Abstract.
Questions remain as to whether an unnoticed Ebola outbreak occurred in Guinea before the 2014–2016 epidemic. To address this, we used a highly sensitive and specific Luminex-based assay for Ebola virus (EBOV) antibody detection to screen blood samples collected in the framework of the Demographic Health Survey performed in 2012 in Guinea. One sample (GF069) of 1,483 tested was positive at very high immunoglobulin G titer to Zaire EBOV in Guinée Forestière. Thus, at least 2 years before the 2014 EVD outbreak in Guinea, Zaire EBOV was circulating in rural areas of this country.
Between 1976 and 2017, a total of 26 Ebola virus (EBOV) outbreaks have been reported in humans in Africa with a fatality rate between 25% and 90%.1,2 Twenty-four of the 26 EBOV disease outbreaks occurred in Central Africa, whereas two occurred in West Africa, in Côte d’Ivoire in 1994 and in Guinea, in 2014. Since 1994, the frequency of outbreaks has increased.1,3 Modeling studies suggest that the regions most at risk of a future Ebola outbreak encompass 22 countries across Central and West Africa including Guinea.1
In the present study, we aimed at investigating on the circulation of EBOV in human population in Guinea before 2014. To that end, we used a recently developed serological screening tool based on the Luminex® technology that detects simultaneously multiple targets with sensitivity and specificity equal or superior to conventional enzyme-linked immuno sorbent assay (ELISA).4 With this method, we screened blood samples collected in the framework of the 2012 Demographic and Health Survey (DHS) conducted in Guinea jointly by the National Institute of Statistics of Guinea and by MEASURE DHS, ICF International Calverton, Maryland.
Samples were collected between June and October 20125 as dried blood spots (DBS) on Whatman 903 filter paper. Blood samples were drawn from approximately 9,000 individuals aged between 15 and 59 years throughout the country. In this pilot screening, we included a subset of these samples collected from participants aged 18 years or older and originating from all the administrative regions of Guinea (Figure 1) to test for antibodies to Zaire EBOV. To reconstitute plasma from DBS, we proceeded as previously described.4 Reconstituted plasma samples from DBS were heat-inactivated at 56°C for 30 minutes in BSL-3 and further incubated overnight at 37°C with continuous shaking for a complete release of immunoglobulins.
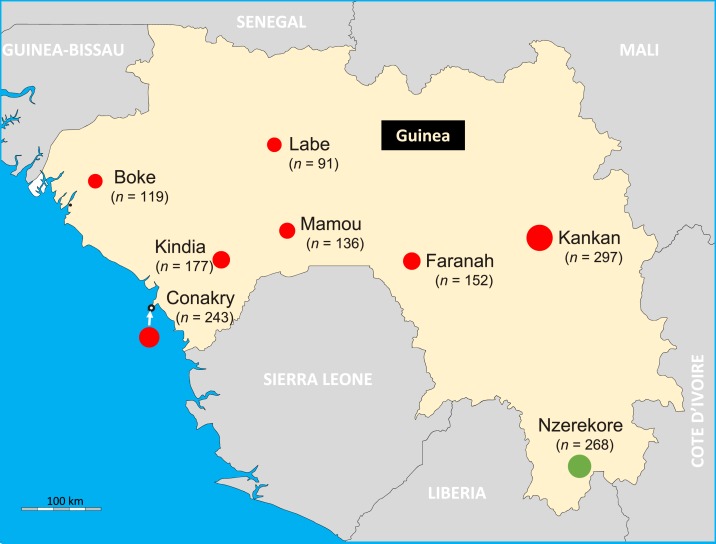
Figure 1.
Sample collection sites in Guinea. The figure shows the geographical map of Guinea and its surrounding countries. The circles indicate sites where the samples have been collected and are proportional to the number of samples tested for each site. The circle in green indicated the site where the Ebola virus antibody positive sample has been collected. This figure appears in color at www.ajtmh.org.
To detect antibodies to EBOV present in human samples, we used the Luminex® approach that we have previously validated.4 To that end, we coupled nucleoprotein (NP), glycoproteins (GP) (strains Mayinga and Kissidougou-Makona), and viral protein-40 (VP40) to Luminex microsphere beads. The results were expressed as median fluorescence intensity for 100 beads. The cutoff values were determined with receiver operating characteristic curves analysis from a panel of EBOV negative and positive plasma samples consisting of European donors who have not traveled in areas where EBOV outbreak has been reported and of survivors of the 2014 West African EBOV outbreak in Guinea. To determine the positivity of a sample, we defined an algorithm in our previous work where we used sera from survivors of the Guinean Ebola outbreak and thus of truly infected persons.4 We defined an algorithm specifying that a sample was considered positive for immunoglobulin G (IgG) to EBOV if it reacted simultaneously and repeatedly with NP and GP proteins as was also observed in a recent report from survivors of the first Ebola outbreak in DRC.9
Overall, we screened 1,483 samples from the 2012 DHS in Guinea. The 1,483 samples were homogenously distributed throughout the country, from the Savannah to the forested areas of Nzérékoré in the borders of Liberia (Figure 1). Of the 1,483 samples, 838 (56.5%) originated from rural areas of Guinea, whereas 645 (43.5%) from urban cities.
By serology, 154/1483 samples (10.4%) reacted with at least one EBOV antigen, ranging by collection site from 4.1% in Conakry to 19.4% in Guinée Forestière (Table 1). Taken individually by antigen, 9.3% of samples presented IgG antibodies directed against EBOV GP and 9.3% to VP40 proteins in Guinée Forestière. Samples collected from other areas of Guinea also presented IgG antibodies directed against EBOV GP and VP40 proteins, roughly between 1% and 5%. Nine samples (0.6%) reacted against EBOV NP.
Table 1.
Reactivity profile of a subset of the 2012 DHS samples on Zaire EBOV proteins stratified by regions of Guinea
| Antigens | Basse Guinée, n reactive (%) | Conakry, n reactive (%) | Guinée Forestière, n reactive (%) | Haute Guinée, n reactive (%) | Moyenne Guinée, n reactive (%) | Total (%) |
|---|---|---|---|---|---|---|
| N = 253 | N = 243 | N = 268 | N = 449 | N = 270 | N = 1,483 | |
| NP | 2 (0.79) | 1 (0.41) | 3 (1.12) | 3 (0.67) | 0 (0.0) | 9 (0.61) |
| GPk* | 10 (3.95) | 3 (1.23) | 10 (3.73) | 15 (3.34) | 11 (4.07) | 49 (3.30) |
| GPm† | 15 (5.93) | 5 (2.06) | 25 (9.33) | 26 (5.79) | 10 (3.70) | 81 (5.5) |
| VP40 | 9 (3.56) | 3 (1.23) | 25 (9.33) | 22 (4.90) | 3 (1.11) | 62 (4.2) |
| NP+GP | 0 (0.0) | 0 (0.0) | 1 (0.37) | 0 (0.0) | 0 (0.0) | 1 (0.07) |
| GP+VP40 | 1 (0.40) | 0 (0.0) | 1 (0.37) | 4 (0.89) | 1 (0.37) | 7 (0.47) |
| NP+GP+VP40 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| At least one antigen | 27 (10.7) | 10 (4.1) | 52 (19.4) | 50 (11.1) | 15 (5.6) | 154 (10.4) |
DHS = Demographic and Health Survey; EBOV = Ebola virus; GP = glycoproteins; NP = nucleoprotein; VP40 = viral protein-40.
Glycoprotein from Kissidougou-Makona strain of EBOV.
Glycoprotein from Mayinga strain of EBOV.
Applying our algorithm, based on survivor’s sera, to the present work, we observed that, of the 1,483 samples screened, one (GF069) fulfilled the criteria of positivity. The sample GF069 was collected from an adult male, living in a rural area of Guinée Forestière, in the Prefecture of Nzérékoré (Figure 1). The sample GF069 tested five times EBOV NP+GP positive (January to December 2017) by two different laboratory technicians on four different batches of protein-coupled Luminex® beads. This sample remained positive when diluted down to 1/6,000 (Table 2). Moreover, EBOV positive status of the sample GF069 was confirmed by a commercial ELISA assay from Alpha Diagnostics used in the report by Rimoin et al.9
Table 2.
Reactivity of the sample GF069 at different dilutions on EBOV antigens
| Plasma dilution | Mean MFI/100 beads (SD)* | ||||
|---|---|---|---|---|---|
| NP | GPk† | GPm‡ | VP40 | ||
| Nonreactive (N = 217) | 1,000 | 57 (76) | 49 (51) | 55 (69) | 124 (148) |
| GF069 (N = 5) | 1,000 | 2,331 (341) | 3,941 (820) | 4,831 (583) | 112 (51) |
| GF069 (N = 2) | 2,000 | 1,531 (130) | 2,572 (301) | 2,795 (80) | 55 (4) |
| GF069 (N = 2) | 4,000 | 787 (22) | 1,213 (13) | 1,381 (31) | 26 (0) |
| GF069 (N = 2) | 6,000 | 714 (46) | 1,078 (157) | 1,389 (179) | 22 (2) |
| GF069 (N = 2) | 8,000 | 482 (17) | 703 (1) | 928 (75) | 15 (1) |
| GF069 (N = 2) | 10,000 | 429 (109) | 639 (66) | 826 (24) | 13 (3) |
EBOV = Ebola virus; GP = glycoproteins; MFI = median fluorescence intensity; NP = nucleoprotein; VP40 = viral protein-40.
Cutoff values (MFI): NP = 600; GP = 400 and VP40 = 650.
Glycoprotein from Kissidougou-Makona strain of EBOV.
Glycoprotein from Mayinga strain of EBOV.
Our objective in the present study was to investigate on unnoticed circulation of EBOV in the Guinean population before the largest 2014 outbreak ever seen. Our work on a subset of samples from the 2012 DHS showed evidence of IgG antibodies to EBOV in an adult male villager from Guinée Forestière. Our findings indicate that at least 2 years before the onset of the largest EBOV outbreak ever seen, Zaire EBOV was circulating in rural areas of Guinea. To reach this conclusion, we used a stringent definition of positivity validated on a large panel of EBOV disease survivors’ samples. The 10.4% of samples that reacted with at least one antigen (Table 1) could be the result of cross-reactions with distantly related filoviruses, a differential kinetics of antibodies to these different proteins or false positive reactions. From these serology data, it is not possible to date the EBOV infection in the individual it was detected, but a few studies reported prevalence and kinetics of antibodies to EBOV in some survivors and pauci-symptomatic individuals.6 In a follow-up study of survivors from the 1999–2000 Sudan EBOV outbreak, Sobarzo et al.7 identified IgG antibodies to NP, VP40, VP30, and to whole virus antigens in 120 survivors on samples collected 6 months to 10 years after viral infection. Another study, conducted in Gabon,8 showed also the maintenance of antibodies in survivors up to 9–12 years after infection. A recent study conducted in the DRC showed the presence of antibodies to NP and GP 40 years after infection on patients who survived the first Ebola outbreak in Yambuku, in 1976.9 Interesting enough, this study used recombinant NP, GP, and VP40 proteins ELISA to detect antibodies to Zaire EBOV in the 14 patients included. Of these, six were confirmed EVD cases at the time of the outbreak, and all the six were simultaneously NP and GP positive, thus independently validating our algorithm.
In Guinea, beside the 2014/2016 EBOV outbreak, there is no other officially recognized Ebola outbreak reported. However, in a seroepidemiological survey performed in 1982/1983 in Madina-Oula district (area of Kindia, basse Guinée administrative region), a study by Boiro et al.10 reported 8% seroprevalence of antibodies to EBOV determined by ELISA using whole virus antigens. The survey was conducted in the framework of a disease outbreak characterized by symptoms of a hemorrhagic fever with an overall case fatality rate of 38%. This 1982/1983 outbreak was not confirmed by virological methods and is not listed among the 26 EBOV disease outbreaks officially reported thus far. To the best of our knowledge, the report by Boiro et al. is the unique pre-2014/2016 hemorrhagic fever outbreak with a suspicion of Ebola mentioned in Guinea.
Whether IgG antibodies from GF069 is a result of a zoonotic infection from wildlife or from a human-to-human transmission could not be determined from our study. We can also not exclude the possibility that the person from whom the sample was collected was a returnee from areas or countries where an Ebola outbreak has occurred and where he has been infected and survived. Nevertheless, we showed here the presence of high titers IgG antibodies to GP and NP proteins of Zaire EBOV in a sample from a villager of a forest zone of Guinea, at least 2 years before the 2014 outbreak.
References
- 1.Pigott DM, Golding N, 2014. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife 3: e04395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pigott DM, et al. 2016. Updates to the zoonotic niche map of Ebola virus disease in Africa. ELife 5: pii: e16412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baize S, et al. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371: 1418–1425. [DOI] [PubMed] [Google Scholar]
- 4.Ayouba A, Toure A, Butel C, Keita AK, Binetruy F, Sow MS, Foulongne V, Delaporte E, Peeters M, 2017. Development of a sensitive and specific serological assay based on luminex technology for detection of antibodies to Zaire Ebola virus. J Clin Microbiol 55: 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DHS Program , 2012. Guinea DHS 2012—Final Report Available at: https://dhsprogram.com/publications/publication-FR280-DHS-Final-Reports.cfm. Accessed February 2, 2018.
- 6.Glynn JR, et al. 2017. Asymptomatic infection and unrecognised Ebola virus disease in Ebola-affected households in Sierra Leone: a cross-sectional study using a new non-invasive assay for antibodies to Ebola virus. Lancet Infect Dis 17: 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobarzo A, et al. 2013. Persistent immune responses after Ebola virus infection. N Engl J Med 369: 492–493. [DOI] [PubMed] [Google Scholar]
- 8.Becquart P, et al. 2010. High prevalence of both humoral and cellular immunity to Zaire Ebola virus among rural populations in Gabon. PLoS One 5: e9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rimoin AW, et al. 2018. Ebola virus neutralizing antibodies detectable in survivors of the Yambuku, Zaire outbreak 40 years after infection. J Infect Dis 217: 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boiro I, Lomonossov NN, Sotsinski VA, Constantinov OK, Tkachenko EA, Inapogui AP, Balde C, 1987. Clinico-epidemiologic and laboratory research on hemorrhagic fevers in Guinea. Bull Soc Pathol Exot 80: 607–612. [PubMed] [Google Scholar]