, a total of 84 genes associated with osteogenesis were compared between the control and 3-mM SIN-1 group in hypertrophic chondrocytes. A scatter plot of 84 genes indicated that 40 genes were differentially expressed by 3-fold or greater (Fig. 3). In total, 34 genes were downregulated (Table II), which were mostly associated with collagen, gene expression regulation (transcription factors), skeletal development, bone mineral metabolism and cell adhesion. A total of 6 genes were upregulated (Table III), which were mostly associated with collagen, skeletal development and cell adhesion molecules.
Figure 3.
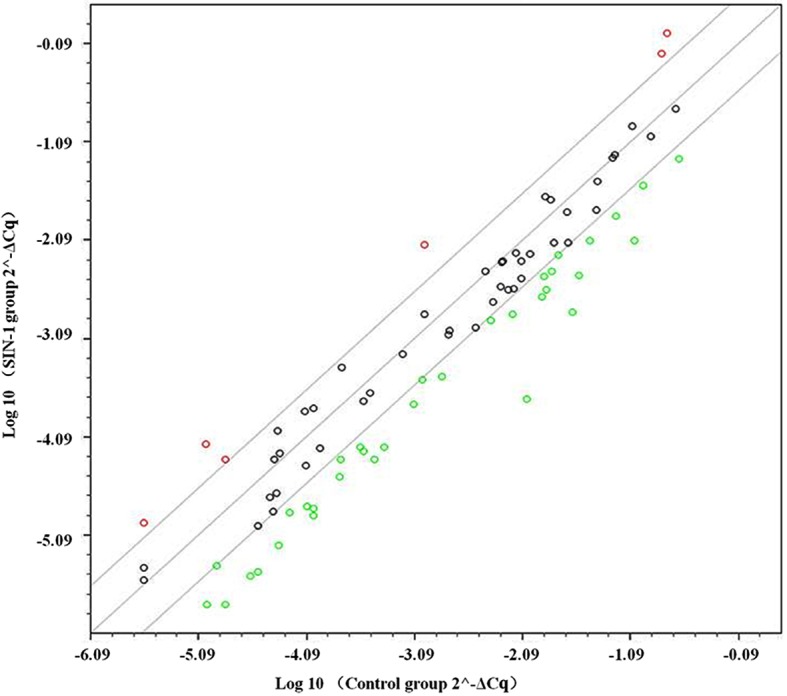
A scatter plot of 84 genes associated with osteoarthritis. Genes whose expression levels were altered by ≥3-fold were deemed differentially expressed. Upregulated genes are marked with a red circle, whereas downregulated genes are marked with a green circle. Black circles represent genes whose expression was unchanged. SIN-1, 3-morpholinosydnonimine.
Table II.
Significantly downregulated genes in hypertrophic chondrocytes treated with 3-morpholinosydnonimine.
| No. | ID | Symbol | Description | Fold regulation |
|---|---|---|---|---|
| 1 | NM_013465 | Ahsg | α-2-HS-glycoprotein | −6.19 |
| 2 | NM_007553 | BMP2 | Bone morphogenetic protein 2 | −6.11 |
| 3 | NM_173404 | BMP3 | Bone morphogenetic protein 3 | −8.52 |
| 4 | NM_009758 | BMPR1a | Bone morphogenetic protein receptor, type 1A | −4.17 |
| 5 | NM_007560 | BMPR1b | Bone morphogenetic protein receptor, type 1B | −7.16 |
| 6 | NM_007643 | Cd36 | Cluster of differentiation 36 antigen | −7.26 |
| 7 | NM_009866 | Cdh11 | Cadherin 11 | −3.05 |
| 8 | NM_009893 | Chrd | Chordin | −5.28 |
| 9 | NM_009925 | Col10a1 | Collagen, type X, α 1 | −4.03 |
| 10 | NM_016685 | Comp | Cartilage oligomeric matrix protein | −5.17 |
| 11 | NM_007802 | Ctsk | Cathepsin K | −3.14 |
| 12 | NM_010197 | Fgf1 | Fibroblast growth factor 1 | −7.89 |
| 13 | NM_010206 | Fgfr1 | Fibroblast growth factor receptor 1 | −11.16 |
| 14 | NM_010207 | Fgfr2 | Fibroblast growth factor receptor 2 | −3.34 |
| 15 | NM_010228 | Flt1 | FMS-like tyrosine kinase 1 | −7.41 |
| 16 | NM_010512 | Igf1 | Insulin-like growth factor 1 | −3.48 |
| 17 | NM_008396 | Itga2 | Integrin α 2 | −4.76 |
| 18 | NM_008402 | Itgav | Integrin α V | −7.52 |
| 19 | NM_010578 | Itgb1 | Integrin β 1 (fibronectin receptor β) | −4.20 |
| 20 | NM_019471 | MMP10 | Matrix metalloproteinase 10 | −45.25 |
| 21 | NM_008610 | MMP2 | Matrix metalloproteinase 2 | −3.66 |
| 22 | NM_013599 | MMP9 | Matrix metalloproteinase 9 | −4.66 |
| 23 | NM_008689 | Nfkb1 | Nuclear factor of κ light polypeptide gene enhancer in B cells 1, p105 | −3.66 |
| 24 | NM_011077 | Phex | Phosphate regulating gene with homologies to endopeptidases on the X chromosome | −4.44 |
| 25 | NM_008539 | Smad1 | Mothers against decapentaplegic homolog 1 (Drosophila) | −3.89 |
| 26 | NM_010754 | Smad2 | Mothers against decapentaplegic homolog 2 (Drosophila) | −3.10 |
| 27 | NM_016769 | Smad3 | Mothers against decapentaplegic homolog 3 (Drosophila) | −4.63 |
| 28 | NM_008540 | Smad4 | Mothers against decapentaplegic homolog 4 (Drosophila) | −4.17 |
| 29 | NM_009367 | Tgfb2 | Transforming growth factor, β 2 | −5.31 |
| 30 | NM_009370 | Tgfbr1 | Transforming growth factor, β receptor I | −16.00 |
| 31 | NM_009371 | Tgfbr2 | Transforming growth factor, β receptor II | −5.70 |
| 32 | NM_013693 | Tnf | Tumor necrosis factor | −9.19 |
| 33 | NM_011613 | Tnfsf11 | Tumor necrosis factor (ligand) superfamily, member 11 | −4.14 |
| 34 | NM_011693 | Vcam1 | Vascular cell adhesion molecule 1 | −6.68 |
Differentially expressed genes that were deemed differentially expressed by 3-fold or greater are presented.
Table III.
Significantly upregulated genes in hypertrophic chondrocytes treated with 3-morpholinosydnonimine.
| No. | ID | Symbol | Description | Fold regulation |
|---|---|---|---|---|
| 1 | NM_007542 | Bgn | Biglycan | 5.82 |
| 2 | NM_007555 | Bmp5 | Bone morphogenetic protein 5 | 7.26 |
| 3 | NM_007743 | Col1a2 | Collagen, type I, α 2 | 4.08 |
| 4 | NM_031163 | Col2a1 | Collagen, type II, α 1 | 7.26 |
| 5 | NM_010544 | Ihh | Indian hedgehog | 3.32 |
| 6 | NM_024449 | Sost | Sclerostin | 4.26 |
Differentially expressed genes that were deemed differentially expressed by 3-fold or greater are presented.
mRNA levels of 9 associated genes in experimental hypertrophic chondrocytes
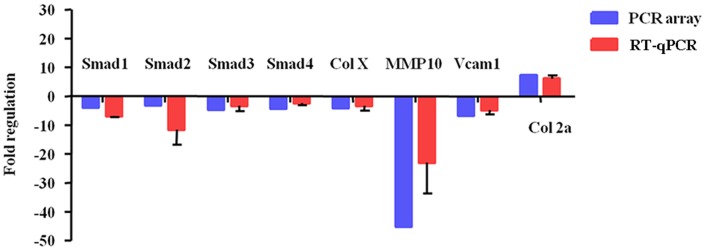
To further confirm these PCR array data, 8 differentially expressed genes (Smad1-4, Col X, MMP10, Vcam1, and Col2a1) were validated in hypertrophic chondrocytes treated with 0 or 3 mM SIN-1. The RT-qPCR results demonstrated that 3 mM SIN-1 induced Smad1-4, Col X, MMP10, and Vcam1 downregulation and Col2a1 upregulation (Fig. 4). These results are consistent with and support the reliability of the PCR array findings.
Figure 4.
A histogram presenting mRNA expression levels of 8 selected genes in hypertrophic chondrocytes treated with 3-morpholinosydnonimine. GAPDH served as an internal control. Data are expressed as the mean ± standard deviation. PCR, polymerase chain reaction; RT-qPCR, reverse transcription-quantitative PCR; Smad, mothers against decapentaplegic homolog; Col X, type X collagen; MMP, matrix metalloproteinase; Vcam1, vascular cell adhesion molecule 1; Col 2a, collagen, type II, α.
Transforming growth factor (TGF)-β signaling cascades are the key pathways in hypertrophic chondrocytes treated with SIN-1
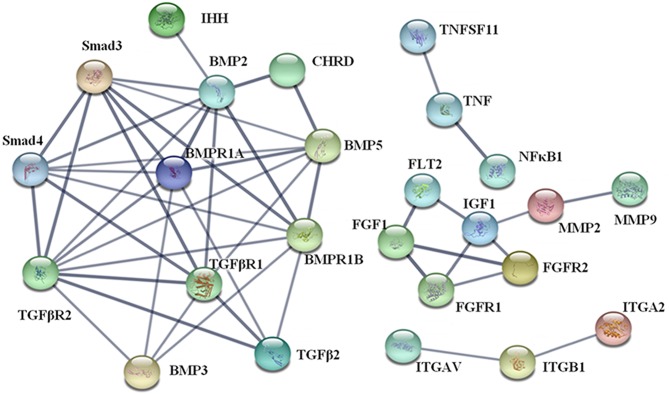
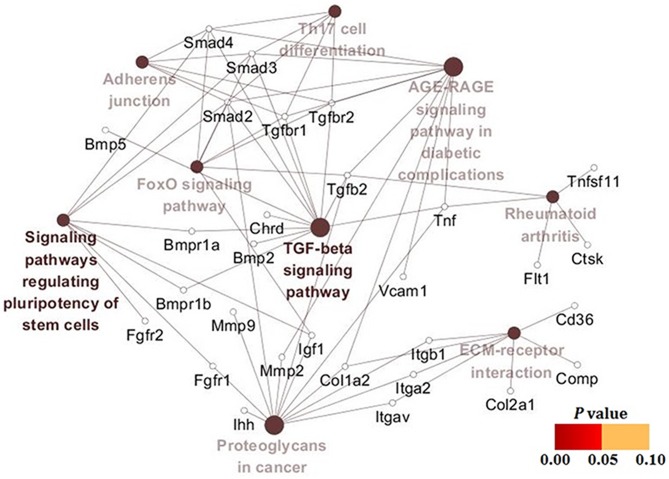
The results of PPI analysis demonstrated that there were four clusters in the PPI network (Fig. 5). The most important cluster included TGF-βs [TGF-β2 and TGF-β receptors (TGF-βr1 and−2)], bone morphogenic proteins (BMPs; BMP2,−3, and−5, and BMP receptors 1A and 1B) and mothers against decapentaplegic homologs (Smads; Smad3 and−4), which were beneficial for the elucidation of biological systems participating in the oxidative stress in hypertrophic chondrocytes. KEGG pathway enrichment results (Table IV) also demonstrated that TGF-β signaling cascades were the key pathways in hypertrophic chondrocytes treated with SIN-1, indicating that TGF-β/Smad signaling and BMP/Smad signaling were potential pathways associated with the above process. Once again, the gene-pathway network analysis yielded results similar to the above findings (Fig. 6).
Figure 5.
Network of protein-protein interactions for differentially expressed genes.
Table IV.
Results on top five GO functions and KEGG pathway enrichment analysis of differentially expressed genes.
| Function and pathway | ID | Description | Count in gene set | False discovery rate |
|---|---|---|---|---|
| Biological process | GO: 0001501 | Skeletal system development | 20 | 2.03×10−20 |
| (GO) | GO: 0009888 | Tissue development | 27 | 3.18×10−19 |
| GO: 0001503 | Ossification | 16 | 2.27×10−18 | |
| GO: 0071495 | Cellular response to endogenous stimulus | 21 | 4.40×10−18 | |
| GO: 0071363 | Cellular response to growth factor stimulus | 17 | 5.39×10−18 | |
| Molecular function | GO: 0046332 | Mothers against decapentaplegic homolog binding | 10 | 5.84×10−14 |
| (GO) | GO: 0005515 | Protein binding | 32 | 4.67×10−13 |
| GO: 0005160 | Transforming growth factor β receptor binding | 8 | 1.02×10−11 | |
| GO: 0005126 | Cytokine receptor binding | 10 | 1.09×10−9 | |
| GO: 0019838 | Growth factor binding | 8 | 1.58×10−9 | |
| Cellular component | GO: 0005615 | Extracellular space | 20 | 2.99×10−13 |
| (GO) | GO: 0044421 | Extracellular region part | 26 | 2.01×10−10 |
| GO: 0005576 | Extracellular region | 27 | 3.32×10−10 | |
| GO: 0031012 | Extracellular matrix | 12 | 3.32×10−10 | |
| GO: 0005578 | Proteinaceous extracellular matrix | 11 | 1.35×10−9 | |
| KEGG pathways | 4350 | Transforming growth factor-β signaling pathway | 13 | 1.50×10−20 |
| 5200 | Pathways in cancer | 17 | 2.04×10−19 | |
| 4390 | Hippo signaling pathway | 12 | 1.87×10−15 | |
| 5205 | Proteoglycans in cancer | 12 | 1.53×10−13 | |
| 4151 | Phosphoinositide 3-kinase-protein kinase B signaling pathway | 12 | 2.49×10−11 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 6.
Analysis of signaling pathways for differentially expressed genes.
Discussion
The oxidative stress in hypertrophic chondrocytes is regulated by multiple intricate signal transduction pathways. The crosstalk among these signals has a fine balance in normal hypertrophic chondrocytes (16). Free radicals, called ‘the second messengers,’ may adjust the expression of relevant target proteins through nuclear transcription factors and regulate chondrocyte growth, differentiation, and maturation (17,18). In the normal physiological state, there is a dynamic balance between the free-radical-generating system and the scavenging system (19). Once excessive free radicals are produced or ingested, the original balanced state is disrupted and then causes cell damage (20). On the basis of previous research findings that implicated oxidative damage in the hypertrophy of chondrocytes in OA and KBD (14,21), the present study was designed to profile altered gene expression of the intricate signaling network associated with oxidative stress to mimic cellular responses within an articular joint affected by OA or KBD.
In the present study, the TGF-β (TGF-β1, −2 and -3, and TGF-βr1,−2 and -3) and Smad (Smad1-5) families of genes. A total of 7 genes were downregulated in hypertrophic chondrocytes following treatment with SIN-1, including TGF-β2, TGF-βr1, TGF-βr2, Smad1, Smad2, Smad3, and Smad4. Smads are the downregulated gene targets of the TGF-β family of genes. TGF-β transduces signals from the cell membrane to the nucleus via its receptors and Smad proteins (22). In a European patient cohort, a single nucleotide polymorphism in the intron region of the Smad3 gene has been demonstrated to be associated with hip and knee OA (23). TGF-β may stimulate chondrocyte matrix production and has been demonstrated to promote cartilage repair to alleviate OA (24). TGF-β/Smad3 signals could repress chondrocyte hypertrophic differentiation, and are essential for maintaining articular cartilage (22). TGF-β3 and phosphorylated Smad2 levels were significantly reduced in two murine models of osteoarthritis, accompanied by a loss of proteoglycans (25). In analyses of PPIs, KEGG pathway enrichment and gene-pathway network in the present study, it was demonstrated that TGF-β signaling cascades were the key pathways in hypertrophic chondrocytes treated with SIN-1. In accordance with previous results, the present study indicates that these differentially expressed TGF-β- and Smad-family genes may provide novel approaches for drug targeting in OA and other osteoarthroses.
BMPs are members of the pleiotropic TGF-β superfamily and are key regulators of skeletal development, osteogenesis and bone healing (26,27). BMP signals are mediated by type I and type II receptors (BMPR1a, BMPR1b and BMPR2) (26). BMP signal binding to BMPRs is translocated to the nucleus via Smad signaling (mainly via Smad1,−5 and−8) (28). BMP2 significantly promotes bone formation and stimulates osteoblast differentiation of C2C12 cells in conjunction with specific levels of static stretching force (29). In addition, it has been confirmed that BMP2 stimulates chondrocyte hypertrophy during chondrogenesis of progenitor ATDC5 cells (30), and in the present study it was also demonstrated that BMP2 was downregulated in hypertrophic chondrocytes treated with SIN-1. Therefore, BMP2 is important for the progression of hypertrophy. BMP3 accelerates the differentiation of human mesenchymal stem cells (31), and is associated with rabbit articular cartilage repair (32). The BMP5 gene contains an ‘injury response’ control region, which is activated by multiple types of injury in adult animals (33). In the present study, it was demonstrated that BMP2, BMP3, BMPR1a and BMPR1b were downregulated in hypertrophic chondrocytes treated with SIN-1 and BMP5 was upregulated. Combined with the above discussion on Smads, these data suggest that the BMP/SMAD pathway may also take part in an oxidative stress reaction. The increased expression of BMP5 was probably the result of the ‘injury response’ to oxidative stress.
Matrix metalloproteinases (MMPs) serve important roles in ECM remodeling and degradation. MMPs and their inhibitors (TIMPs) are believed to be associated with the mechanisms underlying KBD (34). In the present study, it was demonstrated that the mRNA expression levels of MMP2, -9 and -10 were downregulated in hypertrophic chondrocytes following treatment with SIN-1. MMP2 and 9 are known to participate in significant degradation of collagen in the muscle of fish (35). In the present study, the mRNA expression levels of Col I and Col II were both increased; thus it was speculated that this increase was caused by downregulation of MMPs. Nevertheless, the mRNA expression level of Col X, which is mainly expressed in hypertrophic chondrocytes, was decreased in the present study. This phenomenon may be directly caused by oxidative stress induced by SIN-1. Combined with the literature, the present data suggest that the alteration of Mmp2,−9, and -10 genes led to a change in ECM, which may further affect the cell differentiation.
Integrins are the major family of ECM receptors, which have a vital role in cell-ECM interactions (36). Integrins are transmembrane heterodimeric glycoproteins consisting of α and β subunits, combining functions of cell adhesion and bidirectional signal transduction (5). In the present study, levels of α2, αv, and β1 integrins were decreased in hypertrophic chondrocytes following treatment with SIN-1. It has been previously reported that the expression of α2 and α5 integrins increased at later stages of OA in rats, and these levels were associated with the severity of OA (37). Additionally α1, α5β1, and αvβ5 integrins have been demonstrated to mediate human chondrocyte adhesion to cartilage (38). Increased expression of Indian hedgehog (Ihh) and sclerostin was also observed, as was decreased expression of Col X. Ihh is expressed in prehypertrophic chondrocytes on growth plates (39), whereas Col X was mainly expressed in hypertrophic chondrocytes. Sclerostin is a novel BMP antagonist, which competes with type I and type II BMP receptors (40). Thus, SIN-1 may first increase sclerostin and further decrease BMP signaling.
In conclusion, the present study indicated that oxidative stress produced by NO donor induced death of hypertrophic chondrocytes is probably mediated by TGF-β/Smad or BMP/Smad pathways, and dysfunctional pathways may then induce abnormal expression of MMPs and collagens. The present results may provide several novel clues to the molecular mechanisms underlying OA and KBD, which may help to find novel diagnostic and therapeutic targets for these debilitating diseases.
There were, however, certain limitations in the present study: i) KBD does not immediately lead to death, and harvesting the cartilage tissue of patients with KBD is traumatic for patients, so it was very difficult to obtain fresh human cartilage; and ii) there are no functional verification experiments to be conducted. Therefore, further research is required.
Acknowledgements
The authors would like to thank Professor Rongqiang zhang from Shaanxi University of Chinese Medicine (Xi'an, China) for guidance in writing the manuscript.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81273006 and 81573102) and the Shaanxi Natural Science Research Project (grant no. 2017JM8122).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC and YH conceived and designed experiments; YH and JC performed the experiments; YH, WY, MZ, YZ, DZ, ZJ, TM, JS and MS analyzed the data; TM, JS and MS also contributed reagents/materials/analysis tools; YH wrote the manuscript; and JC revised it.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tsolis KC, Bei ES, Papathanasiou I, Kostopoulou F, Gkretsi V, Kalantzaki K, Malizos K, Zervakis M, Tsezou A, Economou A. Comparative proteomic analysis of hypertrophic chondrocytes in osteoarthritis. Clin Proteomics. 2015;12:12. doi: 10.1186/s12014-015-9085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreno-Reyes R, Suetens C, Mathieu F, Begaux F, Zhu D, Rivera MT, Boelaert M, Nève J, Perlmutter N, Vanderpas J. Kashin-Beck osteoarthropathy in rural Tibet in relation to selenium and iodine status. N Engl J Med. 1998;339:1112–1120. doi: 10.1056/NEJM199810153391604. [DOI] [PubMed] [Google Scholar]
- 3.Zhang F, Guo X, Duan C, Wu S, Yu H, Lammi M. Identification of differentially expressed genes and pathways between primary osteoarthritis and endemic osteoarthritis (Kashin-Beck disease) Scand J Rheumatol. 2013;42:71–79. doi: 10.3109/03009742.2012.698303. [DOI] [PubMed] [Google Scholar]
- 4.Gao ZQ, Guo X, Duan C, Ma W, Xu P, Wang W, Chen JC. Altered aggrecan synthesis and collagen expression profiles in chondrocytes from patients with Kashin-Beck disease and osteoarthritis. J Int Med Res. 2012;40:1325–1334. doi: 10.1177/147323001204000411. [DOI] [PubMed] [Google Scholar]
- 5.Kim SJ, Kim EJ, Kim YH, Hahn SB, Lee JW. The modulation of integrin expression by the extracellular matrix in articular chondrocytes. Yonsei Med J. 2003;44:493–501. doi: 10.3349/ymj.2003.44.3.493. [DOI] [PubMed] [Google Scholar]
- 6.Peterson JT. The importance of estimating the therapeutic index in the development of matrix metalloproteinase inhibitors. Cardiovasc Res. 2006;69:677–687. doi: 10.1016/j.cardiores.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 7.Schulze-Tanzil G, de Souza P, Merker HJ, Shakibaei M. Co-localization of integrins and matrix metalloproteinases in the extracellular matrix of chondrocyte cultures. Histol Histopathol. 2001;16:1081–1089. doi: 10.14670/HH-16.1081. [DOI] [PubMed] [Google Scholar]
- 8.Pal S, Ganguly KK, Moulik S, Chatterjee A. Modulation of MMPs by cell surface integrin receptor α5β1. Anticancer Agents Med Chem Sep. 2012;12:726–732. doi: 10.2174/187152012802650183. [DOI] [PubMed] [Google Scholar]
- 9.van der Kraan PM, van den Berg WB. Chondrocyte hypertrophy and osteoarthritis: Role in initiation and progression of cartilage degeneration? Osteoarthritis Cartilage. 2012;20:223–232. doi: 10.1016/j.joca.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Chen AF, Davies CM, De Lin M, Fermor B. Oxidative DNA damage in osteoarthritic porcine articular cartilage. J Cell Physiol. 2008;217:828–833. doi: 10.1002/jcp.21562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W, Wei S, Luo M, Yu B, Cao J, Yang Z, Wang Z, Goldring MB, Chen J. Oxidative stress and status of antioxidant enzymes in children with Kashine-Beck disease. Osteoarthritis Cartilage. 2013;21:1781–1789. doi: 10.1016/j.joca.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Konishi K, Watanabe N, Arai T. SIN-1 cytotoxicity to PC12 cells is mediated by thiol-sensitive short-lived substances generated through SIN-1 decomposition in culture medium. Nitric Oxide. 2009;20:270–278. doi: 10.1016/j.niox.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Tian J, Yan J, Wang W, Zhong N, Tian L, Sun J, Min Z, Ma J, Lu S. T-2 toxin enhances catabolic activity of hypertrophic chondrocytes through ROS-NF-jB-HIF-2α pathway. Toxicol In Vitro. 2012;26:1106–1113. doi: 10.1016/j.tiv.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Hunter DJ, Felson DT. Osteoarthritis. BMJ. 2006;332:639–642. doi: 10.1136/bmj.332.7542.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Ueno T, Yamada M, Sugita Y, Ogawa T. N-Acetyl cysteine protects TMJ chondrocytes from oxidative stress. J Dent Res. 2011;90:353–359. doi: 10.1177/0022034510388035. [DOI] [PubMed] [Google Scholar]
- 17.Hinoi E, Takarada T, Fujimori S, Wang L, Iemata M, Uno K, Yoneda Y. Nuclear factor E2 p45-related factor 2 negatively regulates chondrogenesis. Bone. 2007;40:337–344. doi: 10.1016/j.bone.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Morita K, Miyamoto T, Fujita N, Kubota Y, Ito K, Takubo K, Miyamoto K, Ninomiya K, Suzuki T, Iwasaki R, et al. Reactive oxygen species induce chondrocyte hypertrophy in endochondral ossification. J Exp Med. 2007;204:1613–1623. doi: 10.1084/jem.20062525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoshida S, Kuzuya T, Yamashita N, Oe H, Fuji H, Hori M, Tada M, Kamada T. Brief myocardial ischemia affects free radical generating and scavenging systems in dogs. Heart Vessels. 1993;8:115–120. doi: 10.1007/BF01744795. [DOI] [PubMed] [Google Scholar]
- 20.Del Carlo M, Jr, Loeser RF. Nitric oxide-mediated chondrocyte cell death requires the generation of additional reactive oxygen species. Arthritis Rheum. 2002;46:394–403. doi: 10.1002/art.10056. [DOI] [PubMed] [Google Scholar]
- 21.Guo X, Zuo H, Cao CX, Zhang Y, Geng D, Zhang ZT, Zhang YG, von der Mark K, von der Mark H. Abnormal expression of Col X, PTHrP, TGF-beta, bFGF, and VEGF in cartilage with Kashin-Beck disease. J Bone Miner Metab. 2006;24:319–328. doi: 10.1007/s00774-006-0690-3. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Chen L, Xu X, Li C, Huang C, Deng CX. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdes AM, Spector TD, Tamm A, Kisand K, Doherty SA, Dennison EM, Mangino M, Tamm A, Kerna I, Hart DJ, et al. Genetic variation in the SMAD3 gene is associated with hip and knee osteoarthritis. Arthritis Rheum. 2010;62:2347–2352. doi: 10.1002/art.27530. [DOI] [PubMed] [Google Scholar]
- 24.Blom AB, van der Kraan PM, van den Berg WB. Cytokine targeting in osteoarthritis. Curr Drug Targets. 2007;8:283–292. doi: 10.2174/138945007779940179. [DOI] [PubMed] [Google Scholar]
- 25.Davidson Blaney EN, Vitters EL, van der Kraan PM, van den Berg WB. Expression of transforming growth factor-beta (TGFbeta) and the TGFbeta signalling molecule SMAD-2P in spontaneous and instability-induced osteoarthritis: Role in cartilage degradation, chondrogenesis and osteophyte formation. Ann Rheum Dis. 2006;65:1414–1421. doi: 10.1136/ard.2005.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: A critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Long J, Li P, Du HM, Liu L, Zheng XH, Lin YF, Wang H, Jing W, Tang W, Chen WH, et al. Effects of bone morphogenetic protein 2 gene therapy on new bone formation during mandibular distraction osteogenesis at rapid rate in rabbits. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2011;112:50–57. doi: 10.1016/j.tripleo.2010.09.065. [DOI] [PubMed] [Google Scholar]
- 28.Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad-1 and Smad-5 is required for endochondral bone formation. Development. 2009;136:1093–1104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim IS, Song YM, Cho TH, Kim JY, Weber FE, Hwang SJ. Synergistic action of static stretching and BMP-2 stimulation in the osteoblast differentiation of C2C12 myoblasts. J Biomech. 2009;42:2721–2727. doi: 10.1016/j.jbiomech.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Caron MM, Emans PJ, Cremers A, Surtel DA, Coolsen MM, van Rhijn LW, Welting TJ. Hypertrophic differentiation during chondrogenic differentiation of progenitor cells is stimulated by BMP-2 but suppressed by BMP-7. Osteoarthritis Cartilage. 2013;21:604–613. doi: 10.1016/j.joca.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Zhou X, Tao Y, Liang C, Zhang Y, Li H, Chen Q. BMP3 alone and together with TGF-β promote the differentiation of human mesenchymal stem cells into a nucleus pulposus-like phenotype. Int J Mol Sci. 2015;16:20344–20359. doi: 10.3390/ijms160920344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Z, Yang W, Cao Y, Shi Y, Lei C, Du B, Li X, Zhang Q. The functions of BMP3 in rabbit articular cartilage repair. Int J Mol Sci. 2015;16:25934–25946. doi: 10.3390/ijms161125937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guenther CA, Wang Z, Li E, Tran MC, Logan CY, Nusse R, Pantalena-Filho L, Yang GP, Kingsley DM. A distinct regulatory region of the Bmp5 locus activates gene expression following adult bone fracture or soft tissue injury. Bone. 2015;77:31–41. doi: 10.1016/j.bone.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Luo M, Wang W, Zhang Z, He Y, Duance VC, Hughes CE, Caterson B, Cao J. Altered proteolytic activity and expression of MMPs and aggrecanases and their inhibitors in Kashin-Beck disease. J Orthop Res. 2015;33:47–55. doi: 10.1002/jor.22708. [DOI] [PubMed] [Google Scholar]
- 35.Michelin AC, Justulin LA, Jr, Delella FK, Padovani CR, Felisbino SL, Dal-Pai-Silva M. Differential MMP-2 and MMP-9 activity and collagen distribution in skeletal muscle from pacu (Piaractus mesopotamicus) during juvenile and adult growth phases. Anat Rec (Hoboken) 2009;292:387–395. doi: 10.1002/ar.20863. [DOI] [PubMed] [Google Scholar]
- 36.Tian J, Zhang FJ, Lei GH. Role of integrins and their ligands in osteoarthritic cartilage. Rheumatol Int. 2015;35:787–798. doi: 10.1007/s00296-014-3137-5. [DOI] [PubMed] [Google Scholar]
- 37.Almonte-Becerril M, Costell M, Kouri JB. Changes in the integrins expression are related with the osteoarthritis severity in an experimental animal model in rats. J Orthop Res. 2014;32:1161–1166. doi: 10.1002/jor.22649. [DOI] [PubMed] [Google Scholar]
- 38.Kurtis MS, Schmidt TA, Bugbee WD, Loeser RF, Sah RL. Integrin-mediated adhesion of human articular chondrocytes to cartilage. Arthritis Rheum. 2003;48:110–118. doi: 10.1002/art.10704. [DOI] [PubMed] [Google Scholar]
- 39.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 40.Winkler DG, Sutherland MK, Geoghegan JC, Yu C, Hayes T, Skonier JE, Shpektor D, Jonas M, Kovacevich BR, Staehling-Hampton K, et al. Osteocyte control of bone formation via sclerostin, a novel BMP antagonist. EMBO J. 2003;22:6267–6276. doi: 10.1093/emboj/cdg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.