Abstract
To date, different experimental strategies have been developed for the ex vivo expansion of human hematopoietic stem (HSCs) and progenitor (HPCs) cells. This has resulted in significant advances on the use of such expanded cells in transplantation settings. To this day, however, it is still unclear to what extent those stem and progenitor cells generated in vitro retain the functional and genomic integrity of their freshly isolated counterparts. In trying to contribute to the solving of this issue, in the present study we have selected and purified three different hematopoietic cell populations: HSCs (CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells), myeloid progenitor cells (CD34+ CD38+ CD45RA+ CD71‐ Lin‐ cells), and erythroid progenitor cells (CD34+ CD38+ CD45RA‐ CD71+ Lin‐ cells), obtained directly from fresh human umbilical cord blood (UCB) units or generated in vitro under particular culture conditions. We, then, compared their functional integrity in vitro and their gene expression profiles. Our results indicate that in spite of being immunophenotipically similar, fresh and in vitro generated cells showed significant differences, both in functional and genetic terms. As compared to their fresh counterparts, those HSCs generated in our culture system showed a deficient content of long‐term culture‐initiating cells, and a marked differentiation bias toward the myeloid lineage. In addition, in vitro generated HSCs and HPCs showed a limited expansion potential. Such functional alterations correlated with differences in their gene expression profiles. These observations are relevant in terms of HSC biology and may have implications in UCB expansion and transplantation. Stem Cells Translational Medicine 2018;7:602–614
Keywords: Cord blood, Ex vivo expansion, Gene expression profiles, Hematopoietic stem and progenitor cells
Significance Statement.
This study presents evidence indicating that in vitro‐generated hematopoietic stem and progenitor cells differ in functional and genetic terms from their freshly obtained counterparts. These findings have biologic and clinical implications.
Introduction
Over the last three decades, umbilical cord blood (UCB) has become a major source of hematopoietic stem and progenitor cells (HSCs and HPCs, respectively) for hematopoietic transplantation 1, 2. UCB not only contains clinically relevant amounts of primitive hematopoietic cells that possess great proliferation and expansion potentials 3, 4, 5, 6, but such cells can be stored in public or private UCB banks 7, 8, 9. To date, it is estimated that over 40,000 UCB transplants (UCBT) have been performed worldwide. In pediatric patients, UCB transplantation has become a first‐line therapy for specific hematological conditions, with results comparable to those obtained when using adult marrow or mobilized peripheral blood 10. In contrast, most adult patients transplanted with UCB show prolonged periods of neutropenia and thrombocytopenia after transplant, which results in increased morbidity and mortality 11. This seems to be due, at least in part, to the fact that the absolute numbers of HSCs and HPCs in a UCB unit are significantly lower than in a bone marrow or a mobilized peripheral blood unit 12.
In the last few years, different strategies have been explored to improve UCBT in adult patients. Among them, three approaches have already been taken into the clinic, namely, double‐unit transplants, intra bone injection of cells, and the use of ex vivo‐expanded HSCs and HPCs 13, 14, 15, 16, 17, 18. Regarding the use of ex vivo‐expanded cells, significant advances have been reported. Indeed, a variety of experimental strategies has been developed for the expansion of such primitive cells in culture 19, 20, 21, 22. A common feature in all the laboratory systems reported to date is the presence of particular combinations of recombinant stimulatory cytokines that favor hematopoietic growth. These combinations usually include early acting cytokines, such as stem cell factor (SCF), the ligand for FLT‐3 (FLT3‐L), and thrombopoietin (TPO). Interestingly, other elements, such as mesenchymal stromal cells, metal chelators, Notch ligands, or small molecules that favor stem cell self‐renewal (such as UM171, OAC1, SR1, nicotinamide, and Resveratrol), have also been incorporated into some of these systems 23, 24, 25, 26, 27, 28. In fact, some of these molecules have already been tested in clinical settings or are close to being tested. It is noteworthy that when expanded cells have been taken into the clinic, the results obtained so far have been encouraging 29, 30, 31, 32.
In spite of the advances in the development of ex vivo expansion systems and the clinical achievements reported to date, several basic questions regarding the biology of the hematopoietic cells generated in culture are still unanswered. In this regard, it is still unclear to what extent those stem and progenitor cells generated in vitro retain the functional and genomic integrity of their freshly isolated counterparts. In other words, how similar are those HSCs and HPCs generated in vitro to their equivalent cell populations obtained directly from human UCB units? The fact that ex vivo expanded stem and progenitor cells are capable of engrafting and restoring hematopoiesis in immunodeficient mice seems to argue that these cells are similar, in biological terms, to their initially obtained counterparts. However, to the best of our knowledge, this hypothesis has not been conclusively demonstrated. In fact, there is extensive evidence indicating that when primitive hematopoietic cells are cultured in vitro, they experience a variety of phenotypic and functional changes induced by the culture conditions, including a tendency of HSCs to lose their stemness when they enter cell cycle, and an external pressure on HPCs to mature 33, 34, 35. Thus, even though they may retain their original immunophenotype after ex vivo expansion, their genomic and functional integrity may be altered. This notion, in fact, seems to be the reason why expanded cells transplanted into patients are always infused together with an unmanipulated UCB unit.
In trying to contribute to the solving of this issue, in the present study we have purified three different human hematopoietic cell populations: HSCs, myeloid progenitor cells (MPCs), and erythroid progenitor cells (EPCs)—obtained directly from fresh UCB units or generated in vitro under a particular culture strategy (serum‐ and stroma‐free liquid culture supplemented with human recombinant cytokines)—and compared their functional integrity in vitro and their gene expression profiles.
Materials and Methods
Cell Collection and Processing
Human UCB cells, collected according to institutional guidelines, were obtained from normal full‐term deliveries at the Troncoso Hospital (Instituto Mexicano del Seguro Social (IMSS), Mexico City), as described previously by us 36. Informed consent was obtained from every single donor.
Purification of HSCs, MPCs, and EPCs from UCB
Cells expressing the CD34 antigen (CD34+ cells) were enriched from mononuclear cells (MNCs) by negative selection 37, using the EasySep system, according to the manufacturer's instructions (StemCell Technologies Inc. [STI], Vancouver, Canada). Before negative selection, CD34+ cells corresponded to 0.7%–2.1% of MNCs; after negative selection, CD34+ cells corresponded to 67.2%–89.0% of the enriched Lineage‐negative (Lin‐) cell population. From the recovered cells, CD34+ cell subpopulations were purified by Fluorescence Activated Cell Sorting (FACS), using a FACS Aria II (BD; San Jose, California) (Fig. 1A). All the antibodies used were purchased from BD Bioscience, and were selected using the antibodies approved by EuroFlow Guide for Flow Cytometry (2011). Three different cell populations were obtained by standard FACS protocols: HSCs, defined as CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells; MPCs, defined as CD34+ CD38+ CD45RA+ CD71‐ Lin‐ cells; and EPCs, defined as CD34+ CD38+ CD45RA‐ CD71+ Lin‐ cells. These immunophenotypic criteria were based on previous studies 38. For all three cell populations, purity was >98%, as determined by FACS (Fig. 1B). Cell populations obtained in this manner were referred to as “fresh (f)” cells (e.g., fHSCs, fMPCs, fEPCs).
Figure 1.
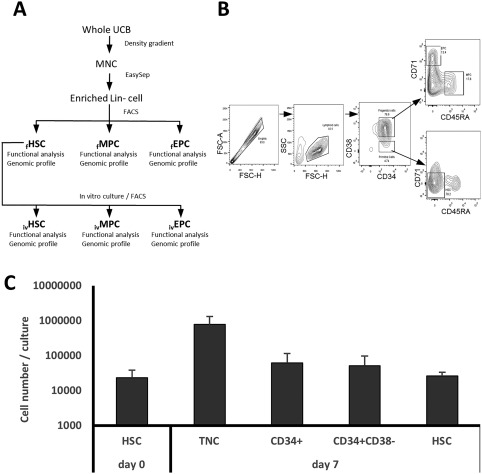
Selection of HSCs, MPCs, and EPCs and in vitro generation of HSCs. (A): Methodological strategy used in the present study for the processing of UCB units and the selection of the three cell populations obtained from fresh UCB units (f) and generated in vitro (iv). (B): FACS strategy for obtaining HSCs, MPCs, and EPCs from a CD34+ cell‐enriched fraction obtained by negative selection. (C): In vitro generation of hematopoietic cells after 7 days of coculture of fHSCs with the OP9 stroma cell line. Values shown correspond to mean ± SD (n = 9). HSC, CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells. Abbreviations: EPCs, erythroid progenitor cells; FACS, fluorescence activated cell sorting; FSC, forward scatter; HSCs, hematopoietic stem cells; MNCs, mononuclear cells; MPCs, myeloid progenitor cells; TNC, total nucleated cells; UCB, umbilical cord blood.
In Vitro Generation of HSCs, MPCs, and EPCs
Fresh HSCs were resuspended in Serum‐Free Expansion Medium (StemSpan SFEM; STI). For the in vitro generation of HSCs, fHSCs (2.3 × 104 cells/ml, in average) were cultured on an adherent cell layer of the OP9 cell line, and the culture medium was supplemented with the following recombinant hematopoietic cytokines: TPO; Flt‐3‐ligand (FL); Steel Factor (SF); Interleukin‐3 (IL‐3); IL‐6; Granulocyte‐Macrophage Colony‐Stimulating Factor (GM‐CSF), and G‐CSF. All cytokines were purchased from Peprotech (Rocky Hill, NJ) and were added at 10 ng/ml. We followed the protocol described in detail in our previous report 36. After 7 days of culture, total nucleated cells were recovered and CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells were purified by FACS.
For the in vitro generation of MPCs and EPCs, fHSCs were cultured in 24‐well tissue culture plates at 37°C in an atmosphere of 5% CO2 in air, at a concentration of 1.1 × 104 cells/ml/well, in average, using the same culture medium supplemented with TPO, FL, SCF, IL‐3, and IL‐6. Culture medium was changed on day 5, and on day 10 of culture total nucleated cells were harvested from the wells, counted, and processed for flow cytometry. MPCs and EPCs were obtained by FACS following the same protocol as for fresh cells (Fig. 1B). All cells generated in culture were referred to as “in vitro generated (iv)” cells (e.g., ivHSCs, ivMPCs, and ivEPCs).
Liquid Suspension Cultures
Hematopoietic stem and progenitor cells were cultured in liquid suspension cultures, in order to assess their proliferation, expansion, and differentiation potentials. Proliferation was defined as the capacity of a primitive cell to divide and give rise to new cells, regardless of their lineage and maturation stage. Expansion was defined as the capacity of a primitive cell to divide and give rise to new primitive cells, biologically similar to the initial cells. Differentiation was defined as the capacity of a primitive cell to divide and give rise to mature cells of specific lineages, identifiable by their morphology. Each one of the cell populations obtained was resuspended in Serum‐Free Expansion Medium (StemSpan SFEM; STI). The culture medium was supplemented with different combinations of the following recombinant hematopoietic cytokines: TPO, FL, SF, IL‐3, IL‐6, GM‐CSF, G‐CSF, and erythropoietin (EPO; R&D, Minneapolis, MN). All cytokines—except for EPO—were added at 10 ng/ml; EPO was added at 3 IU/ml. Cells were cultured in 24‐well tissue culture plates at 37°C in an atmosphere of 5% CO2 in air, at a concentration of 4–12 × 104 cells/ml/well. After 5 days of culture, cells were harvested from the wells, counted, and analyzed for progenitor cell content and morphology. Secondary cultures were initiated with 5–10 × 104 nucleated cells. Cultures were processed in this manner every 5 days.
Hematopoietic Colony Assays
Hematopoietic progenitor cells capable of forming colonies in vitro (colony‐forming cells; CFCs) were assayed in methylcellulose‐based semisolid cultures (MethoCult, STI), as previously described in detail 36.
Long‐Term Culture‐Initiating Cells
The numbers of more primitive (pre‐CFC) cells, that is, those cells capable of initiating and sustaining long‐term hematopoietic growth in culture, were determined right after each cell population was isolated. Such long‐term culture initiating cells (LTC‐ICs) were assessed based on the method established by Eaves and colleagues 39, as described previously by us 40.
Morphologic Analysis
Hematopoietic cell populations obtained from UCB, as well as cells obtained from liquid cultures, were examined on slide preparations stained with Wright‐Giemsa stain. Approximately 300 cells per slide were scored, following standard morphology criteria.
Statistics
Results shown correspond to mean ± SD of the indicated number of samples. Statistical analysis was performed by using ANOVA‐test.
Microarrays and Gene Expression Analysis
The quality of the RNA obtained from the cells was evaluated by capillary electrophoresis (Agilent 2100 Bioanalyzer; Agilent Technologies). Only RNA samples with an RNA Integrity Number greater than 6 were further processed for microarray studies. One hundred picograms of RNA from each experimental condition were evaluated in the Gene Chip Human transcriptome Array (HTA) 2.0 (Affymetrix, Santa Clara, CA). The cDNA synthesis, amplification, and gene expression profiling were done with WT Pico Reagent Kit for fresh samples (Affymetrix). Wash and stain processes were performed with the GeneChip Hybridization wash and stain kit in the GeneChip Fluid Station 450 (Affymetrix). The probe arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix). Array signal intensities were analyzed with the Affymetrix expression console. Raw data probes were normalized using Signal Space Transformation‐Robust Multichip Analysis for the background correction and quantile algorithm. To define the differential expression profile within the different cell populations, Affymetrix Transcriptome Analysis Console (TAC) software was used. Genes with fold change >2 or <–2 and with an ANOVA p‐value <.05 were considered significantly altered between the conditions (in vitro generated populations vs. freshly obtained populations). All data were uploaded in GEO (ID: GSE107497).
Enrichment Analysis
The Gene set enrichment analysis and Over‐Representation Analysis (ORA) were made using GeneTrail2 (PMID: 26787660) 41. Genes from TAC analysis with a fold‐change >2 and <–2, and ANOVA p‐value <.05 were used for the ORA with the following parameters: a false discovery rate of 0.05, and two genes per category as minimum number for all analysis.
Real Time RT‐PCR
To validate the microarray results, cDNA was generated using High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific), from RNA of cells from three different UCB samples, following the manufacturer's protocol. Samples were different from those used for gene microarrays. The following genes were selected for qPCR validation: HSCs: CDC6, KCNQ5, RUNX3 and GBP4; MPCs: VCAN, CCNB1, KCNE3 and RUNX3; EPCs: CCNB1, KCNE3, PIEZO2 and SELP. Primers design and probes selection were made using Universal Probe Library System Assay designed by Roche for qPCR. Quantitative PCR experiments were performed in a Light Cycler 480 (Roche Life Science). Primers for each gene were synthesized at T4 Oligo Company. Fold change of gene expression was determined with the 2−ΔΔCT method using GAPDH as the housekeeping gene. Statistical analysis was performed using Student's t test. For sequence primer details see Supporting Information Table S1.
Results
In Vitro Generation of HSCs, MPCs, and EPCs
We first assessed our culture conditions as experimental systems for the ex vivo generation of human hematopoietic stem and progenitor cells. Following our previous report 36, we generated HSCs in a coculture system in which fHSCs were plated on stromal cells of the OP9 cell line, and the culture medium was supplemented with a cytokine mixture that included TPO, SCF, FL, IL‐3, IL‐6, GM‐CSF, and G‐CSF. Cultures were initiated with 2.3 × 104 CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells. After 7 days of culture, 77.6 × 104 nucleated cells, in average, were generated, which represented a 33.7‐fold increase in total cell number (Fig. 1C). Of those cells, 8.0% corresponded to CD34+ cells and 6.6% to CD34+ CD38‐ cells, indicating a 2.6‐ and a 2.2‐fold increase in the respective cell numbers (Fig. 1C). Interestingly, 26,400 cells (3% of the total cells generated in culture), in average, corresponded to CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells. This represented a 1.13‐fold increase in cells with the HSC immunophenotype, as compared to day 0 (Fig. 1C).
In terms of the in vitro generation of myeloid and erythroid progenitors, it was not possible to determine the fold‐increase of such cell populations based on their immunophenotype, since the cultures were initiated with CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells (HSC immunophenotype). However, we were able to determine the number of cells giving rise to myeloid and erythroid colonies, both before and after fHSC culture for generation of progenitor cells (as described in “Materials and Methods” section). After fHSCs were cultured for 10 days in liquid suspension cultures supplemented with TPO, FL, SCF, IL‐3, and IL‐6, a 6.5‐fold expansion in erythroid CFC numbers was observed, whereas the numbers of myeloid CFCs were increased almost 64‐fold (not shown). Taken together, the above data indicate that the culture conditions that we used in the present study favored the in vitro generation of HSCs, as well as that of myeloid and erythroid progenitors.
Each one of the cell populations analyzed in this study, including those freshly obtained from UCB units and those generated in vitro, was assessed in terms of both its functional integrity in vitro (i.e., LTC‐IC and CFC content, proliferation, expansion, and differentiation potentials), as well as its gene expression profile.
In Vitro Assessment of HSCs
LTC‐IC and CFC Content
As a first approach into the functional characterization of HSCs, we determined their content of LTC‐ICs and CFCs. In the fHSC population, the frequency of LTC‐IC corresponded to 1.85% (1 LTC‐IC per 54 cells). This was a significant enrichment, considering that the frequency of LTC‐IC in the MNC fraction was 1 per 9,506 cells, and in the Lin‐ cell fraction, the frequency was 1 per 670 cells (not shown). In terms of CFCs, we found that 26.6% of CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells were capable of forming colonies in semisolid cultures (Fig. 2A). Out of those CFCs, 52% corresponded to myeloid CFCs (colonies containing granulocytes and/or macrophages), 46% corresponded to erythroid CFCs (colonies containing erythroid cells), and 2% corresponded to multipotent CFCs (colonies containing both myeloid and erythroid cells). When we analyzed ivHSCs, we found that the content of LTC‐IC was 0.12% (1 LTC‐IC per 825 cells), which was significantly reduced, as compared to fHSCs. The frequency of CFCs was 24.6%, which was very similar to that of fHSCs. Interestingly, 91% of the CFCs corresponded to myeloid progenitors, only 8% to their erythroid counterparts and 1% to multipotent progenitors (Fig. 2A). These numbers were very different to those observed for fHSCs.
Figure 2.
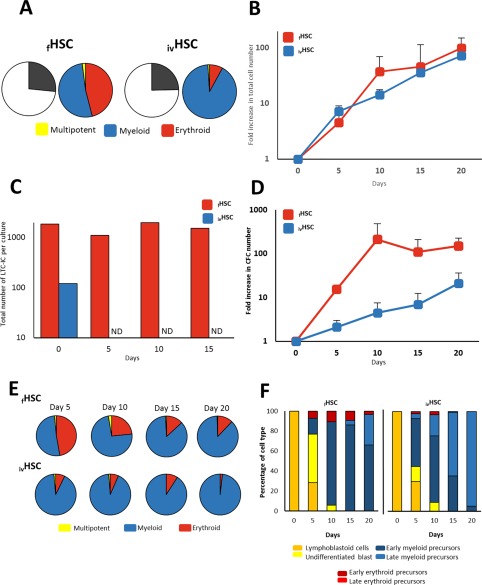
In vitro functional characterization of HSCs. (A): CFC content in the cell fractions corresponding to fHSCs and ivHSCs right after they were obtained by fluorescence activated cell sorting and before culture. Graphics represent the mean frequency (%; n = 9) of myeloid, erythroid, and multipotent progenitors. (B): Proliferation kinetics of fHSCs and ivHSCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in total nucleated cell number, as compared to day 0. (C): Kinetics of LTC‐ICs throughout liquid culture. Values (means; n = 5) represent absolute cell numbers. ND, not detected. (D): Expansion kinetics of fHSCs and ivHSCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in CFC number, as compared to day 0. (E): Frequency of myeloid, erythroid, and multipotent progenitors (as percentage of total CFC; n = 9) throughout liquid cultures of fHSCs and ivHSCs. (F): Frequency (%; n = 3) of the different hematopoietic cell types, recognized by their morphology, generated in cultures of fHSCs and ivHSCs. Abbreviations: CFCs, colony‐forming cells; HSCs, hematopoietic stem cells; LTC‐ICs, long‐term culture initiating cells.
HSC Growth in Liquid Suspension Cultures
We next assessed the proliferation, expansion and differentiation potentials of HSCs. As shown in Figure 2B, both fHSCs and ivHSCs showed similar proliferation patterns throughout the 20‐day culture period, reaching their highest levels (99‐ and 72‐fold increase, respectively) by day 20 of culture. On most of the time points analyzed, the total nucleated cell numbers were higher in cultures of fHSCs; however, such differences were not statistically significant. In terms of their expansion potentials (i.e., LTC‐IC and CFC levels throughout the culture period), we observed that the absolute numbers of LTC‐ICs in cultures of fHSCs were sustained throughout the culture period (Fig. 2C). In contrast, the levels of LTC‐ICs in cultures of ivHSCs abruptly declined, so this type of cell was undetectable from day 5 of culture (Fig. 2C). In terms of progenitor cells, the levels of CFCs were always higher in cultures of fHSCs as compared to ivHSCs. Although such differences were not statistically significant, the tendency was clear (Fig. 2D). It is noteworthy that throughout the culture period there was a marked tendency toward the production of progenitors giving rise to colonies containing granulocytes and/or macrophages. Indeed, by day 20 of culture, myeloid CFCs corresponded to almost 90% of the CFCs in cultures of fHSCs, and to almost 100% of CFCs in cultures of ivHSCs (Fig. 2E). Erythroid progenitors became a minority, whereas multipotent progenitors were detected only in cultures of fHSCs, and at extremely low levels. In keeping with this observation, the differentiation patterns of both fresh and in vitro generated HSCs showed a marked tendency toward the production of myeloid precursors. Such a tendency seemed to be more pronounced in ivHSCs. As shown in Figure 2F, all of the cells observed on day 20 in cultures of ivHSCs consisted of myeloid precursors (the majority of them were late precursors), whereas in cultures of fHSCs, a small proportion of early erythroid precursors was still observed on that same day. No late erythroid precursors were observed, most likely due to the absence of EPO in liquid cultures.
Global Gene Expression Profiles of HSCs
We next performed a global comparison of the gene expression profiles from both fresh and in vitro generated HSCs. By using Affymetrix Gene Chip HTA 2.0, we assessed the expression of over 18,000 genes and clearly observed significant differences in such profiles (Fig. 3A). We found 541 genes differentially expressed (Fig. 3B). Of these, 243 were upregulated in ivHSCs (including CDC6, BRIP1, TOP2A, KCNQ5, DNMT3B, and ANGPT1; Supporting Information Table S2) and 298 were downregulated (including IL1RL1, GNG4, ICAM1, SETBP1, GBP2, SMAD3, and CXCL8; Supporting Information Table S2). KEGG and GO analyses indicated that NFkB, TNF, JAK‐STAT, HIF, and PI3K‐AKT signaling pathways were significantly upregulated in ivHSCs, and that cell cycle, apoptosis, leukocyte activation, myeloid differentiation, and chromatin remodeling were among the most upregulated biological processes (Fig. 3C). No pathways or processes were found to be downregulated at significant levels (not shown). In order to validate these results, we selected four genes (CDC6, KCNQ5, RUNX3, and GBP4) and quantitated their levels by qRT‐PCR (Supporting Information Fig. S1).
Figure 3.
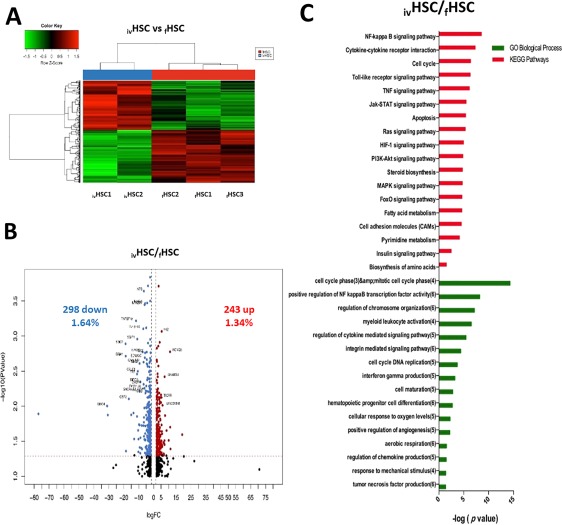
Differentially expressed genes between ivHSCs and fHSCs. (A): Hierarchical cluster showing the genes differentially expressed between ivHSCs and fHSCs. Samples are in columns, genes are in rows. Both cell populations were compared using the TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (B): The volcano plot shows 541 genes differentially expressed (243 genes upregulated ‐red‐ and 298 downregulated ‐blue‐ in ivHSCs). The analysis was made using TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (C): Most highly represented biologic processes/pathways that were upregulated in ivHSCs as compared to fHSCs. Functional annotation in Gene Ontology categories was made using GeneTrail2, and were classified in biological processes and KEGG categories. Abbreviation: HSCs, hematopoietic stem cells.
In Vitro Assessment of MPCs
LTC‐IC and CFC Content
Following a similar approach to the one used for HSCs, we determined the content of LTC‐ICs and CFCs in the MPC population. LTC‐ICs were not detected in any of the MPC populations analyzed (fresh or in vitro generated; not shown). This is in keeping with the fact that within the hematopoietic hierarchy, LTC‐IC corresponds to a cell stage earlier than the committed progenitor stage 39. In terms of CFCs, we found that their frequencies were very similar in fresh and in in vitro generated MPCs (21.4% and 20.6% of the cells in each cell fraction, respectively; Fig. 4A). Among those CFCs, the vast majority (93%–99%) corresponded to cells producing colonies containing granulocytes and/or macrophages, and only 1%–7% corresponded to small erythroid colonies (Fig. 4A). No mixed colonies were observed. These results are also in keeping with the fact that MPCs correspond to a cell fraction enriched for myeloid progenitors.
Figure 4.
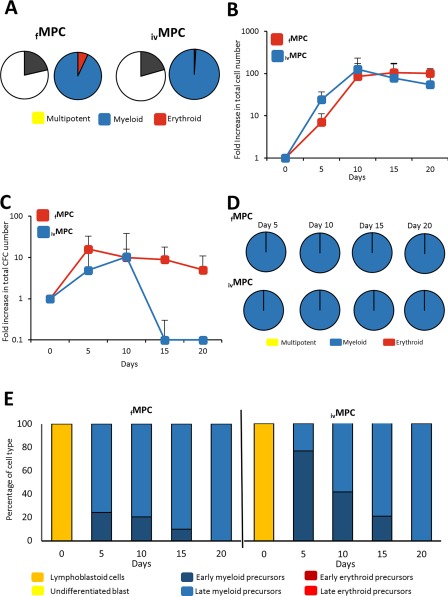
In vitro functional characterization of MPCs. (A): CFC content in the cell fractions corresponding to fMPCs and ivMPCs right after they were obtained by fluorescence activated cell sorting and before culture. Graphics represent the mean frequency (%; n = 9) of myeloid, erythroid, and multipotent progenitors. (B): Proliferation kinetics of fMPCs and ivHSCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in total nucleated cell number, as compared to day 0. (C): Expansion kinetics of fMPCs and ivMPCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in CFC number, as compared to day 0. (D): Frequency of myeloid, erythroid, and multipotent progenitors (as percentage of total CFC; n = 9) throughout liquid cultures of fMPCs and ivMPCs. (E): Frequency (%; n = 3) of the different hematopoietic cell types, recognized by their morphology, generated in cultures of fMPCs and ivMPCs. Abbreviations: CFCs, colony‐forming cells; HSCs, hematopoietic stem cells; MPCs, myeloid progenitor cells.
MPC Growth in Liquid Suspension Cultures
The proliferation potentials of both MPCs were similar, reaching a 105‐ and 126‐fold‐increase in total cell number, respectively (Fig. 4B). From day 10 of culture, the levels of total cells remained relatively constant. In contrast to the kinetics of total cells, CFCs from fMPCs and ivMPCs showed very different kinetics (Fig. 4C). At all time points analyzed, except for day 10, CFC levels were higher in cultures of fMPCs; in fact, by day 15, CFC levels in ivMPCs were extremely low. It is noteworthy that from day 5 of culture, only colonies containing granulocytes and/or macrophages were observed; no erythroid or mixed colonies were detected (Fig. 4D). As shown in Figure 4E, at culture onset, MPCs consisted of lymphoblastoid cells; however, and in keeping with the fact that only granulocyte/macrophage colonies were present in culture, from day 5 of culture only myeloid precursors were observed.
Global Gene Expression Profiles of MPCs
When we compared the gene expression profiles of both fresh and in vitro generated MPCs (Fig. 5A), we found 1,086 genes differentially expressed (Fig. 5B). Of these, 859 were upregulated in ivMPCs (including VCAN, MRC1, FGL2, ADAMDEC1, and RNASE6; Supporting Information Table S3) and 227 were downregulated (including SKIL, CCR7, MED12L, SETBP1, and CD38; Supporting Information Table S3). KEGG and GO analyses indicated that Lysosome, Phagosome, JAK‐STAT, PI3K‐AKT, Toll‐like receptor signaling, and CAMs signaling pathways were significantly upregulated in ivMPCs, and that myeloid leukocyte migration, granulocyte chemotaxis, TNF family production, myeloid leukocyte‐mediated immunity, and phagosome maturation were among the most upregulated biological processes (Fig. 5C). No pathways or processes were found to be downregulated at significant levels (not shown). In order to validate these results, we selected four genes (VCAN, CCNB1, KCNE3, and RUNX3) and quantitated their levels by qRT‐PCR (Supporting Information Fig. S1).
Figure 5.
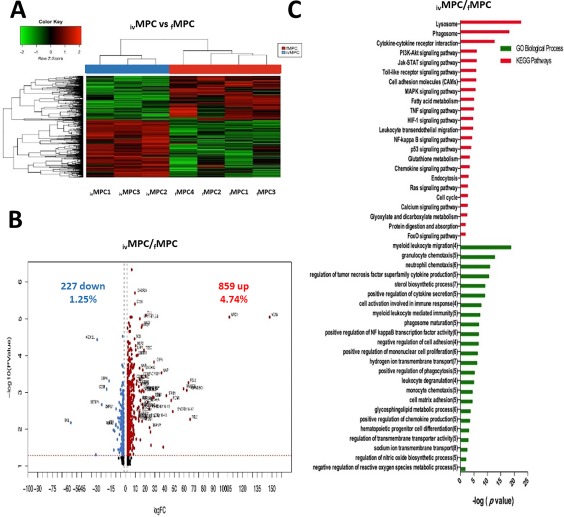
Differentially expressed genes between ivMPCs and fMPCs. (A): Hierarchical cluster showing the genes differentially expressed between ivMPCs and fMPCs. Samples are in columns, genes are in rows. Both cell populations were compared using the TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (B): The volcano plot shows 1,086 genes differentially expressed (859 genes upregulated ‐red‐ and 227 downregulated ‐blue‐ in ivMPCs). The analysis was made using TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (C): Most highly represented biologic processes/pathways that were upregulated in ivMPCs as compared to fMPCs. Functional annotation in Gene Ontology categories was made using GeneTrail2, and were classified in biological processes and KEGG categories. Abbreviation: MPCs, myeloid progenitor cells.
In Vitro Assessment of EPCs
LTC‐IC and CFC Content
As expected, we were unable to detect LTC‐ICs in the EPC population (not shown), which is in keeping with the hierarchical organization of hematopoiesis and with the results obtained for MPCs. Thirty five percent of the cells in the fEPC fraction were capable of forming colonies in semisolid cultures and the vast majority (95%) corresponded to colonies containing exclusively erythroid cells (Fig. 6A). For ivEPCs, the levels of CFCs (24%) were lower than in fEPCs, and more than 95% of them also formed erythroid colonies (Fig. 6A). Myeloid colonies consisted of small granulocyte and/or macrophage clusters. No multipotent (mixed) colonies were detected.
Figure 6.
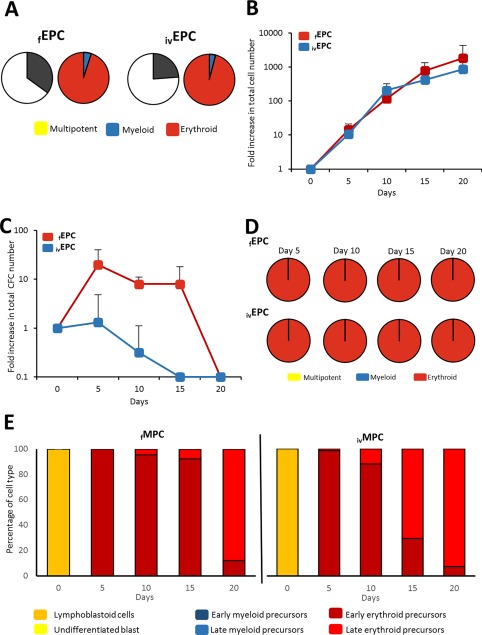
In vitro functional characterization of EPCs. (A): CFC content in the cell fractions corresponding to fEPCs and ivEPCs right after they were obtained by fluorescence activated cell sorting and before culture. Graphics represent the mean frequency (%; n = 9) of myeloid, erythroid, and multipotent progenitors. (B): Proliferation kinetics of fEPCs and ivEPCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in total nucleated cell number, as compared to day 0. (C): Expansion kinetics of fEPCs and ivEPCs throughout liquid culture. Values (mean ± SD; n = 5) represent the increments in CFC number, as compared to day 0. (D): Frequency of myeloid, erythroid, and multipotent progenitors (as percentage of total CFC; n = 9) throughout liquid cultures of fEPCs and ivEPCs. (E): Frequency (%; n = 3) of the different hematopoietic cell types, recognized by their morphology, generated in cultures of fEPCs and ivEPCs. Abbreviations: CFCs, colony‐forming cells; EPCs, erythroid progenitor cells; MPCs, myeloid progenitor cells.
EPC Growth in Liquid Suspension Cultures
Both fEPCs and ivEPCs showed very high proliferation potentials in liquid cultures, reaching higher levels of total cells than the ones observed for HSCs or MPCs (Fig. 6B). The growth patterns were similar in both erythroid cell populations; however, it is noteworthy that on days 15 and 20 of culture, total nucleated cell numbers were much higher in cultures of fEPCs, although such differences were not statistically significant. In contrast, expansion kinetics were very different. Indeed, whereas a significant increase in CFCs was observed in cultures of fEPCs, particularly during the first 15 days of culture, there was no expansion in cultures of ivEPCs (Fig. 6C). From day 5 to day 20 of culture, only colonies containing erythroid cells were observed (no myeloid or mixed colonies were observed; Fig. 6D), and in keeping with this, from day 5 of culture only erythroid precursors were detected in culture (Fig. 6E). During the first days of culture the majority of the erythroid cells corresponded to early precursors (proerythroblasts and basophilic erythroblasts), whereas by day 20, the vast majority corresponded to late erythroblasts (polychromatic and orthochromatic erythroblasts).
Global Gene Expression Profiles of EPCs
We, then, compared the gene expression profiles of both fresh and in vitro generated EPCs (Fig. 7A), and found 438 genes differentially expressed (Fig. 7B). Of these, 196 were upregulated in ivEPCs (including SIGLEC6, EPX, PPBP, RAP27B, and HBBP1; Supporting Information Table S4) and 242 were downregulated (including GBP4, SKIL, THBS1, CRHBP, and CSF1; Supporting Information Table S4). KEGG and GO analyses indicated that PI3K‐Akt, NF‐kB, Jak‐STAT, chemokine signaling, and TNF signaling pathways were significantly upregulated in ivEPCs, and that sterol biosynthesis, cell matrix adhesion, cellular response to oxygen levels, negative regulation of cell growth, cell maturation, and several metabolic routes were among the most upregulated biological processes (Fig. 7C). No pathways or processes were found to be downregulated at significant levels (not shown). In order to validate these results, we selected four genes (CCNB1, KCNE3, PIEZO2, and SELP) and quantitated their levels by qRT‐PCR (Supporting Information Fig. S1).
Figure 7.
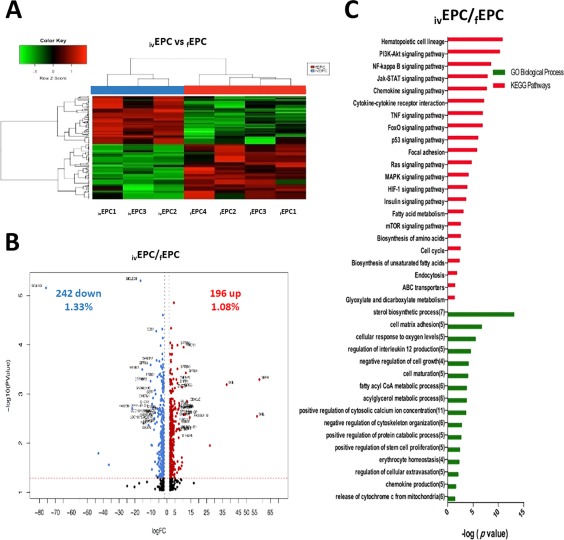
Differentially expressed genes between ivEPCs and fEPCs. (A): Hierarchical cluster showing the genes differentially expressed between ivEPCs and fEPCs. Samples are in columns, genes are in rows. Both cell populations were compared using the TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (B): The volcano plot shows 438 genes differentially expressed (196 genes upregulated ‐red‐ and 242 downregulated ‐blue‐ in ivEPCs). The analysis was made using TAC Affymetrix software, considering a fold change of 2.0 and p < .05. (C): Most highly represented biologic processes/pathways that were upregulated in ivEPCs as compared to fEPCs. Functional annotation in Gene Ontology categories was made using GeneTrail2, and were classified in biological processes and KEGG categories. Abbreviation: EPCs, erythroid progenitor cells.
Discussion
Great interest on the ex vivo generation of primitive hematopoietic cells, including self‐renewing stem cells and multipotent and committed progenitors, has grown over the last few years, due to both biologic and clinical reasons. This is particularly true for UCB‐derived HSCs and HPCs, since UCB transplantation has become a major therapeutic approach for a wide variety of hematologic diseases 2, 12. In this regard, different culture systems and experimental approaches have been developed, and significant achievements have already been reported 19, 20, 21, 22, 23, 24, 25, 26, 27, 28. To date, however, it is still unclear to what extent those stem and progenitor cells generated in vitro retain the functional and genomic integrity of their freshly isolated counterparts. This yet unanswered question possesses both academic and practical implications. In trying to contribute to answering the above question, in the present study we obtained fresh human UCB units and isolated HSCs, MPCs, and EPCs. We, then, determined their content of LTC‐ICs and CFCs, assessed their in vitro proliferation, expansion, and differentiation capacities under particular culture conditions, and determined their global gene expression profiles. Immunophenotipically similar cell populations were generated in vitro, by using specific culture systems, and then were analyzed in the same manner. Thus, a comparative functional and genomic study of both fresh and in vitro generated cell populations was performed.
It has long been recognized that the in vitro growth of hematopoietic cells depends on the input cell population and the culture conditions used 20, 34. Based on our previous report 36, in the present study, we generated HSCs by using a culture system in which fHSCs were cultured with the OP9 stromal cell line and the culture medium was supplemented with a combination of seven different early‐ and late‐acting hematopoietic cytokines. In our previous report, the input cell population consisted of CD34+ CD38‐ Lin‐ cells, and we observed a >20‐fold increase in the number of such cells after culture. Herein, our input cell population consisted of CD34+ CD38‐ CD45RA‐ CD71‐ Lin‐ cells, and after the culture period, we did not observe a significant increase in such a cell number, but a maintenance. In contrast, we did observe a significant increase in the levels of myeloid and erythroid CFCs. On the other hand, by culturing fHSCs in a stroma‐free liquid culture system supplemented with five early‐acting cytokines, we achieved the generation of significant numbers of CD34+ CD38+ CD45RA+ CD71‐ Lin‐ and CD34+ CD38+ CD45RA‐ CD71+ Lin‐ cells. Taken together, these results indicate that our culture systems were efficient in generating committed myeloid and EPCs and were able to sustain HSCs over input levels.
In spite of their identical immunophenotypes, it was clear that fresh and in vitro generated cells showed significant differences in terms of their biology. This was particularly evident for the HSC populations. Indeed, the frequency of LTC‐ICs in the ivHSC population was significantly reduced, as compared to fHSCs, and their levels were undetectable after 5 days of culture, whereas LTC‐IC levels in cultures of fHSCs were sustained throughout the culture period. It was also evident that whereas fHSCs showed a “balanced” frequency of myeloid and erythroid CFCs, the vast majority of the CFCs detected in the ivHSC population corresponded to myeloid progenitors. Work by different groups, including those of Muller‐Sieburg and colleagues, Eaves and colleagues, and Suda and colleagues, has demonstrated that among murine HSCs, some clones are biased toward the production of myeloid cells, some are biased toward the production of lymphoid cells, whereas others show a balanced capacity toward the production of both myeloid and lymphoid cells 42, 43, 44, 45. Although the presence of such HSC subpopulations is less clear in humans, our results seem to suggest that whereas most of the HSCs obtained from fresh UCB units correspond to a balanced myeloid‐erythroid subtype, the majority of the HSCs generated in our culture system are biased toward the myeloid lineage. Interestingly, and in keeping with previous reports 38, a marked tendency toward the myeloid lineage was observed once HSCs were put into culture, regardless of whether HSCs were fresh or generated in vitro.
In all three populations analyzed (i.e., HSCs, MPCs, and EPCs), both fresh and in vitro generated cells showed a similar proliferation capacity (ability to divide and produce new cells, regardless of their lineage and maturation stage). However, their expansion potentials (ability of primitive cells to divide and give rise to more primitive cells) were clearly different. Although the reason for this is not clear, one possibility is that there is an increased proliferation activity at the level of the precursor cells generated in vitro; thus, although the levels of CFCs in cultures of in vitro generated cells were reduced, as compared to their fresh counterparts, the total numbers of cells generated in culture were similar.
Global gene expression analysis of each one of the cell populations included in the present study also demonstrated significant differences between fresh and in vitro generated cells. The latter showed consistent increased expression of particular signaling pathways, such as JAK‐STAT, PI3K‐AKT, NFkB, and HIF‐1, and processes such as cell cycle, apoptosis, and myeloid differentiation, which is in keeping with their proliferation, expansion, and differentiation patterns in culture. In a recent study by Papayannopoulou and colleagues, they observed that when CD34+ cells from adult mobilized peripheral blood were cultured for 7 days in the presence of a combination of recombinant cytokines and small molecules, those cells generated in culture that showed reduced expression of Rap‐1, Ras, and PI3K‐Akt showed a higher hematopoietic performance in vivo, as compared to those with a higher expression 46. It is interesting that in the present study, in all cell populations generated after culture, the levels of PI3K‐Akt were increased. This was, in fact, a consistent finding. So, this may be one of the reasons for the reduced expansion potential of such cells, as compared to their fresh counterparts.
In the last few years, several groups have reported the use of small molecules, including UM171, OAC1, SR1, and Resveratrol, among others, that have been shown to favor the self‐renewal activity of HSCs when used in combination with early‐acting recombinant hematopoietic cytokines 27, 28, 31, 47, 48. Indeed, evidence exists that a particular combination of small molecules and cytokines favors the expansion of CD34+ and CD34+CD38‐ cells by enhancing the Notch pathway—which favors retention of stemness—and by inhibiting the Wnt pathway, which results in blocking of the differentiation process 49. In the present study, we did not include any of those small molecules as part of the culture system. Thus, it would be interesting to include them in future studies and to perform similar analyses to the one described here.
It is noteworthy that in the present study, the functional integrity of the cells analyzed was assessed by using in vitro systems only; no in vivo systems were used. Thus, we are not certain as to the functional in vivo capacities of the fresh and in vitro generated cells included herein. It should also be considered that in the present study, our main criteria for the identification, selection, and purification of the different cell populations was their immunophenotype. And to this purpose, we followed well‐defined, standard criteria. However, it has been shown that the relationship of stem cell immunophenotype and function is altered when primitive hematopoietic cells are cultured in serum‐free liquid suspension cultures 33, 35, 50. Thus, it is possible that when selecting the cell populations generated in vitro, we were gating‐out cells that possess functional capacities related to HSCs and progenitor cells, but whose immunophenotypes do not correspond to those considered as standard phenotypes of HSCs and HPCs. This, of course, is a very important issue that should be taken into account when developing protocols for the ex vivo expansion of primitive hematopoietic cells.
In summary, in the present study we have selected and purified three different cell populations (HSCs, MPCs, and EPCs) obtained directly from fresh human UCB units or generated in vitro under specific culture conditions. We, then, compared their functional integrity in vitro and their gene expression profiles. Our results indicate that in spite of being immunophenotipically similar, fresh and in vitro generated cells showed significant differences, both in functional and genetic terms. In general, those HSCs generated in culture showed a deficient content of LTC‐ICs, and a marked tendency toward the myeloid lineage. In addition, in vitro generated HSCs and HPCs showed limited expansion potentials. Such functional alterations correlated with changes in their gene expression profiles. Our results, however, need to be taken with caution, since in the present study we used a single ex vivo culture strategy (serum‐ and stroma‐free liquid culture supplemented with different combinations of recombinant stimulatory cytokines). Thus, we are not certain as to the functional integrity and gene expression profiles of the same cell populations generated under different culture conditions (e.g., in the presence of stroma cells and/or small molecules such as UM171, OAC1, SR1, nicotinamide, and Resveratrol).
Nevertheless, our observations may have clinical implications in UCB transplantation. Indeed, in all clinical trials reported so far on the use of expanded cells for UCBT, such cells have been infused together with non‐manipulated cells and in the vast majority of cases, it has been the latter the ones that remain long‐term in patients. It is, thus, possible that, as suggested by the results of the present study, expanded cells do not retain the complete functional and genomic integrity of non‐manipulated cells, so that they are at disadvantage when competing under physiological conditions with their fresh counterparts.
Author Contributions
R.D.‐M.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, final approval; P.F.‐G.: data analysis and interpretation, final approval; J.C.‐N., I.M.‐G., and L.A.‐P.: collection and assembly of data, final approval; A.H.‐M.: financial support, data analysis and interpretation, final approval; F.O.B.‐A. and A.C.‐T.: collection and assembly of data, data analysis and interpretation, final approval; O.B.‐O.: provision of study patients, final approval; H.M.: conception and design, financial support, data analysis and interpretation, manuscript writing, final approval.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Supporting Information Figure S1
Supporting Information Figure 1
Supporting Information Tables
Acknowledgments
This study was supported by Grant FIS/IMSS/PROT/PRIO/14/032 from the Mexican Institute of Social Security (IMSS) to H.M. Roberto Dircio‐Maldonado was a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and he received fellowships from CONACYT (fellowship no. 445062) and from the Mexican Institute of Social Security (IMSS; fellowship no. 038–2012). We thank Luis Enrique Torres‐Galeana and Ignacio Valencia‐Plata for excellent technical assistance. We would also like to thank the staff at the Flow Cytometry Core Facility (Coordinacion de Investigacion en Salud, IMSS) for their technical assistance and support.
This article was published online on 26 April 2018. The logo for the Cord Blood Association incorrectly appeared on the article and has now been removed because the authors are not members of the association. This notice is included in the online version to indicate this version was corrected on 7 May 2018.
References
- 1. Gluckman E. History of cord blood transplantation. Bone Marrow Transplant 2009;44:621–626. [DOI] [PubMed] [Google Scholar]
- 2. Ballen KK, Gluckman E, Broxmeyer HE. Umbilical cord blood transplantation: The first 25 years and beyond. Blood 2013;122:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Broxmeyer HE, Hangoc G, Cooper S et al. Growth characteristics and expansion of human umbilical cord blood and estimation of its potential for transplantation in adults. Proc Natl Acad Sci USA 1992;89:4109–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lansdorp PM, Dragowska W, Mayani H. Ontogeny‐related changes in proliferative potential of human hematopoietic cells. J Exp Med 1993;178:787–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vormoor J, Lapidot T, Pflumio F et al. Immature human cord blood progenitors engraft and proliferate to high levels in severe combined immunodeficient mice. Blood 1994;83:2489–2497. [PubMed] [Google Scholar]
- 6. Mayani H. Biological differences between neonatal and adult human hematopoietic stem/progenitor cells. Stem Cells Dev 2010;19:285–298. [DOI] [PubMed] [Google Scholar]
- 7. Navarrete C, Contreras M. Cord blood banking: A historical perspective. Br J Haematol 2009;147:236–245. [DOI] [PubMed] [Google Scholar]
- 8. Murphy A, McKenna D, McCullough J. Cord blood banking and quality issues. Transfusion 2016;56:645–652. [DOI] [PubMed] [Google Scholar]
- 9. Mayani H. Umbilical Cord Blood: Lessons learned and lingering challenges after more than 20 years of basic and clinical research. Arch Med Res 2011;42:645–651. [DOI] [PubMed] [Google Scholar]
- 10. Rocha V, Kabbara N, Ionescu I et al. Pediatric related and unrelated cord blood transplantation for malignant diseases. Bone Marrow Transplant 2009;44:653–659. [DOI] [PubMed] [Google Scholar]
- 11. Ooi J. Cord blood transplantation in adults. Bone Marrow Transplant 2009;44:661–666. [DOI] [PubMed] [Google Scholar]
- 12. Stanevsky A, Goldstein G, Nagler A. Umbilical cord blood transplantation: Pros, cons and beyond. Blood Rev 2009;23:199–204. [DOI] [PubMed] [Google Scholar]
- 13. Kelly SS, Parmar S, de Lima M et al. Overcoming the barriers to umbilical cord blood transplantation. Cytotherapy 2010;12:121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Delaney C, Ratajczak MZ, Laughlin M. Strategies to enhance umbilical cord blood engraftment in adult patients. Expert Rev Hematol 2010;3:273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brunstein CG, Setubal DC, Wagner JE. Expanding the role of umbilical cord blood transplantation. Br J Haematol 2007;137:20–35. [DOI] [PubMed] [Google Scholar]
- 16. Haspel RL, Ballen KK. Double cord blood transplants: Filling a niche? Stem Cell Rev 2006;2:81–86. [DOI] [PubMed] [Google Scholar]
- 17. Frassoni F, Gualandi F, Podesta M et al. Direct intrabone transplant of unrelated cord blood cells in acute leukaemia: A phase I/II study. Lancet Oncol 2008;9:831–839. [DOI] [PubMed] [Google Scholar]
- 18. Brunstein CG, Barker JN, Weisdorf DJ et al. Intra BM‐injection to enhance engraftment after myeloablative umbilical cord blood transplantation with two partially HLA‐matched units. Bone Marrow Transplant 2009;43:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dahlberg A, Delaney C, Bernstein ID. Ex vivo expansion of human hematopoietic stem and progenitor cells. Blood 2011;117:6083–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flores‐Guzmán P, Fernández‐Sánchez V, Mayani H. Ex vivo expansion of cord blood‐derived hematopoietic stem and progenitor cells: Basic principles, experimental approaches, and impact in regenerative medicine. Stem Cells Translational Medicine 2013;2:830–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pineault N, Abu‐Khader A. Advances in umbilical cord blood stem cell expansion and clinical translation. Exp Hematol 2015;43:498–513. [DOI] [PubMed] [Google Scholar]
- 22. Baron F, Ruggeri A, Nagler A. Methods of ex vivo expansion of human cord blood cells: Challenges, successes, and clinical implications. Expert Rev Hematol 2016;9:297–314. [DOI] [PubMed] [Google Scholar]
- 23. Peled T, Mandel J, Goudsmid RN et al. Pre‐clinical development of cord blood‐derived progenitor cell graft expanded ex vivo with cytokines and the polyamine copper chelator tetraethylenepentamine. Cytotherapy 2004;6:344–355. [DOI] [PubMed] [Google Scholar]
- 24. Robinson SN, Ng J, Niu T et al. Superior ex vivo cord blood expansion following co‐culture with bone marrow‐derived mesenchymal stem cells. Bone Marrow Transplant 2006;37:359–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Flores‐Guzman P, Flores‐Figueroa E, Montesinos JJ et al. Individual and combined effects of mesenchymal stromal cells and recombinant stimulatory cytokines on the in vitro growth of primitive hematopoietic cells from human umbilical cord blood. Cytotherapy 2009;11:886–896. [DOI] [PubMed] [Google Scholar]
- 26. Boitano AE, Wang J, Romeo R et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science 2010;329:1345–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fares I, Chagraoui J, Gareau Y et al. Cord blood expansion. Pyrimidoindole derivatives are agonists of human hematopoietic stem cell self‐renewal. Science 2014;345:1509–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang X, Lee M‐R, Cooper S et al. Activation of OCT4 enhances ex vivo expansion of human cord blood hematopoietic stem and progenitor cells by regulating HOXB4 expression. Leukemia 2016;30:144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Delaney C, Heimfeld S, Brashem‐Stein C et al. Notch‐mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med 2010;16:232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Lima MR, McNiece I, Robinson SN et al. Cord blood engraftment with ex vivo mesenchymal‐cell coculture. N Engl J Med 2012;367:2305–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner JE, Brunstein CG, Boitano AE et al. Phase I/II trial of StemRegenin‐1 expanded umbilical cord blood hematopoietic stem cells supports testing as a stand‐alone graft. Cell Stem Cell 2016;18:144–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anand S, Thomas S, Hyslop T et al. Transplantation of ex vivo expanded umbilical cord blood (NiCord) decreases early infection and hospitalization. Biol Blood Marrow Transplant 2017;23:1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dorrell C, Gan OI, Pereira DS et al. Expansion of human cord blood CD34+ CD38‐ cells in ex vivo culture during retroviral transduction without a corresponding increase in SCID‐repopulating cell (SRC) frequency: Dissociation of SCR phenotype and function. Blood 2000;95:102–110. [PubMed] [Google Scholar]
- 34. Douay L. Experimental culture conditions are critical for ex vivo expansion of hematopoietic cells. J Hematother Stem Cell Res 2001;10:341–346. [DOI] [PubMed] [Google Scholar]
- 35. McKenzie JL, Gan OI, Doedens M et al. Reversible cell surface expression of CD38 on CD34‐positive human hematopoietic repopulating cells. Exp Hematol 2007;35:1429–1436. [DOI] [PubMed] [Google Scholar]
- 36. Fernandez‐Sanchez V, Pelayo R, Flores‐Guzman P et al. In vitro effects of stromal cells expressing different levels of Jagged‐1 and Delta‐1 on the growth of primitive and intermediate CD34+ cell subsets from human cord blood. Blood Cells Mol Dis 2011;47:205–213. [DOI] [PubMed] [Google Scholar]
- 37. Wognum AW, Eaves AC, Thomas TE. Identification and isolation of hematopoietic stem cells. Arch Med Res 2003;34:461–475. [DOI] [PubMed] [Google Scholar]
- 38. Mayani H, Dragowska W, Lansdorp PM. Characterization of functionally distinct subpopulations of CD34+ cord blood cells in serum‐free long‐term cultures supplemented with hematopoietic cytokines. Blood 1993;82:2664–2672. [PubMed] [Google Scholar]
- 39. Miller CL, Eaves CJ. Long‐term culture initiating‐cell assays for human and murine cells In: Klug CA, Jordan CT, eds. Hematopoietic Stem Cell Protocols. Totowa, NJ: Humana Press, 2002:123–141. [DOI] [PubMed] [Google Scholar]
- 40. Flores‐Guzman P, Fernandez‐Sanchez V, Valencia‐Plata I et al. Comparative in vitro analysis of different hematopoietic cell populations from human cord blood: In search of the best option for clinically oriented ex vivo cell expansion. Transfusion 2013;53:668–678. [DOI] [PubMed] [Google Scholar]
- 41. Stöckel D, Kehl T, Trampert P et al. Multi‐omics enrichment analysis using the GeneTrail2 web service. Bioinformatics 2016;32:1502–1508. [DOI] [PubMed] [Google Scholar]
- 42. Muller‐Sieburg CE, Cho RH, Karlsson L et al. Myeloid‐biased hematopoietic stem cells have extensive self‐renewal capacity but generate diminished lymphoid progeny with impaired IL‐7 responsiveness. Blood 2004;103:4111–4118. [DOI] [PubMed] [Google Scholar]
- 43. Muller‐Sieburg CE, Sieburg HB. The GOD of hematopoietic stem cells: A clonal diversity model of the stem cell compartment. Cell Cycle 2006;5:394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dykstra B, Kent D, Bowie M et al. Long‐term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell 2007;1:218–229. [DOI] [PubMed] [Google Scholar]
- 45. Ema H, Morita Y, Suda T. Heterogeneity and hierarchy of hematopoietic stem cells. Exp Hematol 2014;42:74–82. [DOI] [PubMed] [Google Scholar]
- 46. Psatha N, Georgolopoulos G, Phelps S et al. Brief report: A differential transcriptomic profile of ex vivo expanded adult human hematopoietic stem cells empowers them for engraftment better than their surface phenotype. Stem Cells Translational Medicine 2017;6:1852–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Heinz N, Ehrnstrom B, Schambach A et al. Comparison of different cytokine conditions reveals resveratrol as a new molecule for ex vivo cultivation of cord blood‐derived hematopoietic stem cells. Stem Cells Translational Medicine 2015;4:1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fares I, Chagraoui J, Lehnertz B et al. EPCR expression marks UM171‐expanded CD34+ cord blood stem cells. Blood 2017;129:3344–3351. [DOI] [PubMed] [Google Scholar]
- 49. Wang L, Guan X, Wang H et al. A small‐molecule/cytokine combination enhances hematopoietic stem cell proliferation via inhibition of cell differentiation. Stem Cell Res Ther 2017;8:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Danet GH, Lee HW, Luongo JL et al. Dissociation between stem cell phenotype and NOD/SCID repopulating activity in human peripheral blood CD34+ cells after ex vivo expansion. Exp Hematol 2001;29:1465–1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1
Supporting Information Figure 1
Supporting Information Tables


