Abstract.
Estero Real virus (ERV) was isolated in 1980 from Ornithodoros tadaridae ticks collected in El Estero Real, Sancti Spiritus, Cuba. Antigenic characterization of the isolate based on serological methods found a relationship with Abras and Zegla viruses and, consequently, the virus was classified taxonomically within the Patois serogroup. Given the fact that genetic characterization of Patois serogroup viruses has not yet been reported and that ERV is the only virus within the Patois serogroup isolated from ticks, we recently conducted nearly complete genome sequencing in an attempt to gain further insight into the genetic relationship of ERV with other Patois serogroup viruses and members of Peribunyaviridae family (Bunyavirales order). With the exception of ERV, our sequencing and phylogenetic studies revealed the close relationship of the Patois serogroup viruses to each other, forming a clear divergent clade from other members of the Orthobunyavirus genus (Peribunyaviridae family). Notably, our analysis also revealed that ERV forms a monophyletic clade that is closely related to species of the Orthonairovirus genus (Nairoviridae family) in all the genome segments. In light of these findings, we believe that the taxonomic classification of ERV should be revised.
INTRODUCTION
The Bunyavirales order contains more than 300 viruses that have been reclassified by the International Committee on Taxonomy of Viruses (ICTV) in nine families, which includes Hantaviridae, Feraviridae, Fimoviridae, Jonviridae, Nairoviridae, Peribunyaviridae, Phasmaviridae, Phenuiviridae, and Tospoviridae.1 Based on this new classification, four of the nine virus families, including Hantaviridae, Nairoviridae, Peribunyaviridae, and Phenuiviridae, contain viruses that are pathogenic in humans. The family Peribunyaviridae is the largest with more than 170 named viruses subdivided into 18 serogroups on the basis of serological reactivity.2 Among them, the Patois serogroup is one of the least studied group of viruses despite their frequent isolation during ecological investigations. Viruses in this serogroup have been isolated from mosquitoes in South, Central, and North America, and rodents have been incriminated as the main reservoir host.3,4 Currently, at least seven viruses have been grouped within the Patois serogroup: Pahayokee virus (PAHV), Shark River virus (SRV), Patois virus (PATV), Zegla virus (ZEGV), Abras virus (ABRV), Babahoyo virus (BABV), and Estero Real virus (ERV). The PATV and ZEGV were originally isolated from mosquitoes collected in Panama in 1961,5 and initial antigenic characterization of these viruses identified them as members of the Group C viruses; however, subsequent studies reclassified them as members of the Patois serogroup.6 Both PATV and ZEGV were frequently isolated along the tropical Gulf coast of southeastern Mexico as part of ecological investigations conducted during 1963–1968.7,8 These studies also revealed the presence of hemagglutination inhibition and neutralizing antibodies to PATV in sera of people residing at the study site and in terrestrial wild mammals, including cotton rats, opossums, and raccoons.8 The PAHV and SRV were isolated in 1963 and 1964, respectively, from Culex (Melanoconion) species captured in the Everglades National Park in Florida6; however, SRV-like viruses have also been isolated from sentinel hamsters in Mexico and Guatemala providing evidence about their vast geographic distribution.8 ABRV and BABV were isolated from Culex spp. mosquitoes and from the blood of sentinel hamsters collected in the coastal areas of Ecuador and found to be closely related by complement fixation but distinct to other described members of the Patois serogroup.9 Last, ERV was isolated from argasid Ornithodoros tadaridae ticks collected from leaves of a palm tree colonized by bats in El Estero Real in Cuba in April 1980 and, to this date, it constitutes the only Patois serogroup virus isolated from ticks.10 Because of the unusual association of a Patois serogroup virus with ticks and the limited information available about the genetic relationship of this group of viruses with other members of the Bunyavirales order, we initiated a coordinated effort to determine the complete genome nucleotide (nt) sequences of six of the Patois serogroup viruses described to date. We report here our phylogenetic analyses which confirm that the Patois serogroup viruses formed a divergent clade from other members of the Orthobunyavirus genus (Peribunyaviridae family) and, more importantly, we provide evidence that ERV is an orthonairovirus.
MATERIALS AND METHODS
Virus strains and RNA extraction.
Prototype Patois serogroup viruses were obtained from the World Reference Center for Arboviruses and Emerging Viruses and propagated in African green monkey kidney (Vero) cells and maintained at 37°C. The cell cultures were examined daily for evidence of cytopathic effect (CPE). On the appearance of CPE or 10 days after virus inoculation, cell culture supernatants were harvested and clarified by centrifugation, and viral RNA was extracted as previously described11 using Trizol reagent (Invitrogen, Carlsbad, CA).
Next generation sequencing.
Following RNA extraction, viral RNA was fragmented and a sequence library was prepared and sequenced on a HiSeq 1000 using the 2 × 50 paired-end protocol. Reads were quality-filtered and assembled using the de novo assembly program ABySS.12 Host reads were filtered out before the de novo assembly. The longest contigs were selected and reads were mapped back to the contigs using bowtie213 and then visualized with the Integrated Genomics Viewer14 to verify that the assembled contigs were correct. Total reads ranged from 1.5 to 12 million and the percentage of reads mapping to the virus genome in each sample ranged from 12% to 33%. Additional details are available on request. The complete genome sequences of Patois segroup viruses obtained in this study were deposited in the GenBank. Accession numbers are listed in Table 1.
Table 1.
Viruses included in this study
| Virus name | Abbreviation | Strain | Collection date | Source of isolate | Location | Accession numbers |
|---|---|---|---|---|---|---|
| Abras virus | ABRV | 75V1183 | 8/21/1974 | Culex (Mel) paracrybda. | Naranjal, Ecuador | MH017269, MH017275, MH017287 |
| Babahoyo virus | BABV | 75V2858 | 2/28/1975 | Culex (Mel) ocossa | Abras, Ecuador | MH017270, MH017276, MH017282 |
| Patois virus | PATV | 63A49 | 1963 | Culex spp | Veracruz, Mexico | MH017273, MH017277, MH017283 |
| Shark River virus | SRV | 64U80 | 1964 | sentinel hamster | Mexico | MH017271, MH017278, MH017284 |
| Zegla virus | ZEGV | BT5012 | 6/23/1961 | Sigmodon hispidus | Almirante, Panama | MH017272, MH017279, MH017285 |
| Estero Real | ERV | K329 | 04/26/1980 | Ornithodoros tadaridae | El Estero real, Sancti Spiritus, Cuba | MH017274, MH017280, MH017286 |
Genome characterization.
Virus genomes were evaluated regarding size, open reading frames (ORF) descriptions, 5′ and 3′ noncoding regions, and conserved motifs with Geneious 9.1.2 (Biomatters, Auckland, New Zealand). We submitted data to the TOPCONS webserver for identification of transmembrane regions and signal peptide, and we submitted data to the NetNglyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) for identification of glycosylation sites. The annotations of protein domains in the M segment were performed with InterProScan in Geneious 9.1.2 (Biomatters) and Conserved Domain Database.15
Phylogenetic analysis.
Maximum likelihood (ML) phylogenetic trees were reconstructed using alignments of complete coding sequences of all RNA segments of the Patois serogroup viruses and representative orthobunyaviruses (S, M, and L) available in the GenBank database (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignments were carried out using the combinations of PROMALS3D and Gblocks method.16,17 The phylogenies were inferred using IQ-TREE version 1.4.3 software under the best-fit model based on Bayesian Information Criterion determined by Modelfinder with 1,000 replicates.18,19 In all cases, statistical supports for individual nodes were estimated using the bootstrap value. The phylogenetic trees were visualized using the FigTree software v.1.4.2.
Reassortment event analysis.
To identify potential reassortment events, the data were analyzed for evidence of distinct phylogenetic topologies based on the depicted trees at the nt level, as described previously. All genes in a single sequence were concatenated and a multiple alignment was performed using the program Muscle 3.7 as described previously.20 Potential reassortment events were then analyzed using the RDP, GENECONV, Bootscan, MaxChi, Chimera, SiScan, and 3Seq methods implemented in RDP4.21 Common program settings for all methods were used to perceive sequences as linear, to require phylogenetic evidence, to refine breakpoints, and to check alignment consistency. The highest acceptable P value was set at 0.05, after considering Bonferroni correction for multiple comparisons. All method-specific program settings remained at their default values.
RESULTS
Genome organization of Patois serogroup viruses.
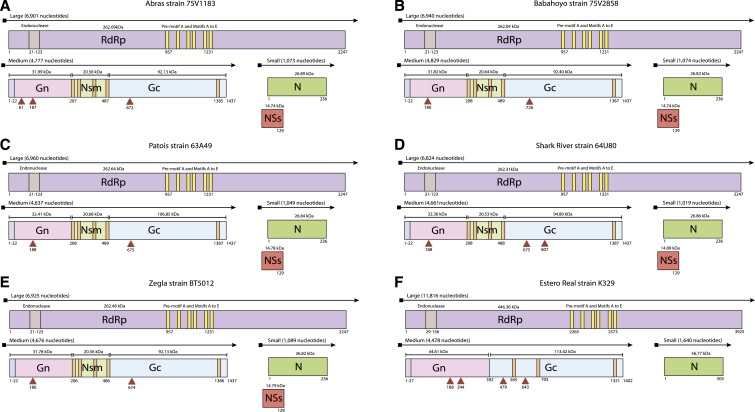
We obtained nearly complete sequences for the S, M, and L genome segments of viruses designated as members of Patois serogroup (Table 1), except for PAHV. Other than ERV, the S segment of all other Patois serogroup isolates ranged from 1,019 to 1,089 nt and presented two ORF encoding the nucleocapsid (N) protein and the nonstructural NSs protein. The N protein is 236 amino acids with a predicted mass of 26.9 kDa, whereas the ORF of NSs gene starts at an ATG codon 35 nt upstream from the N ORF start codon, and encodes a protein of 129 amino acids (∼14.8 kDa) (Figure 1A–E). Surprisingly, we found that the S segment of ERV is 1,640 nt in length and encodes for a unique protein of 505 amino acids (56.7 kDa protein; Figure 1F). Blast search analyses revealed a close association of this protein to the N protein of members of the genus Orthonairovirus but no association with the N protein of other Patois serogroup viruses.
Figure 1.
Genome organization of Patois serogroup. (A) Abras virus, (B) Babahoyo virus, (C) Patois virus, (D) Shark River virus, (E) Zegla virus, and (F) Estero Real virus. This figure appears in color at www.ajtmh.org.
With regard to the M segment sequence, we found that the M segment of the Patois serogroup viruses ranged from 4,637 to 4,829 nt, encoded the viral glycopolyprotein precursor (GPC) of 1,433 to 1,437 amino acids in length, and contained the characteristic N-terminal signal peptide that is common to bunyaviruses. The M polyprotein is cleaved into two different structural proteins, glycoprotein n-terminus (Gn) and glycoprotein c-terminus (Gc), and a nonstructural protein (nonstructural protein m; NSm). The glycoproteins are predicted to contain transmembrane regions (TMDs): a single TMD in Gn (close to the C-terminus), three TMD domains in NSm, and a single domain close to the C-terminus of Gc (Figure 1). The asparagine sites predicted to be N-glycosylated in the GPC are also observed (Figure 1).
By contrast, we found that the M segment of ERV has 4,478 nt and encodes a GPC of 1,402 amino acids in length (molecular weight = 158 kDa) that also contains an N-terminal signal peptide, three TMD, and four sites of N-glycosylation. A RKLL cleavage motif for SKI-I/S1P protease, which is conserved among GPCs of orthonairoviruses,22 was identified between amino acid positions 389 to 392 (Figure 1F). In addition, we identified the Zinc finger region, which is important to the viral RNA binding site and probably is involved in viral assembly.23 Therefore, the GPC shows a topology similar to members of the Hughes serogroup (Hughes orthonairovirus species group).22
Analyses of the L segment sequences revealed that viruses in the Patois serogroup possess an L segment ranging from 6,824 to 6,960 nt in length that codes for an RNA dependent RNA polymerase (RdRp) of 2,247 amino acids, with a predicted molecular weight of ∼262 kDa. Surprisingly, the L segment of ERV is 11,816 nt in length and encodes an RdRp of 3,925 aa, with a predicted molecular weight of ∼446.36 kDa. However, all the proteins contain conserved polymerase activity domains consisting of pre-motif A and motifs A through E, which are highly conserved in negative sense RNA viral polymerases and common to all other Bunyavirales characterized so far24 (Figure 1). In addition, the ERV RdRp contains a characteristic catalytic ovarian tumor-like cysteine protease motif that is common to all orthonairoviruses.22
Phylogenetic relationship of Patois serogroup viruses with other orthobunyaviruses.
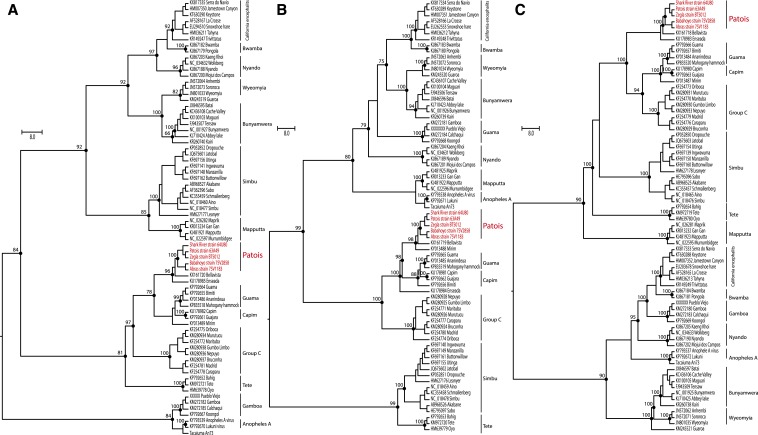
The phylogenetic trees that were generated based on amino acid sequences of 72 orthobunyaviruses produced 14 monophyletic groups, but with different topologies in each segment. Thirteen of the well-supported clades corresponded to already established serogroups: Bunyamwera, Wyeomyia, California encephalitis, Bwamba, Nyando, Simbu, Mapputta, Gamboa, Koongol, Tete, Group C, Guama, and Capim (Figure 2). Furthermore, we assigned a divergent clade as a proposed new “group,” containing the six viruses of the Patois serogroup (Figure 2). The phylogenetic analysis based on all segment sequences reveals that the members of the Patois serogroup form a monophyletic clade with Bellavista virus. This observation suggests that these viruses might have the same evolutionary origin. In addition, the Patois serogroup viruses were also closely related to Guama, Capim, and Group C groups.
Figure 2.
Maximum likelihood phylogenetic trees of Patois serogroup within Orthobunyavirus genus. The nucleoprotein (A), glycoprotein (B), and RdRP (C), all based on LG + I + G4 model amino acids substitution model. Phylogenies are midpoint rooted and with proportional branches for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per amino acid substitutions/site, and the principal bootstrap support levels were indicated. The Patois virus strains sequenced in this study are highlighted in light grey and in red (online). This figure appears in color at www.ajtmh.org.
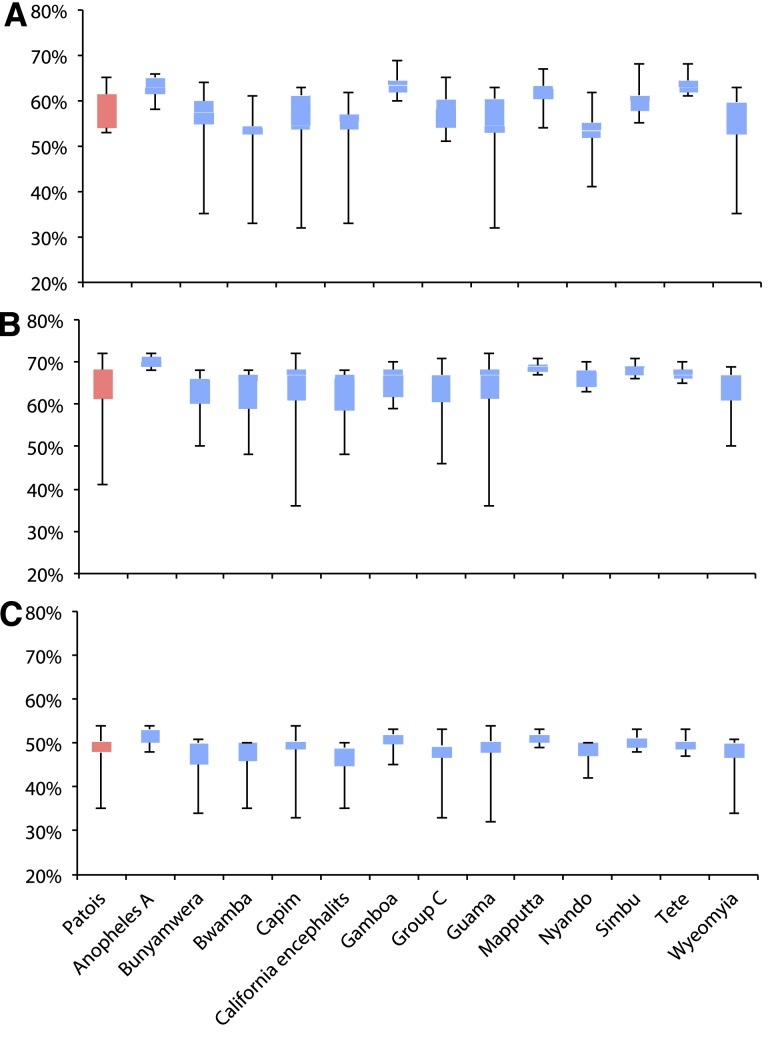
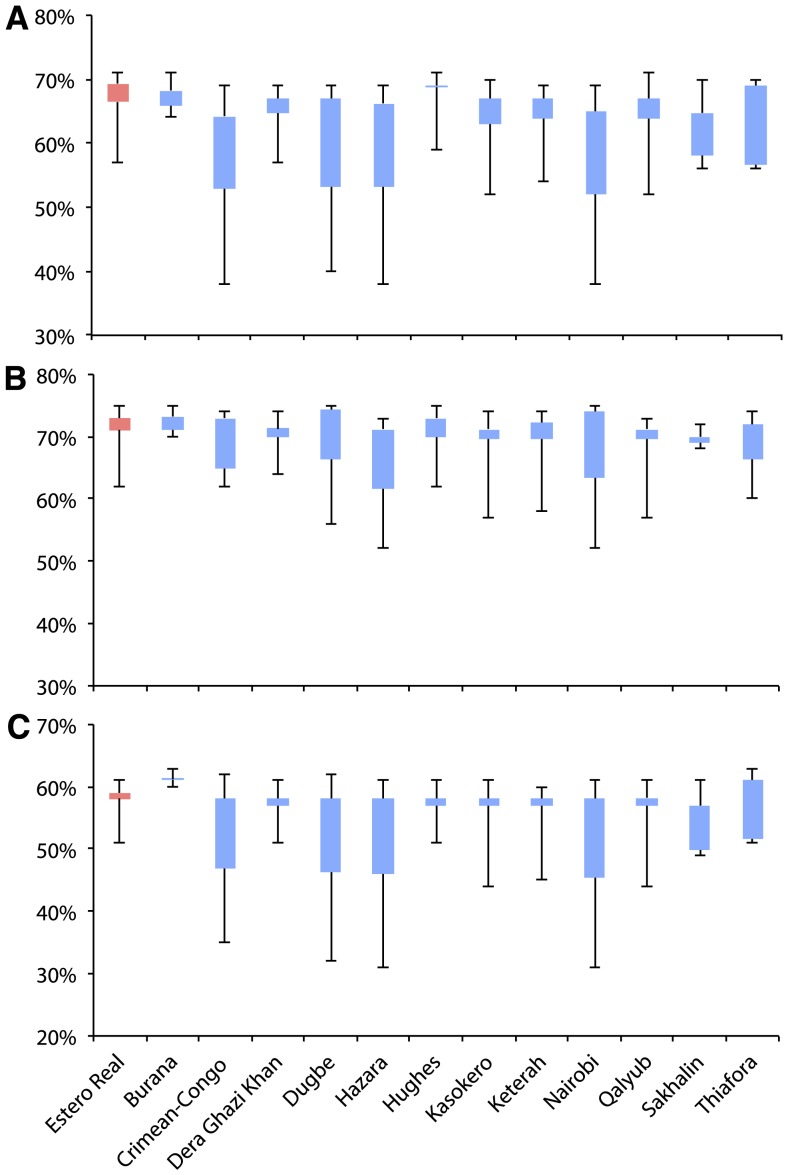
We next conducted pairwise amino acid sequence distance analysis based on the N, GPC, and RdRp sequences of Patois serogroup and representative orthobunyaviruses, and results of an inter-clade analysis are shown in Figure 3. The genetic distances of the Patois serogroup viruses to other orthobunyaviruses was estimated to be ∼57%, ∼60%, and ∼48% for the N, GPC, and RdRp, respectively. Lower levels of genetic diversity were observed among the members of Patois serogroup viruses when compared with those observed with other orthobunyavirus clades. This fact was observed in the p-distance intra-clade analysis, based on all three genes (N, GPC, and RdRp), in both nt and amino acid identity comparisons (Table 2).
Figure 3.
Pairwise genetic similarities (amino acid p-distance) of Patois serogroup compared with other orthobunyavirus groups based on the coding sequencing of N protein (A), glycopolyprotein (B), and RdRp (C). This figure appears in color at www.ajtmh.org.
Table 2.
Nucleotide and amino acid identity comparisons for members of Patois serogroup
| PATV | PATV-like | ABRV | BABV | SRV | ZEGV | ||
|---|---|---|---|---|---|---|---|
| PATV | S | – | 94.49 | 76.59 | 76.59 | 95.76 | 98.31 |
| M | – | 81.75 | 70.49 | 70.65 | 89.42 | 80.71 | |
| L | – | 82.65 | 76.54 | 76.17 | 82.53 | 82.40 | |
| ABRV | S | 75.81 | 77.36 | – | 98.58 | 77.97 | 77.12 |
| M | 68.42 | 69.35 | – | 84.02 | 70.28 | 70.29 | |
| L | 76.54 | 76.57 | – | 92.41 | 76.96 | 76.50 | |
| BABV | S | 75.95 | 77.22 | 99.02 | – | 77.97 | 77.12 |
| M | 69.19 | 69.40 | 74.38 | – | 70.93 | 70.63 | |
| L | 76.17 | 74.73 | 92.41 | – | 77.28 | 76.57 | |
| SRV | S | 89.87 | 86.22 | 76.93 | 76.23 | – | 94.92 |
| M | 80.30 | 74.82 | 69.44 | 70.12 | – | 79.39 | |
| L | 82.53 | 83.66 | 76.96 | 77.28 | – | 83.36 | |
| ZEGV | S | 97.05 | 89.59 | 77.07 | 76.93 | 88.89 | – |
| M | 74.59 | 80.11 | 69.01 | 69.08 | 74.13 | – | |
| L | 82.40 | 94.37 | 76.50 | 76.57 | 83.36 | – |
ABRV = Abras virus; BABV = Babahoyo virus; PATV = Patois virus; SRV = Shark River virus; ZEGV = Zegla virus.
Phylogenetic relationship of ERV with other orthonairoviruses.
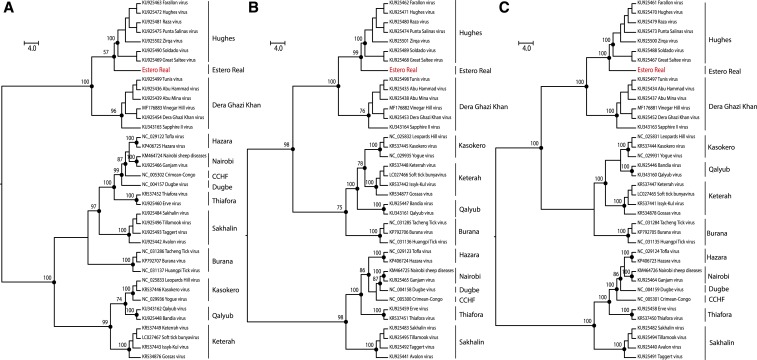
Because blast search analysis revealed the close association of ERV to other orthonairoviruses, we conducted phylogenetic analyses based on amino acid sequences of ERV and 37 complete coding sequences of orthonairoviruses. The ML trees showed 13 well-supported monophyletic groups, but with different topologies in each segment. Twelve of the well-supported clades corresponded to already established orthonairoviruses species.22 The phylogenetic analysis reveals that ERV forms a monophyletic clade and is genetically more closely related to the species of Hughes serogroup (H. orthonairovirus species group) in all segments (Figure 4). In addition, the p-distance intra-clade analysis, based on the N, GPC, and RdRP sequences of ERV and representative orthonairoviruses, indicates that ERV is a distinct species within the genus (Figure 5). The findings further confirm the close association of ERV with orthonairovirus but not with the Patois serogroup viruses.
Figure 4.
Maximum likelihood phylogenetic trees of ERV within Nairoviridae family. The nucleoprotein (A) and glycoprotein (B) based on LG + I + G4 model amino acids substitution model and RdRP (C) based on LG + F + I + G4 model amino acids substitution model. Phylogenies are midpoint rooted and with proportional branches for clarity of presentation. The scale bar indicates evolutionary distance in numbers of substitutions per amino acids substitutions/site, and the principal bootstrap support levels were indicated. Estero Real virus sequenced in this study is highlighted in light grey and in red (online). This figure appears in color at www.ajtmh.org.
Figure 5.
Pairwise genetic similarities (amino acid p-distance) of ERV compared with other orthonairovirus groups based on the coding sequencing of N protein (A), glycopolyprotein (B), and RdRp (C). This figure appears in color at www.ajtmh.org.
Reassortment analyses.
To identify the potential reassortment events among viruses of the Patois serogroup, we inspected the nt ML phylogenies for discordances in clade clustering between the S, M, and L trees (Figure 2), together with Sliding-window RDP4 analyses using concatenated full genomes of 72 orthobunyaviruses available in GenBank. These analyses indicate that the ZEGV BT5012 strain is a potential reassortant that contains the S and L segment from PATV, and an M segment of unknown origin (Supplemental Figure 1). Based on tree topologies and additional analysis to identify potential reassortment events using the RDP4 program,25 we did not observe evidence of reassortment with ERV.
DISCUSSION
In this report, we present the complete coding sequences and genomic characteristics of six of the seven members of the Patois serogroup within the family Peribunyaviridae. All strains exhibit similar genomic characteristics as other orthobunyaviruses. We found that the RdRp contains the endonuclease and main motifs that are conserved for the polymerase activity (pre-motif A and motifs A through E), and are highly conserved in negative sense RNA viral polymerases.24 Our analysis also revealed that, with the exception of ERV, all members of the Patois serogroup viruses sequenced in this study contain putative ORFs for the NSs protein, which has a start codon before the pre-N start codon, as previously described for Enseada and Brazoran viruses.26,27 The NSs protein has been detected in most orthobunyaviruses that infect vertebrates and is known to be an important virulence factor.28–31 Because infection with Patois serogroup viruses has not yet been associated with human illness, it would be interesting to determine whether the NSs of these viruses have similar virulence properties as those described for other orthobunyaviruses.
Our phylogenetic analysis revealed that, with the exception of ERV, the Patois serogroup viruses are most closely genetically related to Bellavista virus, which has not been associated with human illness. Although the Patois serogroup viruses have not been confirmed to cause human disease, they share a common ancestor with Capim, Guama, and Group C serogroup viruses. Both Guama and Group C viruses have been associated with human disease and are a significant cause of febrile illness in Latin America.
We also conducted pairwise amino acid sequence distance analysis of the Patois serogroup viruses and observed a divergence of ∼57%, ∼60%, and ∼48% for the N, GPC, and RdRp, respectively. When these viruses were compared with the amino acid distance from other serogroups, we noted that the degree of evolutionary divergence is equivalent to those observed with other orthobunyavirus serogroups. The Patois serogroup viruses exhibit a higher amino acid distance within the Orthobunyavirus genus for the N and GPC, whereas the distance rates for the RdRp were lower. This means that these viruses are under the same selective pressure, with a more conserved polymerase protein and more plastic N and GPC, as previously reported for other orthobunyaviruses.26
The natural reassortment process in segmented viruses is a form of genetic exchange that has the potential to provide host/vector range shifts and changes in pathogenicity and/or virulence, as well as an important mechanism for the emergence of new viral species. Reassortment events have been suggested as the driving force in the evolution of bunyaviruses because they have been detected among members of the Group C, Simbu, and Bunyamwera serogroups.32 Within the Patois serogroup, the first evidence of potential reassortment events was based on the results of serological assays and the observation that SRV and PAHV have almost identical L and S RNA oligonucleotide fingerprints, but different M RNA fingerprints.33 Here, based on the topology of the phylogenetic trees obtained from complete genome sequences of each segment, combined with a RDP4 analysis, we have confirmed that the ZEGV strain BT5012 is a natural reassortant of PATV. The S and L segments of this virus were provided by PATV, whereas the M segment is of unknown origin (Figure 2). This finding challenges the current ICTV classification which designates ZEGV as a distinct species (Zegla orthobunyaviruses)1 and suggests that the virus should be considered instead as a member of the Patois serogroup (Patois orthobunyavirus species group). Interestingly, ZEGV and PATV were isolated from the hispid cotton rat (Sigmodon hispidus) captured near Almirante city in Panama in June, 1961.5 The potential implications for this reassortment phenomenon in the Patois serogroup need further investigation.
The most important finding from this study is the observation that ERV is closely related genetically to members of the Orthonairovirus genus and not to the Patois serogroup viruses. The genome organization of ERV shares the same characteristics of viruses within the Orthonairovirus genus and phylogenetic analyses clearly corroborated these findings by placing ERV in a distinct clade but closely related to species of H. orthonairovirus in all segments analyzed, suggesting that its current placement with other members of the Patois serogroup viruses should be revised.
In summary, our study provides new insights into the genetic diversity and evolution of the Patois serogroup viruses described to date (with the exception of PAHV) and provides genetic evidence that the ERV taxonomic status does not belong with this group of viruses. It also highlights the need for future studies to determine the epidemiology, geographic distribution, and public health impact of ERV. Furthermore, the sequences generated in this study could be used to develop diagnostic tests and evaluate the potential public health impact of viruses included in the Patois serogroup.
Supplementary Material
Acknowledgments:
This study was supported by the National Institutes of Health (NIH) grants number F31 AI124662-01 (JAS) and R24 AI120942 (PVA), WMS was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo-FAPESP grants no. 12/24150-9 and 17/13981-0); MJF was supported by the FAPESP grant no. 16/01414-1; MRTN was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico grants number 302032/2011-8 and 200024/2015-9).
Note: Supplemental figure appears at www.ajtmh.org.
REFERENCES
- 1.Adams MJ, et al. 2017. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol 162: 2505–2538. [DOI] [PubMed] [Google Scholar]
- 2.Calisher CH, 1996. Chapter 1, Elliot RM, ed. The Bunyaviridae. New York, NY: Springer 1–17. [Google Scholar]
- 3.Scherer WF, Dickerman RW, Ordonez JV, Seymour C, Kramer LD, Jahrling PB, Powers CD, 1976. Ecologic studies of Venezuelan encephalitis virus and isolations of Nepuyo and Patois viruses during 1968–1973 at a marsh habitat near the epicenter of the 1969 outbreak in Guatemala. Am J Trop Med Hyg 25: 151–162. [DOI] [PubMed] [Google Scholar]
- 4.Scherer WF, Dickerman RW, Cupp EW, Ordonez JV, 1985. Ecologic observations of Venezuelan encephalitis virus in vertebrates and isolations of Nepuyo and Patois viruses from sentinel hamsters at Pacific and Atlantic habitats in Guatemala, 1968–1980. Am J Trop Med Hyg 34: 790–798. [DOI] [PubMed] [Google Scholar]
- 5.Srihongse S, Galindo P, Grayson MA, 1966. Isolation of group C arboviruses in Panama including two new members, Patois and Zegla. Am J Trop Med Hyg 15: 379–384. [DOI] [PubMed] [Google Scholar]
- 6.Fields BN, Henderson BE, Coleman PH, Work TH, 1969. Pahayokee and Shark River, two new arboviruses related to Patois and Zegla from the Florida everglades. Am J Epidemiol 89: 222–226. [DOI] [PubMed] [Google Scholar]
- 7.Zarate ML, Geiger RH, Shope RE, Scherer WF, 1968. Intergroup antigenic relationships among arboviruses manifested by a Mexican strain of Patois virus and viruses of the Bunyamwera, C, California, Capim and Guama groups. Am J Epidemiol 88: 273–286. [DOI] [PubMed] [Google Scholar]
- 8.Scherer WF, Anderson K, Dickerman RW, Ordonez JV, 1972. Studies of Patois group arboviruses in Mexico, Guatemala, Honduras, and British Honduras. Am J Trop Med Hyg 21: 194–200. [DOI] [PubMed] [Google Scholar]
- 9.Calisher CH, Gutierrez E, Francy DB, Alava A, Muth DJ, Lazuick JS, 1983. Identification of hitherto unrecognized arboviruses from Ecuador: members of serogroups B, C, Bunyamwera, Patois, and Minatitlan. Am J Trop Med Hyg 32: 877–885. [DOI] [PubMed] [Google Scholar]
- 10.Málková D, Holubová J, Cerný V, Daniel M, Fernández A, de la Cruz J, Herrera M, Calisher CH, 1985. Estero real virus: a new virus isolated from argasid ticks Ornithodoros tadaridae in Cuba. Acta Virol 29: 247–250. [PubMed] [Google Scholar]
- 11.Hontz RD, et al. 2015. Itaya virus, a novel orthobunyavirus associated with human febrile illness, Peru. Emerg Infect Dis 21: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I, 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langmead B, Salzberg SL, 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP, 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchler-Bauer A, et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res 43: D222–D226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei J, Grishin NV, 2014. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods Mol Biol 1079: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talavera G, Castresana J, 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56: 564–577. [DOI] [PubMed] [Google Scholar]
- 18.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS, 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ, 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RC, 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DPMB, Golden M, Khoosal A, Muhire B, 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn JH, et al. 2016. Genomic characterization of the genus Nairovirus (family Bunyaviridae). Viruses 8: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estrada DF, De Guzman RN, 2011. Structural characterization of the Crimean-Congo hemorrhagic fever virus Gn tail provides insight into virus assembly. J Biol Chem 286: 21678–21686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reguera J, Weber F, Cusack S, 2010. Bunyaviridae RNA polymerases (L-protein) have an N-terminal, influenza-like endonuclease domain, essential for viral cap-dependent transcription. PLoS Pathog 6: e1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza WM, Acrani GO, Romeiro MF, Reis O, Tolardo AL, da Silva SP, de Almeida Medeiros DB, Varela M, Nunes MR, Figueiredo LT, 2016. Molecular characterization of Capim and Enseada orthobunyaviruses. Infect Genet Evol 40: 47–53. [DOI] [PubMed] [Google Scholar]
- 27.Lanciotti RS, Kosoy OI, Bosco-Lauth AM, Pohl J, Stuchlik O, Reed M, Lambert AJ, 2013. Isolation of a novel orthobunyavirus (Brazoran virus) with a 1.7 kb S segment that encodes a unique nucleocapsid protein possessing two putative functional domains. Virology 444: 55–63. [DOI] [PubMed] [Google Scholar]
- 28.Elliott RM, Weber F, 2009. Bunyaviruses and the type I interferon system. Viruses 1: 1003–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Léonard VH, Kohl A, Hart TJ, Elliott RM, 2006. Interaction of Bunyamwera orthobunyavirus NSs protein with mediator protein MED8: a mechanism for inhibiting the interferon response. J Virol 80: 9667–9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Knippenberg I, Carlton-Smith C, Elliott RM, 2010. The N-terminus of Bunyamwera orthobunyavirus NSs protein is essential for interferon antagonism. J Gen Virol 91: 2002–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoen A, Weber F, 2015. Orthobunyaviruses and innate immunity induction: alieNSs vs. PredatoRRs. Eur J Cell Biol 94: 384–390. [DOI] [PubMed] [Google Scholar]
- 32.Briese T, Calisher CH, Higgs S, 2013. Viruses of the family Bunyaviridae: are all available isolates reassortants? Virology 446: 207–216. [DOI] [PubMed] [Google Scholar]
- 33.Ushijima H, Clerx-Van Haaster CM, Bishop DH, 1981. Analyses of Patois group bunyaviruses: evidence for naturally occurring recombinant bunyaviruses and existence of immune precipitable and nonprecipitable nonvirion proteins induced in bunyavirus-infected cells. Virology 110: 318–332. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.