Abstract.
Leptospira licerasiae serovar Varillal, a group II intermediate pathogen species/serovar discovered in the Peruvian Amazon city of Iquitos, is commonly recognized in this region by sera from humans (at least 40% seroprevalence) without a known clinical history of leptospirosis. This high frequency of human seroreactivity remains unexplained. To test the hypothesis that the oral route of infection might explain the high rate of human seroreactivity against L. licerasiae, an experimental infection model using Rattus norvegicus was developed, given that rats were one of the original reservoir hosts identified as being colonized by this leptospire. Sprague–Dawley rats were experimentally exposed via mucosa, direct gastric gavage, or parenteral inoculation with nine different isolates of L. licerasiae originally isolated from Peruvian humans, peridomiciliary rodents, and wildlife. As shown by quantitative polymerase chain reaction of kidney tissue, Leptospira infection via these routes of infection was equally successful. Importantly, the data show that L. licerasiae infects R. norvegicus via the oral route, leading to renal colonization. Not only do these findings confirm the infectiousness of group II Leptospira, but also they underscore the potential importance of oral as well as mucosal and transcutaneous routes of Leptospira infection.
INTRODUCTION
Leptospirosis, caused by infectious Leptospira spp., is a reemerging zoonotic disease with growing impact on public health, particularly among impoverished communities in tropical, resource-poor countries.1 It is generally thought that humans contract leptospirosis following mucosal contact with or percutaneous exposure to soil and surface water contaminated by urine from renally colonized mammalian reservoir hosts, including peridomiciliary rodents, dogs, livestock and sylvatic species.1–4
Leptospira licerasiae serovar Varillal was discovered as a new leptospiral species and serovar in the context of a prospective study of human leptospirosis accompanied by animal-trapping studies carried out in the Peruvian Amazon.5 This species was identified as potentially new when pulse-field gel electrophoresis found a novel pattern in Leptospira isolated from humans, Rattus norvegicus and Rattus rattus in the city of Iquitos, Peru.5 16S rDNA sequencing5 followed by whole genome and comparative sequence analysis6,7 demonstrated this new Leptospira to be a group II, intermediately pathogenic Leptospira. Serological,5 carbohydrate composition analysis,8 and genomic analysis of the rfb locus confirmed that L. licerasiae serovar Varillal is a novel serovar, which has the smallest known rfb locus among Leptospira suggesting the possibility that this isolate has a rough lipopolysaccharide.7
There are 22 recognized Leptospira species classified into three large species groups based on DNA–DNA hybridization, later refined by 16S rDNA phylogeny, and phylogenetic analysis of 1,175 proteins comprising the Leptospira core genome.1,9 Group I contains 10 pathogenic species, group II, five so-called intermediately pathogenic species, and group III seven nonpathogenic (“saprophytic”) species. Group I Leptospira—primarily Leptospira interrogans, Leptospira kirschneri, Leptospira borgpetersenii, and Leptospira noguchii—are most commonly implicated in human disease, with symptoms varying in severity ranging from undifferentiated febrile illness to severe disease complicated by varying organ dysfunction including jaundice, hemorrhage, acute kidney injury (oliguric and nonoliguric), refractory shock, with a reported 1 million annual cases with a 5–20% case fatality rate.10 Group II Leptospira, such as Leptospira fainei, Leptospira broomii, and L. licerasiae, do infect mammals.5,11,12 However, in contrast to group I, group II Leptospira grow rapidly in liquid culture, much like saprophytic species, but have rarely been associated with severe disease; lethal infection has not been reported.5,11,13 Leptospira broomii, another group II Leptospira, has been recovered from two severe cases requiring hospitalization.5,11,12,14 Nevertheless, successful experimental infection of rodents with group II Leptospira has not been reported. Hence, despite serological and bacteriological evidence group II Leptospira cause human disease, the pathogenicity of group II Leptospira is still debated.11,12,14
Small animal models of group I Leptospira infection that recapitulate acute human disease have been described using hamsters, guinea pigs, and inbred mouse strains; these models have primarily used serovars of L. interrogans and L. kirschneri, 3,15–18 by contrast, an experimental animal model has not been described for group II Leptospira, despite the isolation of L. fainei from pigs and L. licerasiae from rats and other sylvatic animals and humans.11,19,20 The original report of L. licerasiae described the inability of this species to infect hamsters, guinea pigs, and wild-type and immunodeficient mice in experimental settings.5 Although intraperitoneal injection and conjunctival instillation are the most used routes of challenge infection, the oral/gastric route of infection—which would implicate ingestion as a route of acquiring leptospirosis—has not been carefully examined. One such study concluded that saliva protected against infection by acting as a natural defense against leptospiral infection but did not formally differentiate gastric versus oropharyngeal mucosal sites of infection.21 Given the high seroprevalence of L. licerasiae serovar Varillal among humans in the Peruvian Amazon, we sought to determine whether the oral route of infection might be important for this species or this specie. Hence, the experiments presented here sought to compare infectivity of well-characterized isolates of L. licerasiae in rats (the presumed rodent reservoir) inoculated intraperitoneally or exposed via more biologically relevant (“natural”) routes, i.e., through nasal and conjunctival mucosa, epicutaneously, and the stomach.7
MATERIALS AND METHODS
Ethics.
Experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of California San Diego. All experiments were performed in accordance with standard practices governed by the AAALAC-accredited UC San Diego Animal Care Program.
Bacterial strains and culture conditions.
Leptospira strains were originally isolated in Iquitos, Peru, within the context of a previously published study of human leptospirosis and are available from American Type Culture Collection/ Biodefense and Emerging Infections Research Resources Repository.5 All strains were maintained in liquid Ellinghausen–McCullough–Johnson–Harris (EMJH) (BD, Franklin Lakes, NJ) media as described previously.22 Strains used in the present studies were of unknown passage number given previous experimental inability to infect small animals.5
Experimental rat infections.
Three-week-old, male, specific pathogen-free Sprague–Dawley rats (R. norvegicus) were purchased from Sprague–Dawley (South Easton, MA). Animals were restrained manually for intragastic, intraperitoneal, and intravascular inoculation, whereas those in the intranasal, conjunctival, and epicutaneous groups were anesthetized using an open-drop technique with 1.5% (v/v) isoflurane (VetOne, Boise, ID).
Leptospira cells were counted by dark-field microscopy and adjusted to a final density of 107 cells/mL with sterile EMJH medium. Animals (three per route of infection) were inoculated with a range of 2 × 105 to 1 × 107 Leptospira depending on the route of exposure. Control animals were inoculated with equivalent volumes of sterile EMJH medium. Animals were clinically observed for signs of infection twice daily for up to 2 weeks and then euthanized by inhalation of carbon dioxide in accordance with current American Veterinary Medical Association guidelines.23 Because the isolates had an unknown number of in vitro passages, they were first pooled at equal cell densities for initial infection experiments, then used separately in a second series of experiments to assess differences in infectivity, using one of the inoculation routes. All experiments were repeated at least once to assess reproducibility.
Intraperitoneal inoculations.
A 25G needle (BD), held at a 45-degree angle, was used to inoculate 107 Leptospira directly into the peritoneum. Animals were firmly restrained with the head and body tipped downward; injections were made in the lower right quadrant.
Intragastric inoculations.
The head was gently pulled back to extend the neck and esophagus, an 18-gauge feeding tube was inserted into the esophagus via the mouth, and 107 Leptospira were inoculated directly into the stomach.
Intravenous inoculations.
A 23G needle was used to inject 5 × 106 Leptospira directly into the lateral tail vein. A restraining device was used to immobilize the animals during the injections.
Intranasal inoculations.
Twenty microliters of liquid EMJH medium containing 4 × 105 Leptospira was placed directly into each nostril then gently massaged into the nasal mucosa while the rats were anesthetized with isoflurane.
Conjunctival inoculations.
Ten microliters of liquid EMJH medium containing 2 × 105 Leptospira was placed directly onto the conjunctival surface of each eye. Rats were under anesthesia with isoflurane as described previously.
Epicutaneous inoculations.
Hair (∼0.5 cm2) was shaved off and the skin wiped with 70% (v/v) ethanol. The exposed skin was abraded using a sterile scalpel to remove the top layer of skin. Fifteen microliters of liquid EMJH medium containing 3 × 105 Leptospira was inoculated directly onto the abraded skin.
Necropsy.
Two weeks after inoculation, animals were euthanized and perfused with sterile saline (to eliminate blood from organs to allow for distinguishing organ from blood infection) and kidneys harvested for quantitative polymerase chain reaction (qPCR) and culture. To attempt to reisolate Leptospira from infected animals, sections of left and right kidneys were macerated and inoculated into 10 mL of semisolid EMJH containing 5-fluorouracil (0.01%) and neomycin sulfate (300 μg/mL) supplemented by fetal bovine serum and sodium pyruvate. This material was also inoculated directly into 2 mL of RNAlater (Qiagen, Valencia, CA) and stored at −80°C for subsequent DNA preparation and qPCR.
Nucleic acid testing for presence of Leptospira.
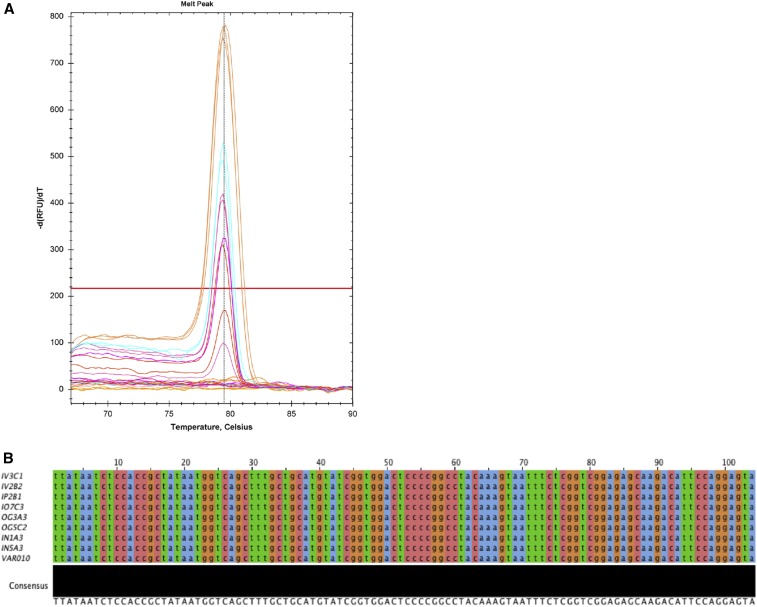
DNA was extracted from kidney samples using the DNeasy Blood and Tissue Kit (Qiagen). For each kidney, two to three separate kidney sections were taken and pooled (18–23 mg total) before DNA extraction. Extraction was performed according to manufacturer’s instructions. DNA concentration was determined by NanoDrop (Thermo Scientific, Wilmington, DE). A qPCR assay using primers against the L. licerasiae lipL32 gene5 was performed to quantify Leptospira burden in renal tissue. Primers sequences were as follows: lipL32_lic F: 5′-CTT ATA ATC TCC ACC GCT ATA AT-3′; lipL32_lic R: 5′-TAC TCC TGG AAT GTC TTG CTC TC-3′. Reaction mixes were prepared using PerfeCTa SYBR Green Supermix (Quanta Biosciences, Gaithersburg, MD) according to manufacturer’s instructions, with primers at a final concentration of 0.2 ng/μL. Reactions were carried out in triplicate on a CFX96 Touch thermal cycler (Bio-Rad, Hercules, CA). Cycling conditions were as follows: 95°C for 120 seconds, followed by 39 cycles of 95°C for 15 seconds, 65.3°C for 30 seconds, 72°C for 60 seconds, and 65°C for 10 seconds (amplification, annealing, and extension). Following amplification, melt curve analysis (65–95°C, 0.2°C increments) was used to confirm the presence of specific amplification products. Reactions with Cq values ≥ 40 were considered negative based on preliminary data when developing the assay (data not shown), as were those lacking the expected melt peak (Tm 79.5°C). A standard curve (108 − 102) was generated using gDNA extracted from cultured L. licerasiae VAR10 and used to determine the number of genome equivalents in infected kidney tissue. Specificity was confirmed by conventional Sanger sequencing of cloned qPCR amplicons (Figure 1).
Figure 1.
Detection of Leptospira licerasiae lipL32 sequences, confirming renal colonization 2 weeks postchallenge. (A) Postamplification melt curve data showing presence of single quantitative polymerase chain reaction (qPCR) product with expected Tm (79.5°C ± 0.1) (B) Gel-purified PCR amplicons from qPCR-positive tissues were cloned, then sequenced by conventional Sanger sequencing. Vector-trimmed sequences were aligned to the full-length L. licerasiae lipL32 gene using the multiple alignment by fast Fourier transform sequence alignment tool.28 Amplicons derived from kidney tissue from all groups were identical to each other and to the VAR010 reference sequence. Groups: intravascular (IV3C1 and IV2B2); intraperitoneal group: (IP2B1); conjunctival group: (IO7C3); intragastric group: (OG3A3 and OG5C2); intranasal group (IN1A3 and IN5A3). This figure appears in color at www.ajtmh.org.
RESULTS AND DISCUSSION
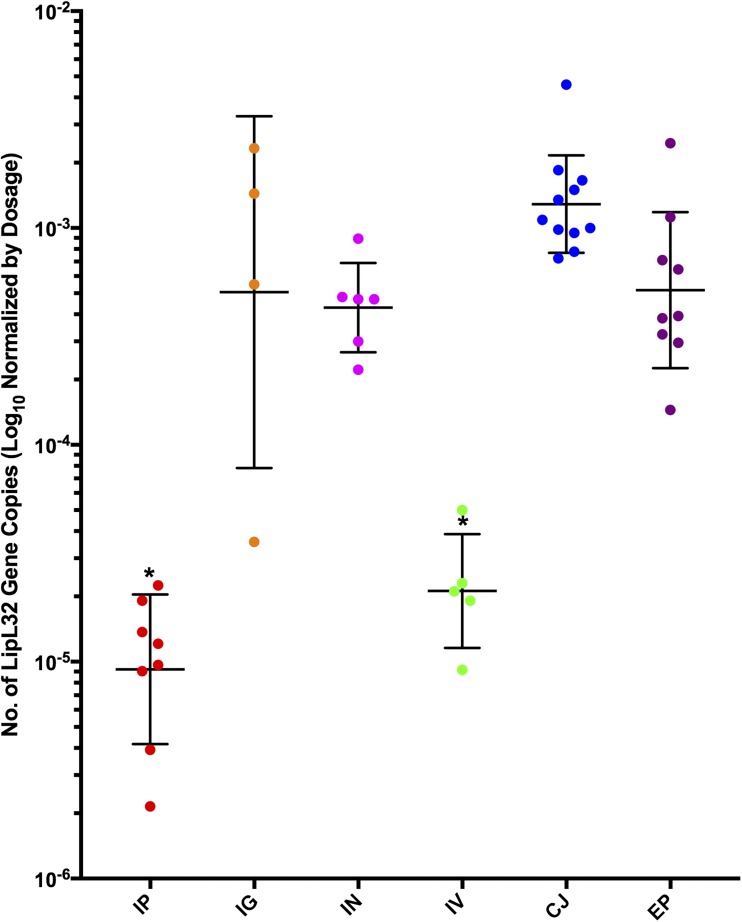
Here we demonstrate that mucosal and direct gastric inoculation of R. norvegicus by the group II intermediately pathogenic L. licerasiae most efficiently leads to renal colonization. Experimental infections were carried out twice, with consistent results. For a total of six animals per exposure route, at least three rats per exposure group were qPCR positive at 2 weeks postinfection (Figure 2). Postamplification melt curve data indicated the presence of a single qPCR product with the expected melt peak (Tm 79.5°C) (Figure 1A). Conventional Sanger sequencing of the PCR products confirmed the presence of L. licerasiae in all qPCR-positive specimens 2 weeks postinoculation (Figure 1).
Figure 2.
Quantification of Leptospira licerasiae in kidney tissue of infected Sprague Dawley (Rattus norvegicus) rats by qPCR. Geometric mean and geometric SD of number of lipL32 gene copies, normalized by inoculation dosage of corresponding route (total number of leptospires). Intraperitoneal 107; Intragastric (IG) 107; Intranasal (IN) 4 × 105; Intravascular (IV) 5 × 106; Conjunctival (CJ) 2 × 105; Epicutaneous (EP) 3 × 105. * Mean number of genome copies significantly different from other routes. This figure appears in color at www.ajtmh.org.
Semisolid cultures to which macerated kidney tissue had been added were checked for Leptospira growth by dark-field microscopy biweekly for up to 3 months. Leptospira were observed 6 weeks postinoculation in cultures inoculated with renal tissue from individual animals challenged by conjunctival and epicutaneous routes. As expected for a presumed reservoir host, and in accordance with the literature, no rats in any of the experimental groups developed observable clinical disease, including hunching, lack of feeding or grooming, weight loss or death.
To test whether L. licerasiae strains differed in their infectivity, follow-up experiments were carried out with groups of three animals being challenged intranasally with individual strains of L. licerasiae. These experiments were based on previous observations that a higher Leptospira burden in kidney tissue was achieved while rats were exposed to fewer spirochetes in different routes of exposure. We found that five of nine strains tested (56%) led to detectable kidney infection at 2 weeks postchallenge (Table 1).
Table 1.
Infection rates of Sprague–Dawley rats following intranasal exposure to individual isolates of Leptospira licerasiae serovar Varillal as determined by quantitative polymerase chain reaction of kidney tissue
| L. licerasiae strains | Isolate source | n | Infected | Percentage |
|---|---|---|---|---|
| MMD4787 | Proechimys sp. | 3 | 0 | 0 |
| MMD3847 | Proechimys brevicauda | 3 | 1 | 33.33 |
| HAI1026 | Human | 3 | 2 | 66.66 |
| VAR10 | Human | 3 | 2 | 66.66 |
| MMD3782 | Metachirus nudicaudatus | 3 | 0 | 0 |
| MMD3795 | P. brevicauda | 3 | 2 | 66.66 |
| CEH008 | Rattus norvegicus | 3 | 0 | 0 |
| CEH006 | R. norvegicus | 3 | 1 | 33.33 |
| MMD4847 | Uroderma magnirostrum* | 3 | 0 | 0 |
| Total | – | 27 | 8 | 29.6 |
CEH indicates peridomestic rodents trapped near houses, MMD indicates wild-caught rodents trapped along a forest transect, VAR and HAI indicate strains isolated from human cases at health centers. The study during which strains were isolated was approved by the Human Subjects Protection Program, University of California San Diego, and the Ethical Committees of Asociacion Benefica PRISMA, Lima, Peru, and Universidad Peruana Cayetano Heredia, Lima, Peru. Animal trapping and use for was approved by the Instituto Nacional de Recursos Naturales of Peru (INRENA), Lima, Peru, and the Institutional Animal Care and Use Committee, University of California San Diego.
To the present, no reports have described successful experimental animal infections by group II Leptospira species. Nevertheless, recent whole-genome comparisons demonstrate that both group I (pathogenic) and group II (intermediate) species of Leptospira are more closely related than either is to the noninfectious Leptospira.7,24 Here, using several recent isolates of L. licerasiae, we demonstrate reproducible renal infection via different routes of exposure, confirming the infectivity of group II Leptospira. Based on the percentage of colonized animals, infection through nasal and gastric mucosa was more efficient compared with intraperitoneal and intravascular inoculation, illustrating the importance of the route of exposure on transmission and in vivo dissemination of group II Leptospira. Moreover, these experiments provide additional evidence that intraperitoneal inoculation may not be optimal for Leptospira infectivity studies and provide a strong foundation for the development of an infection model for group II Leptospira based on the ability for the organisms to colonize the kidney, an important milestone for future gain-of-function genetic studies using this group of organisms.
To test whether route of inoculation affected the ability of Leptospira to establish renal colonization, we compared mean Leptospira burden in kidney tissue relative to dosage for each route. Of nine L. licerasiae strains studied, five were detected in renal tissue 2 weeks after infection. The variation in infectivity among strains could have been due to loss of virulence due to repeated subculture without animal passage that was not previously possible in the absence of an animal infection model. However, because VAR010 and HAI1026 were subjected to more subcultures in vitro than the other strains and both infected readily, this seems unlikely. Notably, five of nine strains that were undetectable at 2 weeks, MMD4787, MMD3782, and MMD4847, were isolated from kidney tissues from species other than R. norvegicus (Spiny rat [Proechimys sp.], brown four-eyed opossum [Metachirus nudicaudatus], and brown tent-making bat [Uroderma magnirostrum] respectively). Accordingly, our observations might be explained by varied host specificity among L. licerasiae strains, though this has yet to be tested experimentally, and any possible genomic differences among these strains would need to be identified. It is also possible that these strains produced only transient infections and were therefore undetectable at the time of euthanasia.
Demonstrating that multiple isolates/strains of L. licerasiae serovar Varillal infects R. norvegicus by the oral route may help to explain the high seroprevalence of L. licerasiae among humans in the Peruvian Amazon. This observation is of significant public health importance because it would predict that ingesting contaminated water might give rise to human infection. Furthermore, we showed that group II Leptospira are able to infect rats and colonize renal tissue, providing a relevant animal model for understanding fundamental host-pathogen interactions and pathogenetic mechanisms of Leptospira. Given previous observations that group II Leptospira lack the CRISPR/Cas9 bacterial defense mechanism against taking up exogenous DNA,7 leading to new avenues of research including genetic manipulation of this group of infectious Leptospira spp. Furthermore, in vivo gain-of-function studies are now possible (based on a recently described replicative expression vector), which will complement traditional knockout/complementation studies, which have been mostly unsuccessful because of the difficulty in generating targeted gene knockouts in pathogenic Leptospira.25–27
REFERENCES
- 1.Ko AI, Goarant C, Picardeau M, 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat Rev Microbiol 7: 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderon A, Rodriguez V, Mattar S, Arrieta G, 2014. Leptospirosis in pigs, dogs, rodents, humans, and water in an area of the Colombian tropics. Trop Anim Health Prod 46: 427–432. [DOI] [PubMed] [Google Scholar]
- 3.Levett PN, 2001. Leptospirosis. Clin Microbiol Rev 14: 296–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharti AR, et al. Peru-United States Leptospirosis Consortium , 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 5.Matthias MA, et al. 2008. Human leptospirosis caused by a new, antigenically unique Leptospira associated with a Rattus species reservoir in the Peruvian Amazon. PLoS Negl Trop Dis 2: e213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ricaldi JN, et al. 2012. Whole genome analysis of Leptospira licerasiae provides insight into leptospiral evolution and pathogenicity. PLoS Negl Trop Dis 6: e1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouts DE, et al. 2016. What makes a bacterial species pathogenic?: Comparative genomic analysis of the genus Leptospira. PLoS Negl Trop Dis 10: e0004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patra KP, Choudhury B, Matthias MM, Baga S, Bandyopadhya K, Vinetz JM, 2015. Comparative analysis of lipopolysaccharides of pathogenic and intermediately pathogenic Leptospira species. BMC Microbiol 15: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morey RE, Galloway RL, Bragg SL, Steigerwalt AG, Mayer LW, Levett PN, 2006. Species-specific identification of Leptospiraceae by 16S rRNA gene sequencing. J Clin Microbiol 44: 3510–3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa F, Hagan JE, Calcagno J, Kane M, Torgerson P, Martinez-Silveira MS, Stein C, Abela-Ridder B, Ko AI, 2015. Global morbidity and mortality of leptospirosis: a systematic review. PLoS Negl Trop Dis 9: e0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petersen AM, Boye K, Blom J, Schlichting P, Krogfelt KA, 2001. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J Med Microbiol 50: 96–100. [DOI] [PubMed] [Google Scholar]
- 12.Levett PN, Morey RE, Galloway RL, Steigerwalt AG, 2006. Leptospira broomii sp. nov., isolated from humans with leptospirosis. Int J Syst Evol Microbiol 56: 671–673. [DOI] [PubMed] [Google Scholar]
- 13.Tsuboi M, Koizumi N, Hayakawa K, Kanagawa S, Ohmagari N, Kato Y, 2017. Imported Leptospira licerasiae infection in traveler returning to Japan from Brazil. Emerg Infect Dis 23: 548–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chappel RJ, Khalik DA, Adler B, Bulach DM, Faine S, Perolat P, Vallance V, 1998. Serological titres to Leptospira fainei serovar hurstbridge in human sera in Australia. Epidemiol Infect 121: 473–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zuerner RL, Alt DP, Palmer MV, 2012. Development of chronic and acute golden Syrian hamster infection models with Leptospira borgpetersenii serovar Hardjo. Vet Pathol 49: 403–411. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Lou XL, Yang HL, Guo XK, Zhang XY, He P, Jiang XC, 2012. Establishment of a leptospirosis model in guinea pigs using an epicutaneous inoculations route. BMC Infect Dis 12: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nally JE, Fishbein MC, Blanco DR, Lovett MA, 2005. Lethal infection of C3H/HeJ and C3H/SCID mice with an isolate of Leptospira interrogans serovar copenhageni. Infect Immun 73: 7014–7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viriyakosol S, Matthias MA, Swancutt MA, Kirkland TN, Vinetz JM, 2006. Toll-like receptor 4 protects against lethal Leptospira interrogans serovar Icterohaemorrhagiae infection and contributes to in vivo control of leptospiral burden. Infect Immun 74: 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perolat P, Chappel RJ, Adler B, Baranton G, Bulach DM, Billinghurst ML, Letocart M, Merien F, Serrano MS, 1998. Leptospira fainei sp. nov., isolated from pigs in Australia. Int J Syst Bacteriol 48: 851–858. [DOI] [PubMed] [Google Scholar]
- 20.Arzouni JP, Parola P, La Scola B, Postic D, Brouqui P, Raoult D, 2002. Human infection caused by Leptospira fainei. Emerg Infect Dis 8: 865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asoh T, Saito M, Villanueva SY, Kanemaru T, Gloriani N, Yoshida S, 2014. Natural defense by saliva and mucosa against oral infection by Leptospira. Can J Microbiol 60: 383–389. [DOI] [PubMed] [Google Scholar]
- 22.Levett PN, 2003. Leptospira and Leptonema. Murray PR, ed. Manual of Clinical Microbiology. Washington, DC: ASM Press, 929–936. [Google Scholar]
- 23.American Veterinary Medical Association , 2013. AVMA Guidelines for the Euthanasia of Animals. Schaumberg, IL: AVMA. [Google Scholar]
- 24.Thibeaux R, Iraola G, Ferres I, Bierque E, Girault D, Soupe-Gilbert ME, Picardeau M, Goarant C, 2018. Deciphering the unexplored Leptospira diversity from soils uncovers genomic evolution to virulence. Microb Genom 4: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pappas CJ, Benaroudj N, Picardeau M, 2015. A replicative plasmid vector allows efficient complementation of pathogenic Leptospira strains. Appl Environ Microbiol 81: 3176–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lourdault K, Cerqueira GM, Wunder EA, Jr., Picardeau M, 2011. Inactivation of clpB in the pathogen Leptospira interrogans reduces virulence and resistance to stress conditions. Infect Immun 79: 3711–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Croda J, Figueira CP, Wunder EA, Jr., Santos CS, Reis MG, Ko AI, Picardeau M, 2008. Targeted mutagenesis in pathogenic Leptospira species: disruption of the LigB gene does not affect virulence in animal models of leptospirosis. Infect Immun 76: 5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Standley DM, 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]