Abstract
A 16-year-old female with Ewing sarcoma, a very rare disease with poor prognosis in women, was admitted to the hospital with abdominal pain. Diagnostic laparotomy revealed the Ewing sarcoma originating from the extramural uterus. Histological examination yielded positive test results for CD99, vimentin, S-100, eosin 5-maleimide and periodic acid-Shiff. EWS-FLI1 type 1 translocation was confirmed. Fibroblast growth factor receptor (FGFR) 4 (c.1162G> A) and HRas proto-oncogene (HRAS; c.182A> G) mutations were also detected. At eight months following complete remission, pelvic lymph node metastasis was confirmed. Epithelial cell adhesion molecule-positive circulating tumor cells (CTCs) were detected in liquid biopsy. FGFR3 (c.1948A> G) and FGFR4 (c.1162G> A) mutations were found in the CTCs. FGFR4 (c.1162G> A) and HRAS (c.182A> G) mutations were confirmed in cell-free circulating tumor DNA. A sequence of EWSR1 gene was also confirmed in the CTCs. To the best of our knowledge, this is the first report of skeletal Ewing sarcoma being detected using multiple noninvasive diagnostic methods to observe genetic translocation and mutation in blood CTCs. It may be used to monitor the therapeutic effect of cancer or predict prognosis. Therefore, liquid biopsy is expected to serve a pivotal role in ultra-precise medicine in the future.
Keywords: Ewing sarcoma, circulating tumor cells, EWS-FLI1 translocation, genomic mutation profiling, liquid biopsy
Introduction
Ewing sarcoma is a very rare malignant tumor usually associated with femur, pelvis, ribs, and scapula (1). Ewing sarcoma originating from the uterus is very rare. According to Loverro et al (2), only 48 cases of Ewing sarcoma have been found in female genitalia until 2015. It is more common in men than in women (55:45). Ewing sarcoma in young females is associated with worse prognosis. It is predominantly seen in Caucasians rather than in Asians or Black teenagers and young adults (3). Ewing sarcoma is associated with primary site pelvic lengthening from the first sign until diagnosis (4). Diagnosis of Ewing sarcoma lesions is made through histological and physical examination, as well as radiologically via X-ray, CT, bone scan, and magnetic resonance imaging (MRI). A screening antibody is used to characterize Ewing sarcoma cells immunohistochemically. Cytogenetic and reverse transcription polymerase chain reaction (PCR) assays are used to determine the presence of characteristic genetic alterations in Ewing sarcoma cells. Molecular genetic testing has revealed that most Ewing sarcoma cases have chromosomes 11 and 22 dislocations involving fusion of EWS gene on chromosome 22 and FLI1 gene on 11 (5). Transmission of Ewing sarcoma is confirmed by standard imaging technique or bone marrow aspirate/biopsy. Metastasis occurs in about 25% of cases diagnosed with Ewing sarcoma (6).
Blood contains intact circulating tumor cells (CTCs) and cell-free circulating tumor DNA (cfDNA), which facilitate precise molecular diagnosis. In addition, blood sampling is relatively noninvasive and easy. CTC and cfDNA carry mutations in tissues of primary carcinomas. Clinical studies are investigating the role of CTC and cfDNA in early diagnosis of cancer, monitoring effects of anticancer therapy, and in prognosis. Liquid biopsy of cancer is expected to become a new paradigm for ultra-precise medicine (7,8). Centrifugal microfluidic system based on a new fluid-assisted separation technology (FAST) has been developed to detect CTCs in the blood based on their size and can be used in a ‘Lab-on-a-disc’ to isolate CTCs in a few ml of blood within a minute at high efficiency (more than 95%). The role of blood CTCs has been investigated in cancer diagnosis based on representative markers (9). Among malignant neoplasms of hepatocellular origin, distant metastasis occurs in half of sarcoma patients with primary lesions with primary metastasis (10). Most sarcomas spread through the vascular system following infiltration of cells from primary carcinoma into blood vessels. These blood CTCs float through the lymphatic system and cause metastasis (11). To investigate the metastatic process in rare cancers such as Ewing sarcoma and identify new therapeutic targets, blood CTCs have been characterized using biomarkers to predict prognosis of patients (12). In the present study, we found Ewing sarcoma as primary carcinoma and blood CTCs of uterine origin in a pediatric patient. Following liquid biopsy, molecular tests including personalized genetic variation analysis were conducted. Cancer mutations were monitored using CTC and cfDNA for comprehensive insight into cancer progress and prognosis.
Case report
Ethics statements and informed consent
The present study was approved by the Institutional Review Board of Chonbuk National University Hospital (Jeonju, Republic of Korea). It was conducted according to the Declaration of Helsinki for biomedical research involving human subjects and Guidelines for Good Clinical Practice. A detailed explanation of this study was provided to all subjects. Written informed consent was obtained from each participant prior to screening.
A 16-year-old female patient visited the hospital with complaints of pain and a feeling of stabbing injury to the underbelly. Abdominal distension occurred starting two weeks ago. No specific personal or family history was detected. Abdominal ultrasonography showed multiple diaphragmatic cysts on a thick wall measuring about 12 cm in size. In addition to a hemoglobin level of 9.6 g/dl (normal range, 12 to 16 g/dl), LFT, BUN, creatinine, and coagulation profiles were within normal range. No tumor markers such as CA125, CA19-9, CEA, AFP, and hCG were detected either (Table I). This is a MRI image taken at the first hospital visit. Pelvic MRI revealed a round mass measuring 12.3×11.6×13.0 cm in size and located in the left pelvic cavity. Multi-septate and focal wall thickening was also detected (Fig. 1). Preoperative laparotomy was performed under general anesthesia to investigate suspected left ovarian tumor, which originated in the posterior wall of the uterus. Complete excision of the mass was done followed by reconstruction of the uterus via myomectomy. Immunohistochemical analysis of the patient's tissues confirmed positive results for CD99, vimentin, S-100, EMA, and PAS (Fig. 2). Molecular genetic testing confirmed the fusion of EWS gene on chromosome 22 and FLI1 gene on chromosome 11, a EWS-FLI1 translocation (Fig. 3). Bone scan and PET were performed preceded by postoperative pelvic MRI, revealing no evidence of metastasis. Patients and caregivers rejected reoperation and radiotherapy. Combination chemotherapy regimens comprising VaC (vincristine, adriamycin, cyclophosphamide) and IE (ifosfamide, etoposide) were administered. Pelvic (lymph node) metastasis was confirmed in the eighth month after complete remission (Fig. 4). Using high-efficiency blood-CTC isolation technique, EpCAM-positive CTCs and vimentin-positive CTCs were detected in the patient's biopsy specimens (Fig. 5). The patient underwent cervical lymph node metastasis five months after transplantation, when EpCAM-positive CTCs and vimentin-positive CTCs were confirmed by liquid phase biopsy (Table II).
Table I.
Clinicopathological features of Ewing sarcoma.
| Characteristics | Feature |
|---|---|
| Sex | Female |
| Age | 16 |
| Primary site | Uterus (extraosseous) |
| Initial symptom | Stabbing pain in lower abdomen |
| Diagnostic evaluation | MRI |
| PET/CT scan | |
| Bone scan | |
| X-ray | |
| Blood chemistry study | |
| Hemoglobin | 9.6 g/dl |
| Liver function test | Normal range |
| BUN | Normal range |
| Creatinine | Normal range |
| Coagulation profile | Normal range |
| CA125 | 25.5 U/ml |
| CA19-9 | 4.16 U/ml |
| CEA | 1.66 ng/ml |
| AFP | 1.1 ng/ml |
| hCG | 0 mIU/m |
| IHC | |
| CD99 | Positive, membranous and cytoplasmic |
| Vimentin | Diffuse, 3+ |
| S-100 | Focal positive |
| EMA | Focal positive |
| PAS | Positive |
MRI, magnetic resonance imaging; CT, computed tomography, PET, positron emission tomography; BUN, blood urea nitrogen; CA125, cancer antigen 125; CA19-9, cancer antigen 19-9; CEA, carcinoembryonic antigen; AFP, alpha-fetoprotein; hCG, human chorionic gonadotropin.
Figure 1.
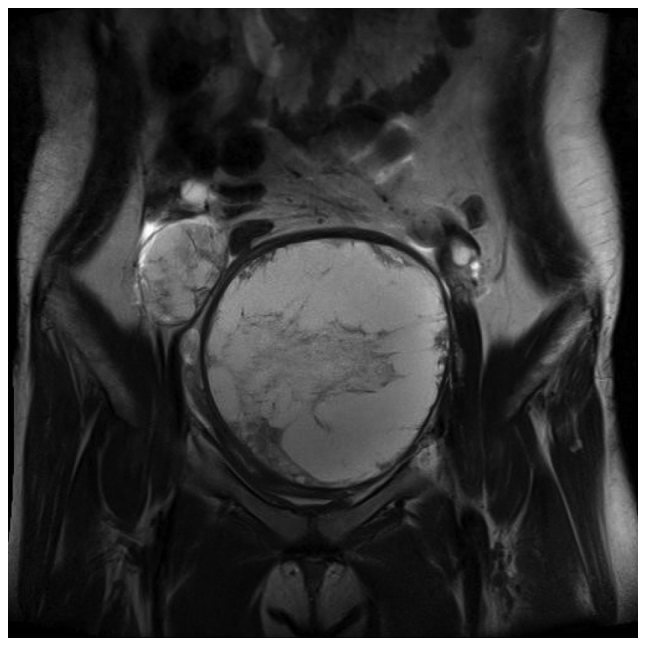
MRI image taken at the first hospital visit. Magnetic resonance image showing a round mass measuring 12.3×11.6×13.0 cm in size with multiple septae and focal wall thickening in the left pelvic cavity. MRI, magnetic resonance image.
Figure 2.
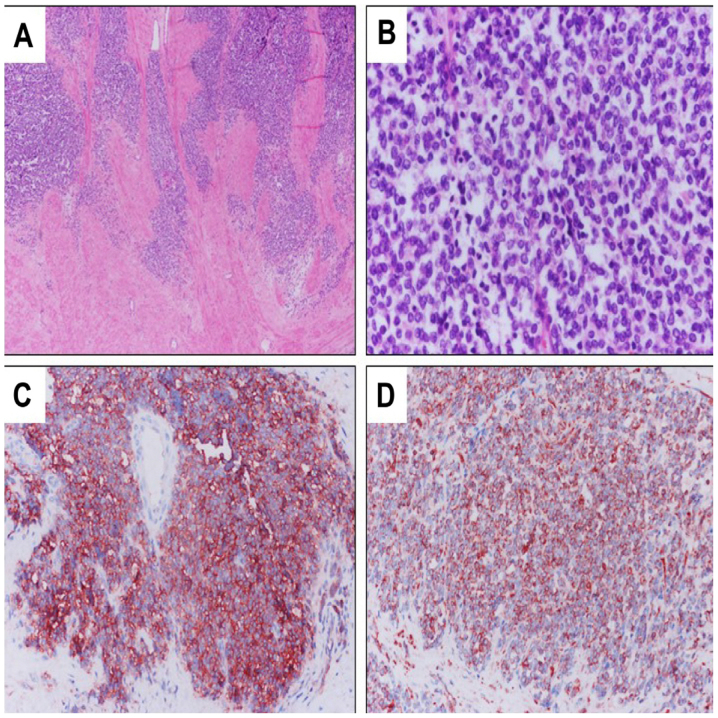
Histological findings of Ewing sarcoma. (A) Under low power (magnification, ×40). (B) Under high power (magnification, ×400). Tumor cells show strong immunoreactivity to (C) cluster of differentiation 99 and (D) vimentin (magnification, ×200).
Figure 3.
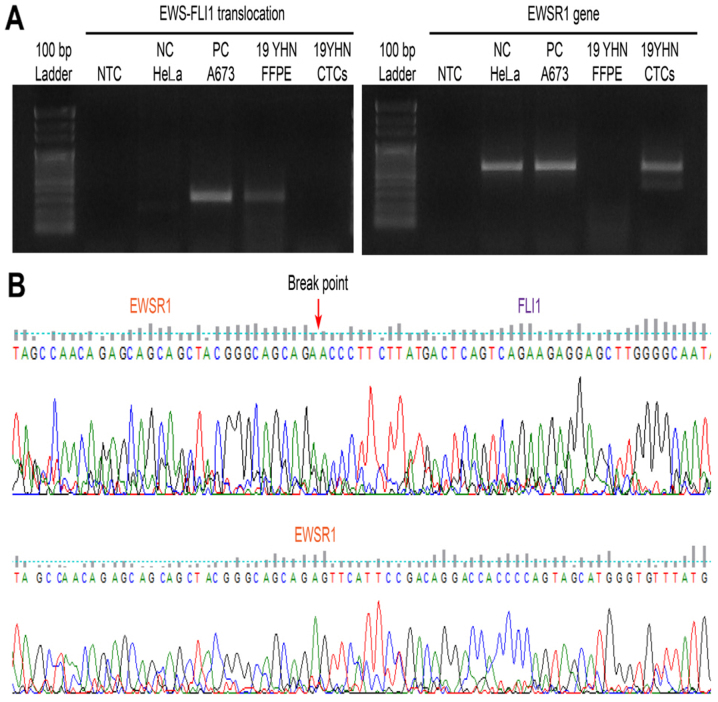
Molecular pathology findings of Ewing sarcoma tissue. (A) EWS-FLI1 fusion transcript was detected in the patient's FFPE DNA. (B) Translocation breakpoint of the EWS-FLI1 fusion transcript was observed in the patient's FFPE sample DNA. FFPE, formalin-fixed paraffin-embedded.
Figure 4.
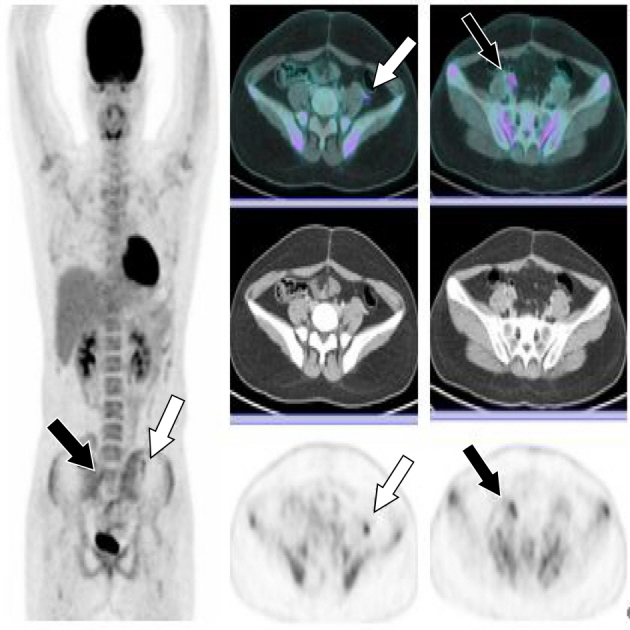
Pelvic (lymph node) metastasis was confirmed in the eighth month after complete remission. FDG-avid focal thickening lesion measuring 1.2 cm long, with an SUVmax of 5.06 in the left paracolic gutter (white arrow); and FDG-avid nodular lesion ~2.0 cm long, with an SUVmax of 5.06 in the right external iliac area (black arrow). SUV, standardized uptake values; FDG, fluorodeoxyglucose.
Figure 5.
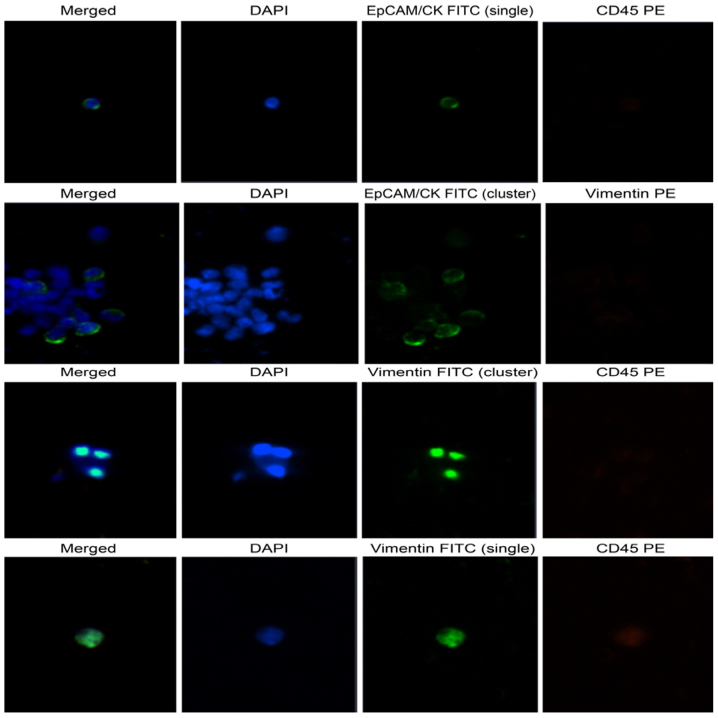
Immunohistochemistry findings of Ewing sarcoma blood-CTC isolation. Observation of CTC captured on membrane using CD-PRIME system through Bioview microscope. EpCAM/CK positive CTC was detected by collecting CTCs of Ewing sarcoma blood with pelvic metastasis. EpCAM/CK cell immunohistochemistry was detected as a green signal in the cytosol. Vimentin was detected as a green and orange signal on the slides. DAPI appeared as a blue signal in the nucleus. Clustering was observed (original magnification, ×100). At the time of cell examination, the patient's blood was sampled at the point when fusion was confirmed at FFPE on June 27, 2017, and vimentin was confirmed. Thereafter, a large number of clusters were found in the second blood CTC. CTCs, circulating tumor cells; EpCAM, epithelial cell adhesion molecule; CK, cytokeratin; FFPE, formalin-fixed paraffin-embedded.
Table II.
Identification of EWS-FLI1 fusion in FFPE and CTCs in Ewing sarcoma patients.
| A673 spiking (no. of cells) | NTC | 0.15 | 0.5 | 1.5 | 5 | 15 | 50 | 150 | 500 | Patient CTCs |
|---|---|---|---|---|---|---|---|---|---|---|
| PBMC (no. of cells) | 2,000 | |||||||||
| ∆∆Ct-1st | ND | 33.821 | 33.224 | 30.651 | 30.364 | 28.16 | 26.707 | 24.18 | 22.884 | ND |
| ∆∆Ct-2nd | ND | 34.292 | 33.346 | 30.531 | 30.276 | 28.058 | 26.454 | 24.206 | 22.937 | ND |
Reverse transcription-quantified PCR was performed after extracting RNA from the patient's CTC. The patient's ΔΔCt value was revealed to be undetermined as NTC. HeLa cells were the negative control. A673 cells were the positive control. EWS-FLI1 fusion was detected in FFPE of Ewing sarcoma, but not in the CTC. After spiking A673 Ewing sarcoma cells on normal PBMCs, the LOD for EWS-FLI1 fusion detection was determined. FFPE, formalin-fixed paraffin-embedded; CTCs, circulating tumor cells; NTC, no template control; PBMC, peripheral blood mononuclear cell; ND, not-detected (undetermined); PCR, polymerase chain reaction; LOD, limit of detection.
Chromosomal translocation analysis using RT-PCR
Ewing sarcoma cell line A673 was used as a negative control and a positive control. RNAs were extracted from CTCs captured from membranes of Ewing sarcoma patients using RNeasy mini kit (Qiagen GmbH, Hilden, Germany) following successful therapy. RNA extraction from formalin-fixed paraffin-embedded (FFPE) slide was performed using RNease FFPE kit (Qiagen GmbH) at the time of Ewing sarcoma diagnosis. SuperScript VILO cDNA Synthesis Kit (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was used for cDNA synthesis, under the following reaction conditions: 25°C for 10 min, 42°C for 120 min, and 85°C for 5 min. To confirm chromosomal translocation, RT-PCR was performed using specific primers and Platinum PCR SuperMix High Fidelity (Invitrogen; Thermo Fisher Scientific, Inc.). RT-PCR was performed under the following conditions: 94°C for 10 min, 45 cycles of 94°C for 1 min, 68°C for 1 min, 72°C for 1 min 30 sec, and 72°C for 5 min (13). The following Ewing sarcoma-specific fusion primers were used for RT-PCR: Forward primer, 5′-TCCTACAGCCAAGCTCCAAGTC-3′; Reverse primer, 5′-ACTCCCCGTTGGTTCCCCTCC-3′. PCR products were subjected to 1.5% agarose gel electrophoresis at 50 volts for 15 min. The breakpoint of EWS-FLI1 translocation was confirmed by RT-PCR and direct sequencing using DNA from FFPE slide of Ewing sarcoma patient. However, EWSR1 gene was sequenced (Cosmogenetech Inc., Seoul, Republic of Korea) and detected in the patient's CTCs, confirming the absence of gene arrangement (Fig. 3).
Immunohistochemical analysis
Enrichment and enumeration of CTCs
A 21G needle was used to collect blood from the patient into a K2 EDTA vacuum tube coated with anticoagulant (BD Biosciences, Franklin Lakes, NJ, USA). The blood was mixed with Ficoll-paque reagent in a 1:1 ratio. After separating mononuclear cells, the CD-OPR-1000 driver and CD-PRIME systems of CD-CTC solo disposable disk were used to isolate CTCs with high efficiency. The CD-CTC Enrichment kit was used for cell enrichment. The membrane on which CTCs were captured was placed on a glass slide (Clinomics, Inc., Ulsan, Republic of Korea) and stained with EpCAM/CK, Pan-CK, CD45, DAPI, vimentin-FITC, and vimentin-PE antibodies using CD-CTC Enumeration kit. Stained CTCs were observed using a Bioview CCBS system (BioView, Ltd., Nes Ziona, Israel). Of a total of 7,440 cells captured on the membrane, 175 (2.73%) tumor cells stained positive for EpCAM/CK and pan-CK. Clustering of CTCs was observed in 11 (6.28%) of these 175 DAPI-stained nuclei (Fig. 5).
Chromosomal translocation analysis using RT-qPCR
CTCs were membrane-captured using CD-PRIME (Clinomics, Inc.). RNAs were extracted from CTCs using QIAamp RNA Blood Mini Kit (Qiagen GmbH). The quality of extracted RNA was measured using Agilent RNA 6000 Pico Kit and 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA). However, RNA integrity number (RIN) value or concentration was not obtained. RNA concentration was measured using a Qubit RNA HS Assay kit and a Qubit Fluorometer (Thermo Fisher Scientific, Inc.). However, it showed a very low concentration. All the extracted RNAs were used for cDNA synthesis using the same cDNA synthesis kit used for RT-PCR. EWS-FLI1 primers (forward primer, 5′-CCAAGTCAATATAGCCAACAG-3′; reverse primer, 5′-GGCCAGAATTCATGTTATTGC-3′) were designed using the EWS-FLI1 fusion primer designed by Lewis et al (14), Power SYBR-Green PCR Master Mix and ABI ViiA 7 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) were used for RT-qPCR. The following PCR program parameters were used: Hold stage at 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 sec and 60°C for 1 min. For melt curve analysis, the following parameters were added: 95°C for 15 sec, 60°C for 1 min, and 95°C for 15 sec. After spiking A673 cells to normal PBMC, the LOD value confirming EWS-FLI1 translocation based on ΔΔCt value in RT-qPCR was found to be a mixing ratio of one A673 cell to 2,000 normal PBMC cells. Using CTCs isolated from patients, the ΔΔCt value was undetermined, indicating the absence of EWS-FLI1 translocation (Table II).
Personal genomic mutation profiling using targeted massively parallel sequencing
DNA was extracted from FFPE slide prepared at the time of laparotomy for the diagnosis of patient's Ewing sarcoma using QIAamp DNA FFPE Tissue kit (Qiagen GmbH). Using CD-PRIME, CTCs were captured from patient's blood when pelvic (lymph node) metastasis occurred at eight months after the diagnosis. DNA was extracted from CTCs using QIAamp DNA Micro kit (Qiagen GmbH). The cfDNA was extracted from the patient's plasma using QIAamp Circulating Nucleic Acid kit (Qiagen GmbH). Using Cancer-PRIME (Clinomics, Inc.), a comprehensive cancer panel, sequencing was performed on an Ion S5 platform (Thermo Fisher Scientific, Inc.) according to the application guide. Mutations of FGFR4 (c.1162G> a; p.G388A) and HRAS (c.182A> G; p.Q61R) were detected in DNA extracted from FFPE. In DNA extracted from CTC, FGFR3 (c.1948A> G; p.K650E) and the same FGFR4 (c.1162G> A; p.G388A) mutation were detected in FFPE DNA. Therefore, the FGFR4 genetic mutation was identified as a germ line mutation. In addition, FGFR3 (c.1948A> G; p.K650E), FGFR4 (c.1162G> A; p.G388A) and HRAS (c.182A> G; p.Q61R) mutations in FFPE DNA and CTC DNA, were also detected in cfDNA (Table III). On the other hand, BRAF (c.1799T> A; p.V600E), CDKN2A (c.1_471del471), or TP53 (c.354_355insCA) mutations were not identified in Ewing sarcoma A673 cells. The CTNNB1 (c.133_135delTCT; p.S45del) or NRAS (c.181C> A; p.Q61K) mutation identified in oncogene mutation profiling of Ewing sarcoma patients reported by Shukla et al (15), was not identified in Ewing sarcoma A673 cells either.
Table III.
Molecular genetics and mutational analysis of Ewing sarcoma.
| Patient | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Mutation | Mutation type | CosmicID | Positive control A673 | FFPE | CTCs | cfDNA |
| FGFR3 | c.1948A>G (p.K650E) | Somatic | COSM719 | √ | √ | ||
| FGFR4 | c.1162G>A (p.G388A) | Germline | – | √ | √ | √ | |
| HRAS | c.182A>G (p.Q61R) | Somatic | COSM499 | √ | √ | ||
| BRAF | c.1799T>A (p.V600E) | Somatic | COSM476 | √ | |||
| CDKN2A | c.1_471del471 (p.0?) | Somatic | COSM12526 | √ | |||
| TP53 | c.354_355insCA (p.A119fs*5) | Not confirmed | COSM26839 | √ | |||
The FGFR4 mutation observed in the primary cancer FFPE sample of the patient was also detected in the CTC and cfDNA. BRAF, CDKN2A, or TP53 mutations identified in A673 Ewing sarcoma cell line were not observed in the CTC of the present case. FFPE, formalin-fixed paraffin-embedded; CTCs, circulating tumor cells; cfDNA, cell-free circulating tumor DNA; FGFR3, fibroblast growth factor receptor 3; FGFR4, fibroblast growth factor receptor 4; HRAS, HRas proto-oncogene; CDKN2A, cyclin-dependent kinase inhibitor 2A; TP53, tumor protein p53.
Discussion
Ewing sarcoma is a very rare cancer. Skeletal Ewing sarcoma is the most common (40%), followed by trunk (32%), extremity limb (26%), head and neck (18%), retroperitoneum (16%), and other sites (9%) (16). Ewing sarcoma of the skeleton originating in the uterus is very rare. Early symptoms of Ewing sarcoma include hemorrhage, pelvic mass, uterine hypertrophy, abdominal pain, tibial pain, and metastasis of the pelvis and lungs (17–19). Diagnosis of Ewing sarcoma is confirmed via histopathological examination, blood tests, MRI, PET/CT, bone scan, bone marrow aspiration, biopsy, and X-ray. Ewing sarcoma cells are small and round in shape and blue in color. Immunohistochemical analysis of the cells reveals positive staining for CD99, Vimentin, and S-100 (20). In the present study, early symptoms of Ewing sarcoma and pathological findings were similar to those of previous reports. Chromosomal translocation has been observed in about 90% of Ewing sarcoma cases (21,22). The EWS-FLI1 fusion gene was also observed in primary cancer tissues in this study. Although not investigated in this study, EWS-FLI1 fusion gene can be observed cytogenetically using FISH for diagnosis (19). EWS is known to play an important role in the molecular diagnosis of sarcoma. Fusion of EWS with chromosome 11q24 FLI1 gene, resulted in EWS gene translocation (23). When EWS-FLI1 fusion gene was observed in primary cancer of the present study, pelvic lymph node metastasis was confirmed radiologically at eight months after complete remission. In this study, CTCs in the blood were used for non-invasive diagnosis of Ewing sarcoma. The detection of CTCs in the blood indicates pelvic lymph node metastasis. EpCAM-positive CTCs are markers of circulating epithelial tumor cells and sarcoma. In the present case, at the time of pelvic lymph node metastasis, EpCAM and vimentin positive CTCs were detected in mesenchymal cancer cell line. EWS-FLI1 fusion did not occur in the patient's CTCs. However, the EWSR1 gene sequence was confirmed in CTC suggesting that Ewing sarcoma-specific fusion gene was no longer expressed following treatment. Therefore, CTCs may be used as a noninvasive method to predict prognosis following biopsy. However, despite the absence of Ewing sarcoma-specific fusion gene in CTCs and detection of primary cancer-derived cells with similar mutational features as the primary cancer in the patient's blood, continuous invasive monitoring of CTCs is needed. This study elucidates the role of blood CTCs derived from primary cancer in diagnostic methods to indicate the prognosis of cancer. Since Ewing sarcoma in the uterus is rare, studies profiling its specific genetic mutations are very limited. Shukla et al (15), studied the profile of oncogene mutations in pediatric solid tumors and found that Ewing sarcoma represented the smallest proportion (19.73%, 75/380) of pediatric solid tumors. Based on sequence analysis, genetic variation was found only in 4% (3/75) of these subjects with Ewing sarcoma (15). The mechanism involved in the release of CTCs and cf DNA into the blood from primary cancers has yet to be elucidated. However, we identified a pathogenic somatic mutation in the patient's blood that closely resembled the genetic variation of CTC and cfDNA in primary cancers because CTCs occur in the pathway of tumor cells (originating from primary cancer tissues into the blood). However, Ewing sarcoma-specific EWS-FLI1 translocation might be associated with a favorable prognosis following treatment. Further studies are needed to validate the role of CTCs and molecular pathologic changes in cfDNA as indicators of cellular metastasis and progression or treatment. In the present study, we used various methods for the diagnosis of Ewing sarcoma, especially skeletal Ewing sarcoma. In particular, liquid biopsy was performed using CTCs and the DNA of circulating free cells in blood. Liquid biopsy represents a relatively noninvasive and clinically significant modality facilitating the monitoring of therapeutic effect and prognosis in cancer, and therefore, is expected to play a pivotal role in ultra-precise medicine.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- FLI1
friend leukemia integration 1 transcription factor
- CTCs
circulating tumor cells
- LFT
liver function test
- BUN
blood urea nitrogen
- CA125
cancer antigen 125
- CA19-9
cancer antigen 19-9
- CEA
carcinoembryonic antigen
- AFP
alpha-fetoprotein
- hCG
human chorionic gonadotropin
- LOD
limit of detection
- EpCAM
epithelial cell adhesion molecule
- FFPE
formalin-fixed paraffin-embedded
- EDTA
ethylenediaminetetraacetic acid
- PET/CT
positron emission tomography-computed tomography
- CK
cytokeratin
- PBMC
peripheral blood mononuclear cell
- TP53
tumor protein p53
- FGFR3
fibroblast growth factor receptor 3
- FGFR4
fibroblast growth factor receptor 4
- HRAS
HRas proto-oncogene
- CDKN2A
cyclin-dependent kinase inhibitor 2A
- CTNNB1
catenin beta 1
- NRAS
NRAS proto-oncogene
- FISH
fluorescence in situ hybridization
Funding
The present study was supported by a grant from the Basic Research Program through the National Research Foundation funded by the Ministry of Science, ICT & Future Planning, Republic of Korea (grant no. 2017R1A2B4012353).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SYL and DHC designed the study. SL performed the experiment. SYL, SL and DHC analyzed the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of Chonbuk National University Hospital (Jeonju, Republic of Korea). Written informed consent was obtained from the participant.
Patient consent for publication
Written informed consent was obtained from the patient for the publication of their data and associated images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Burt M, Karpeh M, Ukoha O, Bains MS, Martini N, McCormack PM, Rusch VW, Ginsberg RJ. Medical tumors of the chest wall. Solitary plasmacytoma and Ewing's sarcoma. J Thorac Cardiovasc Surg. 1993;105:89–96. [PubMed] [Google Scholar]
- 2.Loverro G, Resta L, Di Naro E, Caringella AM, Mastrolia SA, Vicino M, Tartagni M, Schonauer LM. Conservative treatment of Ewing's sarcoma of the uterus in young women. Case Rep Obstet Gynecol. 2015;2015:871821. doi: 10.1155/2015/871821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jawad MU, Cheung MC, Min ES, Schneiderbauer MM, Koniaris LG, Scully SP. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: An analysis of 1631 cases from the SEER database, 1973-2005. Cancer. 2009;115:3526–3536. doi: 10.1002/cncr.24388. [DOI] [PubMed] [Google Scholar]
- 4.Brasme JF, Chalumeau M, Oberlin O, Valteau-Couanet D, Gaspar N. Time to diagnosis of Ewing tumors in children and adolescents is not associated with metastasis or survival: A prospective multicenter study of 436 patients. J Clin Oncol. 2014;32:1935–1940. doi: 10.1200/JCO.2013.53.8058. [DOI] [PubMed] [Google Scholar]
- 5.Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing's sarcoma. PLoS One. 2008;3:e1965. doi: 10.1371/journal.pone.0001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esiashvili N, Goodman M, Marcus RB., Jr Changes in incidence and survival of Ewing sarcoma patients over the past 3 decades: Surveillance epidemiology and end results data. J Pediatr Hematol Oncol. 2008;30:425–430. doi: 10.1097/MPH.0b013e31816e22f3. [DOI] [PubMed] [Google Scholar]
- 7.Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51:2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::AID-CNCR2820511127>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 9.Kim TH, Lim M, Park J, Oh JM, Kim H, Jeong H, Lee SJ, Park HC, Jung S, Kim BC, et al. FAST: Size-selective, clog-free isolation of rare cancer cells from whole blood at a liquid-liquid interface. Anal Chem. 2017;89:1155–1162. doi: 10.1021/acs.analchem.6b03534. [DOI] [PubMed] [Google Scholar]
- 10.Mackall CL, Meltzer PS, Helman LJ. Focus on sarcomas. Cancer Cell. 2002;2:175–178. doi: 10.1016/S1535-6108(02)00132-0. [DOI] [PubMed] [Google Scholar]
- 11.Pennacchioli E, Tosti G, Barberis M, De Pas TM, Verrecchia F, Menicanti C, Testori A, Mazzarol G. Sarcoma spreads primarily through the vascular system: Are there biomarkers associated with vascular spread? Clin Exp Metastasis. 2012;29:757–773. doi: 10.1007/s10585-012-9502-4. [DOI] [PubMed] [Google Scholar]
- 12.Tellez-Gabriel M, Brown HK, Young R, Heymann MF, Heymann D. The challenges of detecting circulating tumor cells in sarcoma. Front Oncol. 2016;6:202. doi: 10.3389/fonc.2016.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aryee DN, Niedan S, Kauer M, Schwentner R, Bennani-Baiti IM, Ban J, Muehlbacher K, Kreppel M, Walker RL, Meltzer P, et al. Hypoxia modulates EWS-FLI1 transcriptional signature and enhances the malignant properties of Ewing's sarcoma cells in vitro. Cancer Res. 2010;70:4015–4023. doi: 10.1158/1538-7445.AM10-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis TB, Coffin CM, Bernard PS. Differentiating Ewing's sarcoma from other round blue cell tumors using a RT-PCR translocation panel on formalin-fixed paraffin-embedded tissues. Mod Pathol. 2007;20:397–404. doi: 10.1038/modpathol.3800755. [DOI] [PubMed] [Google Scholar]
- 15.Shukla N, Ameur N, Yilmaz I, Nafa K, Lau CY, Marchetti A, Borsu L, Barr FG, Ladanyi M. Oncogene mutation profiling of pediatric solid tumors reveals significant subsets of embryonal rhabdomyosarcoma and neuroblastoma with mutated genes in growth signaling pathways. Clin Cancer Res. 2012;18:748–757. doi: 10.1158/1078-0432.CCR-11-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cates JM, Coffin CM. Neurogenic tumors of soft tissue. Pediatr Dev Pathol. 2012;15(1 Suppl):S62–S107. doi: 10.2350/11-03-1003-PB.1. [DOI] [PubMed] [Google Scholar]
- 17.Park JY, Lee S, Kang HJ, Kim HS, Park SY. Primary Ewing's sarcoma-primitive neuroectodermal tumor of the uterus: A case report and literature review. Gynecol Oncol. 2007;106:427–432. doi: 10.1016/j.ygyno.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Fadare O. Uncommon sarcomas of the uterine cervix: A review of selected entities. Diagn Pathol. 2006;1:30. doi: 10.1186/1746-1596-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yi T, Wang P, Lin L, Jiang W. Ewing's sarcoma/peripheral primitive neuroectodermal tumors of the uterus confirmed with fluorescence in situ hybridization in a 29-year-old Chinese female: A case report and published work review. J Obstet Gynaecol Res. 2015;41:478–482. doi: 10.1111/jog.12552. [DOI] [PubMed] [Google Scholar]
- 20.Iwamoto Y. Diagnosis and treatment of Ewing's sarcoma. Jpn J Clin Oncol. 2007;37:79–89. doi: 10.1093/jjco/hyl142. [DOI] [PubMed] [Google Scholar]
- 21.Couturier J. Soft tissue tumors: Ewing's tumors/Primitive neurectodermal tumors (PNET) Atlas Genet Cytogenet Oncol Haematol. 1998;2:148–150. [Google Scholar]
- 22.Turc-Carel C, Aurias A, Mugneret F, Lizard S, Sidaner I, Volk C, Thiery JP, Olschwang S, Philip I, Berger MP, et al. Chromosomes in Ewing's sarcoma. I. An evaluation of 85 cases of remarkable consistency of t(11;22)(q24;q12). Cancer Genet Cytogenet. 1988;32:229–238. doi: 10.1016/0165-4608(88)90285-3. [DOI] [PubMed] [Google Scholar]
- 23.Weiss S, Goldblum J. Extraskeletal Ewing's sarcoma/primitive neuroectodermal tumor family. In: Weiss SW, Goldblum JR, editors. In: Enzinger and Weiss's soft tissue tumors. 5th. Mosby, St Louis, MO: 2007. pp. 963–979. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.