Abstract.
Limited access to nucleic acid tests for hepatitis B virus (HBV) DNA is a significant barrier to the effective management of chronic HBV infection in resource-poor countries. Alternatively, HBV e antigen (HBeAg) may accurately indicate high viral replication. We assessed the diagnostic performance of three commercially available rapid diagnostic tests (RDTs) for HBeAg (SD Bioline, Insight and OneStep) against a quantitative chemiluminescent immunoassay (CLIA, Architect). Using stored sera from adults with chronic HBV infection, we tested RDTs in three groups in Senegal (48 HBeAg-positive, 196 HBeAg-negative, and 117 cases with high HBV DNA (≥ 106 IU/mL)) and one group in France (17 HBeAg-positive East Asians). In Senegal, the sensitivity and specificity for HBeAg detection were 29.8% and 100% for SD Bioline, 31.1% and 100% for Insight, and 42.5% and 98.4% for OneStep, respectively. The lower limits of detection of these RDTs were very high (> 2.5 log10 Paul Ehrlich Institut units/mL). Their low sensitivity was also confirmed in HBeAg-positive Asian samples (35.3–52.9%). The prevalence of HBeAg in highly viremic (≥ 106 IU/mL) Senegalese patients was low: 58.1% using CLIA and 24.5–37.5% using RDTs. Hepatitis B e antigen prevalence was similarly low in a subgroup of 28 Senegalese women of childbearing age with a high viral load (≥ 106 IU/mL). Approximately, half of highly viremic adults do not carry HBeAg in Africa, and HBeAg RDTs had remarkably poor analytical and diagnostic sensitivity. This implies that HBeAg-based antenatal screening, particularly if using the currently available HBeAg RDTs, may overlook most pregnant women at high risk of mother-to-child transmission in Africa.
INTRODUCTION
Viral hepatitis ranks as the seventh leading cause of death worldwide and kills more people than any of the major infectious agents: HIV, tuberculosis, or malaria.1 Consequently, viral hepatitis is now targeted in the UN’s sustainable development goals, and the WHO has adopted a global strategy to eliminate hepatitis as a public health threat by the year 2030.2 Of all hepatitis deaths, the vast majority occur in low- and middle-income countries and about half are attributable to hepatitis B virus (HBV).1 Sub-Saharan Africa (SSA) suffers from the highest HBV prevalence in the world (> 8%).3
To eliminate HBV infection, it is imperative to prevent mother-to-child transmission (MTCT).4,5 Since 2009, WHO recommends the prevention of MTCT by administering hepatitis B vaccine to all neonates within 24 hours of birth.6 However, the birth dose vaccination alone is imperfect in preventing MTCT, particularly in babies born to women with high viral replication, represented by positive hepatitis B e antigen (HBeAg) status and/or high HBV DNA levels.7 Consequently, in resource-rich countries, pregnant women are systematically screened for hepatitis B surface antigen (HBsAg), and subsequently for HBeAg in those found to carry HBsAg to identify high-risk babies who will benefit from hepatitis B immunoglobulin (HBIG) in addition to hepatitis B vaccination at birth. More recently, antenatal screening for HBV DNA has been added to identify highly viremic women (> 106 IU/mL) who have residual risk of MTCT despite both birth dose vaccine and HBIG8 and who thus require antiviral therapy during the third trimester.9
These additional measures to prevent MTCT have rarely been implemented in low- and middle-income countries,10 partly because the access to diagnostic tools to evaluate HBV replication is severely limited. The nucleic acid test to quantify HBV DNA is hardly accessible because of high cost (US$60–200/assay) and the need for a sophisticated laboratory with highly skilled technologists.11 Laboratory-based immunoassays for HBeAg, such as enzyme immunoassay or chemiluminescent immunoassay (CLIA), may be more affordable (US$5–30/assay); however, these are infrequently performed in peripheral laboratories in resource-limited countries. Alternatively, the use of lateral flow immunochromatographic rapid diagnostic tests (RDTs) for HBeAg may largely overcome these limitations because they are inexpensive (US$ < 2), rapid (< 20–30 minutes), easy-to-use, and only require small volumes (75–100 μL) of serum or plasma without any further preparation (Table 1). Indeed, the RDT for HBeAg was recently included in the WHO’s list of essential in vitro diagnostics for primary healthcare.12 However, the diagnostic accuracy of the HBeAg RDTs has been poorly studied.
Table 1.
Characteristics of commercial RDTs evaluated in this study
| Commercial name | Manufacturer (country) | Sample | Time for detection (minutes) | Sensitivity/specificity compared with ELISA reported by the manufacturers | Price (US$, per test) | CE marked | ||
|---|---|---|---|---|---|---|---|---|
| Type | Quantity | Sensitivity | Specificity | |||||
| SD Bioline | Standard Diagnostics Inc. (Korea) | Serum, plasma | 100 μL | 5–20 | 95.5% | 98.6% | 1.3 | No |
| Insight | Tulip Diagnostics Ltd. (India) | Serum | 100 μL | 10–15 | Not reported | Not reported | 0.5 | No |
| OneStep | AMS UK Ltd. (UK) | Serum, plasma | 75 μL | 15 | 94.9% | 99.4% | 0.7 | Yes |
ELISA = enzyme-linked immunosorbent assay; RDT = rapid diagnostic test.
We therefore conducted a case-control study in Senegal, West Africa, to assess retrospectively the diagnostic sensitivity, specificity, and lower detection limit of three commercially available RDTs, in reference to a new quantitative HBeAg CLIA (Architect). We also estimated the prevalence of positive HBeAg using CLIA or RDTs in patients with an HBV DNA level ≥ 106 IU/mL, which is a threshold associated with the risk of immunoprophylaxis failure for MTCT.8 Finally, a small number of HBeAg-positive samples from a cohort of East Asian people were tested with the RDTs to evaluate whether their performance varies between African and Asian patients because of the difference in major genotypes (A/E in West Africa and B/C in East Asia).13
MATERIALS AND METHODS
Study samples.
Senegal has a very high (11%) prevalence of chronic HBV infection,3 and the Institut Pasteur de Dakar (IPD) is one of the few laboratories in the country that routinely performs quantification of HBV DNA. The laboratory has been accredited for the standard NF EN ISO 15189. Every week, the IPD receives 80–100 patients with chronic HBV infection referred by their physicians to be tested for HBV markers such as HBeAg or HBV DNA. Since 2014, serum samples from patients with chronic HBV infection have been systematically stored at −80°C at the IPD without multiple freeze–thaw cycles. From the anonymized list of consecutive samples collected between 2014 and 2016, we excluded duplicate samples from the same individuals. Then, all the samples that had been tested for both HBV DNA and HBeAg at the time of referral were used to make two groups: HBeAg-positive cases (Group 1) and HBeAg-negative controls (Group 2), defined by the reference qualitative HBeAg CLIA. These groups were designed to evaluate the diagnostic sensitivity and specificity of RDTs to detect HBeAg through a case-control study. In January 2017, all these samples were analyzed for HBeAg using RDTs. In addition, Group 1 samples were further quantified for HBeAg (quantitative HBeAg) by the new commercial calibrator (Architect HBeAg Quantitative Calibrators; Abbott, Wiesbaden, Germany) to determine the lower detection limits of the RDTs.
Of the samples that had been tested for HBV DNA but not for HBeAg at the time of blood collection, all those with high HBV DNA levels (≥ 106 IU/mL) were selected to make Group 3. Stored sera were tested by both RDTs and quantitative HBeAg in January 2017, to estimate the prevalence of positive HBeAg using the reference laboratory-based CLIA or RDTs in patients with high viremia (≥ 106 IU/mL).
The study was carried out in accordance with the Declaration of Helsinki and approved by the National Ethics Committee (SEN 17/59). The study was reported in accordance with the Standards for Reporting of Diagnostic Accuracy.14
Reference HBeAg assay.
Hepatitis B e antigen was detected using CLIA (Architect, Abbott) with a lower detection limit of 0.5 Paul Ehrlich Institut units (PEIUs)/mL. Samples positive for HBeAg were quantified using the commercial calibrator (Architect HBeAg Quantitative Calibrators, Abbott), with a reportable range from 0.59 to 700.00 PEIU/mL. When the initial value exceeded 700.00 PEIU/mL, the sample was diluted 10 times and retested according to the manufacturer’s instructions.
Rapid diagnostic tests.
Of five immunochromatographic RDTs that we had identified in November 2016, two were not assessed in the present study: EASY Line (Adaltis, Rome, Italy) because of its shortage and OnSite HBV-5 (CTK Biotech Co, San Diego, CA) because of its high price (US$15/kit) that is related to it being a multi-panel test (HBsAg, antibody to HBsAg [anti-HBsAb], HBeAg, antibody to HBeAg [anti-HBeAb], and antibody to HBV core [anti-HBcAb]). We therefore evaluated three commercially available immunochromatographic RDTs: SD Bioline (Standard Diagnostics Inc., Gyeonggi-do, Republic of Korea); Insight (Tulip Diagnostics Ltd., Goa, India); and OneStep (AMS UK Ltd., Antrim, UK). The characteristics of these kits are summarized in Table 1. These tests are intended to be used for both fresh and frozen samples, and multiple freeze–thaw cycles should be avoided. When a test was invalid (i.e., neither control band nor test band appears), it was repeated until a valid result was obtained. To evaluate the inter-rater agreement, the results of the rapid tests were read and recorded independently by two well-trained laboratory staff who were blinded to the results of the reference test (CLIA). Although only results on which the two readers agreed were included in the analysis to assess the sensitivity and specificity, a subgroup analysis was also conducted for each of the laboratory staff by including the samples with discordant results.
HBV DNA assay.
Hepatitis B virus DNA was measured using commercial real-time polymerase chain reaction (PCR) (AMPLIX; Biosynex, Strasbourg, France), with a lower detection limit of 26 IU/mL.
East Asian samples in Paris, France.
To assess whether diagnostic sensitivity differs between African and Asian samples, we tested the RDTs using HBsAg-positive HBeAg-positive plasma samples from patients originating in East Asia. Of patients with chronic HBV infection followed up at the Infectious Disease Department of Saint Louis Hospital, Paris, France, from 2015 to 2017, we retrospectively selected all East Asian subjects positive for HBeAg. Hepatitis B e antigen was detected using the reference CLIA (Architect, Abbott) and quantified using the commercial calibrator (Architect HBeAg Quantitative Calibrators, Abbott). Plasma HBV DNA levels were measured using Cobas®Ampli Prep/Cobas Taq Man HBV assay (Roche Diagnostics, Meylan, France), with a lower detection limit of 20 IU/mL. Using the plasma samples stored at −80°C, we tested the three RDTs with two laboratory staff independently reading each result.
Sample size.
For the case-control study to determine diagnostic sensitivity and specificity for HBeAg RDTs (Groups 1 and 2), the sample size was determined based on the minimally acceptable sensitivity and specificity of 90% and 98%, respectively. One hundred and ten HBeAg-positive cases were required to show that the sensitivity of RDT is at least higher than 90% at a two-sided significance level of 5% with a power of 80% when the true sensitivity was 97%. Similarly, 188 HBeAg-negative controls were needed to demonstrate that the specificity is at least higher than 98% with a power of 80% when the true specificity was 100%.
Statistical analysis.
Quantitative HBeAg and HBV DNA levels were transformed into logarithmic scale. Comparisons were made between the groups using the χ2 test for categorical variables and the Kruskal–Wallis one-way analysis of variance for continuous variables. The inter-rater agreement of each RDT was determined using the kappa statistic. The analysis of Group 1 and Group 2 allowed us to estimate the sensitivity and specificity of the HBeAg RDTs, respectively, in reference to the CLIA. The differences in sensitivity and specificity between the RDTs were examined by McNemar’s test of paired proportions. To evaluate whether the diagnostic sensitivity varies according to sample storage conditions, Group 1 was further divided into two groups: the samples either collected more than a year or within a year before testing. Group 3 was used to estimate the prevalence of positive HBeAg by CLIA or RDTs in patients with high HBV DNA (≥ 106 IU/mL). Within Group 3, we performed a subgroup analysis to assess the HBeAg prevalence in highly viremic women of childbearing age (15–50 years old).
To determine the lower limit of detection of each RDTs, the proportions successfully detected by each rapid test were plotted at various levels of quantitative HBeAg (log10 PEIU/mL): < 0.5, 0.5–1.0, 1.0–1.5, 1.5–2.0, 2.0–2.5, 2.5–3.0, and ≥ 3.0. The limit of detection was determined as the level above which ≥ 95% of samples were consistently detected. The proportions tested positive by each HBeAg assay (including both RDTs and the reference CLIA) at various levels of HBV DNA (log10 IU/mL) were also plotted graphically. To understand the role of quantitative HBeAg in African patients, the correlation between quantitative HBeAg and age, as well as the correlation between quantitative HBeAg and HBV DNA levels, were assessed using Pearson’s correlation coefficient. Finally, HBeAg-positive Asian samples were used to estimate the sensitivity of the RDTs for HBeAg detection.
RESULTS
Samples in Senegal.
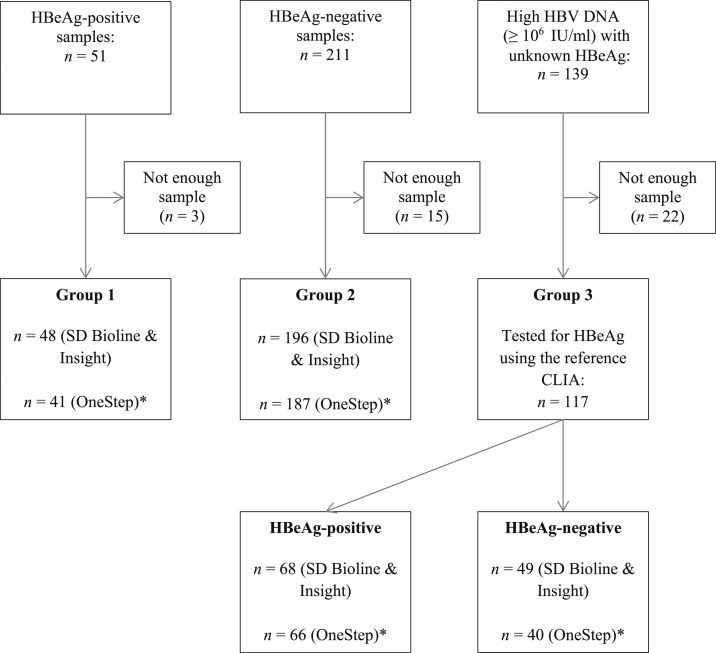
Figure 1 presents the flow diagram of study sample in Senegal. Between 2014 and 2016, there were 51 samples positive for HBeAg (Group 1), 211 samples negative for HBeAg (Group 2), and 139 samples with high HBV DNA (≥ 106 IU/mL) and unknown HBeAg status (Group 3). After excluding the samples with inadequate volume for testing, there were 48, 196, and 117 samples in Groups 1, 2, and 3, respectively. A total of 361 samples were tested by two RDTs: SD Bioline and Insight. Because of the lack of quantity of some samples, the third RDT (OneStep), which was made available later, was tested in 334 samples.
Figure 1.
Flow diagram of study samples in Senegal. CLIA = chemiluminescent immunoassay; HBeAg = hepatitis B e antigen; HBV = hepatitis B virus.
Characteristics of study participants in Senegal are presented in Table 2. Most participants (> 60%) were male in each of the three groups, and the median age was significantly higher in Group 2 (37 years, interquartile range [IQR]: 31–47) than in Group 1 (29, IQR: 21–36) or in Group 3 (33, IQR: 26–43) (P < 0.001). In Group 3, there were 28 samples from women of childbearing age (median: 32 years, IQR: 29–40). Of 117 cases with high viral load (Group 3), 68 (58.1%, 95% confidence interval [CI]: 48.9–66.8) were positive for HBeAg using the reference CLIA, and their median quantitative HBeAg level was 1.9 log PEIU/mL (IQR: 0.9–2.8). In the subgroup of women of childbearing age, 53.6% (15/28) were positive for HBeAg, with median quantitative HBeAg was 1.7 log PEIU/mL (IQR: 1.1–2.7). In Group 1, the median quantitative HBeAg level was 1.5 log PEIU/mL (IQR: 0.4–2.4). The median HBV DNA level was significantly higher in Group 1 (7.7 log IU/mL, IQR: 6.8–8.5) and Group 3 (7.6 log IU/mL, 6.9–8.4) than in Group 2 (3.1 log IU/mL, 2.4–3.9) (P < 0.001).
Table 2.
Characteristics of study samples in Senegal
| Group 1 | Group 2 | Group 3 | ||
|---|---|---|---|---|
| HBeAg-positive cases | HBeAg-negative controls | High HBV DNA (≥ 106 IU/mL) cases | ||
| All | Women of childbearing age | |||
| Total number | 48 | 196 | 117 | 28 |
| Male sex (n, %) | 31 (64.6%) | 129 (65.8%) | 80 (68.4%) | 0 |
| Median age (years, IQR) | 29 (21–36) | 37 (31–47) | 33 (26–43) | 32 (29–40) |
| Positive HBeAg by CLIA (Architect) | 48 (100%) | 0 | 68 (58.1%) | 15 (53.6%) |
| Median HBeAg levels (log10 PEIU/mL, IQR) | 1.5 (0.4–2.4) | N/A | 1.9 (0.9–2.8)* | 1.7 (1.1–2.7)† |
| Median HBV DNA levels (log10 IU/mL, IQR) | 7.7 (6.8–8.5) | 3.1 (2.4–3.9) | 7.6 (6.9–8.4) | 7.4 (6.7–8.3) |
CLIA = chemiluminescent immunoassay; HBeAg = Hepatitis B e antigen; HBV = hepatitis B virus; N/A = not applicable; PEIU = Paul Ehrlich Institut unit; RDT = rapid diagnostic test.
Excluding 49 samples with negative HBeAg by the reference CLIA test.
Excluding 13 samples with negative HBeAg by the reference CLIA test.
Inter-rater agreement of the rapid HBeAg tests.
The inter-rater agreement of the RDTs was evaluated using all three of the Senegalese groups (N = 361 for SD Bioline and Insight; N = 334 for OneStep) (Supplemental Table 1). Discrepancies between the two laboratory staff were observed in 2.2% (8/361) for SD Bioline, 4.7% (17/361) for Insight, and 1.2% (4/334) for OneStep, with Kappa statistics (95% CI) of 0.90 (0.83–0.97), 0.81 (0.73–0.90), and 0.96 (0.92–0.99), respectively. Most discordant results were from the samples positive for HBeAg using the reference CLIA: 62.5% (5/8) for SD Bioline, 88.2% (15/17) for Insight, and 75.0% (3/4) for OneStep.
Sensitivity and specificity for HBeAg detection.
The sensitivity of RDTs for HBeAg detection was estimated using Group 1 and the specificity using Group 2. After excluding discordant results between two examiners (SD Bioline, N = 1; Insight, N = 3; OneStep, N = 2), the sensitivity and specificity were 29.8% (95% CI: 17.3–44.9) and 100% (98.1–100) for SD Bioline; 31.1% (18.2–46.6) and 100% (98.1–100) for Insight; and 42.5% (27.0–59.1) and 98.4% (95.4–99.7) for OneStep, respectively (Table 3). The sensitivity of OneStep was significantly higher than that of SD Bioline (P = 0.008) and Insight (P = 0.03). The samples with false negative results had significantly lower median quantitative HBeAg levels (log PEIU/mL) than those with true positive results: 0.7 (IQR: 0.2–1.6) and 2.8 (2.3–3.1) for SD Bioline (P < 0.001); 0.6 (0.1–1.6) and 2.8 (2.3–3.1) for Insight (P < 0.001); and 0.5 (0.1–1.0) and 2.6 (2.0–2.9) for OneStep (P < 0.001), respectively. Supplemental Table 2 presents the subgroup analysis for the sensitivity and specificity of the RDTs by each of the laboratory staff. This analysis included the discordant results between the two laboratory staff. Similarly low sensitivities were noted irrespective of the reader.
Table 3.
Performance of RDTs for HBeAg detection (Group 1, Group 2 and East Asian samples)
| Senegalese samples* | East Asian samples | |||||
|---|---|---|---|---|---|---|
| Group 1: HBeAg-positive cases (N = 48) | Group 2: HBeAg-negative controls (N = 196) | HBeAg-positive cases (N = 17) | ||||
| Sensitivity | 95% CI | Specificity | 95% CI | Sensitivity | 95% CI | |
| SD Bioline | 29.8% (14/47) | 17.3–44.9 | 100% (196/196) | 98.1–100 | 47.1% (8/17) | 23.0–72.2 |
| Insight | 31.1% (14/45) | 18.2–46.6 | 100% (196/196) | 98.1–100 | 52.9% (9/17) | 27.8–77.0 |
| OneStep | 42.5% (17/40) | 27.0–59.1 | 98.4% (183/186) | 95.4–99.7 | 35.3% (6/17) | 14.2–61.7 |
HBeAg = hepatitis B e antigen; RDT = rapid diagnostic test.
Discordant results on each RDT between two laboratory staff were excluded (SD Bioline, N = 1; Insight, N = 3; OneStep, N = 2).
To evaluate whether the low diagnostic sensitivities of the RDTs were related to the sample storage condition, Group 1 was further divided into two groups according to the duration of sample storage. For the older (> 1 year) and more recently (≤ 1 year) collected samples, the sensitivities were 30.4% (7/23, 95% CI: 13.2–52.9) and 29.2% (7/24, 12.6–51.1) for SD Bioline; 31.8% (7/22, 13.9–54.9) and 30.4% (7/23, 13.2–52.9) for Insight; and 47.8% (11/23, 26.8–69.4) and 35.3% (6/17, 14.2–61.7) for OneStep, respectively.
Lower limit of detection of the RDTs.
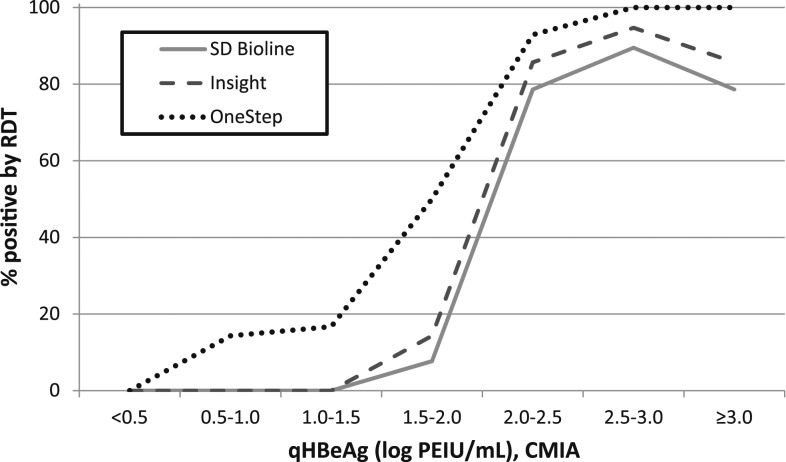
Figure 2 presents the proportion of HBeAg-positive samples successfully detected by each RDT at different quantitative HBeAg levels (log10 PEIU/mL) determined by the quantitative CLIA. This analysis included all samples positive for HBeAg (N = 116; all the samples in Group 1 and 68 HBeAg-positive samples in Group 3). Using the threshold of a ≥ 95% chance to be detected, the limit of detection was estimated at 2.5–3.0 log PEIU/mL for OneStep. SD Bioline and Insight did not detect ≥ 95% of samples even above the highest quantitative HBeAg levels (≥ 3.0 log PEIU/mL). Below the quantitative HBeAg cut-off of 2.0 log PEIU/mL, the performance of the RDT became particularly poor: SD Bioline, Insight, and OneStep detected only 1.6% (1/64), 1.9% (1/54), and 17.2% (10/58) of the HBeAg-positive samples, respectively (Supplemental Table 3).
Figure 2.
Proportion of Hepatitis B e antigen (HBeAg)–positive samples successfully detected by each rapid diagnostic test (RDT) at various quantitative HBeAg levels in Senegal (N = 116).
Hepatitis B e antigen prevalence in highly viremic patients (≥ 106 IU/mL).
The prevalence of HBeAg in highly viremic adult patients (≥ 106 IU/mL) was estimated using Group 3 (N = 117): 58.1% (95% CI: 48.9–66.8) using the reference CLIA, 24.5% (17.3–33.6) using SD Bioline, 30.1% (21.9–39.8) using Insight, and 37.5% (28.6–47.3) using OneStep. In a subgroup of women of childbearing age (N = 28) with high HBV DNA levels (≥ 106 IU/mL), the prevalence of HBeAg was 53.6% (95% CI: 34.3–71.8) using CLIA, 28.6% (14.3–48.9) using SD Bioline, 32.0% (16.0–53.7) using Insight, and 34.8% (17.4–57.4) using OneStep.
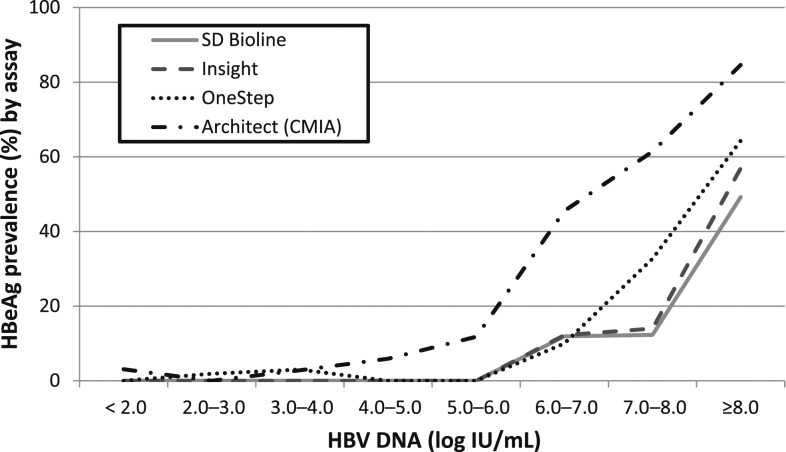
Figure 3 and Supplemental Table 4 present the prevalence of HBeAg at various levels of HBV DNA in Senegalese samples. This analysis included all three groups (Groups 1, 2, and 3; N = 361). The HBeAg prevalence increased with increasing viral load, irrespective of the type of HBeAg assay.
Figure 3.
Prevalence of Hepatitis B e antigen (HBeAg) by different HBeAg assays at various hepatitis B virus (HBV) DNA levels in Senegal (N = 361).
Role of quantitative HBeAg in Senegal.
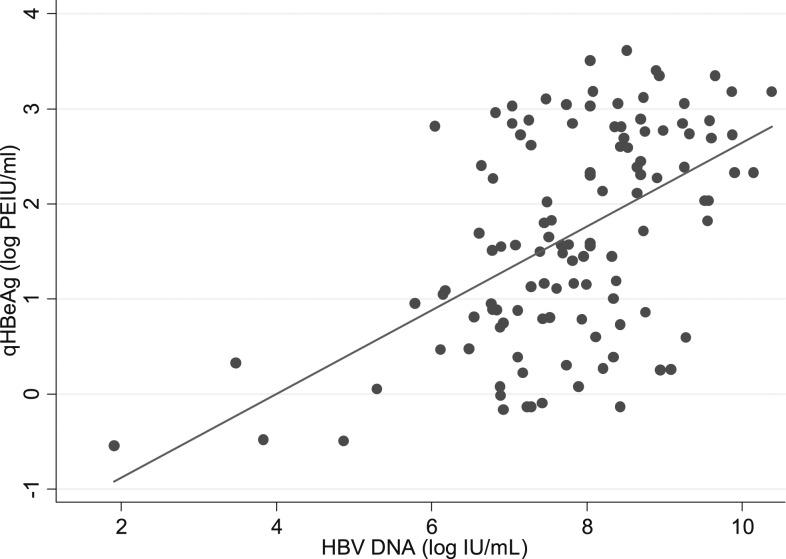
In HBeAg-positive subjects (N = 116: all the samples in Group 1 and 68 HBeAg-positive samples in Group 3), quantitative HBeAg was inversely correlated with age (correlation coefficient: r = −0.37, P < 0.0001) (Supplemental Figure 1) and positively correlated with HBV DNA levels (r = 0.52, P < 0.0001) (Figure 4).
Figure 4.
Correlation between quantitative HBeAg (log PEIU/mL) and hepatitis B virus (HBV) DNA levels (log IU/mL) in HBeAg-positive samples in Senegal (N = 116). HBeAg = hepatitis B e antigen, PEIU = Paul Ehrlich Institut unit.
Asian samples.
In 17 HBeAg-positive Asian samples, the median quantitative HBeAg level was 2.0 log10 PEIU/mL (IQR: −0.4 to 2.7) and the median HBV DNA level was 3.3 log10 IU/mL (IQR: 1.8–7.0). The concordance between the laboratory staff was 100% for all of the RDTs. Sensitivity of the RDTs for HBeAg detection in reference to Architect (CLIA) was 8/17 (47.1%; 95% CI: 23.0–72.2) for SD Bioline, 9/17 (52.9%; 27.8–77.0) for Insight, and 6/17 (35.3%, 14.2–61.7) for OneStep (Table 3).
DISCUSSION
This is the first African study to evaluate the diagnostic accuracy of the commercial RDTs for HBeAg. Using well-characterized serum samples from Senegalese patients, we found good inter-rater agreements (kappa 0.81–0.96) of these RDTs; however, HBeAg was only detected when quantitative HBeAg levels were very high, resulting in poor diagnostic sensitivity ranging from 29.8% to 42.5%. We also found that in Senegal only half of highly viremic (≥ 106 IU/mL) women of childbearing age were positive for HBeAg using the reference assay. In those positive for HBeAg, quantified HBeAg levels were negatively correlated with age and positively correlated with HBV DNA levels. Finally, we confirmed similarly poor diagnostic sensitivity of the RDTs in a small number of Asian samples (range: 35.3–52.9%).
To date, few studies have investigated the performance of HBeAg RDTs. Binax NOW® (Binax Inc., Portland, ME), a rapid test that simultaneously detects HBsAg and HBeAg, has been validated in three studies.15–17 Using sera or whole blood, these studies constantly reported adequate diagnostic sensitivity (80–100%) and specificity (98–100%) for HBeAg detection,15–17 with a lower limit of detection of 2.0 PEIU/mL.15 Advanced Quality™ (InTec Products Inc, Xiamen, China), another RDT for HBeAg, has been evaluated in one study which reported a sensitivity of 64% and a specificity of 97%.18 Unfortunately, these RDTs have subsequently been taken off the market, and are currently unavailable.
To understand the poor diagnostic sensitivity of the RDTs that we evaluated, we determined their lower limits of detection in reference to the new commercial quantitative HBeAg calibrator. The limit of detection of OneStep was estimated to be above 2.5–3.0 log PEIU/mL (i.e., 320–1,000 PEIU/mL), which was much higher than that reported for Binax NOW® (2.0 PEIU/mL) or the reference CLIA (0.5 PEIU/mL). For SD Bioline and Insight, HBeAg detection was not consistently observed even above the level of 3.0 log PEIU/mL. To further investigate whether poor performance of these RDTs is related with the difference in HBeAg epitopes between African HBV genotypes (E and A in Senegal)19 and other genotypes (B and C in Asia, e.g.), we evaluated these RDTs in samples from East Asian patients positive for HBeAg. The sensitivities were also poor (35.3–52.9%) in these Asian samples. However, among the three RDTs, OneStep demonstrated the highest sensitivity in Senegal and the lowest sensitivity in Asian samples. This supports the potential role of HBV genotype in the performance of the RDTs.
Hepatitis B virus DNA has been established to be the single most important marker to identify pregnant women at the highest risk of MTCT.20,21 Consequently, current international guidelines recommend administration of antiviral therapy during pregnancy in HBsAg-positive women with an HBV DNA level > 200,000 IU/mL, irrespective of HBeAg status.22,23 However, this strategy is hardly applicable to low- and middle-income countries where the access to HBV DNA PCR techniques is severely limited.24 Alternatively, HBeAg-based screening without HBV DNA would be appealing if most HBV-infected women with high viral load also carried HBeAg. Indeed, 98% and 86% of highly viremic pregnant women (> 200,000 IU/mL) were positive for HBeAg in Taiwan and the United States, respectively.8,25
Although positive HBeAg is highly correlated with increased HBV DNA levels in HBV-infected young women, emerging basal core promoter (BCP) or precore (PC) variants abolish or reduce HBeAg production without affecting the capacity of the virus to replicate.26 Consequently, there are pregnant women with negative HBeAg who still carry high HBV viral loads by harboring these mutations.27 In our study in Senegal, we found that only 54% of HBV-infected women of childbearing age with high viral load (≥ 106 IU/mL) were positive for HBeAg, suggesting that half of the women with an elevated risk of immunoprophylaxis failure will be missed through HBeAg-based screening. Moreover, by using the HBeAg RDTs which further misclassify because of poor sensitivity, we will overlook > 65% of highly viremic women. A similarly low prevalence of HBeAg in women with high HBV DNA levels has been previously reported from other African studies.28,29 This geographical difference in HBeAg positivity in women with high HBV viral loads might be related to the predominant HBV genotypes in each region, and the difference in the frequency of emerging BCP or PC variant quasispecies by HBV genotype.12,30 To further elucidate the natural history of chronic HBV infection in Africa, additional molecular studies that characterize viral genotype, viral mutations, and HBV serological markers, are warranted.
Until recently, HBeAg could only be semi-quantified through in-house calibration using a reference panel of defined HBeAg concentration supplied from the Paul Ehrlich Institute, Germany.31,32 In our study, we quantified HBeAg using the new commercial calibrator of Abbott. We demonstrated, for the first time in African HBeAg-positive subjects, that its concentration decreases with age. This is not surprising because HBeAg loss is preceded by decreasing levels of quantitative HBeAg,33 and spontaneous loss of HBeAg occurs over time in those who established chronic HBV infection during childhood,5 with the probable emergence of BCP and/or PC variants.26 This is supported by a longitudinal cohort of Gambian children with chronic HBV infection, in which Mendy et al.34 demonstrated a reduction in qualitative HBeAg seropositivity with age in those who have a high viral load (> 2,000 IU/mL): 96% at ages < 5 years, 90% at ages 6–9 years, 86% at ages 10–13 years, and 63% at ages 20–23 years. We also found a good correlation between quantitative HBeAg and HBV DNA levels in HBeAg-positive subjects in Senegal. In low- and middle-income countries, the limited access to nucleic acid tests to measure HBV DNA has been a significant impediment to the effective management of chronic HBV infection.11 As quantitative HBeAg is more affordable than nucleic acid tests, this may be a useful alternative tool to HBV DNA PCR to define treatment eligibility in these settings.
This study has several limitations. First, we could not directly assess the influence of the different HBV genotypes on the performance of the RDTs. Nevertheless, we confirmed that the sensitivity for HBeAg detection was low in both Senegalese and East Asian samples, suggesting that these RDTs may perform poorly irrespective of HBV genotype. Second, we assessed the performance of the RDTs in adults with chronic HBV infection who were referred to the laboratory for HBV DNA measurement, rather than in pregnant women, who are the target population for use of such an RDT. Third, we had limited access to clinical information on the study subjects; we do not know why the physicians requested both HBV DNA and HBeAg for some patients (Groups 1 and 2) and only HBV DNA for the rest (Group 3). Fourth, although we had a high enough number of HBeAg-negative samples, we could not reach the desired number of HBeAg-positive samples originally derived by our sample size calculation. As mentioned earlier, in SSA the prevalence of HBeAg in chronic HBV carriers is much lower than in Asia, and thus, it is extremely difficult to have a large number of HBeAg-positive samples.35 Finally, we could not test matched fresh and frozen samples to evaluate the effect of the storage condition on the test performance. Nevertheless, we found similar diagnostic sensitivities between those stored for longer (> 1 year) and shorter (≤ 1 year) periods, implying that the influence of the storage condition might be minimal for the test performance.
Without having simple, affordable, and reliable diagnostic tools to evaluate HBV viral replication, it is unlikely that the WHO’s global elimination goals can be achieved, especially as the HBV burden is unequally distributed to low- and middle-income countries. In these settings, RDTs may have an important role in scaling up clinical management of HBV infection through decentralized care; however, we found unacceptably poor sensitivity of RDTs for HBeAg. These findings should be further confirmed through a large study. In addition, urgent efforts need to be made to improve these RDTs and to develop and validate other markers of viral replication that are adapted to resource-limited settings.36
Supplementary Material
Supplemental figure and tables
Acknowledgments:
We thank all the study participants.
Note: Supplemental figure and tables appear at www.ajtmh.org.
REFERENCES
- 1.Stanaway JD, et al. 2016. The global burden of viral hepatitis from 1990 to 2013: findings from the Global Burden of Disease Study 2013. Lancet 388: 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO , 2016. Global Health Sector Strategy on Viral Hepatitis 2016–2021. Geneva, Switzerland: World Health Organization, 1–56. [Google Scholar]
- 3.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ, 2015. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 6736: 1–10. [DOI] [PubMed] [Google Scholar]
- 4.Nayagam S, Thursz M, Sicuri E, Conteh L, Wiktor S, Low-Beer D, Hallett TB, 2016. Requirements for global elimination of hepatitis B: a modelling study. Lancet Infect Dis 16: 1399–1408. [DOI] [PubMed] [Google Scholar]
- 5.Shimakawa Y, et al. 2016. Natural history of chronic HBV infection in West Africa: a longitudinal population-based study from the Gambia. Gut 65: 2007–2016. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization , 2009. Hepatitis B vaccines. Wkly Epidemiol Rec 84: 405–419. [PubMed] [Google Scholar]
- 7.Keane E, Funk AL, Shimakawa Y, 2016. Systematic review with meta-analysis: the risk of mother-to-child transmission of hepatitis B virus infection in sub-Saharan Africa. Aliment Pharmacol Ther 44: 1005–1017. [DOI] [PubMed] [Google Scholar]
- 8.Wen WH, Chang MH, Zhao LL, Ni YH, Hsu HY, Wu JF, Chen PJ, Chen DS, Chen HL, 2013. Mother-to-infant transmission of hepatitis B virus infection: significance of maternal viral load and strategies for intervention. J Hepatol 59: 24–30. [DOI] [PubMed] [Google Scholar]
- 9.Visvanathan K, et al. 2016. Managing HBV in pregnancy. Prevention, prophylaxis, treatment and follow-up: position paper produced by Australian, UK and New Zealand key opinion leaders. Gut 65: 340–350. [DOI] [PubMed] [Google Scholar]
- 10.Lemoine M, Thursz MR, 2017. Battlefield against hepatitis B infection and HCC in Africa. J Hepatol 66: 645–654. [DOI] [PubMed] [Google Scholar]
- 11.WHO , 2017. Global Hepatitis Report, 2017. Geneva, Switzerland: World Health Organization, 1–68. [Google Scholar]
- 12.WHO , 2018. World Health Organization Model List of Essential In Vitro Diagnostics, 1st edition. Geneva, Switzerland: World Health Organization, 1–35. [Google Scholar]
- 13.Kramvis A, 2016. The clinical implications of hepatitis B virus genotypes and HBeAg in pediatrics. Rev Med Virol 26: 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossuyt PM, et al. 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology 277: 826–832. [DOI] [PubMed] [Google Scholar]
- 15.Clement F, Dewint P, Leroux-Roels G, 2002. Evaluation of a new rapid test for the combined detection of hepatitis B virus surface antigen and hepatitis B virus e antigen. J Clin Microbiol 40: 4603–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau DT-Y, Ma H, Lemon SM, Doo E, Ghany MG, Miskovsky E, Woods GL, Park Y, Hoofnagle JH, 2003. A rapid immunochromatographic assay for hepatitis B virus screening. J Viral Hepat 10: 331–334. [DOI] [PubMed] [Google Scholar]
- 17.Akanmu AS, Esan OA, Adewuyi JO, Davies AO, Okany CC, Olatunji RO, Babalola T, 2006. Evaluation of a rapid test kit for detection of HBsAg/eAg in whole blood: a possible method for pre-donation testing. Afr J Med Med Sci 35: 5–8. [PubMed] [Google Scholar]
- 18.Mainet-González D, Palenzuela-Gardon DO, Aguilar Rubido JC, 2009. Comparison between an immunochromatographic test with an amplified ELISA for detecting e antigen and anti-e antigen antibodies in chronic hepatitis B. Biotecnol Apl 26: 143–145. [Google Scholar]
- 19.Vray M, Debonne J, Sire J-M, Tran N, Chevalier B, Plantier J-C, Fall F, Vernet G, Mb PS, Simon F, 2006. Molecular epidemiology of hepatitis B virus in Dakar, Sénégal. J Med Virol 78: 329–334. [DOI] [PubMed] [Google Scholar]
- 20.Burk RD, Hwang L-Y, Ho GYF, Shafritz DA, Beasley RP, 1994. Outcome of perinatal hepatitis B virus exposure is dependent on maternal virus load. J Infect Dis 170: 1418–1423. [DOI] [PubMed] [Google Scholar]
- 21.Pan CQ, Duan Z, Bhamidimarri KR, Zou H, Liang X, Li J, Tong MJ, 2012. An algorithm for risk assessment and intervention of mother to child transmission of hepatitis B virus. Clin Gastroenterol Hepatol 10: 452–459. [DOI] [PubMed] [Google Scholar]
- 22.Terrault NA, Bzowej NH, Chang K-M, Hwang JP, Jonas MM, Murad MH, 2016. AASLD guidelines for treatment of chronic hepatitis B. Hepatology 63: 261–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.European Association for the Study of the Liver , 2017. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol 67: 370–398. [DOI] [PubMed] [Google Scholar]
- 24.Andriamandimby SF, Olive M, Shimakawa Y, Rakotomanana F, Razanajatovo IM, Andrianinarivomanana TM, Ravalohery J, Andriamamonjy S, Rogier C, Heraud J-M, 2017. Prevalence of chronic hepatitis B virus infection and infrastructure for its diagnosis in Madagascar: implication for the WHO’s elimination strategy. BMC Public Health 17: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubo A, Shlager L, Marks AR, Lakritz D, Beaumont C, Gabellini K, Corley DA, 2014. Prevention of vertical transmission of hepatitis B. Ann Intern Med 160: 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson AJV, et al. 2010. Serum hepatitis B surface antigen and hepatitis B e antigen titers: disease phase influences correlation with viral load and intrahepatic hepatitis B virus markers. Hepatology 51: 1933–1944. [DOI] [PubMed] [Google Scholar]
- 27.Fujiko M, et al. 2015. Chronic hepatitis B in pregnant women: is hepatitis B surface antigen quantification useful for viral load prediction? Int J Infect Dis 41: 83–89. [DOI] [PubMed] [Google Scholar]
- 28.Ducancelle A, Abgueguen P, Birguel J, Mansour W, Pivert A, Le Guillou-Guillemette H, Sobnangou JJ, Rameau A, Huraux JM, Lunel-Fabiani F, 2013. High endemicity and low molecular diversity of hepatitis B virus infections in pregnant women in a rural district of north Cameroon. PLoS One 8: 10–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimakawa Y, Toure-Kane C, Mendy M, Thursz M, Lemoine M, 2016. Mother-to-child transmission of hepatitis B in sub-Saharan Africa. Lancet Infect Dis 16: 19–20. [DOI] [PubMed] [Google Scholar]
- 30.Dumpis U, Mendy M, Hill A, Thursz M, Hall A, Whittle H, Karayiannis P, 2001. Prevalence of HBV core promoter/precore/core mutations in Gambian chronic carriers. J Med Virol 65: 664–670. [DOI] [PubMed] [Google Scholar]
- 31.Wursthorn K, Zacher BJ, Jaroszewicz J, Darnedde M, Manns M, Wedemeyer H, 2011. Development of a protocol for the quantitative determination of HBeAg using the Elecsys® HBeAg immunoassay. J Viral Hepat 18: 179–183. [DOI] [PubMed] [Google Scholar]
- 32.Maylin S, et al. 2013. Quantification of hepatitis B e antigen between Elecsys HBeAg and Architect HBeAg assays among patients infected with hepatitis B virus. J Clin Virol 56: 306–311. [DOI] [PubMed] [Google Scholar]
- 33.Maylin S, et al. 2012. Kinetics of hepatitis B surface and envelope antigen and prediction of treatment response to tenofovir in antiretroviral-experienced HIV–hepatitis B virus-infected patients. AIDS 26: 939–949. [DOI] [PubMed] [Google Scholar]
- 34.Mendy ME, McConkey SJ, van der Sande MAB, Crozier S, Kaye S, Jeffries D, Hall AJ, Whittle HC, 2008. Changes in viral load and HBsAg and HBeAg status with age in HBV chronic carriers in the Gambia. Virol J 5: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemoine M, et al. 2016. Acceptability and feasibility of a screen-and-treat programme for hepatitis B virus infection in the Gambia: the Prevention of Liver Fibrosis and Cancer in Africa (PROLIFICA) study. Lancet Glob Health 4: e559–e567. [DOI] [PubMed] [Google Scholar]
- 36.Shimakawa Y, Seck A, Nayagam S, Toure-Kane C, Lemoine M, 2018. Screening strategies to prevent mother-to-child transmission of hepatitis B in sub-Saharan Africa. Lancet Gastroenterol Hepatol 3: 222–223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables