Abstract
Arsenic is a known potent risk factor for bladder cancer. Increasing evidence suggests that epigenetic alterations, e.g., DNA methylation and histones posttranslational modifications (PTMs), contribute to arsenic carcinogenesis. Our previous studies have demonstrated that exposure of human urothelial cells (UROtsa cells) to monomethylarsonous acid (MMAIII), one of arsenic active metabolites, changes the histone acetylation marks across the genome that are correlated with MMAIII-induced UROtsa cell malignant transformation. In the current study, we employed a high-resolution and high-throughput liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify and quantitatively measure various PTM patterns during the MMAIII-induced malignant transformation. Our data showed that MMAIII exposure caused a time-dependent increase in histone H3 acetylation on lysine K4, K9, K14, K18, K23, and K27, but a decrease in acetylation on lysine K5, K8, K12, and K16 of histone H4. Consistent with this observation, H3K18ac was increased while H4K8ac was decreased in the leukocytes collected from people exposed to high concentrations of arsenic compared to those exposed to low concentrations. MMAIII was also able to alter histone methylation patterns: MMAIII transformed cells experienced a loss of H3K4me1, and an increase in H3K9me1 and H3K27me1. Collectively, our data shows that arsenic exposure causes dynamic changes in histone acetylation and methylation patterns during arsenic-induced cancer development. Exploring the genomic location of the altered histone marks and the resulting aberrant expression of genes will be of importance in deciphering the mechanism of arsenic-induced carcinogenesis.
Keywords: Arsenic, Cell malignant transformation, Histone acetylation and methylation, LC-MS/MS based histone modification analysis
Introduction
Chronic arsenic (As) exposure in drinking water afflicts >140 million people in 70+ countries and contributes to various chronic diseases, notably multiple types of cancer (Smith and Steinmaus, 2009; Naujokas et al., 2013). Arsenic exposure is an established cause of bladder cancer (BCa) (Steinmaus et al., 2003; Bates et al., 2004; Chen and Ahsan, 2004; Lamm et al., 2004; Marshall et al., 2007; Chen et al., 2010; Melak et al., 2014). Exposure to high level arsenic might be associated with a 50-fold increase in mortality and a significantly higher incidence of BCa, indicating the strong carcinogenic potential of arsenic (Marshall et al., 2007; Steinmaus et al., 2014). However, arsenic does not fall into the classic model of carcinogenesis as it is not efficient at inducing point mutations or initiating and promoting the development of tumors in experimental animals (Jacobson-Kram and Montalbano, 1985; Jongen et al., 1985; Simeonova and Luster, 2000). Mounting evidence from in vitro, animal, and human studies shows that As is a strong epigenetic regulator that can alter DNA methylation (Broberg et al., 2014; Kile et al., 2014; Liu et al., 2014; Seow et al., 2014; Argos et al., 2015; Rojas et al., 2015), histone modification (Chu et al., 2011; Chervona et al., 2012; Ge et al., 2013; Howe et al., 2016; Tauheed et al., 2017), and microRNA (miRNA) expression (Ngalame et al., 2014; Wang et al., 2014; Ren et al., 2015; Humphries et al., 2016).
Histone posttranslational modifications (PTMs) have rapidly emerged as important regulators of gene expression. Dynamic changes in histone modifications have been broadly associated with chromatin regulation, which in turn affects DNA related cellular processes and plays a central role in various physiological and pathological conditions including cancer (Bannister and Kouzarides, 2011). Comparing histone modification profiles between cancer and normal states, particularly across multiple stages of malignant transformation of human cells that exhibit different genetic characteristics, can provide insights into understanding cancer initiation, progression, and response to therapy. The nature of histone PTM marks and their importance in carcinogenesis and cancer recurrence have made them prime candidates for the study of disease progression and drug development.
Our previous studies have showed that exposure of human urothelial cells (UROtsa cells) to monomethylarsonous acid (MMAIII) can alter histone acetylation patterns on gene-regulatory regions across the genome, which resulted in the deregulation of the expression of key tumor control genes and regulated gene networks during the development and progression of malignant transformation in urinary bladder cells (Zhu et al., 2017). The precise balance of histone acetylated and deacetylated states, which is a dynamic process and represents important features of gene regulation associated with MMAIII-induced cell transformation, has yet to be determined. In addition, studies have indicated that arsenic could also induce changes in other histone modifications (e.g. histone methylations) (Zhou et al., 2008; Jensen et al., 2009; Jo et al., 2009; Herbert et al., 2014; Liu et al., 2015). In the current study, we employed high-resolution liquid chromatography tandem mass spectrometry (LC-MS/MS) to identify and quantitatively measure a variety of histone variants and explore the dynamic combinatorial nature of histone PTMs in MMAIII-treated UROtsa cells. LC-MS/MS provides a sensitive and accurate method for examining the combinatorial nature of histone PTMs in a high-throughput fashion. Site-specific quantification of lysine acetylation/methylation on the N-terminal tails of H3 and H4 was performed to elicit the role of histone acetylation and methylation in regulating nucleosome architecture. We also explored whether the altered histone modifications could be manifested in samples collected from a human population exposed to arsenic.
Methods and Materials
Cell culture and treatment
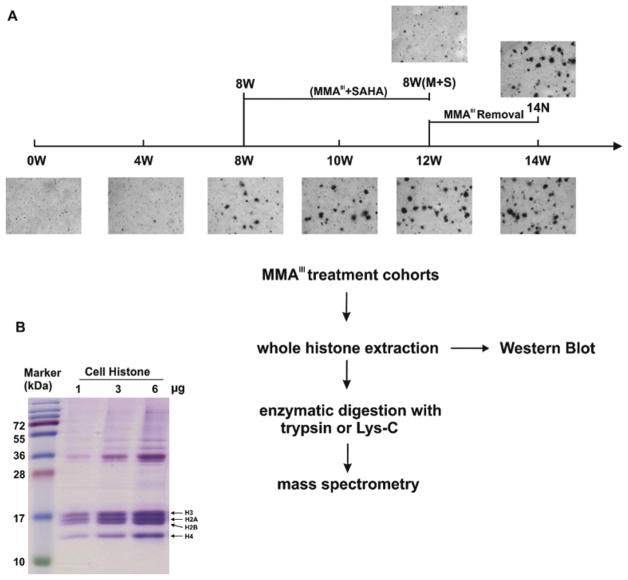
Cell samples collected from our previous study were used (Zhu et al., 2017). The procedure of cell culture and treatment was described in detail previously. In brief, human UROtsa cells, were maintained in DMEM with 5% fetal bovine serum, 100 IU/ml penicillin, and 100 μg/ml streptomycin. UROtsa cells were treated and continuously cultured in a medium enriched with 50 nM MMAIII for four weeks (4W), eight weeks (8W), 10 weeks (10W), 12 weeks (12W), and 14 weeks (14W), while passage-matched controls were maintained in a MMAIII-free medium. Culture medium was refreshed every three days with freshly prepared MMAIII solution to ensure the continuous presence of MMAIII. After eight weeks of the initial MMAIII treatment, an aliquot of MMAIII-treated cells was treated with 50 nM SAHA (histone deacetylase inhibitor, HDACi), and the cells were then continuously cultured in a medium enriched with both MMAIII and SAHA for four weeks [8W (M+S)]. Following 12 weeks of MMAIII treatment, an aliquot of MMAIII-treated cells was cultivated under standard cell culture conditions for an additional two weeks after removal of MMAIII (14N). Cells used in this study, with normal karyotype and negative for mycoplasma, were tested for viability, morphology, and growth curve analysis on a regular basis. Our previous analysis, e.g. gene expression profiles, did not identify any significant difference of all control cells no matter the length of culture and the passage times (data not shown). Therefore, the following experiment and the subsequent analysis were performed on MMAIII treated cells at each time point including the 0W (untreated samples).
Human population study
In 2010, we conducted an epidemiological investigation in arsenic-exposed villages of Wuyuan County, Inner Mongolia, China (Chen et al., 2017), where a very high percentage of residents were exposed to arsenic through contaminated drinking water (Guo et al., 2003). The protocols were carried out in accordance with guidelines approved by the Internal Review Boards (IRB) of Wenzhou Medical University, China and the University at Buffalo. Detailed information including the subjects’ profiles, arsenic exposure analysis, and blood collection methods, were described in our previous study (Chen et al., 2017). In the current study, buffy coat samples from 10 participants of low iAs exposure (10–30 μg/l iAs in drinking water) and 11 participants of high iAs exposure (>150 μg/l iAs in drinking water) were used for immunoblot analysis due to their high abundance of histone proteins. Detailed information regarding the samples are listed in supplementary materials Table S.1.
In vitro colony assay and nude mice xenograft assay
The stages of cell transformation and the malignant status of transformed cells were tested by colony formation in soft agar and by nude mice xenograft assay, respectively, and were performed previously as described (Zhu et al., 2017). The animal study was approved by the Institutional Animal Care and Use Committee (MED19014).
Histone extraction
Total histone protein was prepared from 8×106 cells using an EpiQuik Histone Extraction Kit (OP-0006; EpiGentek) according to the manufacturer's instructions, and histone protein concentration was measured by the Bradford Dye (500-0006, Bio-rad). A modified protocol was applied for histone extraction of human buffy coat samples. Briefly, 200–300 μl buffy coat samples were incubated with 10 ml hypotonic Tris-HCl solution (10 mM Tris, PH 8.0) for two hours at 4 °C. A protease inhibitor cocktail (P2714; Sigma) was used to prevent protein degradation after lysing the cells. At the end of the incubation, centrifugation was performed for 20 min at 10,000 rpm, 4 °C. The pellet was then mixed with lysis buffer from the EpiQuik Histone Extraction Kit (OP-0006; EpiGentek) and the remaining extraction steps were completed following the manufacturer’s protocol.
Histone acetylation and methylation analysis
Extracted histones were subjected to immunoblot analysis for H3/H4 acetylation and methylation marks using antibodies to acetyl-Histone H3 Lys4 (ABE223; Millipore), acetyl-Histone H3 Lys14 (04-1044; Millipore), acetyl-Histone H3 Lys18 (AB1191; Abcam), acetyl-Histone H3 Lys23 (07-360; Millipore), acetyl-Histone H4 Lys5 (07-327; Millipore), acetyl-Histone H4 Lys8 (61103; Active Motif), acetyl-Histone H4 Lys12 (07-595; Millipore), acetyl-Histone H4 Lys16 (07-329; Millipore), monomethyl-Histone H3 Lys4 (07-436; Millipore), monomethyl-Histone H3 Lys9 (ABE101-S; Millipore), unmethylated-Histone H3 Lys9 (MABE263; Millipore), monomethyl-Histone H3 Lys27 (07-448; Millipore), total Histone H3 (AB18521; Abcam) and total Histone H4 (AB17036; Abcam). Each measured protein was normalized to total H3 or H4. The chemiluminescent blots were imaged with a ChemiDoc MP imager (Bio-Rad) and quantified using Image software (NIH, Bethesda, MD). At the same time, an EpiQuik Histone H3 Modification Multiplex Assay Kit (P-3100; EpiGentek) was used to detect and quantify various H3 methylations simultaneously in an ELISA-like format following the manufacturer’s recommended protocols.
Sample preparation for LC-MS/MS based histone modification analysis
The isolated histone protein was prepared for LC-MS/MS analysis as previously described (Lott et al., 2015). Briefly, for each sample, 100 μg of total histone protein was reduced using a final concentration of 5 mM DTT (Dithiothreitol) kept in the dark at 56 °C for 30 min and then alkylated using a final concentration of 20 mM IAM (iodoacetamide) kept in the dark at 37 °C for 30 min. The protein mixture was then precipitated by stepwise addition of six volumes of acetone with continuous vortex and then incubated overnight at −20 °C. After centrifugation at 20,000 g force at 4 °C, the supernatant was removed and the pellet was washed with cold methanol and then allowed to air-dry. Digestion was performed in Tris-FA(formic acid) buffer (50 mM Tris, adjust pH value to 8.5 by formic acid) containing enzyme (trypsin or Lys-C) at an enzyme/substrate ratio of 1:20 (w/w). Total digestion was controlled at a volume of 100 μl, and the sample was incubated at 37 °C overnight, with vortex at 500 rpm in an Eppendorf Thermomixer® (Hamburg, Germany). Digestion was terminated by adding 1% (v/v) formic acid, after centrifuging at 20,000 g force for 30 min at 4 °C. The supernatant was obtained for nanoLC Orbitrap lumos mass spectrometry, 4 μl of each sample was loaded into the system.
Histone modification analysis by mass spectrometry
A Dionex UltiMate 3000 Nano-LC system (Thermo Scientific) was applied to load and separate the complex peptide mixture for highly sensitive identification. The nano-LC/nanospray setup, featuring a low void volume and high chromatographic reproducibility, enabled us to achieve comprehensive separation of the complex histone peptide mixture. Mobile phases A and B were 0.1% formic acid in 1% acetonitrile and 0.1% formic acid in 88% acetonitrile, respectively. Samples were loaded onto a large ID trap (300 μm inner diameter × 5 mm, packed with Zorbax 5μm C18 material) with 1% loading mobile phase B (0.05% Tris-FA in 88% acetonitrile) and 99% loading mobile phase A (0.05% Tris-FA in 1% acetonitrile) at a flow rate of 10 μl/min. A series of nanoflow gradients were used to back-flush the trapped samples onto the nano-LC column (75 μm inner diameter × 100 cm, packed with Pepmap® 3 μm C18 material). The nano-LC column was heated to 52 °C to improve both chromatographic resolution and reproducibility. A gradient profile consisted of the following steps for resolving the complex peptide mixture: a linear increase from 3 to 8% B for 5 min, 8–27% B for 117 min, 27–45% B for 10 min, 45–98% B for 20 min, and finally, isocratic at 98% B for 20 min. The nano-LC was coupled to a high-resolution Orbitrap Lumos Tribrid mass spectrometer (Thermo Fisher Scientific, San Jose, CA) with CID (collision-induced dissociation) activation, and the instrument was operated in data-dependent product ion mode. A three-second scan cycle was used, including an MS1 survey scan (m/z 400–1600) that was performed at a resolution of 120,000 with an AGC (automatic gain control) target of 5×105, maximum injection time = 50, followed by MS2 scans with CID activation, charge states of 2–7 were fragmented; dynamic exclusion was used for sensitive identification. A duration time of 45 s with data-dependent mode was employed. The isolation device was the front-end quadrupole with an isolation window = 1.2 units. n-trap CID was used for CID fragmentation, with collision energy of 30%, top speed mode for cycles, 35 ms injection time, and AGC set at 1x104, while the dual-cell ion trap was used as the detector.
Database searching and acetylation calculation
All raw data was processed using Proteome Discoverer 1.4 (Thermo Scientific), incorporating the SEQUEST algorithm with the specified protein amino acid sequences. For identification of lysine acetylation (+42.011 Da), differential modifications of single, double, and triple acetylation of lysine residues on the histone peptide mixture were examined for CID spectra. Also, carbamidomethylation of cysteines (+57.021 Da) was set as a fixed modification, and a variable modification of methionine oxidation (+15.995 Da) was allowed. Confirmation of the most probable assignment was obtained by manual inspection of b and y ions for putatively identified acetylated peptides. The relative quantification of different acetylated peptides was calculated by extracting ion currents of the precursors obtained by the Orbitrap Lumos mass spectrometry. For each identified acetylated peptide, the extracting ion currents were extracted in a narrow m/z window (0.005 units) around the monoisotopic m/z for each available charge state. The peptide-filtering criteria included Delta Cn scores of >0.1 and Xcorr scores of >1.9 and 2.3, respectively, for doubly and triply charged peptides. Stringent cutoffs were put in place for the Delta Cn scores and Xcorrs. The peak area under the curve for each precursor at each charge state was calculated using Qualbrowser (Thermo Scientific) and then the percentage of each product was calculated. The calculation of different acetylation types of the same molecular weight was based on the spectra count information generated by Sequest. The peak area of a peptide at 0W was designated as 100%, and the relative quantity of each modified acetylation was calculated by dividing the area of the same peptide MS signal peaks at a particular time point by the total area. A sum-intensity method was used to aggregate the quantitative data from peptide level to protein level. All different modified forms of a histone peptide were considered for acetylation calculation. The averages of all these ratios were used to estimate the relative acetylation quantification at the specific lysine residue (Tu et al., 2013; Lott et al., 2015).
Statistical analysis
The Western-blot data were obtained from at least three independent sets of experiments. One-way ANOVA was used for multiple comparisons and P < 0.05 was considered to indicate significant differences among the experimental groups.
Results
Workflow to assess acetylation changes at N-terminal of H3/H4 tails
The experimental workflow, shown in Fig. 1A, combined high-resolution LC-MS/MS profiling and immunoblot approaches to examine the impact of arsenic exposure on histone modifications at the N-terminal of H3/H4 tails in UROtsa cells. An in vitro malignant transformation model of human bladder epithelial cells, induced by chronic MMAIII exposure, was established as previously described (Zhu et al., 2017). Briefly, the study showed that exposure to MMAIII at a dose of 50 nM for 12 weeks was able to induce the malignant transformation of UROtsa cells (Fig. 1A, supplementary materials Fig. S.1 and Table S.2). Significant alterations of gene-regulatory networks that facilitate malignant transformation began to occur at eight weeks when cells transitioned from a reversible to an ultimately irreversible malignant transformation (Zhu et al., 2017). In addition, administration of the pharmaceutical histone deacetylase inhibitor, suberoylanilide hydroxamic acid (SAHA), resulted in the inhibition of MMAIII-induced malignant transformation (Ge et al., 2013). We utilized a high-resolution LC-MS/MS approach for sensitive and accurate quantification of histone PTM profiles in UROtsa cells during the transformation course upon MMAIII/SAHA treatment. Western blot was used to verify the identified changes in lysine acetylation. Purified whole histone proteins from UROtsa cells obtained from an EpiQuik Total Histone Extraction Kit were used for both mass spectrometry and Western blot analysis (Fig. 1B, supplementary materials Fig. S.2).
Figure 1.
Arsenic exposure causes lysine acetylation changes in opposite directions at the N-terminal of H3/H4 tails
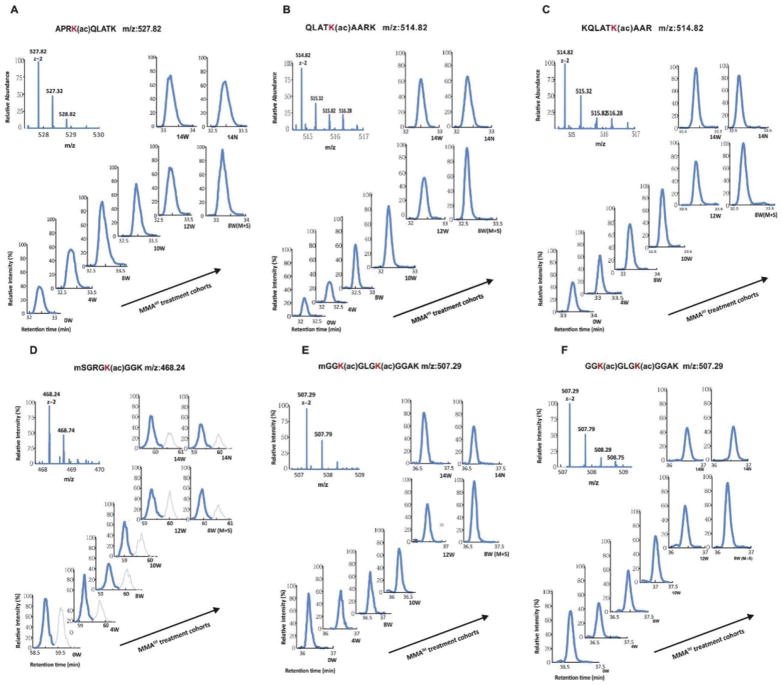
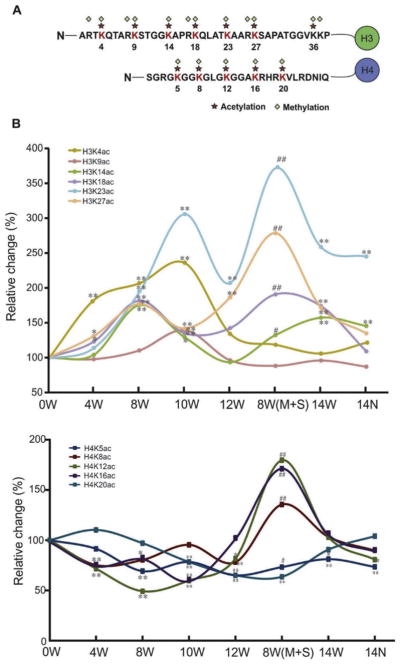
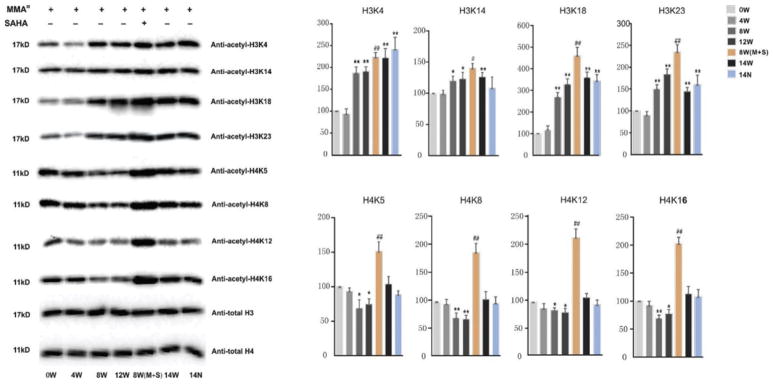
Both Lys-C and trypsin digestion of extracted histones produced fragments of suitable sizes. Peptides derived from digestion were then subjected to LC-MS/MS analysis. Small diagnostic MS/MS fragmentation of acetylated lysine residues generated a highly specific diagnostic fragment and was used to quantify the levels of acetylation at the N-terminal of histone H3/H4 tails. Histones are highly modified proteins with each peptide containing several acetylation acceptor sites, thus resulting in many possible modified forms for a given peptide. The detected peptides with a high Xcoor score and recognizable abundant peak with areas over 108 are listed in Table 1. The relative modified ratios of histone peptides were obtained by dividing the extracted normalized peak area of all modified forms for a given peptide sequence by their corresponding peak area at 0W without MMAIII treatment. A chromatogram of six representative peptides on H3 or H4 after Lys-C or trypsin digestion is shown in Fig. 2. Peptides eluted at different times and MS/MS targeting was required throughout the elution. We observed a time-dependent alteration in histone acetylation over the course of MMAIII treatment (Fig. 3). Interestingly, histone acetylation changes on H3 and H4 N-terminal tails occurred in an inverse manner, especially during the critical window for MMAIII-induced UROtsa cells’ malignant transformation of four to 12 weeks: MMAIII exposure increased acetylation on lysine sites K4, K9, K14, K18, K23, and K27 at the N-terminal of H3 tails, but decreased acetylation on lysine sites K5, K8, K12, and K16 at the N-terminal of H4 tails. However, many histone acetylation sites did not show an obvious change but instead reverted to the baseline status and were comparable to 0W after UROtsa cells underwent malignant transformation and subsequently possessed the properties of cancer. These differential changes of lysine acetylation on H3 and H4 were largely verified by Western blot using total histones extracted from UROtsa cells. As shown in Fig. 4, the acetylation levels on H3 lysine K4, K14, K18, and K23 sites at the N-terminal of histone H3 tails were significantly increased in a time-dependent manner over the time-course of MMAIII treatment, while the acetylation levels on H4 lysine K5, K8, K12, and K16 at the N-terminal of histone H4 tails were altered in the opposite direction, in which they were decreased during the 12 weeks of MMAIII exposure.
Table 1.
Representative acetylated peptides from Histone H3 and H4 post Lys-C or trypsin proteolytic digestion identified by LC-MS/MS.
| Protein Accession | Peptide | Sequence | m/z (Da) | Charge | Acetylated site(s) |
|---|---|---|---|---|---|
| H3 | LysC-1 | mARTK(ac)QTARK | 637.86 | 2 | H3K4 |
| LysC-2 | mARTKQTARK(ac)STGGK | 839.94 | 2 | H3K9 | |
| LysC-3 | QTARK(ac)STGGK(ac)APRK | 523.96 | 3 | H3K9, H3K14 | |
| LysC-4 | STGGK(ac)APRKQLATK | 742.93 | 2 | H3K14 | |
| LysC-5 | STGGK(ac)APRK(ac)QLATK | 763.93 | 2 | H3K14, H3K18 | |
| LysC-6 | APRK(ac)QLATK | 527.82 | 2 | H3K18 | |
| LysC-7 | STGGKAPRK(ac)QLATK | 742.93 | 2 | H3K18 | |
| LysC-8 | QLATK(ac)AARK | 514.82 | 2 | H3K23 | |
| LysC-9 | AARK(ac)SAPATGGVKK | 692.39 | 2 | H3K27 | |
| LysC-10 | QLATK(ac)AARK(ac)SAPATGGVK | 613.68 | 2 | H3K23, H3K27 | |
| LysC-11 | AARK(ac)SAPATGGVK | 433.24 | 3 | H3K27, H3K36 | |
| Tryp-1 | STGGK(ac)APR | 815.44 | 2 | H3K14 | |
| Tryp-2 | KQLATK(ac)AAR | 514.82 | 2 | H3K23 | |
| Tryp-3 | K(ac)QLATK(ac)AAR | 535.82 | 2 | H3K18, H3K23 | |
| Tryp-4 | K(ac)SAPATGGVK | 957.54 | 2 | H3K27 | |
| H4 | LysC-1 | mSGRGK(ac)GGK | 468.24 | 2 | H4K5 |
| LysC-2 | mSGRGKGGK(ac)GLGK | 645.84 | 2 | H4K8 | |
| LysC-3 | mGGK(ac)GLGK(ac)GGAK | 507.29 | 2 | H4K8, H4K12 | |
| LysC-4 | mSGRGK(ac)GGK(ac)GLGK(ac) | 687.84 | 2 | H4K5, H4K8, H4K12 | |
| LysC-5 | GGKGLGK(ac)GGAK | 486.28 | 2 | H4K12 | |
| LysC-6 | GLGK(ac)GGAK(ac)RHRK | 674.90 | 2 | H4K12, H4K16 | |
| LysC-7 | GGKGLGKGGAK(ac) | 486.28 | 2 | H4K16 | |
| LysC-8 | GGAK(ac)RHRK(ac)VLRDNIQGITK | 744.44 | 3 | H4K16, H4K20 | |
| LysC-9 | RHRK(ac)VLRDNIQGITK | 626.05 | 3 | H4K20 | |
| LysC-10 | GLGKGGAKRHRK(ac) | 653.89 | 2 | H4K20 | |
| Tryp-1 | GK(ac)GGK(ac)GLGK | 885.51 | 2 | H4K5, H4K8 | |
| Tryp-2 | GKGGK(ac)GLGK | 843.49 | 2 | H4K8 | |
| Tryp-3 | GGK(ac)GLGK(ac)GGAK | 507.29 | 2 | H4K8, H4K12 | |
| Tryp-4 | GLGK(ac)GGAK(ac)R | 927.53 | 2 | H4K12, H4K16 | |
| Tryp-5 | GLGKGGAK(ac)R | 885.51 | 2 | H4K16 |
Figure 2.
Figure 3.
Figure 4.
SAHA treatment of UROtsa cells induces global changes in histone acetylation
SAHA, a histone deacetylase inhibitor, has been widely used in clinical trials for cancer therapies. We previously reported that co-treatment of UROtsa cells with MMAIII and SAHA globally increased both H3 and H4 acetylation levels. Accordingly, SAHA also inhibited MMAIII-induced cell transformation, suggesting that histone acetylation dysregulation plays a key role in arsenic-induced carcinogenesis (Ge et al., 2013). To further examine the impact of SAHA on histone acetylation, UROtsa cells were co-treated with MMAIII and 50 nM SAHA for four weeks after eight weeks of initial MMAIII treatment; during that time, the cells are undergoing an irreversible phase of malignant transformation. As shown in Figs. 3 and 4, the most vivid response of SAHA on acetylation regulation was elicited on lysine sites at histone N tails: SAHA induced a global increase in histone acetylation on both H3 and H4. These data demonstrate that SAHA can reverse MMAIII-induced changes in histone acetylation and suggest that HDACi treatment may have a direct effect on the physiology of these cells, at least in part, by modifying epigenetic PTM marks.
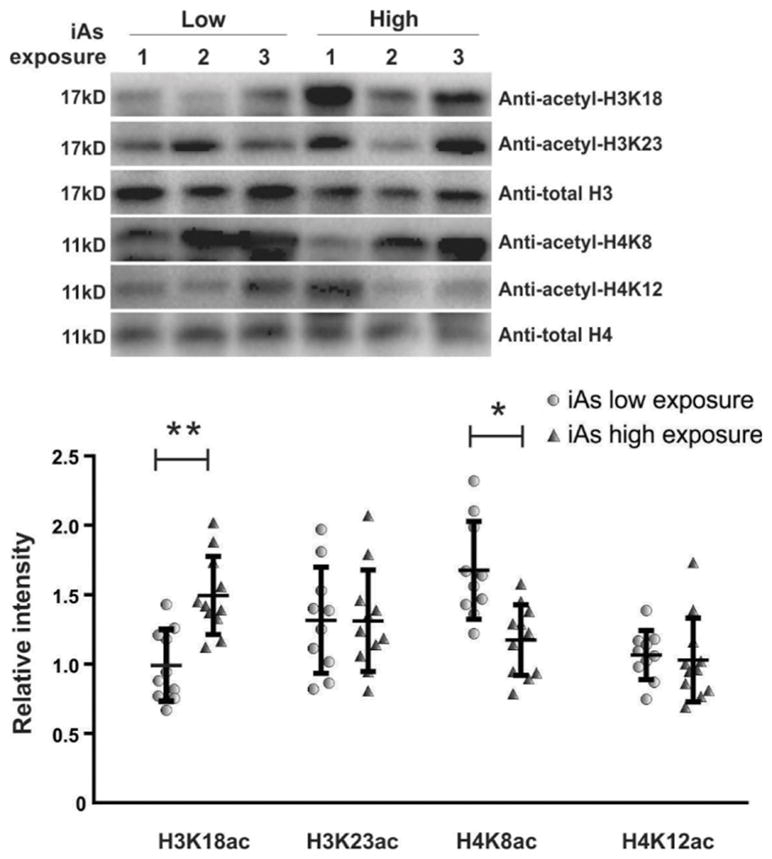
Histone acetylation alterations in peripheral human blood leukocytes of an arsenic-exposed population
In light of our in vitro data, we wondered if epigenetic histone acetylation signatures, coincident with iAs exposure, were altered in a similar manner in a human population exposed to arsenic. We performed a pilot analysis on 21 leukocytes samples collected from a population exposed to different concentrations of iAs in their drinking water in Wuyuan County, China (Chen et al., 2017). Western blot was used to examine the level of H3K18ac, H3K23ac, H4K8ac and H4K12ac markers in these samples. We found that H3K18ac was significantly increased in leukocytes of people exposed to high iAs (165 μg/l) while H4K8ac was substantially decreased (Fig. 5). However, there was a high variability between individuals for the histone acetylation level of the other two markers, H3K23ac and H4K12ac; therefore, no change tendency was observed for these two markers.
Figure 5.
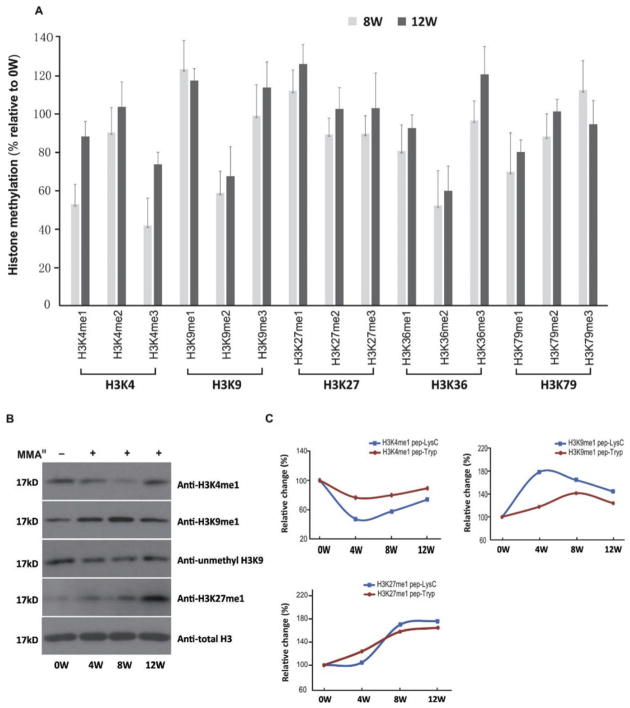
Histone methylation patterns were altered in accordance with histone acetylation dysregulation during malignant transformation
To examine other epigenetic markers on H3 that are modified along with histone acetylation due to arsenic exposure, we assessed histone methylation changes via multiple analytical tools, including ELISA assay, LC-MS/MS, and Western blot. In accordance with the alterations in histone acetylation, chronic arsenic exposure induced histone methylation dysregulation in a time-dependent manner promoting malignant transformation of UROtsa cells across the critical time window of four to 12 weeks. As shown in Fig. 6A, hypomethylation of H3K4me1, as well as hypermethylation of H3K9me1 and H3K27me1 were identified by an EpiQuik Histone H3 Modification Multiplex Assay Kit, and were further verified by immunoblot (Fig. 6B). In the LC-MS/MS analysis, after Lys-C and trypsin digestion, typical peptides that contained methylated sites, which exhibited changed methylation patterns, were chosen and identified (Table 2). As shown in Fig. 6C, the peptides that contain the methylated lysine sites H3K4me1, H3K9me1, and H3K27me1, showed a similar trend of altered methylation levels as results from EpiQuik Histone H3 Modification Multiplex Assay Kit and Western-blot.
Figure 6.
Table 2.
Representative methylated peptides from Histone H3 post Lys-C or trypsin proteolytic digestion identified by LC-MS/MS.
| Peptide | Sequence | m/z (Da) | Charge | Methylated site(s) |
|---|---|---|---|---|
| LysC-1 | MAR(me)TK(me)QTARKSTGGK | 886.96 | 2 | H3K4me1, H3R2me2 |
| LysC-2 | MARTKQTARK(me)STGGK | 846.95 | 2 | H3K9me1 |
| LysC-3 | QLATKAARK(me) | 521.82 | 2 | H3K27me1 |
| Tryp-1 | MARTK(me)QTAR | 618.77 | 2 | H3K4me1 |
| Tryp-2 | QTAR(me)K(me)STGGK | 585.30 | 2 | H3K9 me1, H3R8me1 |
| Tryp-3 | AARK(me)SAPATGGVK | 682.35 | 2 | H3K27me1 |
Discussion
Our previous studies have shown that changes in H3 and H4 acetylation following chronic exposure to low doses of MMAIII coincide with and contribute to MMAIII-induced cell malignant transformation (Ge et al., 2013; Zhu et al., 2017). Here, we report the results of a comprehensive screening of histone acetylation and methylation on individual lysine sites of H3 and H4 by high-resolution-MS. We validate that chronic MMAIII exposure causes histone acetylation changes in opposite directions on histones H3 and H4, with increase in H3 acetylation but decrease in H4 acetylation. This trend is also observed in a human population exposed to arsenic-contaminated drinking water. H3K18ac was remarkably increased in the leukocytes of people exposed to high iAs compared to low iAs, while H4K8ac was significantly decreased. MMAIII also altered histone methylation patterns: the transformed cells experienced a loss of H3K4me1, and an increase in H3K9me1 and H3K27me1. Together, our data show that arsenic exposure leads to dynamic histone acetylation and methylation changes during arsenic-induced cancer development.
Exposure to arsenic interferes with numerous regulatory processes that maintain genomic fidelity and chromosomal transmission integrity, including those responsible for epigenetic imprinting (Reichard and Puga, 2010). The acetylation status of H3 and H4 is broadly associated with changes in chromatin condensation and patterns of transcriptional activation and repression (Herbert et al., 2014). Recent studies indicated arsenic exposure may induce histone PTMs by disrupting the regulatory function of histone deacetylase SIRT1 (Herbert et al., 2014), histone acetyltransferase hMOF (Liu et al., 2015), and histone acetyltransferase MYST1 (Jo et al., 2009). These changes alter cellular stress responses to arsenic toxicity by impairing regulation of tumor suppressor gene-mediated signaling pathways and substrates involved in cell cycle regulation and apoptosis. As we showed previously, arsenic-induced aberrant regulation of PTMs, such as histone acetylation, can be enriched at gene promoter-specific regions across the genome, and are associated with differential binding patterns in a large number of critical genes that act as upstream regulators of networks with known functions in cancer development and progression (Zhu et al., 2017).
PTMs dictate the higher-order chromatin structure and are important regulators of chromatin function and gene expression (Lin and Garcia, 2012). Histone modifications play vital roles in many fundamental biological processes by rearranging the structure and composition of chromatin. In eukaryotes, such chromatin restructuring events can help partition the genome into distinct domains such as euchromatin and heterochromatin and result in DNA transcription, repair, and replication. Moreover, some histone modifications may also participate in chromosome condensation, indicating their importance in the cell cycle and cell mitosis. Histone lysine modification systems, along with DNA methylation and small noncoding RNA, collectively make up mechanisms referred to as “epigenetic” controls, which are known to affect gene expression patterns and phenotypes in a heritable manner (Ren et al., 2011). Dysregulation of histone PTMs is known to be associated with various human diseases including cancer. Transcriptional machinery components and factors as well as histone modifying enzymes can bind to different histone PTMs in an ordered manner and manipulate DNA expression (Sterner and Berger, 2000). Mass spectrometry (MS) has been a powerful tool for studying histone PTMs as it allows accurate quantification of specific modifications of various combinations in an unbiased manner and in a high-throughput fashion. (Villar-Garea et al., 2008). To understand the biological functions of the global dynamics of histone acetylation in arsenic-induced carcinogenesis, we applied Nano-LC/high-resolution-MS to elucidate the histone modification patterns in UROtsa cells under malignant transformation induced by chronic MMAIII treatment. We developed a targeted Nano-LC/high-resolution-MS method for the site-specific quantification of lysine acetylation in the N-terminal region of H3 and H4 by combining chemical derivatization at the peptide level with digestion using Lys-C and trypsin. Our finding that chronic MMAIII exposure increased acetylation at the H3 N-tail but decreased acetylation at the H4 N-tail showed a complex relationship between arsenic and histone acetylation modifications.
In general, increased histone acetylation is associated with an open status of chromatin, which translates to increased gene-expression activity, while reduction of histone acetylation down-regulates chromatin accessibility and is associated with transcriptional repression. Therefore, the opposing changes of H3 and H4 acetylation may lead to both up- and down-regulation of gene expression. In functional studies, the effects of H4 acetylation is often found to be inversely correlated with the effects of H3 acetylation such as in the binding of transcription factors, expression of genes, or remodeling of chromatin (Kurdistani et al., 2004). There is an increasing body of evidence which suggest that H3- and H4- acetylation may have opposing roles in regulating nucleosome architecture. Moreover, H4-acetylation may act to counteract the effects of acetylated H3, suggesting that distinct aspects of nucleosome dynamics might be independently controlled by individual histones (Gansen et al., 2015). Previously, we reported that the treatment of HDACi, e.g., SAHA, which is clinically used for the treatment of cutaneous T cell lymphoma, could block the MMAIII-induced cell malignant transformation. Here we showed that SAHA treatment caused a global increase in both histone H3 and H4 acetylation. However, whether or not this indicates that decreased histone H3 acetylation plays a more critical role in MMAIII-induced cell malignant transformation than increased histone H4 acetylation remains to be determined.
In addition, to histone acetylation, histone methylation also plays an important role in epigenetic regulation. Histone methylation mainly occurs on the side chains of lysines and arginines, with an added level of complexity when compared to acetylation: lysines may be mono-, di- or tri-methylated, whereas arginines may be mono-, symmetrically or asymmetrically di-methylated. Methylated lysine sites on histones can bind to other regulatory proteins for the regulation of gene activity, e.g., HP1 (Lehnertz et al., 2003). Gender-specific patterns of association were observed between iAs exposure and several histone marks (Chervona et al., 2012). In the present study, altered histone methylation patterns were observed in arsenic-transformed human bladder cells. The transformed cells experienced a loss of H3K4me1 and an increase in H3K9me1 and H3K27me1. H3K4me1 is a well-established feature of enhancers and promoters and is associated with poised enhancers (Ferrari et al., 2014). H3K9me1 is a marker of heterochromatin, the condensed and transcriptionally inactive state of chromatin. H3K27me1 can directly influence gene expression: H3K27me1 is found to be enriched at the transcriptional start site of active genes, coinciding with a general increase in gene activation and decrease in DNA methylation (Barski et al., 2007). Given the critical role of H3K27 methylation in the balance of gene activity, it is not surprising to find anomalies of this system in cancer. Generally, high levels of histone acetylation and H3K4 methylation are detected in promoter regions of active genes, whereas elevated levels of H3K9 and H3K27 methylation correlates with gene repression (Barski et al., 2007).
In conclusion, acetylation on H3 and H4 were found to be associated with arsenic exposure and were altered differentially. In addition, H3 methylation levels were also changed as a result of exposure to arsenic. The human sample analysis is merely a pilot study, and a more dedicated research with larger sample size is warranted to not only validate our findings but also to examine the application of our in vitro findings in human population investigations.
Supplementary Material
Highlights.
Quantitatively measure histone modifications by a high-resolution LC-MS/MS.
MMAIII induced a time-dependent change of histone H3 and H4 acetylation patterns.
MMAIII altered acetylation patterns on histone H3 and H4 in an opposite direction.
Histone acetylation pattern was similar in human as in cell exposed to arsenic.
Arsenic reduced H3K4me1 and increased H3K9me1 and H3K27me1 methylation levels.
Acknowledgments
We thanks all villagers who participated in the human epidemiological study and all members in the investigation team. This study was supported by National Institutes of Health (NIH) grant ES022629 (to X.R.).
Abbreviations
- ac
Acetyl/Acetylated
- AGC
Automatic Gain Control
- BCa
Bladder Cancer
- CID
Collision-Induced Dissociation
- DTT
Dithiothreitol
- IAM
Iodoacetamide
- iAs
Inorganic Arsenic
- ID
Inner Diameter
- H
Histone
- HDACi
Histone Deacetylase Inhibitor
- K
Lysine
- LC-MS/MS
Liquid Chromatography tandem-Mass Spectrometry
- Lys-c
Lysyl Endopeptidase
- me
Methylation
- MMAIII
Monomethylarsonous acid
- PTM
Histones Posttranslational Modification
- SAHA
Suberoylanilide Hydroxamic Acid
- Tryp
Trypsin
Footnotes
Competing financial interests
All authors have no competing financial interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, Paul-Brutus R, Gamble MV, Harper KN, Parvez F, Rahman M, Rakibuz-Zaman M, Slavkovich V, Baron JA, Graziano JH, Kibriya MG, Ahsan H. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect. 2015;123:64–71. doi: 10.1289/ehp.1307884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bates MN, Rey OA, Biggs ML, Hopenhayn C, Moore LE, Kalman D, Steinmaus C, Smith AH. Case-control study of bladder cancer and exposure to arsenic in Argentina. Am J Epidemiol. 2004;159:381–389. doi: 10.1093/aje/kwh054. [DOI] [PubMed] [Google Scholar]
- Broberg K, Ahmed S, Engstrom K, Hossain MB, Jurkovic Mlakar S, Bottai M, Grander M, Raqib R, Vahter M. Arsenic exposure in early pregnancy alters genome-wide DNA methylation in cord blood, particularly in boys. J Dev Orig Health Dis. 2014;5:288–298. doi: 10.1017/S2040174414000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Chiou HY, Hsu LI, Hsueh YM, Wu MM, Wang YH, Chen CJ. Arsenic in drinking water and risk of urinary tract cancer: a follow-up study from northeastern Taiwan. Cancer Epidemiol Biomarkers Prev. 2010;19:101–110. doi: 10.1158/1055-9965.EPI-09-0333. [DOI] [PubMed] [Google Scholar]
- Chen X, Guo X, He P, Nie J, Yan X, Zhu J, Zhang L, Mao G, Wu H, Liu Z, Aga D, Xu P, Smith M, Ren X. Interactive Influence of N6AMT1 and As3MT Genetic Variations on Arsenic Metabolism in the Population of Inner Mongolia, China. Toxicol Sci. 2017;155:124–134. doi: 10.1093/toxsci/kfw181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ahsan H. Cancer burden from arsenic in drinking water in Bangladesh. Am J Public Health. 2004;94:741–744. doi: 10.2105/ajph.94.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Hall MN, Arita A, Wu F, Sun H, Tseng HC, Ali E, Uddin MN, Liu X, Zoroddu MA, Gamble MV, Costa M. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol Biomarkers Prev. 2012;21:2252–2260. doi: 10.1158/1055-9965.EPI-12-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Ren X, Chasse A, Hickman T, Zhang L, Yuh J, Smith MT, Burlingame AL. Quantitative mass spectrometry reveals the epigenome as a target of arsenic. Chem Biol Interact. 2011;192:113–117. doi: 10.1016/j.cbi.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari KJ, Scelfo A, Jammula S, Cuomo A, Barozzi I, Stutzer A, Fischle W, Bonaldi T, Pasini D. Polycomb-dependent H3K27me1 and H3K27me2 regulate active transcription and enhancer fidelity. Mol cell. 2014;53:49–62. doi: 10.1016/j.molcel.2013.10.030. [DOI] [PubMed] [Google Scholar]
- Gansen A, Toth K, Schwarz N, Langowski J. Opposing roles of H3- and H4-acetylation in the regulation of nucleosome structure--a FRET study. Nucleic Acids Res. 2015;43:1433–1443. doi: 10.1093/nar/gku1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Gong Z, Olson JR, Xu P, Buck MJ, Ren X. Inhibition of monomethylarsonous acid (MMA(III))-induced cell malignant transformation through restoring dysregulated histone acetylation. Toxicology. 2013;312:30–35. doi: 10.1016/j.tox.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Guo X, Fujino Y, Chai J, Wu K, Xia Y, Li Y, Lv J, Sun Z, Yoshimura T. The prevalence of subjective symptoms after exposure to arsenic in drinking water in Inner Mongolia, China. J Epidemiol. 2003;13:211–215. doi: 10.2188/jea.13.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert KJ, Holloway A, Cook AL, Chin SP, Snow ET. Arsenic exposure disrupts epigenetic regulation of SIRT1 in human keratinocytes. Toxicol Appl Pharmacol. 2014;281:136–145. doi: 10.1016/j.taap.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Howe CG, Liu X, Hall MN, Slavkovich V, Ilievski V, Parvez F, Siddique AB, Shahriar H, Uddin MN, Islam T, Graziano JH, Costa M, Gamble MV. Associations between Blood and Urine Arsenic Concentrations and Global Levels of Post-Translational Histone Modifications in Bangladeshi Men and Women. Environ Health Perspect. 2016;124:1234–1240. doi: 10.1289/ehp.1510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries B, Wang Z, Yang C. The role of microRNAs in metal carcinogen-induced cell malignant transformation and tumorigenesis. Food Chem Toxicol. 2016;98:58–65. doi: 10.1016/j.fct.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson-Kram D, Montalbano D. The reproductive effects assessment group's report on the mutagenicity of inorganic arsenic. Environ Mutagen. 1985;7:787–804. doi: 10.1002/em.2860070515. [DOI] [PubMed] [Google Scholar]
- Jensen TJ, Wozniak RJ, Eblin KE, Wnek SM, Gandolfi AJ, Futscher BW. Epigenetic mediated transcriptional activation of WNT5A participates in arsenical-associated malignant transformation. Toxicol Appl Pharmacol. 2009;235:39–46. doi: 10.1016/j.taap.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo WJ, Ren X, Chu F, Aleshin M, Wintz H, Burlingame A, Smith MT, Vulpe CD, Zhang L. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol Appl Pharmacol. 2009;241:294–302. doi: 10.1016/j.taap.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongen WM, Cardinaals JM, Bos PM, Hagel P. Genotoxicity testing of arsenobetaine, the predominant form of arsenic in marine fishery products. Food Chem Toxicol. 1985;23:669–673. doi: 10.1016/0278-6915(85)90155-3. [DOI] [PubMed] [Google Scholar]
- Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, Cardenas A, Wright RO, Christiani DC. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9:774–782. doi: 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Lamm SH, Engel A, Kruse MB, Feinleib M, Byrd DM, Lai S, Wilson R. Arsenic in drinking water and bladder cancer mortality in the United States: an analysis based on 133 U.S. counties and 30 years of observation. J Occup Environ Med. 2004;46:298–306. doi: 10.1097/01.jom.0000116801.67556.8f. [DOI] [PubMed] [Google Scholar]
- Lin S, Garcia BA. Examining histone posttranslational modification patterns by high-resolution mass spectrometry. Methods Enzymol. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wu D, Zhao L, Yang Y, Ding J, Dong L, Hu L, Wang F, Zhao X, Cai Y, Jin J. Arsenic Trioxide Reduces Global Histone H4 Acetylation at Lysine 16 through Direct Binding to Histone Acetyltransferase hMOF in Human Cells. PloS One. 2015;10:e0141014. doi: 10.1371/journal.pone.0141014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Y, Zhang W, Zhang X, Lioyd-Jones DM, Baccarelli AA, Ning H, Fornage M, He K, Liu K, Hou L. Blood methylomics in response to arsenic exposure in a low-exposed US population. J Expo Sci Environ Epidemiol. 2014;24:145–149. doi: 10.1038/jes.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Mukhopadhyay S, Li J, Wang J, Yao J, Sun Y, Qu J, Read LK. Arginine methylation of DRBD18 differentially impacts its opposing effects on the trypanosome transcriptome. Nucleic Acids Res. 2015;43:5501–5523. doi: 10.1093/nar/gkv428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall G, Ferreccio C, Yuan Y, Bates MN, Steinmaus C, Selvin S, Liaw J, Smith AH. Fifty-year study of lung and bladder cancer mortality in Chile related to arsenic in drinking water. J Nati Cancer Inst. 2007;99:920–928. doi: 10.1093/jnci/djm004. [DOI] [PubMed] [Google Scholar]
- Melak D, Ferreccio C, Kalman D, Parra R, Acevedo J, Perez L, Cortes S, Smith AH, Yuan Y, Liaw J, Steinmaus C. Arsenic methylation and lung and bladder cancer in a case-control study in northern Chile. Toxicol Appl Pharmacol. 2014;274:225–231. doi: 10.1016/j.taap.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, Suk WA. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect. 2013;121:295–302. doi: 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngalame NN, Tokar EJ, Person RJ, Xu Y, Waalkes MP. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci. 2014;138:268–277. doi: 10.1093/toxsci/kfu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard JF, Puga A. Effects of arsenic exposure on DNA methylation and epigenetic gene regulation. Epigenomics. 2010;2:87–104. doi: 10.2217/epi.09.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Gaile DP, Gong Z, Qiu W, Ge Y, Zhang C, Huang C, Yan H, Olson JR, Kavanagh TJ, Wu H. Arsenic responsive microRNAs in vivo and their potential involvement in arsenic-induced oxidative stress. Toxicol Appl Pharmacol. 2015;283:198–209. doi: 10.1016/j.taap.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ Health Perspect. 2011;119:11–19. doi: 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobna Z, Rubio-Andrade M, Styblo M, Garcia-Vargas G, Fry RC. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci. 2015;143:97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow WJ, Kile ML, Baccarelli AA, Pan WC, Byun HM, Mostofa G, Quamruzzaman Q, Rahman M, Lin X, Christiani DC. Epigenome-wide DNA methylation changes with development of arsenic-induced skin lesions in Bangladesh: a case-control follow-up study. Environ Mol Mutagen. 2014;55:449–456. doi: 10.1002/em.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonova PP, Luster MI. Mechanisms of arsenic carcinogenicity: genetic or epigenetic mechanisms? J Environ Pathol Toxicol Oncol. 2000;19:281–286. [PubMed] [Google Scholar]
- Smith AH, Steinmaus CM. Health effects of arsenic and chromium in drinking water: recent human findings. Annu Rev Public Health. 2009;30:107–122. doi: 10.1146/annurev.publhealth.031308.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C, Ferreccio C, Acevedo J, Yuan Y, Liaw J, Duran V, Cuevas S, Garcia J, Meza R, Valdes R, Valdes G, Benitez H, VanderLinde V, Villagra V, Cantor KP, Moore LE, Perez SG, Steinmaus S, Smith AH. Increased lung and bladder cancer incidence in adults after in utero and early-life arsenic exposure. Cancer Epidemiol Biomarkers Prev. 2014;23:1529–1538. doi: 10.1158/1055-9965.EPI-14-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmaus C, Yuan Y, Bates MN, Smith AH. Case-control study of bladder cancer and drinking water arsenic in the western United States. Am J Epidemiol. 2003;158:1193–1201. doi: 10.1093/aje/kwg281. [DOI] [PubMed] [Google Scholar]
- Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauheed J, Sanchez-Guerra M, Lee JJ, Paul L, Ibne Hasan MOS, Quamruzzaman Q, Selhub J, Wright RO, Christiani DC, Coull BA, Baccarelli AA, Mazumdar M. Associations between post translational histone modifications, myelomeningocele risk, environmental arsenic exposure, and folate deficiency among participants in a case control study in Bangladesh. Epigenetics. 2017;12:484–491. doi: 10.1080/15592294.2017.1312238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C, Li J, Jiang X, Sheflin LG, Pfeffer BA, Behringer M, Fliesler SJ, Qu J. Ion-current-based proteomic profiling of the retina in a rat model of Smith-Lemli-Opitz syndrome. Mol Cell Proteomics. 2013;12:3583–3598. doi: 10.1074/mcp.M113.027847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Garea A, Israel L, Imhof A. Analysis of histone modifications by mass spectrometry. Curr Protoc Protein Sci. 2008;Chapter 14(Unit 14):10. doi: 10.1002/0471140864.ps1410s51. [DOI] [PubMed] [Google Scholar]
- Wang Z, Humphries B, Xiao H, Jiang Y, Yang C. MicroRNA-200b suppresses arsenic-transformed cell migration by targeting protein kinase Calpha and Wnt5b-protein kinase Calpha positive feedback loop and inhibiting Rac1 activation. J Biol Chem. 2014;289:18373–18386. doi: 10.1074/jbc.M114.554246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sun H, Ellen TP, Chen H, Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29:1831–1836. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang J, Chen X, Tsompana M, Gaile D, Buck M, Ren X. A time-series analysis of altered histone H3 acetylation and gene expression during the course of MMAIII-induced malignant transformation of urinary bladder cells. Carcinogenesis. 2017;38:378–390. doi: 10.1093/carcin/bgx011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.