Abstract
The aim of the present study was to identify the differentially expressed genes (DEGs) in esophageal squamous-cellcarcinoma (ESCC) and provide potential therapeutic targets. The microarray dataset GSE20347 was downloaded from the Gene Expression Omnibus (GEO) database, and included 17 tissue samples and 13 normal adjacent tissue samples from patients with ESCC. A total of 22,277 DEGs were identified. A heat map for the DEGs was constructed with the Morpheus online tool and the top 200 genes (100 upregulated and 100 downregulated) were selected for further bioinformatics analysis, including analysis of gene ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, protein-protein interaction networks and Spearman's correlation tests. The results of the GO analysis indicated that the upregulated DEGs were most significantly enriched in membrane-bounded vesicles in the cellular component (CC) category, but were not significantly enriched in any GO terms of the categories biological process (BP) or molecular function (MF); furthermore, the downregulated DEGs were most significantly enriched in regulation of DNA metabolic processes, nucleotide binding and chromosomes in the categories BP, MF and CC, respectively. The KEGG analysis indicated that the downregulated DEGs were enriched in the regulation of cell cycle pathways. The top 10 hub proteins in the protein-protein interaction network were cyclin-dependent kinase 4, budding uninhibited by benzimidazoles 1, cyclin B2, heat shock protein 90AA1, aurora kinase A, H2A histone family member Z, replication factor C subunit 4, and minichromosome maintenance complex component 2, −4 and −7. These proteins are mainly involved in regulating tumor progression. The genes in the four top modules were mainly implicated in regulating cell cycle pathways. Secreted Ly-6/uPAR-related protein (SLURP) was the hub gene, and SLURP and its interacting genes were most enriched in the chromosomal part in the CC category, organelle organization in the BP category and protein binding in the MF category, and were involved in pathways including DNA replication, cell cycle and P53 signaling. The expression of SLURP-1 in fifteen patients with esophageal carcinoma was detected using quantitative polymerase chain reaction analysis, and the results indicated that SLURP-1 expression was significantly decreased in the tumor samples relative to that in normal adjacent tissues. These results suggest that several hub proteins and the hub gene SLURP-1 may serve as potential therapeutic targets, and that gene dysfunction may be involved in the tumorigenesis of ESCC.
Keywords: esophageal squamous-cell carcinoma, differentially expressed genes, bioinformatics analysis, microarray
Introduction
Esophageal cancer (EC) has the sixth highest mortality and the eighth highest incidence rate worldwide (1). Its incidence rate in China is the highest in the world (2). The primary histological type is esophageal squamous-cell carcinoma (ESCC), which accounts for ~90% of all EC cases in China. Patients with ESCC are usually diagnosed at an advanced stage, and their 5-year survival rate is therefore low (~10-20%) (3,4). It has been reported that smoking and alcohol consumption are major causative factors of ESCC, as they promote gene mutations associated with processes including tumor initiation, progression and even metastasis. Thus, it is important to understand the molecular mechanisms of the tumorigenesis process to identify targets and develop novel treatments for ESCC.
Various genes, mRNAs and micro (mi)RNAs have been reported to form a network regulating the tumorigenesis and development of EC. Numerous studies have indicated that certain genes act as tumor suppressors, and that several genes inhibit cancer cell migration, invasion and tumor progression in ESCC (5–8). High-throughput sequencing technologies, including microarrays, which are able to detect changes in the expression of a vast amount of genes, have been widely used in cancer diagnosis and cancer research. In a previous study, numerous differentially expressed genes (DEGs) were detected in the tumor tissues of patients with ESCC relative to those in normal tissue or normal epithelial cells by microarrays (9). These hundreds of DEGs are involved in signalling pathways in ESCC, which encompass biological processes (BP), molecular functions (MF) and cellular components (CC). Hu et al (10) examined DEGs in tumor and matched normal adjacent tissue samples from patients with ESCC using microarrays. However, the regulatory roles of these DEGs, including the pathways in their interaction network, have remained to be elucidated (10).
Therefore, in the present study, bioinformatics methods were used to analyze the DEGs and their interaction networks. Original data were downloaded from the Gene Ontology (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The DEGs were identified from tumor tissues of patients with ESCC compared with those in matched normal adjacent tissues. The 200 top DEGs were then selected for further bioinformatics analysis, including analysis of Gene Ontology (GO) terms, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, protein-protein interaction (PPI) networks and Spearman's correlation tests (11). In general, the present study may help identify potential therapeutic targets and provide valuable information to further illuminate the molecular mechanisms of ESCC.
Materials and methods
Microarray data
Gene expression profiles of GSE20347 were downloaded from the GEO repository collated by Hu et al (10). These data were based on the AgilentGPL571 platform (Affymetrix Human Genome U133A 2.0 Array, HG-U133A_2; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA), which included 13 samples of normal adjacent esophageal tissues (with the ID nos. GSM509787-GSM509803) and 17 samples of tumor tissues (with the ID nos. GSM509804-GSM509820) from patients with ESCC. Total RNA had been extracted using the PureLink Micro-to-Midi RNA Purification System (Invitrogen; Thermo Fisher Scientific, Inc.) and was detected by Affymetrix HG-U133A 2.0 gene expression arrays (Affymetrix; Thermo Fisher Scientific, Inc.). R (Bioconductor; http://www.bioconductor.org/) was used for background correction and normalization of the data.
Identification of DEGs
The raw data files used for analysis included TXT files (Agilent platform). The files were used to create heat maps with the Morpheus online software (https://software.broadinstitute.org/morpheus/). The data were classified into two groups, namely the normal and tumor groups. The top 200 DEGs (100 upregulated and 100 downregulated genes) were screened by their signal-to-noise ratio (SNR).
GO and pathway enrichment analysis of DEGs
The top 200 DEGs were analyzed using with the database for annotation, visualization, integrated discovery (DAVID) online tool (https://david.ncifcrf.gov/; DAVID bioinformatics resources 6.8 of the National Institute of Allergy and Infectious Diseases/National Institutes of Health), which may be used for the high-throughput functional analysis of genes (12). The database includes GO and KEGG pathway analyses. GO is a useful tool for identifying characteristic biological information by using high-throughput genome transcriptome data (13) and was used in the present study for GO enrichment analysis in the categories BP, cellular component (CC) and MF. KEGG pathway analysis was also performed to gain insight into the signaling pathways regulated by the DEGs (14–16).
PPI network and module analysis
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://version10.string-db.org/; version 10.0) database is a powerful online tool that overlays 9.6 million proteins from 2,034 organisms and 184 million interactions. The top 200 DEGs were analyzed by STRING for the PPI. Subsequently, interactions with a combined score of >0.4 (the value that was considered statistically significant) were selected to construct the PPI network using Cytoscape software 3.4.0 (http://www.cytoscape.org). Finally, the plug-in Molecular Complex Detection (MCODE) of Cytoscape was used to construct the modules of the PPI network. The top modules with an MCODE score of >3 and a node number of >4 were selected for further pathway analysis with KEGG.
Spearman correlation test of the top 200DEGs in patients with ESCC
To further determine DEGs that has the most connections to the other top DEG, a Spearman's correlation test was performed in Excel 2007 (Microsoft Corp., Redmond, WA, USA). Subsequently, the genes were screened with the following settings: 0.95<Pearson correlation coefficient (PCC) <1 and −1<PCC<-0.95. The gene with the highest correlation was selected as the hub gene to construct the co-expression network in Cytoscape. Finally, GO enrichment analyses in the categories BP, CC and MF for the hub gene and their associated genes were further performed by (Biological Networks Gene Ontology) BiNGO tool (17) and the KEGG analysis was performed with the ClueGO plug-in of Cytoscape.
Sample collection
ESCC and normal tissue samples were collected from the Panyu Central Hospital and the Third Affiliated Hospital of Southern Medical University (Guangzhou, China) from June 30, 2015 to May 2, 2017. All patients were male, with a mean age of 62.20±5.98 years, and diagnosed by clinical pathology (14 cases were squamous carcinoma and 1 was adenocarcinoma). All samples were stored at −80°C after collection. A total of 30 samples, which included 15 tumor samples and 15 normal adjacent tissue samples (>3 cm from the tumor tissue) as controls, were used to detect the gene expression of SLURP by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR assay
Total RNA was extracted from all samples using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. First-strand complementary (c)DNA was synthesized with 1 µg total RNA per sample using the All-in-One™ First-Strand cDNA Synthesis kit (Gene Copoeia, Inc., Rockville, MD, USA). Subsequently, the cDNA sample was amplified using All-in-One qPCR mix (Gene Copoeia, Inc.) in a final volume of 20 µl in an ABI Vii7 dx reactor (Applied Biosystems; Thermo Fisher Scientific, Inc.). The amplifications were performed as follows: Initial incubation for 2 min at 50°C, denaturation for 30 sec at 95°C, and 45 cycles of 95°C for 5 sec and 65.6°C for 34 sec. The experiments were performed in triplicate and quantified using melting curve analysis. β-actin was used as an endogenous reference control. The relative gene expression levels were calculated using the 2−ΔΔCq method (18). The primer pairs for SLURP-1 and β-actin were as follows: SLURP-1 forward, 5′-GCTCCTGTGTGGCCACCGAC-3′ and reverse, 5′-GAGCCAGGCCCCGTCAGAGA-3′; β-actin forward, 5′-ACTCTTCCAGCCTTCCTTCC-3′ and reverse, 5′-GCGGCGCAATACGAATGCCCC-3′.
Statistical analysis
P<0.05 was considered to indicate a statistically significant difference. The P-value was adjusted by using the false discovery rate (FDR) method for multiple hypothesis testing. FDR <0.05 was established as the threshold (15–17). The data are expressed as mean ± standard deviation. Independent t-tests were used to analyze the PCR results and performed using SPSS software (version 16.0; SPSS, Inc., Chicago, IL, USA).
Results
Identification of DEGs
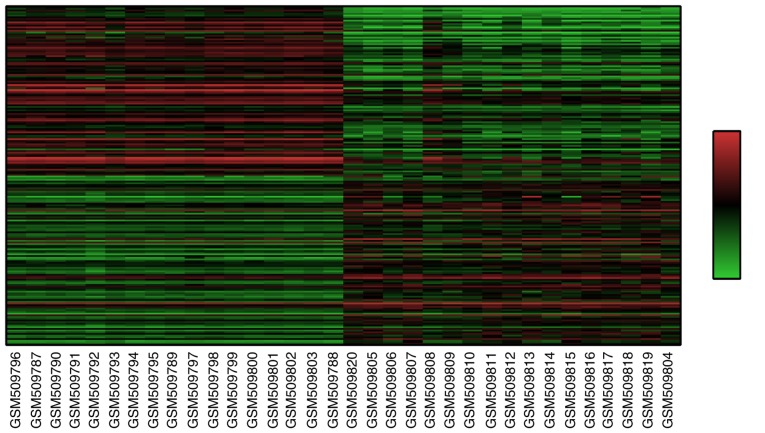
A total of 22,277 DEGs were identified. The expression of these genes was analyzed with the Morpheus online tool to form a heat map (top 100 upregulated and 100 downregulated genes), which were selected according to their SNR value. The heat map is presented in Fig. 1.
Figure 1.
Heat map of the top 200 differentially expressed genes (100 upregulated and 100 downregulated genes). GSM509796 to GSM509788: Normal tissue of patients with esophageal squamous-cell carcinoma; GSM509820 to GSM509804: Tumor tissue of patients with esophageal squamous-cell carcinoma. Red indicates high expression levels and green low expression levels.
GO term enrichment and KEGG pathway analysis of DEGs
GO term enrichment analysis indicated that the upregulated DEGs were most significantly enriched in membrane-bound vesicles in the category CC, but no significant enrichment was identified in the categories BP and MF. The downregulated DEGs were most significantly enriched in the regulation of DNA metabolic processes, nucleotide binding and chromosomes in the categories BP, MF and CC, respectively (Table I). The KEGG analysis indicated that the downregulated DEGs were enriched in the regulation of cell cycle pathways (Table II).
Table I.
GO analysis of differentially expressed genes in esophageal squamous cell carcinoma.
| A, Upregulated genes in the category cellular component | |||
|---|---|---|---|
| GO term | Function | N (%) | P-value |
| GO:0031988 | Membrane-bound vesicle | 33 (0.25) | 4.02×10−8 |
| GO:0070062 | Extracellular exosome | 29 (0.22) | 1.27×10−7 |
| GO:1903561 | Extracellular vesicle | 29 (0.22) | 1.45×10−7 |
| GO:0043230 | Extracellular organelle | 29 (0.22) | 1.46×10−7 |
| GO:0044421 | Extracellular region part | 30 (0.23) | 3.16×10−5 |
| B, Downregulated genes | |||
| GO term | Function | N (%) | P-value |
| Biological process | |||
| GO:0006259 | DNA metabolic process | 17 (0.11) | 1.81×10−7 |
| GO:0051276 | Chromosome organization | 17 (0.11) | 1.33×10−6 |
| GO:0010564 | Regulation of cell cycle process | 12 (0.08) | 7.36×10−6 |
| GO:1903047 | Mitotic cell cycle process | 12 (0.08) | 2.68×10−5 |
| Cellular component | |||
| GO:0005694 | Chromosome | 20 (0.13) | 2.80×10−8 |
| GO:0044427 | Chromosomal part | 19 (0.12) | 4.98×10−8 |
| GO:0098687 | Chromosomal region | 11 (0.07) | 1.35×10−6 |
| GO:0000228 | Nuclear chromosome | 14 (0.09) | 1.78×10−6 |
| GO:0000793 | Condensed chromosome | 8 (0.05) | 1.00×10−5 |
| GO:0044454 | Nuclear chromosome part | 12 (0.08) | 3.14×10−5 |
| Molecular function | |||
| GO:0000166 | Nucleotide binding | 27 (0.17) | 6.38×10−6 |
| GO:1901265 | Nucleoside phosphate binding | 27 (0.17) | 6.38×10−6 |
| GO:0035639 | Purine ribonucleoside triphosphate binding | 23 (0.15) | 1.03×10−5 |
| GO:0032550 | Purine ribonucleoside binding | 23 (0.15) | 1.08×10−5 |
| GO:0001883 | Purine nucleoside binding | 23 (0.15) | 1.09×10−5 |
| GO:0032549 | Ribonucleoside binding | 23 (0.15) | 1.12×10−5 |
| GO:0001882 | Nucleoside binding | 23 (0.15) | 1.18×10−5 |
| GO:0032555 | Purine ribonucleotide binding | 23 (0.15) | 1.41×10−5 |
| GO:1901363 | Heterocyclic compound binding | 44 (0.28) | 1.48×10−5 |
| GO:0017076 | Purine nucleotide binding | 23 (0.15) | 1.50×10−5 |
| GO:0032553 | Ribonucleotide binding | 23 (0.15) | 1.62×10−5 |
| GO:0005524 | ATP binding | 20 (0.13) | 1.70×10−5 |
| GO:0036094 | Small molecule binding | 27 (0.17) | 1.97×10−5 |
| GO:0097159 | Organic cyclic compound binding | 44 (0.28) | 2.02×10−5 |
| GO:0032559 | Adenyl ribonucleotide binding | 20 (0.13) | 2.15×10−5 |
| GO:0030554 | Adenyl nucleotide binding | 20 (0.13) | 2.28×10−5 |
GO, Gene Ontology.
Table II.
Kyoto Encyclopedia of Genes and genomes pathway analysis of differentially expressed genes in esophageal squamous cell carcinoma.
| Expression | Term | Function | N (%) | P-value |
|---|---|---|---|---|
| Downregulated | cfa04110 | Cell cycle pathway | 8 (0.05) | 1.62×10−5 |
PPI network and top module construction
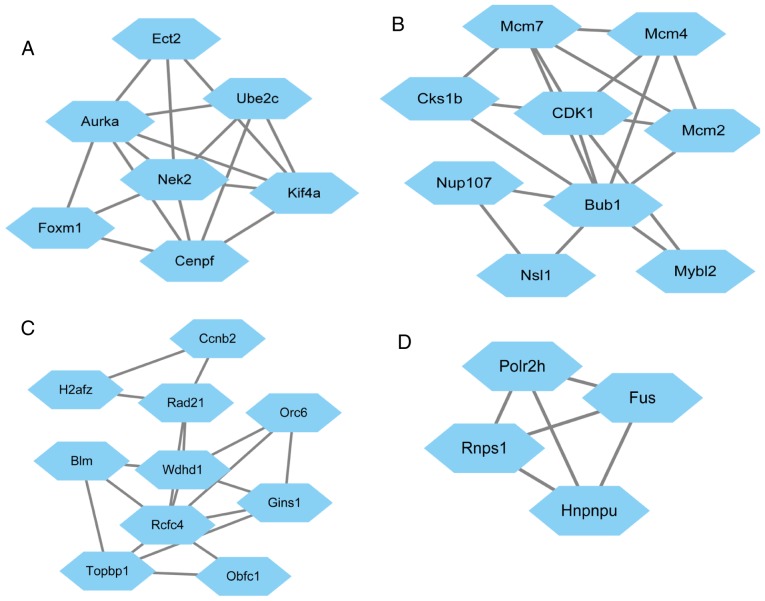
Based on the STRING database, the PPI network of the top 200 DEGs was constructed with Cytoscape software (Fig. 2). In the PPI networks, nodes with a high degree of connectivity were defined as hub proteins. The top 10 hub proteins included cyclin-dependent kinase 4 (CDK4), budding uninhibited by benzimidazoles 1 (BUB1) and cyclin B2 (CCNB2). The degree of connectivity of CDK4 was 30, and it was therefore the most highly connected node. The network consisted of 110 nodes and 262 edges. The top four significant modules were selected for further pathway enrichment analysis (MCODE score, >3; number of nodes, >4) and the results indicated that the genes were significantly enriched in cell cycle pathways (Table III).
Figure 2.
Top 4 modules from the protein-protein interaction networks. (A) Module 1. (B) Module 2. (C) Module 3. (D) Module 4.
Table III.
The enriched pathway of module 2.
| Pathway | P-value | False discovery rate | Nodes |
|---|---|---|---|
| Cell cycle | 1.71×10−6 | 9.35×10−4 | Cyclin-dependent kinases regulatory subunit 1, Cyclin-dependent kinase 1, MCM7, Nuclear pore complex protein Nup107, MCM4, MCM2, Mitotic checkpoint serine/threonine-protein kinase BUB1, KAT8 regulatory NSL complex subunit 1, Myb-related protein B |
MCM, DNA replication licensing factor MCM.
Spearman correlation analysis of the top 200 DEGs
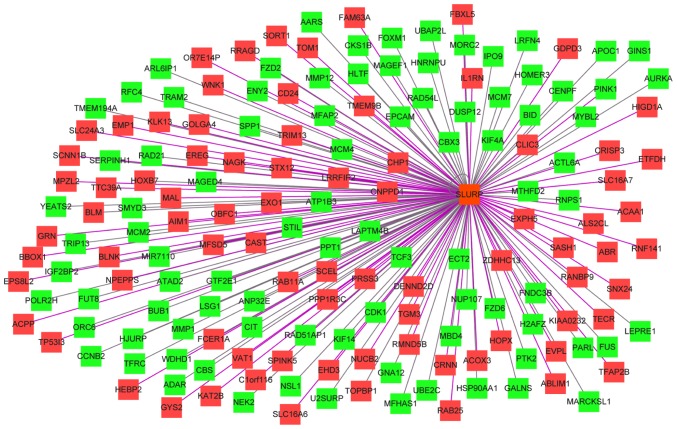
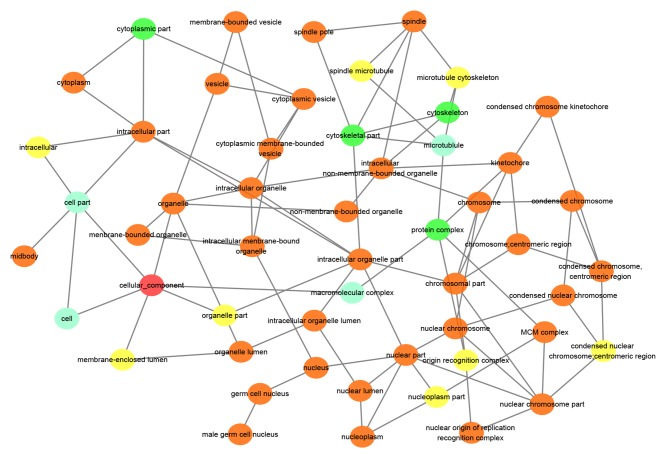
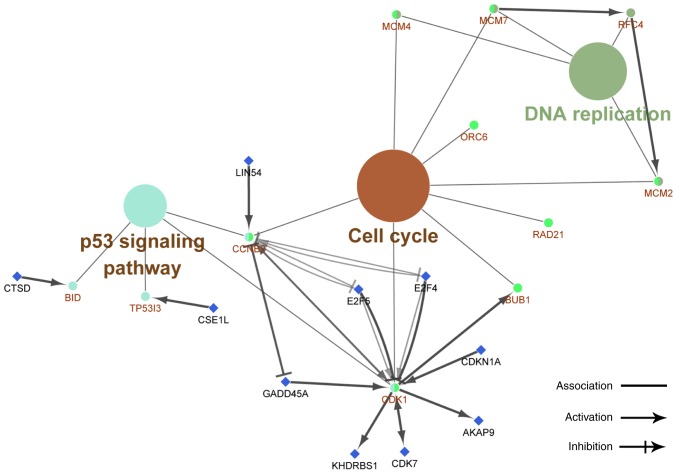
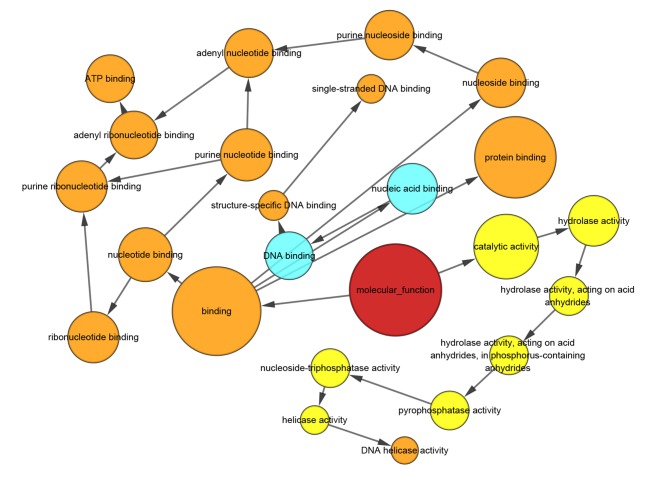
The results of the Spearman correlation analysis indicated that the most connected gene was secreted LY6/PLAUR domain (SLURP). SLURP was selected as a hub gene and further analysis was performed on SLURP and its associated genes (Fig. 3). The results of the bioinformatics analysis suggested that these genes were most significantly enriched in the chromosomal part, organelle organization and protein binding in the categories CC, BP and MF, respectively (Figs. 4–6). KEGG pathway analysis revealed that the genes were involved in DNA replication, cell cycle and P53 signaling pathways (Fig. 7).
Figure 3.
Co-expression network of the hub gene secreted LY6/PLAUR domain and its connected genes. The red nodes indicate a positive and the green nodes indicate a negative correlation. The colors of the connection lines from black to purple represent a low to high value of the correlation coefficient, respectively.
Figure 4.
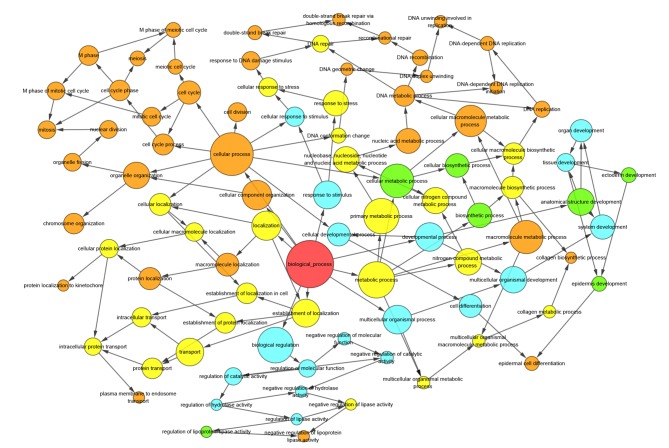
Gene Ontology enrichment analysis in the category cellular component for the hub gene secreted LY6/PLAUR domain and its connected genes. Arrows indicate regulation. MCM, DNA replication licensing factor MCM. The red circle represents the category of interest. Light blue, green, yellow and orange circles represent P>0.05, P<0.05, P<0.01 and P<0.01, respectively. P<0.05 was considered as significant.
Figure 6.
Gene Ontology enrichment analysis in the category biological processes for the hub gene secreted LY6/PLAUR domain and its connected genes. Arrows indicate regulation. The red circle represents the category of interest. Light blue, green, yellow and orange represent P>0.05, P<0.05, P<0.01 and P<0.001, respectively.
Figure 7.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of the hub gene secreted LY6/PLAUR domain and its connected genes. The results revealed that they are involved in DNA replication, cell cycle and P53 signaling pathways. The large colored indicate nodes with a P<0.05: Turquiose, 0.001; green, 8.097E−5; brown, 1.236E−6. The small circles represent the proteins that are in the pathway denoted by the node. The small blue squares indicated the proteins that are regulated by the proteins in the pathways.
PCR analysis of SLURP
The results indicated that the expression of the hub gene SLURP-1 was significantly decreased in the tumor samples relative to that in the normal adjacent tissues in patients with esophageal carcinoma (P<0.05; Fig. 8).
Figure 8.
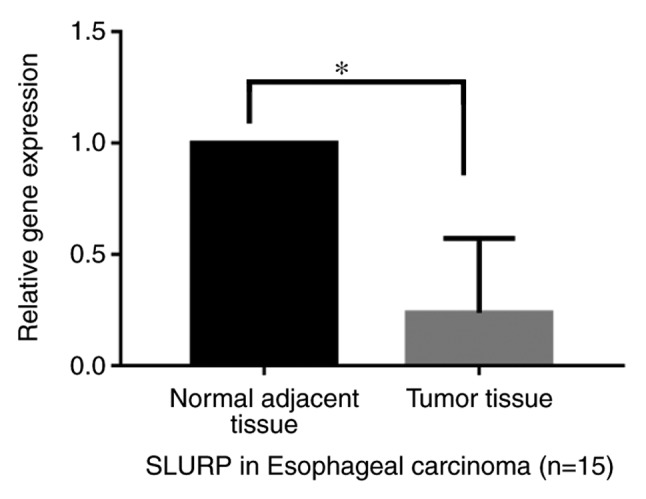
Gene expression of SLURP-1 in tumor tissues relative to normal adjacent tissues of patients with esophageal carcinoma. Fifteen patients with esophageal carcinoma were assessed by using reverse transcription-quantitative polymerase chain reaction analysis. The results indicated that SLURP-1 was significantly decreased in the tumor samples relative to that in normal adjacent tissues. *P<0.05 vs. control. SLURP, secreted LY6/PLAUR domain.
Discussion
ESCC is caused by external factors leading to gene mutations. Thus, understanding the underlying molecular mechanisms is of pivotal importance for ESCC diagnosis and treatment. In the present study, a dataset downloaded from the GEO database was analyzed and the bioinformatics tools Morpheus, DAVID and STRING were used to obtain DEGs, hub proteins, hub genes and major deregulated pathways in ESCC.
A total of 22,277 DEGs were identified from the dataset GSE20347. To better understand the interactions of DEGs, the top 200 DEGs were selected for further analysis. The results of the GO term enrichment revealed that these DEGs were most highly involved in membrane-bound vesicles, DNA metabolic processes, nucleotide binding and chromosomes. KEGG analysis indicated that the DEGs were enriched in cell cycle pathways. These results suggest that the DEGs may be mainly involved in regulating the cell cycle (19,20) and organogenesis, which are closely associated with tumorigenesis (21) and tumor progression (22–29). The results from a study by Lin et al (30) confirmed that the dysregulation of genes that regulate the G1/S transition is common in ESCC. Reduced expression of the protein p21WAF1/Cip1 was reported to predict a shorter overall survival time of patients with ESCC (27,28). The present results indicated that the deregulation of certain genes is involved in ESCC, and that various DEGs may be associated with the genesis of ESCC.
The analysis of PPI networks indicated that the top 10 hub proteins included CDK4, BUB1, CCNB2, heat shock protein (HSP)90AA1, aurora kinase (AURK)A, H2A histone family member Z (H2AFZ), replication factor C subunit 4 (RFC4), as well as minichromosome maintenance complex component 7 (MCM7), MCM4 and MCM2. These proteins are closely associated with the cell cycle, tumorigenesis (31), transferase signaling pathway (32,33), transforming growth factor β-mediated cell cycle control (34,35), embryonic development (36), DNA-dependent ATPase activity (37) and DNA unwinding enzymes (38–40). CDK4 was the node with the highest degree of interaction and was the most connected hub protein, interacting with 30 genes in the regulatory network. This result was consistent with that by Su et al (41), which also indicated that CDK4 was the most significantly upregulated gene by analyzing 5 mRNA expression datasets of EC tissues/cell lines from GEO. A recent study revealed that CDK4 had a negative association with EC-related gene 4, which has a tumor suppressor function in ESCC (42). CDK4/6 inhibitor-SHR6390 was reported to exert an antitumor effect against ESCC (43). AURKA, MCM7 and MCM4 were closely associated with cell proliferation and migration in ESCC (44–46). BUB1-related protein kinase was significantly higher in cancerous tissue than in adjacent normal tissue, and after radiochemotherapy, it was significantly decreased in the tissue of patients with ESCC (47). HSP90A and CCNB1 protein were reported to be associated with tumor malignancy and prognosis in patients with ESCC (48). It was demonstrated that abnormal levels of H2AF may be associated with poor survival of ESCC patients (49). However, as the involvement of RFC4 and MCM2 in ESCC has been rarely investigated, further study is necessary. Analysis of Hub protein functions indicated that these proteins have a key role in the regulation of ESCC, including the genesis development and progression of tumors, and that these hub proteins may serve as therapeutic targets in ESCC.
The present study used the plug-in MCODE to construct the modules. The results indicated that the functions of genes in the top 4 modules were mainly associated with cell cycle pathways. Li et al (50) indicated that overall, the DNA repair pathways were significantly associated with a risk of ESCC. Roncalli et al (51) studied cell cycle-associated genes in patients with EC, and their results demonstrated that these were significantly associated lymph node metastasis and unfavorable survival rates. These results indicate that the pathways associated with the top modules mainly regulate tumor progression in ESCC and that certain genes in theses pathways may serve as potential prognostic biomarkers.
In the present study, Spearman's correlation test was used to analyze the correlation between the top 200 DEGs. The results revealed that SLURP was the hub gene that was most highly connected to the other genes. The results of the bioinformatics analysis indicated that this hub gene and its associated genes are significantly enriched in the chromosomal part, organelle organization and protein binding in the GO categories CC, BP and MF, respectively. These genes are also involved in DNA replication, cell cycle and P53 signaling pathways. The expression of SLURP-1 was the assessed in 15 patients with EC, and the results revealed that SLURP-1was significantly decreased in the tumor samples relative to that in the normal adjacent tissues. SLURP-1 has been reported to participate in signal transduction, immune activation and cell adhesion to exert its antitumor activity (52). This was consistent with the results of recent studies, which indicated that DEGs screened by RNA-sequencing data or The Cancer Genome Atlas analysis have important roles in regulating growth, invasion and metastasis of tumors as well as immune responses, and the DEGs included collagen type I α 1, matrix metallopeptidases, keratin 4, cysteine-rich secretory protein2 and 3, mucin 21 and cyclin D1 (53,54). The present results indicated that the hub gene SLURP-1 may have a key role in regulating the tumorigenesis of ESCC and that it may serve as a potential biomarker in tumor diagnosis.
In conclusion, the present study identified DEGs and the hub proteins (CDK4, BUB1, CCNB2, HSP90AA1, AURKA, H2AFZ, RFC4, MCM7, MCM4 and MCM2) and a hub gene (SLURP) in ESCC. These genes are primarily involved in regulating the tumorigenesis and progression of ESCC. The hub proteins and gene may be considered as candidate therapeutic targets and may provide information for further studies on the molecular biological functions and mechanisms of ESCC. However, the present study had certain limitations, including the fact that only GEO 1 dataset of microarray data was used and that the sample size was relatively small.
Figure 5.
Gene Ontology enrichment analysis for the hub gene secreted LY6/PLAUR domain and its connected genes in the category molecular function. Arrows indicate regulation. The red circle represents the category of interest. Light blue, yellow and orange circles represent P>0.05, P<0.01 and P<0.001, respectively.
Acknowledgements
The authors would like to thank Professor Edward I. Wong (Milton International Education Group, Hong Kong) for revising the manuscript.
Funding
This work was funded by grants of the Technical New Star of Zhujiang, Pan Yu district, Guangzhou (grant no. 2013-special-15-6.10), the Science and Technology program of Pan Yu (grant no. 2015-Z03-09), the Traditional Chinese Medicine Bureau of Guangdong Province (grant no. 20161186) and the Science and Technology Program of Guangzhou (grant no. 201804010012).
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Authors' contributions
XC and SC performed the analysis of the data and writing of the paper. BL, XZ, WL and HL collected the samples and performed the PCR. XC performed the data analysis of GEO.
Ethical approval and consent to participate
This study was approved by the Ethics Committee of Panyu Central Hospital (ethical approval no. 20180001). All patients provided informed consent to participate in this study.
Patient consent for publication
All patients gave consent for the publication on their data in the current study.
Competing interests
The authors declared they have no competing conflicts.
References
- 1.Chen W, Zheng R, Zhang S, Zeng H, Xia C, Zuo T, Yang Z, Zou X, He J. Cancer incidence and mortality in China, 2013. Cancer Lett. 2017;401:63–71. doi: 10.1016/j.canlet.2017.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer. 2005;113:456–463. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 3.Yang Q, Wang YX, He M, Li J, Qi Z, Zhu SC, Qiao XY. Factors affecting on long-time survival in patients with stageIII thoracic esophageal carcinoma after esophagectomy. Zhonghua Zhong Liu Za Zhi. 2016;38:530–537. doi: 10.3760/cma.j.issn.0253-3766.2016.07.010. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 4.Yu S, Zhang W, Ni W, Xiao Z, Wang X, Zhou Z, Feng Q, Chen D, Liang J, Fang D, et al. Nomogram and recursive partitioning analysis to predict overall survival in patients with stage IIB-III thoracic esophageal squamous cell carcinoma after esophagectomy. Oncotarget. 2016;7:55211–55221. doi: 10.18632/oncotarget.10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi Y, Lu X, Chen J, Jiao C, Zhong J, Song Z, Yu X, Lin B. Downregulated miR-486-5p acts as a tumor suppressor in esophageal squamous cell carcinoma. Exp Ther Med. 2016;12:3411–3416. doi: 10.3892/etm.2016.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osako Y, Seki N, Kita Y, Yonemori K, Koshizuka K, Kurozumi A, Omoto I, Sasaki K, Uchikado Y, Kurahara H, et al. Regulation of MMP13 by antitumor microRNA-375 markedly inhibits cancer cell migration and invasion in esophageal squamous cell carcinoma. Int J Oncol. 2016;49:2255–2264. doi: 10.3892/ijo.2016.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo J, Wang D, Shen H, Liu F, Han J, Zhang X. MicroRNA-153 inhibits tumor progression in esophageal squamous cell carcinoma by targeting SNAI1. Tumour Biol. 2016 Oct 13; doi: 10.1007/s13277-016-5427-x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 8.Jing C, Ma G, LiX, Wu X, Huang F, Liu K, Liu Z. MicroRNA-17/20a impedes migration and invasion via TGF-β/ITGB6 pathway in esophageal squamous cell carcinoma. Am J Cancer Res. 2016;6:1549–1562. [PMC free article] [PubMed] [Google Scholar]
- 9.Hu N, Wang C, Clifford RJ, Yang HH, Su H, Wang L, Wang Y, Xu Y, Tang ZZ, Ding T, et al. Integrative genomics analysis of genes with biallelic loss and its relation to the expression of mRNA and micro-RNA in esophageal squamous cell carcinoma. BMC Genomics. 2015;16:732. doi: 10.1186/s12864-015-1919-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu N, Clifford RJ, Yang HH, Wang C, Goldstein AM, Ding T, Taylor PR, Lee MP. Genome wide analysis of DNA copy number neutral loss of heterozygosity (CNNLOH) and its relation to gene expression in esophageal squamous cell carcinoma. BMC Genomics. 2010;11:576. doi: 10.1186/1471-2164-11-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garner E, Wallace JS, Argoty GA, Wilkinson C, Fahrenfeld N, Heath LS, Zhang L, Arabi M, Aga DS, Pruden A. Metagenomic profiling of historic Colorado Front Range flood impact on distribution of riverine antibiotic resistance genes. Sci Rep. 2016;6:38432. doi: 10.1038/srep38432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. GenomeBiol. 2003;4:P3. [PubMed] [Google Scholar]
- 13.Torto-Alalibo T, Purwantini E, Lomax J, Setubal JC, Mukhopadhyay B, Tyler BM. Genetic resources for advanced biofuel production described with the Gene Ontology. Front Microbiol. 2014;5:528. doi: 10.3389/fmicb.2014.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y. KEGG-PATH: Kyotoencydopedia of genes and genomes-based pathway analysis using a path analysis model. MolBiosyst. 2014;10:2441–14447. doi: 10.1039/c4mb00287c. [DOI] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: Kyotoencyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–9344. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, Li Z, Han X. Long Noncoding RNA HOTAIR Controls Cell Cycle by Functioning as a Competing Endogenous RNA in Esophageal Squamous Cell Carcinoma. Transl Oncol. 2016;9:489–497. doi: 10.1016/j.tranon.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang HF, Alshareef A, Wu C, Jiao JW, Sorensen PH, Lai R, Xu LY, Li EM. miR-200b induces cell cycle arrest and represses cell growth in esophageal squamous cell carcinoma. Carcinogenesis. 2016;37:858–869. doi: 10.1093/carcin/bgw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao J, Ge MH, Ling ZQ. Fbxw7 tumor suppressor: A vital regulator contributes to human tumorigenesis. Medicine (Baltimore) 2016;95:e2496. doi: 10.1097/MD.0000000000002496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa C, Santos M, Martínez-Fernández M, Lorz C, Lázaro S, Paramio JM. Deregulation of the pRb-E2F4 axis alters epidermal homeostasis and favors tumor development. Oncotarget. 2016;7:75712–75728. doi: 10.18632/oncotarget.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambot MA, Peny MO, Fayt I, Haot J, Noël JC. Overexpression of 27-kDa heat shock protein relates to poor histological differentiation in human oesophageal squamous cell carcinoma. Histopathology. 2000;36:326–330. doi: 10.1046/j.1365-2559.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- 24.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL, et al. CCircular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, Qiu M, Chen Y, Wang J, Xia W, Mao Q, Yang L, Li M, Jiang F, Xu L, Yin R. Long noncoding RNA, tissue differentiation-inducing nonprotein coding RNA is upregulated and promotes development of esophageal squamous cell carcinoma. Dis Esophagus. 2016;29:950–958. doi: 10.1111/dote.12436. [DOI] [PubMed] [Google Scholar]
- 26.Zhi H, Zhang J, Hu G, Lu J, Wang X, Zhou C, Wu M, Liu Z. The deregulation of arachidonic acid metabolism-related genes in human esophageal squamous cell carcinoma. Int J Cancer. 2003;106:327–333. doi: 10.1002/ijc.11225. [DOI] [PubMed] [Google Scholar]
- 27.Mao Y, Li L, Liu J, Wang L, Zhou Y. MiR-495 inhibits esophageal squamous cell carcinoma progression by targeting Akt1. Oncotarget. 2016;7:51223–51236. doi: 10.18632/oncotarget.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng LS, Yang XZ, Wen YF, Mail SJ, Wang MH, Zhang MY, Zheng XF, Wang HY. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging (Albany NY) 2016;8:1236–1249. doi: 10.18632/aging.100980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Zhang C, Wang C, Li L, Li C, Li Q, Zhang M, Wu Q. Miz-1 promotes the proliferation of esophageal cancer cells via suppression of p21 and release of p21-arrested cyclin D1. Oncol Rep. 2016;35:3532–3540. doi: 10.3892/or.2016.4731. [DOI] [PubMed] [Google Scholar]
- 30.Lin DC, Shi ZZ, Xue LY, Chen W, Xu X, Han YL, Lv N, Wang MR. Expression of cell cycle related proteins cyclin D1, p53 and p21WAF1/Cip1 in esophageal squamous cell carcinoma. Yi Chuan. 2010;32:455–460. doi: 10.3724/SP.J.1005.2010.00455. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 31.Lang E, Zelenak C, Eberhard M, Bissinger R, Rotte A, Ghashghaeinia M, Lupescu A, Lang F, Qadri SM. Impact of cyclin-dependent kinase CDK4 inhibition on eryptosis. Cell Physiol Biochem. 2015;37:1178–1186. doi: 10.1159/000430241. [DOI] [PubMed] [Google Scholar]
- 32.Breit C, Bange T, Petrovic A, Weir JR, Müller F, Vogt D, Musacchio A. Role of intrinsic and extrinsic factors in the regulation of the mitotic checkpoint kinase Bub1. PLoS One. 2015;10:e0144673. doi: 10.1371/journal.pone.0144673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takashima S, Saito H, Takahashi N, Imai K, Kudo S, Atari M, Saito Y, Motoyama S, Minamiya Y. Strong expression of cyclin B2 mRNA correlates with a poor prognosis in patients with non-small cell lung cancer. Tumour Biol. 2014;35:4257–4265. doi: 10.1007/s13277-013-1556-7. [DOI] [PubMed] [Google Scholar]
- 34.Chen JT, Younusi A, Cao L, Tian Z, Zhou YJ, Song XH. Potential role of heat-shock proteins in giant cell tumors. Genet Mol Res. 2015;14:19144–19154. doi: 10.4238/2015.December.29.24. [DOI] [PubMed] [Google Scholar]
- 35.Kim SR, Kim KB, Chae YC, Park JW, Seo SB. H3S10 phosphorylation-mediated transcriptional regulation by Aurora kinase A. Biochem Biophys Res Commun. 2016;469:22–28. doi: 10.1016/j.bbrc.2015.11.063. [DOI] [PubMed] [Google Scholar]
- 36.Kusakabe M, Oku H, Matsuda R, Hori T, Muto A, Igarashi K, Fukagawa T, Harata M. Genetic complementation analysis showed distinct contributions of the N-terminal tail of H2A.Z to epigenetic regulations. Genes Cells. 2016;21:122–135. doi: 10.1111/gtc.12327. [DOI] [PubMed] [Google Scholar]
- 37.Arai M, Kondoh N, Imazeki N, Hada A, Hatsuse K, Matsubara O, Yamamoto M. The knockdown of endogenous replication factor C4 decreases the growth and enhances the chemosensitivity of hepatocellular carcinoma cells. Liver Int. 2009;29:55–62. doi: 10.1111/j.1478-3231.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 38.Ishibashi Y, Kinugasa T, Akagi Y, Ohchi T, Gotanda Y, Tanaka N, Fujino S, Yuge K, Kibe S, Yoshida N, et al. Minichromosome maintenance protein 7 is a risk factor for recurrence in patients with Dukes C colorectal cancer. Anticancer Res. 2014;34:4569–4575. [PubMed] [Google Scholar]
- 39.Watanabe E, Ohara R, Ishimi Y. Effect of an MCM4 mutation that causes tumours in mouse on human MCM4/6/7 complex formation. J Biochem. 2012;152:191–198. doi: 10.1093/jb/mvs060. [DOI] [PubMed] [Google Scholar]
- 40.Razavi SM, Jafari M, Heidarpoor M, Khalesi S. Minichromosome maintenance-2 (MCM2) expression differentiates oral squamous cell carcinoma from pre-cancerous lesions. Malays J Pathol. 2015;37:253–258. [PubMed] [Google Scholar]
- 41.Su P, Wen S, Zhang Y, Li Y, Xu Y, Zhu Y, Lv H, Zhang F, Wang M, Tian Z. Identification of the key genes and pathways in esophageal carcinoma. Gastroenterol Res Pract. 2016;2016:2968106. doi: 10.1155/2016/2968106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Wang W, Li X, Gao T. Association of ECRG4 with PLK1, CDK4, PLOD1 and PLOD2 in esophageal squamous cell carcinoma. Am J Transl Res. 2017;9:3741–3748. [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J, Li Q, Yuan J, Wang J, Chen Z, Liu Z, Li Z, Lai Y, Gao J, Shen L. CDK4/6 inhibitor-SHR6390 exerts potent antitumor activity in esophageal squamous cell carcinoma by inhibiting phosphorylated Rb and inducing G1 cell cycle arrest. J Transl Med. 2017;15:127. doi: 10.1186/s12967-017-1231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Li X, Li C, He C, Ren B, Deng Q, Gao W, Wang B. Aurora-A modulates MMP-2 expression via AKT/NF-κB pathway in esophageal squamous cell carcinoma cells. Acta Biochim Biophys Sin (Shanghai) 2016;48:520–527. doi: 10.1093/abbs/gmw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qiu YT, Wang WJ, Zhang B, Mei LL, Shi ZZ. MCM7 amplification and overexpression promote cell proliferation, colony formation and migration in esophageal squamous cell carcinoma by activating the AKT1/mTOR signaling pathway. Oncol Rep. 2017;37:3590–3596. doi: 10.3892/or.2017.5614. [DOI] [PubMed] [Google Scholar]
- 46.Choy B, LaLonde A, Que J, Wu T, Zhou Z. MCM4 and MCM7, potential novel proliferation markers, significantly correlated with Ki-67, Bmi1, and cyclin E expression in esophageal adenocarcinoma, squamous cell carcinoma, and precancerous lesions. Hum Pathol. 2016;57:126–135. doi: 10.1016/j.humpath.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tanaka K, Mohri Y, Ohi M, Yokoe T, Koike Y, Morimoto Y, Miki C, Tonouchi H, Kusunoki M. Mitotic checkpoint genes, hsMAD2 and BubR1, in oesophageal squamous cancer cells and their association with 5-fluorouracil and cisplatin-based radiochemotherapy. Clin Oncol (R Coll Radiol) 2008;20:639–646. doi: 10.1016/j.clon.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Huang T, Chen S, Han H, Li H, Huang Z, Zhang J, Yin Q, Wang X, Ma X, Dai P, et al. Expression of Hsp90α and cyclin B1 were related to prognosis of esophageal squamous cell carcinoma and keratin pearl formation. Int J Clin Exp Pathol. 2014;7:1544–1552. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang K, Li L, Zhu M, Wang G, Xie J, Zhao Y, Fan E, Xu L, Li E. Comparative analysis of histone H3 and H4 post-translational modifications of esophageal squamous cell carcinoma with different invasive capabilities. J Proteomics. 2015;112:180–189. doi: 10.1016/j.jprot.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Li WQ, Hu N, Hyland PL, Gao Y, Wang ZM, Yu K, Su H, Wang CY, Wang LM, Chanock SJ, et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis. 2013;34:1536–1542. doi: 10.1093/carcin/bgt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roncalli M, Bosari S, Marchetti A, Buttitta F, Bossi P, Graziani D, Peracchia A, Bonavina L, Viale G, Coggi G. Cell cycle-related gene abnormalities and product expression in esophageal carcinoma. Lab Invest. 1998;78:1049–1057. [PubMed] [Google Scholar]
- 52.Pettersson A, Nylund G, Khorram-Manesh A, Nordgren S, Delbro DS. Nicotine induced modulation of SLURP-1 expression in human colon cancer cells. Auton Neurosci. 2009;148:97–100. doi: 10.1016/j.autneu.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 53.Fu JH, Wang LQ, Li T, Ma GJ. RNA-sequencing based identification of crucial genes for esophageal squamous cell carcinoma. J Cancer Res Ther. 2015;11:240–425. doi: 10.4103/0973-1482.160122. [DOI] [PubMed] [Google Scholar]
- 54.Cancer Genome Atlas Network; Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.