Summary
NMDA-type glutamate receptors (NMDARs) are ligand-gated ion channels that mediate excitatory neurotransmission in the central nervous system (CNS). Here, we describe functional and single channel properties of triheteromeric GluN1/GluN2A/GluN2C receptors, which contain two GluN1, one GluN2A, and one GluN2C subunits. This NMDAR has three conductance levels and opens in bursts similar to GluN1/GluN2A receptors, but with single channel open time and open probability reminiscent of GluN1/GluN2C receptors. The deactivation time course of GluN1/GluN2A/GluN2C receptors is intermediate to GluN1/GluN2A and GluN1/GluN2C receptors and is not dominated by GluN2A or GluN2C. We show that triheteromeric GluN1/GluN2A/GluN2C receptors are the predominant NMDARs in cerebellar granule cells, and propose that co-expression of GluN2A and GluN2C in cerebellar granule cells occludes cell-surface expression of diheteromeric GluN1/GluN2C receptors. This new insight to neuronal GluN1/GluN2A/GluN2C receptors highlights the complexity of NMDAR signaling in the CNS.
Keywords: cerebellar granule cells, excitatory neurotransmission, ligand-gated ion-channel, ionotropic glutamate receptors
ETOC BLURB
Bhattacharya et al. show that the NMDA receptor GluN2C subunit is preferentially incorporated into triheteromeric GluN1/GluN2A/GluN2C receptors in cerebellar granule cells. Triheteromeric GluN1/GluN2A/GluN2C receptors have single channel properties that cannot be predicted from the composite subunits.
Introduction
N-methyl-D-aspartate receptors (NMDARs) are members of the ionotropic glutamate receptor family that bind the co-agonists glutamate and glycine to mediate a Ca2+-permeable component of excitatory synaptic transmission. NMDARs are tetrameric complexes that contain two GluN1 and two GluN2 or GluN3 subunits, and are involved in many critical processes in the central nervous system (CNS), including circuit development and memory formation (Cotman et al., 1987; Daw et al., 1993; Kentros et al., 1998; Lisman, 2003; Traynelis et al., 2010; Tsien et al., 1996). The four different GluN2 subunits (GluN2A-D) endow NMDARs with divergent pharmacological and functional properties (Cull-Candy et al., 2001; Monyer et al., 1994; Monyer and Sprengel, 1992; Paoletti and Neyton, 2007; Traynelis et al., 2010; Vicini et al., 1998; Watanabe et al., 1992). Moreover, the GluN2 subunits are expressed differentially across the CNS (Akbarian et al., 1996; Monyer et al., 1994; Standaert et al., 1999).
Most studies of recombinant NMDARs have focused on diheteromeric receptors that contain two copies of GluN1 and two copies of the same GluN2 subunit. However, two or more GluN2 subunits are expressed in most neurons (Buller et al., 1994; Cull-Candy et al., 2001; Landwehrmeyer et al., 1995), and many native NMDARs appear to be triheteromeric assemblies of two GluN1 and two different GluN2 subunits (Al-Hallaq et al., 2007; Luo et al., 1997; Rauner and Köhr, 2011; Sheng et al., 1994; Tovar et al., 2013). Moreover, there is emerging evidence that triheteromeric receptors exhibit distinct functional properties compared to diheteromeric receptors (Hansen et al., 2014; Hatton and Paoletti, 2005; Stroebel et al., 2014; Tovar and Westbrook, 1999), underscoring their relevance in physiology and development. For example, the best-studied triheteromeric receptor complex contains GluN2A and GluN2B, and shows pharmacological properties distinct from diheteromeric receptors, with intermediate agonist potencies and a submaximal degree of inhibition at saturating concentrations for some modulators (Hansen et al., 2014; Hatton and Paoletti, 2005; Stroebel et al., 2014). However, there is no comprehensive functional description of any other recombinant or native triheteromeric NMDARs.
Cerebellar granule cells are known to express both GluN2A and GluN2C after receiving mossy fiber synaptic input (> P10 age in mice), and their NMDAR properties suggest they contain the GluN2C subunit (Akazawa et al., 1994; Cathala et al., 2000; Lu et al., 2006; Takahashi et al., 1996). However, the existence of triheteromeric GluN1/GluN2A/GluN2C receptors in these cells has not been explicitly demonstrated. The GluN2A and GluN2C subunits are also co-expressed in the cortex of rodent olfactory bulb, spinal cord, and locus coeruleus (Akazawa et al., 1994; Allgaier et al., 2001; Sun et al., 2000; Sundström et al., 1997), as well as the suprachiasmatic nucleus (Clark and Kofuji, 2010; O’Hara et al., 1995), brainstem nuclei (Guthmann and Herbert, 1999), hypothalamus (Al-Ghoul et al., 1997), thalamus (Wenzel et al., 1997), and retinal ganglion cells (Lagréze et al., 2000). Despite these reports of GluN2A and GluN2C co-expression in the CNS by detecting mRNA expression and subunit-selective antibody labeling, there is only a single description of glutamate and glycine potencies in Xenopus oocytes co-expressing GluN1, GluN2A, and GluN2C, in which the authors report an intermediate EC50 with variable Hill slopes (Wafford et al., 1993). However, this study provides no data from neuronal NMDA receptors nor any quantitative analysis of the proportion of receptors with different stoichiometries, complicating interpretation of receptor subtypes. Gradual incorporation of GluN2C subunits into synaptic receptors with age in cerebellar granule cells has been demonstrated (Cathala et al., 2000), and it has been proposed that co-assembly of GluN2C with GluN2A in HEK cells can result in MK801 binding affinities similar to homogenized tissue (Chazot et al., 1994). These studies point to the existence of functional triheteromeric GluN1/GluN2A/GluN2C receptors, but do not address the relative abundance of diheteromeric and triheteromeric GluN2C-containing NMDARs in neurons.
Previous findings from our laboratory have shown that the GluN2C-selective positive allosteric modulator PYD-106 is sensitive to subunit stoichiometry, acting only on diheteromeric GluN1/GluN2C receptors with two copies of the GluN2C subunit and not on triheteromeric GluN1/GluN2A/GluN2C receptors (Kaiser et al., 2018; Khatri et al., 2014). Thus, evaluation of the response of neuronal receptors to this compound and other GluN2C modulators (Mullasseril et al., 2010) could provide information about the relative proportion of native receptors that contain one or two GluN2C subunits. In addition, excision of outside-out patches that contain only a single receptor allows unambiguous determination of the functional properties or receptors with a single copy of GluN2A and GluN2C. We show that GluN2C in neurons overwhelmingly exists as a triheteromeric GluN1/GluN2A/GluN2C receptor, which has unique single channel properties that affect neuronal function. These data shed new light on critical aspects of GluN2C function and expression in the presence of GluN2A, which advances our understanding of triheteromeric NMDARs in central neurons.
Results
GluN2C subunits exist as triheteromeric NMDARs in cerebellar granule cells
To determine the subunit stoichiometry of GluN2C-containing NMDARs in neurons, we exploited two previously described subunit-selective allosteric modulators of GluN2C function that show differential sensitivity to GluN2 subunit composition. Pyrrolidinones, including PYD-106, selectively potentiate recombinant diheteromeric GluN1/GluN2C, but not triheteromeric GluN1/GluN2A/GluN2C NMDARs (Khatri et al., 2014). By contrast, GluN2C/2D-selective positive allosteric modulators such as CIQ act on both di- and triheteromeric GluN2C-containing NMDARs (Mullasseril et al., 2010). To evaluate the expression of diheteromeric GluN1/GluN2C NMDARs and triheteromeric NMDARs that contain only one GluN2C subunit, we compared the effects of PYD-106 and CIQ on cerebellar granule cell current responses evoked by brief pressure-applied pulses of NMDA in acute cerebellar slices from wild type mice. As a control, we confirmed the activity of the CIQ and PYD-106 we used on recombinant NMDARs (Table S1, Supplemental Information). Granule cells in P16-28 cerebellum express both GluN2A and GluN2C subunits (Akazawa et al., 1994; Cathala et al., 2000; Lu et al., 2006; Takahashi et al., 1996). The glycine site agonist D-cycloserine (DCS) was used as a co-agonist in these experiments because it acts as a partial GluN1 agonist at GluN2A-, GluN2B- and GluN2D-containing NMDARs (Hood et al., 1989; Watson et al., 1990), but is more efficacious than glycine at GluN1/GluN2C NMDARs (Supplemental Table S2; (Dravid et al., 2010; Sheinin et al., 2001). Granule cell responses to pressure-applied NMDA in the continuous presence of 30 μM DCS were not potentiated by 50 μM PYD-106 (Figure 1A-D, F; 84±3.8% of DCS alone, n=8 animals, p=0.01), although PYD-106 can potentiate responses of recombinant GluN1/GluN2C in DCS (Khatri et al., 2014). PYD-106 also had no effect on NMDA responses recorded from granule cells with glycine as the co-agonist (n=3 animals, data not shown). In contrast, the GluN2C/2D-selective positive allosteric modulator CIQ (20 μM), which is active at both diheteromeric GluN1/GluN2C and triheteromeric GluN1/GluN2A/GluN2C receptors (Mullasseril et al., 2010), significantly enhanced granule cell responses (Figure 1C,D; 141±6.7% of DCS, n=6 animals, p=0.03). These data confirm that GluN2C-containing NMDARs are expressed in granule cells, and suggest that functional NMDARs carry only a single GluN2C subunit, raising the possibility that diheteromeric receptors that contain GluN1/GluN2C are rarely expressed on the cell surface of neurons in vivo when GluN2A is expressed in the same cell. While our results do not directly address whether diheteromeric GluN1/GluN2A receptors exist once neurons express GluN2C, cerebellar granule cells retain a fast component of synaptic transmission at P21-40 that is compatible with GluN1/GluN2A receptors (Cathala et al., 2000; Takahashi et al., 1996).
Figure 1.
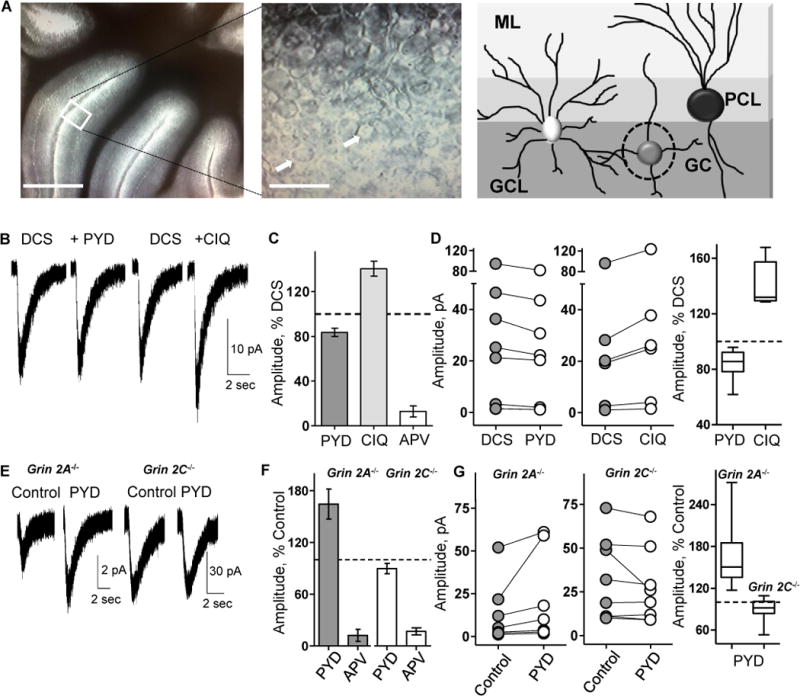
PYD-106 does not potentiate NMDA evoked currents in wild type mouse granule cells. A. Left panel shows acute slice of cerebellum maintained in ACSF (scale 0.5 mm); inset shows a photomicrograph of granule cells (scale 30 μm). White arrows indicate granule cells. Right panel shows cerebellar architecture with molecular layer (ML), the Purkinje cell layer (PCL) and the granule cell layer (GCL). The dashed circle indicates a granule cell (GC). B. Representative current responses to a pulse of NMDA with 30 μM DCS, DCS plus 50 μM PYD-106 (PYD), DCS wash before CIQ application, and DCS plus 20 μM CIQ in the bath are shown. C. The mean±SEM current response amplitudes in PYD-106, CIQ, and 200 μM DL-APV (APV) were expressed as percentage of the mean response in DCS. D. Left panel shows amplitude for DCS and PYD-106 current responses to NMDA (n=8 slices). Middle panel shows response amplitude in DCS before CIQ application and CIQ responses (n=6 slices, p=0.03). Right panel shows comparison of NMDA responses in DCS and PYD-106 and CIQ (horizontal line is median, box 25-75 percentile, whiskers are 5-95 percentile). E. Representative current responses to NMDA/glycine in Grin2A−/− and Grin2C−/− slices for control and 50 μM PYD-106 are shown. F. The current responses in PYD-106 and 200 μM DL-APV were expressed as percent of control. G. Left panel shows individual responses for control and PYD-106 in Grin2A−/− slices (n=8, p=0.0078). Middle panel shows response amplitude for control and PYD-106 in Grin2C−/− slices (n=8, p=0.058). Right panel shows comparison of control and PYD-106 current responses as box-and-whisker plots. Wilcoxon Matched Pair Signed Rank test was used for each experiment.
To investigate whether functional diheteromeric GluN1/GluN2C receptors can be assembled at the plasma membrane in neurons lacking GluN2A, we evaluated granule cell responses in cerebellar slices prepared from Grin2A−/− mice. Granule cell responses to pressure- applied NMDA and glycine in Grin2A−/− slices were potentiated by 50 μM PYD-106 (Figure 1E-G; 164±17% of control, n=8 animals, p=0.008). Since PYD-106 only potentiates diheteromeric GluN1/GluN2C receptors (Figure 3E), these data suggest that diheteromeric GluN1/GluN2C NMDARs are expressed on the cell surface in the absence of GluN2A. To confirm that the NMDAR response was not mediated by compensatory GluN1/GluN2B/GluN2C receptors, which might render GluN2C-containing triheteromeric receptors PYD-sensitive, we expressed GluN1/GluN2B/GluN2C receptors in oocytes and confirmed that they were insensitive to PYD-106 (Figure S1).
Figure 3.
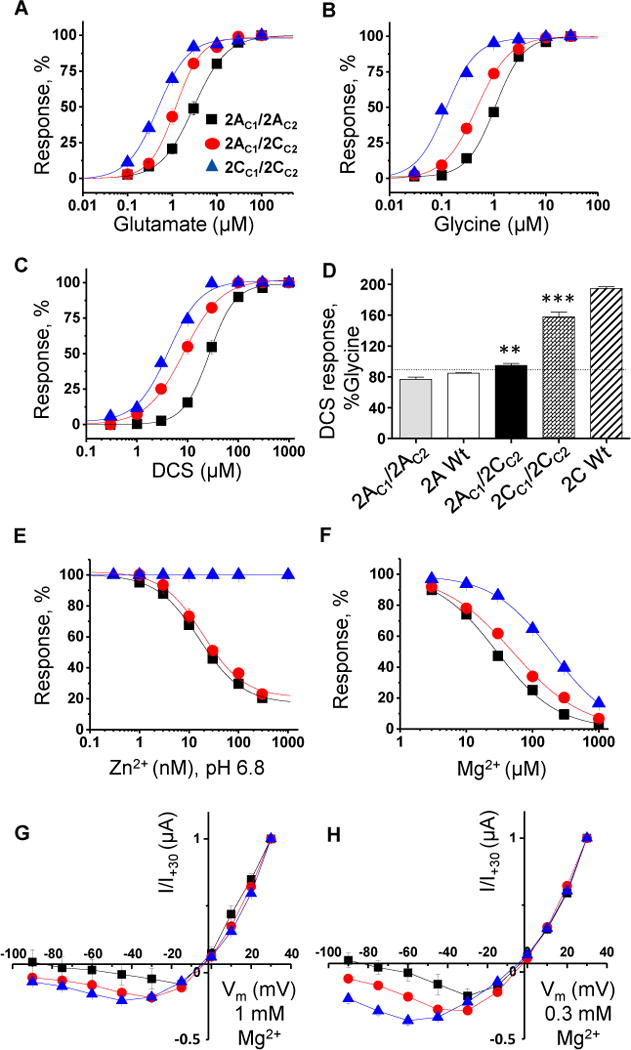
Pharmacological profile of triheteromeric GluN1/GluN2A/GluN2C receptors. A, B, C, D. Glutamate, glycine, and DCS concentration-response data for GluN1/GluN2AC1/GluN2CC2 (2AC1/2CC2), GluN1/GluN2AC1/GluN2AC2 (2AC1/2AC2), and GluN1/GluN2CC1/GluN2CC2 (2CC1/2CC2) receptors in Xenopus oocytes. DCS maximal response at 1000 μM was compared to that of glycine response at 30 μM in the presence of 100 μM glutamate for triheteromeric receptors and wild type GluN1/GluN2A (2A Wt) and GluN1/GluN2C (2C Wt) receptors. The statistical evaluation given in Table S2. E, F. Concentration-response curves for Zn2+ at pH 6.8, and Mg2+. G, H. Current-voltage relationships of NMDAR responses in different concentrations of extracellular Mg2+ (see Table S3).
As a negative control, we evaluated the sensitivity of NMDAR responses in granule cells from Grin2C −/− mice. As expected, granule cell responses in cerebellar slices were not altered by 50 μM PYD-106, confirming the specificity of this compound (Figure 1E-G; 90±7% of control, n=8 animals, p=0.058, Wilcoxon Signed Rank test). All granule cell recordings were terminated by applying 200 μM of the NMDAR antagonist DL-APV, confirming that the current responses were mediated by NMDARs.
GluN2C subunits form functional triheteromeric but not diheteromeric NMDARs in cultured cortical neurons
To assess whether the apparent preference of GluN2C to co-assemble with GluN2A is specific for cerebellar granule cells, we repeated the experiments using stoichiometry-selective modulators in cultured cortical neurons. We transiently expressed either GFP alone (control) or GluN2C and GFP in cortical neurons that endogenously express GluN2A and GluN2B subunits, but not GluN2C or GluN2D (Mizuta et al., 1998). We subsequently evaluated the effects of PYD-106 (50 μM) and CIQ (20 μM) on current responses to pressure-applied NMDA and glycine. PYD-106 did not potentiate NMDAR current responses in either GFP control or GluN2C-transfected cortical neurons (Figure 2A-C, 83±0.6% and 89±1.6% of control, respectively; paired t-tests, p=0.034, n=5, p<0.001, n=5). In the same cortical neurons, CIQ potentiated NMDAR-mediated currents in GluN2C-transfected neurons to 145±9.4% of control (Figure 2B, paired t-test, p=0.04, n=5), but had no effect in control cortical neurons (Figure 2C, 96±4.6% of baseline, p=0.4, n=4). These data demonstrate that GluN2C was not endogenously expressed, and that recombinant GluN2C was incorporated into functional NMDARs. DL-APV (400 μM) inhibited the current responses to 3.4±0.7% of control, confirming these were NMDAR-mediated responses. These data suggest that surface-expressed GluN2C is almost exclusively incorporated into triheteromeric receptors when introduced into cultured cortical neurons.
Figure 2.
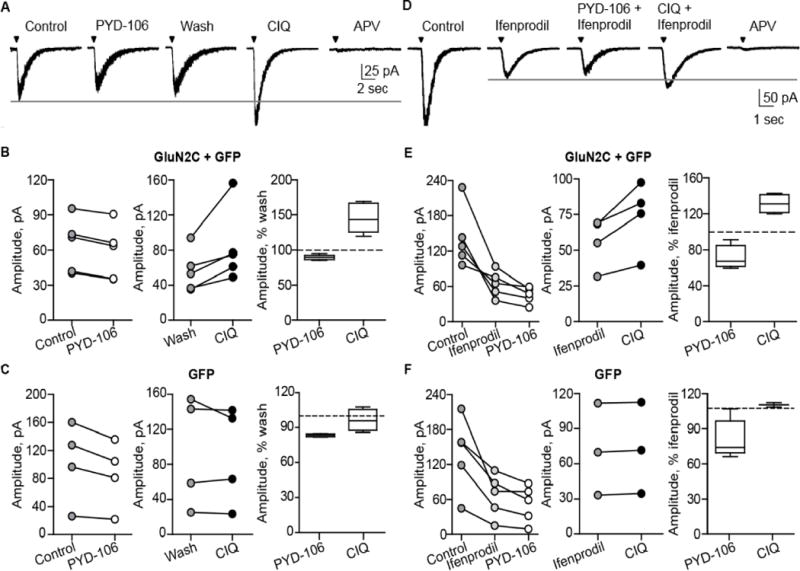
PYD-106 does not potentiate NMDA/glycine-evoked currents in cultured cortical neurons. A, D. NMDAR current responses were evoked by pressure-applied NMDA/glycine (1 mM/0.5 mM) in cultured cortical neurons transfected with GFP or GluN2C and GFP. Current responses were recorded by whole-cell voltage clamp in the presence of 50 μM PYD-106, 10 μM CIQ, 3 μM ifenprodil, or 400 μM DL-APV. B, C. Comparison of NMDAR responses for control vs. PYD-106 followed by CIQ in neurons transfected with GluN2C and GFP or GFP alone; the experiment was terminated with 400 μM DL-APV. E, F. Comparison of NMDAR responses for control and PYD-106 followed by CIQ in the presence of ifenprodil in neurons transfected with GluN2C and GFP or GFP alone. The experiment was terminated in DL-APV and ifenprodil. Responses from the experiments shown (B, C, E, F) with the horizontal lines as the median, boxes showing 25-75 percentile, and whiskers showing 5-95 percentile.
To further explore the failure of PYD-106 to potentiate NMDAR-mediated currents in GluN2C-transfected cortical neurons, we tested if PYD-106 or CIQ could potentiate NMDAR currents when currents mediated by GluN2B-containing receptors were blocked. We reasoned that blocking GluN2B-containing receptors, the majority of native receptors in these cells (Mizuta et al., 1998), might reveal a small current mediated by diheteromeric GluN1/GluN2C receptors. If so, then PYD-106 should potentiate the residual ifenprodil-insensitive current. CIQ potentiated NMDAR current responses in GluN2C-transfected cortical neurons in the presence of ifenprodil (131±5% of baseline, paired t-test, n=4, p=0.03), in contrast to PYD-106 (Figure 2D-F, 72±6% of baseline, paired t-test, n=5, p=0.04). These data further suggest that surface-expressed GluN2C subunits, similar to native receptors in cerebellar granule cells, may only be present as triheteromeric receptors when they are co-expressed with GluN2A or GluN2B.
PYD-106 does not potentiate NMDARs with exon 5-containing GluN1 subunits (Khatri 2014). Therefore, the lack of PYD-106 potentiation could be due to inclusion of alternatively spliced GluN1 exon 5 in cortical neurons. To test for exon 5-containing NMDARs, we examined spermine potentiation of native GluN2B-mediated current responses, as exon 5 also blocks spermine potentiation (Zhang et al., 1994). Spermine potentiated NMDAR currents to 142±7.2% of baseline responses (paired t-test, n=3, p=0.005, Figure S2A, B), indicating that a large population of NMDARs do not contain GluN1 exon 5. These findings are consistent with the idea that recombinant GluN2C is not functionally expressed as diheteromeric NMDAR assemblies at the cell surface in cultured cortical neurons, but that GluN2C preferentially resides in triheteromeric receptors.
Activation of triheteromeric GluN1/GluN2A/GluN2C NMDARs
Our neuronal studies raise the possibility that in cerebellar granule cells, and perhaps other neurons, the majority of GluN2C is present in triheteromeric assemblies with either GluN2A or GluN2B. However, there is virtually no information on the functional properties of GluN2C in triheteromeric NMDARs. To assess the functional properties of GluN1/GluN2A/GluN2C receptors, we co-injected cRNA encoding GluN2A modified to contain coiled-coil domains and an ER retention signal (GluN2AC1), chimeric GluN2C with the sequence encoding the intracellular C-terminal replaced with that of our modified GluN2A (GluN2CC2), and GluN1 into Xenopus oocytes. The resulting GluN1/GluN2AC1/GluN2CC2 receptors produced current responses activated by glutamate and glycine that were detectable 2-3 days post-injection. In addition to triheteromeric GluN1/GluN2AC1/GluN2CC2 receptors, we expect a small percentage of the measured current response arose from diheteromeric GluN1/GluN2AC1 or GluN1/GluN2CC2 receptors that reached the cell surface despite the absence of masking of the ER retention signal by heterodimeric coiled-coil formation. To evaluate the fraction of total current that arose from diheteromeric receptors, we introduced two agonist binding site mutations to each GluN2 subunit that eliminate glutamate binding; these mutant subunits were referred to as GluN2AC1(RK+TI) and GluN2CC2(RK+TI). NMDA receptors require simultaneous binding of two glutamate molecules for channel gating (Benveniste and Mayer, 1991; Clements and Westbrook, 1991; Hansen et al., 2014). Oocytes co-injected with GluN1, GluN2AC1, and GluN2CC2(RK+TI) could produce GluN1/GluN2AC1, GluN1/GluN2CC2(RK+TI), and GluN1/GluN2AC1/GluN2CC2(RK+TI). Of these, only GluN1/GluN2AC1 will be active, and thus the current response can be expressed as a percentage of the current response of oocytes expressing GluN1/GluN2AC1/GluN2CC2 to estimate the percentage current from GluN1/GluN2AC1 receptors that escaped the ER. Similarly, we co-injected GluN1, GluN2AC1(RK+TI), and GluN2CC2 and determined the current response as a fraction of current through GluN1/GluN2AC1/GluN2CC2 to estimate the percentage of the response potentially arising from diheteromeric GluN1/GluN2CC2 that escaped the ER. We restricted our analysis to experiments with escape currents below 10% for the GluN1/GluN2AC1/GluN2CC2(RK+TI) and 2% for the GluN1/GluN2AC1(RK+TI)/GluN2CC2 (Figure S3). On average, the experiments included here showed a summed escape current of 10%.
Using the above strategy, we evaluated agonist potency at GluN1/GluN2AC1/GluN2CC2 receptors. Analysis of glutamate concentration-response curves in the presence of 30 μM glycine for GluN1/GluN2AC1/GluN2CC2 yielded a glutamate EC50 value of 1.3 μM, which is intermediate compared to GluN1/GluN2AC1/GluN2AC2 (2.9 μM) and GluN1/GluN2CC1/GluN2CC2 (0.5 μM) (Figure 3A, Table 1). Analysis of the glycine concentration-response curves generated in the presence of 100 μM glutamate yielded a glycine EC50 value of 0.48 μM for GluN1/GluN2AC1/GluN2CC2, which is intermediate to those for GluN1/GluN2AC1/GluN2AC2 (1.1 μM) and GluN1/GluN2CC1/GluN2CC2 (0.14 μM) (Figure 3B, Table 1). There was no significant difference in the Hill slope for diheteromeric or triheteromeric receptors for either glycine (Hill slope range 1.2-1.6) or glutamate (Hill slope range 1.4-1.7; One-way ANOVA with Tukey’s multiple comparison test), suggesting the response of oocytes co-injected with GluN1, GluN2AC1, GluN2CC2 reflected a single population of receptors (Table 1). Similar results were obtained when a mutation was introduced that removed Mg2+ block for GluN2C-containing receptors, allowing escape current from diheteromeric GluN1/GluN2A receptors to be blocked by addition of Mg2+ (Table 1), confirming that the escape current did not contribute to the intermediate phenotype.
Table 1.
Pharmacological properties of GluN1/GluN2A/GluN2C
| Co-agonists | GluN2AC1/GluN2AC2 EC50 (95% CI) μM maximum b% | N | GluN2AC1/GluN2CC2 EC50 (95% CI) μM maximum b% | N | GluN2CC1/GluN2CC2 EC50 (95% CI) μM maximum b% | N | GluN2AC1/GluN2CC2(NKa) EC50 (95% CI) μM maximum b% | N |
|---|---|---|---|---|---|---|---|---|
| Glutamate | 2.9 (2.1, 3.7) | 14 | 1.3 (0.9, 1.7) | 11 | 0.50 (0.40, 0.60) | 10 | 0.86 (0.73, 1.0) | 8 |
| Glycine | 1.1 (0.97, 1.2) | 15 | 0.48 (0.40, 0.56) | 10 | 0.14 (0.12, 0.16) | 9 | 0.60 (0.50, 0.60) | 9 |
| DCS | 29 (22, 36) | 14 | 10 (8, 13) | 9 | 4.8 (3.9, 5.8) | 8 | 13 (11, 15) | 8 |
| Mg2+ | 26 (25, 32) | 18 | 62 (50, 79) | 11 | 200 (180, 220) | 14 | – | – |
| Zn2+ (pH 7.3) | 0.050 (0.040, 0.063) | 10 | 0.040 (0.037, 0.045) | 10 | NE | 10 | 0.045 (0.042, 0.047) | 12 |
| 57±2.0 c | 55±1.0 c | 57±0.4 c | ||||||
| Zn2+ (pH 6.8) | 0.018 (0.017, 0.020) | 13 | 0.020 (0.018, 0.028) | 14 | NE | 9 | 0.019 (0.017, 0.022) | 14 |
| 21±1.7 c | 23±1.7 c | 27±1.6 c |
The mutation N624K was introduced into GluN2CC2 (see Methods) and responses recorded in 1 mM extracellular Mg2+ to reduce leak current generated from expression of any GluN1/GluN2AC1/GluN2AC1 receptors that reach the surface, which are sensitive to Mg2+ (see Supplemental Figure S3). For Zn2+ concentration-response experiments, 0.1 mM Mg2+ was used to minimize possible effects of Mg2+ on Zn2+ binding.
The mean fitted EC50 value to 2 significant figures is given with the 95% confidence interval (CI) determined from the log EC50, followed by the mean ± SEM fitted maximal response in saturating ligand as a percent of control; no overlap in CI was considered statistically significant.
The fitted response in saturating Zn2+ agonist is given as a percent of control. – indicates no detectable effect. Hill slopes ranged from 0.6-2.8.
Effect of endogenous modulators on GluN1/GluN2A/GluN2C NMDARs
Endogenous modulators exert important effects on circuit function, but their actions on triheteromeric GluN1/GluN2A/GluN2C NMDARs are not known. Here, we determined the effects of two endogenous ions, Zn2+ and Mg2+, on GluN1/GluN2A/GluN2C NMDARs. Extracellular Zn2+ plays important roles in synaptic transmission in the brain through NMDAR modulation (Guilarte et al., 1994; Smart et al., 2004; Ueno et al., 2002; Westbrook and Mayer, 1987), and can inhibit GluN2A-containing NMDARs at concentrations in the low nanomolar range by increasing proton-mediated inhibition of NMDARs at physiological pH. This inhibition is voltage-independent and reflects Zn2+ binding to a high affinity binding site in the GluN2A amino-terminal domain (Choi and Lipton, 1999; Erreger and Traynelis, 2008; Low et al., 2000; Paoletti et al., 1997; Paoletti et al., 2000; Romero-Hernandez et al., 2016; Traynelis et al., 1998). Zn2+ only acts at GluN2C-containing NMDARs via a low affinity binding site within the pore (Paoletti et al., 1997; Williams, 1996). Because of the interaction between Zn2+ and proton inhibition, we analyzed Zn2+ concentration-response curves at pH 8.4, 7.3 and 6.8. To avoid voltage-dependent low affinity inhibition by Zn2+, recordings were performed at -20 mV and the highest concentration of Zn2+ used was 300 nM. Zn2+ did not show any inhibition at alkaline pH (8.4) for triheteromeric receptors activated by glutamate and glycine concentrations of 50 μM (data not shown). The IC50 values of Zn2+ inhibition at triheteromeric GluN1/GluN2AC1/GluN2CC2 were 40 nM at pH 7.3 and 20 nM at pH 6.8, not significantly different from IC50 values at GluN1/GluN2AC1/GluN2AC2 (Table 1, Figure 3E). The extent of inhibition at saturating Zn2+ showed no significant difference compared to GluN1/GluN2AC1/GluN2AC2 at all pH levels (Table 1). These data suggest that the presence of a single GluN2A subunit in the triheteromeric NMDAR assembly is enough for Zn2+ to exert its full inhibitory effect.
Extracellular Mg2+ produces a voltage-dependent channel block of NMDARs that enables the receptor to act as a coincidence detector of activity and synaptic glutamate release (Traynelis 2010). The IC50 value for Mg2+ at −60 mV (62 μM) was intermediate for triheteromeric GluN1/GluN2AC1/GluN2CC2 receptors compared to GluN1/GluN2AC1/GluN2AC2 (26 μM) and GluN1/GluN2CC1/GluN2CC2 (200 μM) receptors (Figure 3F, Table 1). We analyzed the voltage-dependence of Mg2+ inhibition on NMDAR responses to 100 μM glutamate and 30 μM glycine in the presence of 0, 0.1, 0.3, and 1.0 mM Mg2+. The current responses of GluN1/GluN2AC1/GluN2CC2 receptors were intermediate to those of GluN1/GluN2AC1/GluN2AC2 and GluN1/GluN2CC1/GluN2CC2 receptors at corresponding voltages (Figure 3G, H, Table S3). These data suggest that the pore, as expected, is influenced by both GluN2A and GluN2C subunits. The negative modulators for GluN2A (TCN-201, Bettini et al., 2010) and for GluN2C/GluN2D (DQP-1105, Acker et al., 2011) show unique sensitivity at GluN1/GluN2A/GluN2C receptors (Table S1).
Deactivation time course of triheteromeric GluN1/GluN2A/GluN2C NMDARs
We determined the deactivation time course for triheteromeric GluN1/GluN2AC1/GluN2CC2 as well as diheteromeric GluN1/GluN2A and GluN1/GluN2C NMDARs in outside-out patches excised from oocytes. We compared the deactivation kinetics of responses to brief (5 ms) pulses of saturating glutamate (1 mM) in the continuous presence of glycine (100 μM). The time constant for glutamate deactivation (τ) of diheteromeric GluN1/GluN2A (81±8 ms; n=6 from 4 frogs) was approximately 6-fold lower than the corresponding τ of diheteromeric GluN1/GluN2C (Figure 4A-D; 477±40 ms; n=8 from 4 frogs). Glutamate deactivation of triheteromeric GluN1/GluN2AC1/GluN2CC2 was 301±15 ms (n=24 from 4 frogs), which is intermediate to the respective diheteromeric receptors (Figure 4). Thus, neither GluN2A nor GluN2C appear to dominate the deactivation kinetics of triheteromeric GluN1/GluN2AC1/GluN2CC2, in contrast to the deactivation time course of triheteromeric GluN1/GluN2A/GluN2B receptors, which is dominated by the GluN2A subunit through inter-subunit allosteric interactions (Hansen et al., 2014; Sun et al., 2017).
Figure 4.
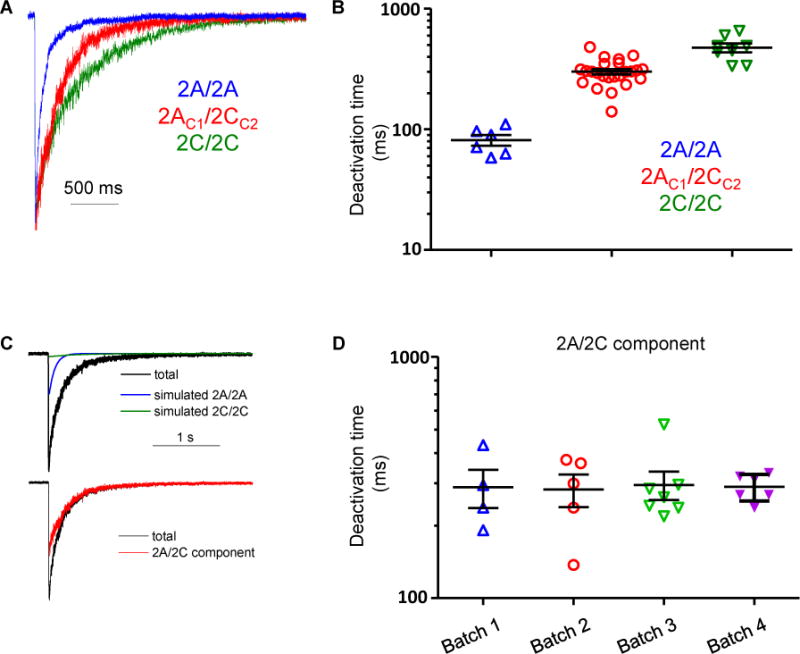
Glutamate deactivation time courses of diheteromeric and triheteromeric NMDARs. A. Normalized representative current responses to brief (<5 ms) application of 1 mM glutamate in the continuous presence of 100 μM glycine from outside-out patches excised from oocytes expressing of GluN1/GluN2A/GluN2A (blue), GluN1/GluN2AC1/GluN2CC2 (red), or GluN1/GluN2C/GluN2C (green). B. Deactivation time constants for GluN1/GluN2A/GluN2A and GluN1/GluN2C/GluN2C receptors were obtained by fitting the time course with a single exponential function. Deactivation time constants for GluN1/GluN2AC1/GluN2CC2 receptors were obtained by fitting the response time course with the sum of three exponential functions. C. The total response from GluN1/GluN2AC1/GluN2CC2 receptors is shown with the GluN1/GluN2A/GluN2A and GluN1/GluN2C/GluN2C receptor components, which were simulated using the deactivation time constant determined from oocytes expressing GluN1/GluN2A/GluN2A and GluN1/GluN2C/GluN2C receptors. The amplitudes of the diheteromeric contribution to the time course were set as the escape current measured from GluN1/GluN2AC1/GluN2AC1 and GluN1/GluN2CC2/GluN2CC2 receptors. The GluN1/GluN2AC1/GluN2CC2 receptor component was isolated by subtracting the GluN1/GluN2A/GluN2A and GluN1/GluN2C/GluN2C receptor components from the total current. D. Summary of deactivation time constants for the GluN1/GluN2AC1/GluN2CC2 receptor component determined from 4 different batches of oocytes.
Open probability of GluN1/GluN2A/GluN2C NMDARs
To determine the open probability of agonist-bound triheteromeric NMDARs, we expressed GluN2AC1 and GluN2CC2 subunits with GluN1-A652C in oocytes, and bath applied 0.2 mM methanethiosulfonate ethylammonium (MTSEA) in the presence of 100 μM glutamate and 30 μM glycine (Wood et al., 1995; Yuan et al., 2005). The open probability (Popen) for GluN1/GluN2AC1/GluN2CC2 (0.03±0.004, n=12, p<0.001) and GluN1/GluN2AC1/GluN2CC2(NK) (0.04±0.06, n=13, p<0.001) was an order of magnitude lower than GluN1/GluN2AC1/GluN2AC2 (0.3±0.02, n=10). However, Popen for GluN1/GluN2AC1/GluN2CC2 was not significantly different from GluN1/GluN2CC1/GluN2CC2 (0.005±0.001, n=11; one-way ANOVA with Tukey’s Test). Popen for wild type GluN1/GluN2A (0.3±0.01, n=4) and GluN1/GluN2C (0.007±0.001, n=6) receptors were also similar to that of GluN1/GluN2AC1/GluN2AC2 and GluN1/GluN2CC1/GluN2CC2 receptors (Figure S4A-E, Table S4). These data suggest that unlike GluN1/GluN2A/GluN2B triheteromers (Sun et al., 2017), GluN2A is not dominant in terms of the open probability of GluN1/GluN2AC1/GluN2CC2 receptors.
Single channel properties of GluN1/GluN2A/GluN2C NMDARs
We determined how single channel properties of triheteromeric GluN1/GluN2A/GluN2C differed from diheteromeric GluN1/GluN2A and GluN1/GluN2C receptors using outside-out patches pulled from HEK 293 cells transiently transfected with GluN2AC1, GluN2CC2, and GluN1. Unitary NMDAR currents were evoked by 1 mM glutamate and 100 μM glycine. Patches were determined to contain a single channel when no double openings were observed and analysis of opening frequency suggested a low probability (p<0.001) of a multichannel recording (Colquhoun and Hawkes, 1990; Dravid et al., 2008) (Figure 5). Records were idealized using time course fitting, and analysis of the total open time during the recording determined that GluN1/GluN2AC1/GluN2AC2 receptors had an open probability of 0.17±0.065 and a mean open time of 2.8±0.56 ms (n=3). Diheteromeric GluN1/GluN2CC1/GluN2CC2 receptors had an open probability of 0.049±0.025 and mean open time of 0.70±0.060 ms (n=7).
Figure 5.
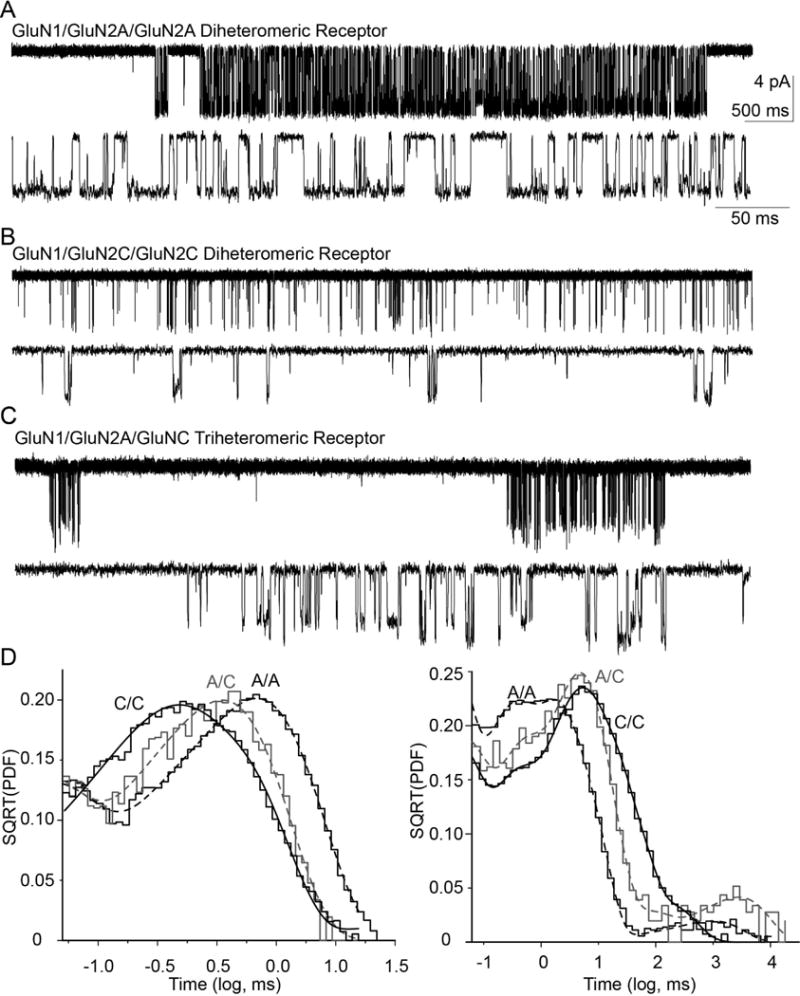
Representative data for single channel recordings from outside-out patches. A, B, C. Unitary currents recorded at −80 mV are shown on compressed and expanded scale for (A) GluN1/GluN2AC1/GluN2AC2, (B) GluN1/GluN2CC1/GluN2CC2, and (C) GluN1/GluN2AC1/GluN2CC2. Openings of diheteromeric GluN1/GluN2AC1/GluNAC2 and triheteromeric GluN1/GluN2AC1/GluN2CC2 NMDARs channels are clustered into bursts, separated by inactive periods. By contrast, openings of diheteromeric GluN1/GluN2CC1/GluN2CC2 NMDARs show no burst structure. D. Representative open duration histogram from a one channel outside-out patch for GluN1/GluN2AC1/GluN2AC2 (31,655 events), GluN1/GluN2CC1/GluN2CC2 (3,004 events), and GluN1/GluN2AC1/GluN2CC2 (2,737 events) NMDARs fitted by three exponential components. Representative shut duration histograms from GluN1/GluN2AC1/GluN2AC2, GluN1/GluN2CC1/GluN2CC2, and GluN1/GluN2AC1/GluN2CC2 NMDARs from single patch recordings fitted with six exponential components.
We subsequently analyzed the unitary currents in patches with a single active channel obtained from cells co-transfected with cDNAs encoding (1) GluN1, GluN2AC1, GluN2AC2, (2) GluN1, GluN2AC1, GluN2CC2, or (3) GluN1, GluN2CC1, GluN2CC2. Three different receptor populations could be present in cells co-transfected with GluN1, GluN2AC1, GluN2CC2, which include GluN1/GluN2AC1/GluN2AC1 and GluN1/GluN2CC2/GluN2CC2, as well as triheteromeric GluN1/GluN2AC1/GluN2CC2. We hypothesized that single channel properties of the triheteromeric receptors would be distinct from those observed for GluN2A and GluN2C receptors, and thus searched for patches with different properties from diheteromeric channels. From this analysis we identified 7 patches containing a single active channel with a unique open time and complex multi-conductance levels that were distinct from diheteromeric receptors, which we conclude were from GluN1/GluN2AC1/GluN2CC2 channels in the patch. Because we restricted analyses to patches with a single active channel, the complex patterns could not have arisen from a mixture of channels with different stoichiometry.
The open probability of triheteromeric receptors calculated from the overall open time was 0.015±0.0092, 11-fold lower than GluN1/GluN2AC1/GluN2AC2 (p<0.0001, see Table 2) but similar to GluN1/GluN2CC1/GluN2CC2 (p=0.13, Table 2). The mean open time for the triheteromeric receptors was 0.84±0.07 ms, significantly different from GluN1/GluN2AC1/GluN2AC2 (p<0.0001, Table 2) but similar to GluN1/GluN2CC1/GluN2CC2 receptors (p=0.8, Table 2). Open duration histograms for di- and triheteromeric NMDARs were fitted with the sum of three exponential components and shut duration histograms were fitted with the sum of six exponential components (Table 2). This analysis revealed differences in stability of the open state and the stability of the pre-gating steps (inferred from three briefest shut time components). Both diheteromeric GluN1/GluN2AC1/GluN2AC2 and triheteromeric GluN1/GluN2AC1/GluN2CC2 channels opened in prolonged bursts separated by long closed times (Figure 5A-C, Table 2), which is apparent from the closed time histograms (Figure 5D). By contrast, there is no prominent slow component in the GluN1/GluN2CC1/GluN2CC2 histogram that separates open periods into clearly defined bursts of openings, and thus no burst analysis is possible (Figures 5).
Table 2.
Single channel properties for GluN1/GluN2A/GluN2C
| Open duration | GluN2AC1/GluN2AC2 | GluN2CC1/GluN2CC2 | GluN2AC1/GluN2CC2 |
|---|---|---|---|
| Tau 1, ms (area) | 0.037±0.004 (23.7±0.8 %) | 0.034±0.005 (15.1±2.8 %) | 0.084±0.05 ms (29.4±3.5 %) |
| Tau 2, ms (area) | 0.303±0.03 (7.3±1.7%) | 0.406±0.05 (49.1±5.5%) | 0.547±0.12 (26.8±10.6%) |
| Tau 3, ms (area) | 3.35±0.69 (68.9±2.2%) | 1.02±0.11 (35.6±6.6%) | 1.63±0.25 (43.7±10.7%) |
| Mean open time, ms | 2.83±0.56 | 0.70±0.055* | 0.84±0.070* |
| Overall Open probability | 0.17±0.065 | 0.049±0.025* | 0.015±0.0092* |
| Intervals | 57,369 | 72,210 | 18,824 |
| Closed duration | GluN2AC1/GluN2AC2 | GluN2CC1/GluN2CC2 | GluN2AC1/GluN2CC2 |
|
| |||
| Tau 1, ms (area) | 0.026±0.0 (35.9±1.6 %) | 0.024±0.002 (26.6±3.5 %) | 0.027±0.003 ms (30.8±4.5 %) |
| Tau 2, ms (area) | 0.212±0.006 (22.4±1.2%) | 0.24±0.02 (10.3±1.3 %) | 0.26±0.04 (14.4±0.78 %) |
| Tau 3, ms (area) | 0.96±0.05 (16.9±0.8%) | 2.89±0.58 (19.1±2.0%) | 3.29±0.99 (19.3±3.3%) |
| Tau 4, ms (area) | 3.39±0.40 (21.1±1.6%) | 11.9±1.5 (29.1±5.3%) | 12.8±4.7 (27.9±3.6%) |
| Tau 5, ms (area) | 8.24±2.2 (3.1±0.5%) | 47.8±8.8 (13.4±3.3%) | 61.2±25.4 (4.8±1.4%) |
| Tau 6, ms (area) | 1754±485 (0.6±0.2%) | 349±164 (1.6±0.7%) | 2853±399 (2.8±0.9%) |
| Intervals | 59,449 | 71,410 | 18,678 |
| Burst Properties | GluN2AC1/GluN2AC2 | GluN2CC1/GluN2CC2 | GluN2AC1/GluN2CC2 |
|
| |||
| Burst length, ms | 654±95 | ND | 209±36 |
| Burst Open Probability | 0.60 | ND | 0.15 |
| Opens in Burst | 99.9% | ND | 99.2% |
| Number Opens/Burst | 504±139 | ND | 64±17 |
| Number of Bursts | 309 | ND | 967 |
| Tcrit (ms) | 41±6.2 | ND | 127±15 |
| Chord conductance | GluN2AC1/GluN2AC2 | GluN2CC1/GluN2CC2 | GluN2AC1/GluN2CC2 |
|
| |||
| γHIGH (area) | 75±5.5 pS (92±0.4%) | – | 75±3.0 pS (29±1.2%) |
| γ MEDIUM (area) | 58±5.0 pS (8±0.4%) | 47±1.8 pS (78±2.2%) * | 59±2.3 pS (66±1.2%) |
| γ LOW (area) | – | 29 ±1.7 pS (22±2.2%) * | 44±1.2 pS (5±0.05%) * |
| Number of currents | 104,014 | 83,536 | 20,116 |
| Number of patches | 3 | 7 | 7 |
Values are mean+SEM. Time constants and relative areas for maximum likelihood fits of the open and shut duration distributions to the sum or 3 to 6 exponential functions. Chord conductances were calculated assuming a reversal potential of 0 mV. Burst properties were calculated using a Tcrit determined for each patch to optimally separate the two slowest components of the shut duration histogram. ND indicates not determined.
p<0.05, significantly different from GluN1/GluN2AC1/GluN2AC2, ANOVA, Tukey post hoc, Holm-Bonferroni correction for family wise error. Power to detect an effect size of 1 was 0.9.
Diheteromeric GluN1/GluN2AC1/GluN2AC2 NMDARs opened to two levels with chord conductances of 75 pS (92%) and 58 pS (8%), assuming a reversal potential of 0 mV. Diheteromeric GluN1/GluN2CC1/GluN2CC2 NMDARs also opened to two sublevels, but with lower chord conductances of 47 pS (78%) and 29 pS (22%). By contrast, the triheteromeric GluN1/GluN2AC1/GluN2CC2 receptors had three clear levels with chord conductances of 75 pS (29%), 59 pS (66%), and 44 pS [5%, Table 2, Figure 6A-D, (Cheffings and Colquhoun, 2000)]. Comparison of the chord conductance levels for the triheteromeric and diheteromeric receptors (Table 2), suggested that the highest conductance state for the triheteromeric receptor (75 pS) is similar to the highest conductance state of diheteromeric GluN1/GluN2AC1/GluN2AC2 (75 pS). The intermediate chord conductance for triheteromeric receptors (59 pS) was not significantly different from the lowest conductance level of GluN1/GluN2AC1/GluN2AC2 (58 pS), but was different from the highest conductance level for GluN1/GluN2CC1/GluN2CC2 (47 pS). The lowest conductance state for the triheteromeric receptor (44 pS) was similar to the highest conductance state of diheteromeric GluN1/GluN2CC1/GluN2CC2 (47 pS). The lowest diheteromeric GluN1/GluN2CC1/GluN2CC2 conductance level (29 pS) appears unique (ANOVA for all, Tukey post-test, p < 0.05, Holm-Bonferroni correction for family wise error). These data raise the possibility that different conductance levels of the triheteromeric receptor are differentially influenced by GluN2A and GluN2C subunits.
Figure 6.
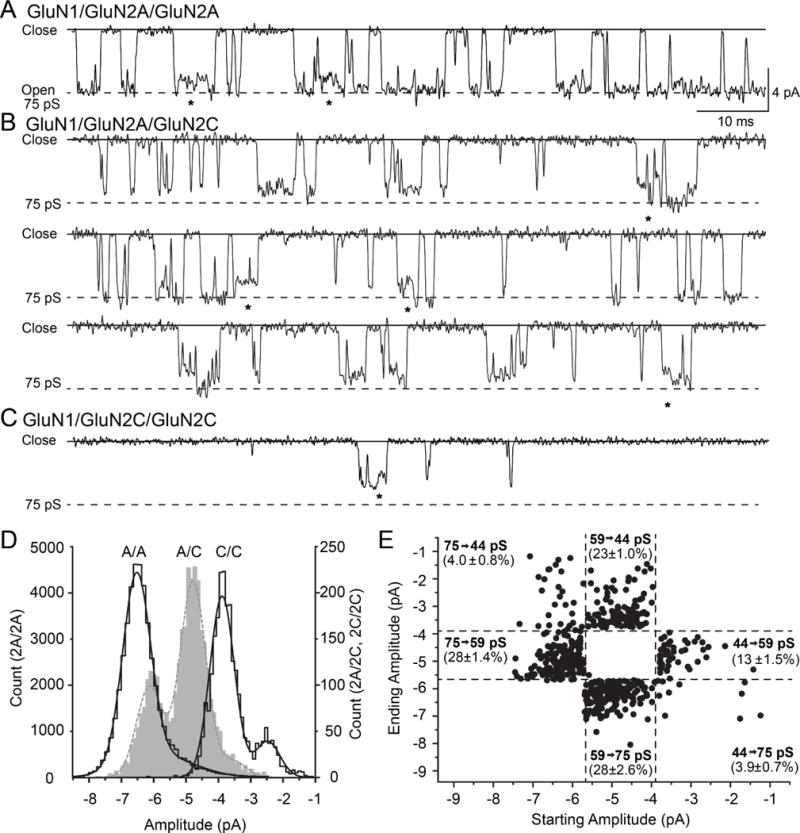
An expanded view of single channel recordings shows multiple conductance levels. A, B, C. Unitary currents recorded from (A) GluN1/GluN2AC1/GluN2AC2, (B) GluN1/GluN2AC1/GluN2CC2, and (C) GluN1/GluN2CC1/GluN2CC2 NMDARs illustrate multiple conductance levels. Direct sublevel transitions are indicated by *. D. Fitted amplitude histograms from single channel analysis of a representative patch with a GluN1/GluN2AC1/GluN2AC2 (A/A; black line, left axis), GluN1/GluN2AC1/GluN2CC2 (A/C; gray, broken line, right axis), or GluN1/GluN2CC1/GluN2CC2 receptor (C/C; black line, right axis) were fitted by the sum of 2-3 Gaussian distributions (smooth lines). E. All contiguous openings with a direct transition between two sublevels with different amplitudes and durations greater than 2.5 filter rise times were identified from the idealized record, and a scatter plot made of the first initial amplitude and the second amplitude. The ACRIT values (broken lines) were calculated as described in the methods to determine which conductance level each amplitude was assigned. The relative proportion of all direct transitions (determined by ACRIT) is given for all possible transitions between sublevels in all patches. When multiple direct transitions occurred, only the first two were analyzed.
We identified 6,733 direct transitions between two different conductance states for diheteromeric GluN1/GluN2CC1/GluN2CC2. We observed an asymmetry in transitions between sub-conductance levels previously, as previously described for GluN1/GluN2C receptors (Dravid 2008). Direct transitions from the high to low diheteromeric GluN1/GluN2CC1/GluN2CC2 sub-conductance states occurred in 65±1.2% of the consecutive openings, whereas 35±1.2% of the direct transitions were from the low to high sub-conductance state.
We identified 1,783 direct transitions between the three different conductance levels in our triheteromeric receptor recordings. When openings showed direct sublevel transitions, 32±1.6% of all openings were to the 75 pS level, 51±2.0% were to the 59 pS state, and 17±2.1% were to the 44 pS level. Thus, the triheteromeric receptors that show direct sublevel transitions open 3-fold more often to the lowest (44 pS) conductance level than when channels directly open to and close from a single level (Table 2). In contrast to direct transitions that include the 75 pS level, an apparent asymmetry exists for transitions between the two lowest conductance levels, with almost twice as many transitions occurring between 59 to 44 pS than from 44 to 59 pS (Figure 6). This asymmetry suggests that the lowest levels share properties with GluN1/GluN2C channels (Dravid et al., 2008), and thus provides clues about how the two subunits contribute to the pore properties of triheteromeric receptors as the various subunits undergo conformational changes that precede channel activation.
Discussion
In spite of their abundant and ubiquitous expression in the CNS, pharmacological and physiological properties of triheteromeric NMDARs that contain two distinct GluN2 subunits largely remain unknown (Al-Hallaq et al., 2007; Luo et al., 1997; Rauner and Köhr, 2011; Sheng et al., 1994). Lack of tractable approaches to express triheteromeric NMDARs of known subunit composition in heterologous systems is a principal reason for the deficit of experimentation on triheteromeric receptors (Javitt and Zukin, 1991; Paoletti and Neyton, 2007; Traynelis et al., 2010; Tu et al., 2010; VanDongen, 2008). Nevertheless, studies continue to highlight important roles of triheteromeric receptors in CNS function (Gray et al., 2011; Rauner and Köhr, 2011; Soares et al., 2013; Tovar et al., 2013). A recently developed system that controls stoichiometry of triheteromeric assemblies on the cell surface has provided a means to overcome the stumbling block of specific triheteromeric NMDAR expression, allowing a detailed investigation of their function (Hansen et al., 2014).
We have evaluated the subunit composition of native triheteromeric NMDARs, and used bioengineered receptors that form triheteromeric assemblies to study receptors that contain both GluN2A and GluN2C. The most important finding of this study is that most GluN2C subunits appear to be expressed in tetramers with GluN2A in cerebellar granule cells. Similarly, all transfected GluN2C subunits in cultured cortical neurons appear to be expressed in tetrameric assemblies with GluN2A or GluN2B. This raises the possibility that diheteromeric GluN2C receptors may be rare or absent on the surface of central neurons when GluN2A or GluN2B are expressed. In addition, we show that receptor function differs for triheteromeric GluN1/GluN2A/GluN2C receptors compared to diheteromeric GluN1/GluN2A and GluN1/GluN2C receptors. In some cases (e.g. agonist potency), intermediate phenotypes might have been predicted. By contrast, there is no data in the literature from which to predict the interesting combination of biophysical properties displayed by GluN1/GluN2A/GluN2C channels that we describe here. We will discuss implications of these two findings in turn.
Functional GluN2C subunits preferably express with GluN2A-containing NMDARs
An important aspect of NMDAR biology is whether GluN2C subunits express on the neuronal surface as diheteromeric GluN1/GluN2C assemblies or prefer to associate with GluN2A as triheteromeric receptors. Although granule cells express GluN2A and GluN2C after P14 (Akazawa et al., 1994), NMDAR responses were not potentiated by PYD-106, which acts only on NMDARs containing two GluN2C subunits. However, CIQ potentiated responses, indicating the presence of functional GluN2C-containing NMDARs. A possible explanation of this result is that GluN2C-containing NMDARs on the neuronal surface contain only one GluN2C subunit. Further supporting this conclusion, PYD-106 potentiated granule cell responses in slices from Grin2A−/− mice, suggesting that NMDARs that contain two GluN2C subunits are functionally expressed in the absence of GluN2A. Hence, diheteromeric GluN2C-containing receptors appear capable of travelling to the surface neurons, but GluN2C preferentially expresses with GluN2A when present, limiting expression of diheteromeric GluN2C-containing NMDARs on the cell surface. Notably, PYD-106 did not potentiate NMDAR responses from cultured cortical neurons transfected with GluN2C, suggesting this may be a general property of GluN2C in neurons. These findings provide a new picture of GluN2C biology, suggesting GluN2C may only exist, at least in terms of functionality, as a triheteromeric receptor in the brain.
Our findings are consistent with single channel recordings from granule cells in cerebellar slices from wild type rodents, which show conductance patterns similar to our triheteromeric receptors (Farrant et al., 1994). Furthermore, single channel conductance levels from granule cells in slices obtained from Grin2A−/− mice almost certainly arise from GluN1/GluN2C receptors (Takahashi et al., 1996), consistent with our data showing PYD-106 sensitivity of NMDAR responses in Grin2A−/− neurons. Takahashi et al. (1996) conclude that different GluN2 subunits form distinct NMDARs in cerebellar granule cells, and do not exclude the possibilities that multiple GluN2 subunits may contribute to individual NMDARs in situ. This is consistent with our explicit demonstration of different channel properties in receptors that contain GluN1/GluN2A/GluN2A, GluN1/GluN2A/GluN2C, and GluN1/GluN2C/GluN2C. We also note that multiple studies of cultured cerebellar granule cells (Cull-Candy et al., 1988; Howe and Cull-Candy, 1988; Howe et al., 1991) show larger conductances than we describe for GluN1/GluN2A/GluN2C receptors here, which likely reflect expression of GluN2B early in development, prior to expression of GluN2C (Iijima et al., 2008).
The synaptic time course is known to vary in granule cells as a function of development, which has been interpreted as a change in NMDAR subunit composition with time. There is an initial acceleration of the EPSC time course (early postnatal development P7-P14, Takahashi et al., 1996, Lu et al., 2006) that correlates with inclusion of GluN1 alternative exon5 (Prybylowski et al., 2000) and is absent in mutant mice lacking GluN2A, suggesting this reflects a switch from GluN2B to GluN2A. Multiple lines of evidence, including altered Mg2+ sensitivity and mRNA expression, suggest that a subsequent slowing of the deactivation time course in older animals reflects incorporation of GluN2C into the NMDAR (Akazawa et al. 1994; Takahashi et al., 1996; Cathala et al., 2000; Iijima et al., 2008). The relative abundance of low conductance channels in cerebellar granule cells (Farrant et al., 1994; Takahashi et al., 1996) that resemble GluN2C-containing NMDARs (Dravid et al., 2008; Stern et al., 1992) supports our interpretation that the intermediate deactivation time course for GluN1/GluN2A/GluN2C receptors accounts for the slowing of the synaptic NMDAR current (Cathala et al., 2000; Lu et al., 2006). Our neuronal recordings at P20 also support expression of triheteromeric receptors at this age, although we cannot rule out expression of diheteromeric GluN2C receptors later than P28, the oldest age we recorded from.
Two potential explanations for the lack of diheteromeric GluN1/GluN2C receptors on the surface of P28 cerebellar granules cells and cultured cortical neurons are that GluN2C preferentially assembles with GluN2A, or that GluN1/GluN2C diheteromers assemble, but do not traffic efficiently to the cell surface. Consistent with reduced trafficking efficiency, GluN2C surface expression was reduced compared to GluN2A in a heterologous system and cultured granule cells, while total protein levels were similar (Lichnerova et al., 2014). The differential regulation of GluN2A and GluN2C trafficking is not well understood, but two binding partners have been identified for GluN2C, 14-3-3 and sorting nexin 27; these proteins promote and restrict GluN2C surface expression, respectively, and do not interact with GluN1 or GluN2A (Cai et al., 2011; Chen and Roche, 2009; Chung et al., 2015). In addition, a truncated GluN2C lacking the C-terminal domain is not expressed on the cell surface, whereas GluN2A and GluN2B lacking the C-terminal domain do traffic to the cell surface, albeit less efficiently than the full-length protein (Lichnerova et al., 2014). Thus, GluN2C likely contains additional regulatory elements that restrict surface expression, and perhaps, inclusion of GluN2A in the tetramer overrides restrictions imposed by GluN2C and enhances surface expression of triheteromers compared to GluN1/GluN2C (Cai et al., 2011; Chen and Roche, 2009; Lichnerova et al., 2014).
Triheteromeric GluN1/GluN2A/GluN2C receptors have distinct properties
The data presented here shed light on unique properties of triheteromeric GluN1/GluN2A/GluN2C NMDARs that are distinct from diheteromeric GluN1/GluN2A or GluN1/GluN2C NMDARs. For many agonists, potentiators, and inhibitors as well as endogenous ligands, GluN1/GluN2A/GluN2C NMDARs had intermediate properties compared to diheteromeric receptors (Table 1, Supplemental Table S1). Interestingly, we found that Zn2+ had a GluN1/GluN2A-like effect on GluN1/GluN2A/GluN2C triheteromeric receptors, suggesting GluN2A dominates the response to this endogenous modulator, similar to the effect of Zn2+ on GluN1/GluN2A/GluN2B NMDARs (Hansen et al., 2014).
We used a single molecule approach to unequivocally record from triheteromeric receptors. Our analysis of single channel unitary currents identified three conductance levels for the triheteromeric GluN1/GluN2A/GluN2C NMDARs, which were clearly distinct from those observed in GluN1/GluN2A or GluN1/GluN2C receptors. The multiple levels in receptors containing three different subunits supports the idea that, like AMPARs (Jin et al., 2003; Rosenmund et al., 1998), individual subunits in the NMDA receptor can also influence unique conductance levels. The single channel properties observed here are consistent with those identified in cerebellar granule neurons (Farrant et al., 1994; Takahashi et al., 1996), suggesting that the heterologous expression system faithfully reproduces native channel properties. Lastly, it appears that the triheteromeric receptors have unique burst properties that capture some aspects of GluN2A function (prominent burst-like activation), yet maintain low open probability and mean open time similar to GluN1/GluN2C NMDARs (Table 2). Taken together, this work reveals the properties of GluN1/GluN2A/GluN2C receptors in both heterologous and native systems and provides important insight into the roles of GluN2C-containing NMDARs in brain physiology.
In summary, we have revealed unique functional properties of GluN1/GluN2A/GluN2C receptors that form a major portion of NMDARs in areas of the adult brain. Using pharmacological probes that can detect subunit composition, we have provided functional data suggesting that GluN2A and GluN2C subtypes are preferred partners for cell surface expression in cerebellar granule cells. These insights create a new precedent for considering the role of GluN2C as a modulatory subunit, and emphasize the need to study receptors that have subunit composition matching that in the central nervous system.
Star methods
Contacts for reagent and resource sharing
Further information and requests for resources and reagents should be directed to the Lead Contact, Dr. Stephen Traynelis (strayne@emory.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAIL
All procedures involving the use of animals were reviewed and approved by the Emory University IACUC. The subjects of this study were randomly selected male and female C57BL/6J mice (The Jackson Laboratory, 16-30 days of age, RRID: MGI:5650797). Animals were housed in fully supervised vivarium, attended by regular technical and veterinarian staff for regular animal welfare and husbandry. Each cage had at most four mice housed at the same time with ad libatum access to food and water. Animals from multiple breeder pairs or trios were randomly chosen for experimentation. Grin2A knock-out (Grin2A−/−) mice were generously provided by Dr. S. Nakanishi at Kyoto University (Kadotani et al., 1996), obtained from Dr. Stefano Vicini at Georgetown University, Washington D.C. Grin2A−/− mice were genotyped through Transnetyx (Cordova, TN). Primer sequences used in genotyping are as follows: Grin2A WT Forward Primer: GCCCGTCCAGAATCCTAAAGG, Reverse Primer: GCAAAGAAGGCCCACACTGATA, Reporter 1: CAACCAGCAAGATAATG, Neomycin Forward Primer: GGGCGCCCGGTTCTT, Reverse Primer Sequence: CCTCGTCCTGCAGTTCATTCA, Reporter 1: ACCTGTCCGGTGCCC. Grin2C knock-out (Grin2C−/−) mice were obtained from Dr. Andres Buonanno at NICHD-NIH, Bethesda, MD, (Karavanova et al., 2007), provided by Dr. Shashank Dravid at Creighton University and were genotyped through Transnetyx. Primers used in genotyping are as follows: Grin2C WT ForwardPrimer: GATGTGGGCAGATGTTCTTCCA, Reverse Primer: TCTGAGGCCTGACCATTGAGA, Reporter 1: CCTCCTCTGCTTCTCC, Lac Z Forward Primer: CGATCGTAATCACCCGAGTGT, Reverse Primer: CCGTGGCCTGACTCATTCC and the corresponding Reporter 1: CCAGCGACCAGATGAT.
Cortical neurons were harvested from Sprague-Dawley rat embryos of both sexes at embryonic day 18 (Charles River Labs, RRID:MGI:5651135) under similar care conditions. Defolliculated stage V-VI Xenopus laevis oocytes were obtained from EcoCyte Bioscience (Austin, TX) or dissected from ovaries obtained from a commercial vendor (Xenopus One, Dexter, MI) as mentioned in method details.
METHOD DETAILS
Patch clamp recording from granule cells in cerebellar slices
Acute cerebellar slices were prepared from male and female wild type P15–P28 C57BL/6J mice (Jackson Laboratories) as well as Grin2A−/− and Grin2C−/− C57BL/6J mice. Mice were euthanized using isoflurane overdose, and then transcardially perfused with ice cold sucrose cutting artificial cerebrospinal fluid (aCSF) that contained (in mM) 230 sucrose, 24 NaHCO3, 10 glucose, 3 KCl, 10 MgSO4, 1.25 NaH2PO4, and 0.5 CaCl2 saturated with 95% O2/5% CO2. The brain was removed, placed in ice-cold cutting aCSF, and the cerebellum was dissected and glued to the stage of a vibratome (Leica VT1200S). Brain slices (250-300 μm) were sectioned in ice-cold cutting aCSF. All slices were incubated at room temperature in aCSF with high magnesium and low calcium, which contained (in mM) 130 NaCl, 24 NaHCO3, 10 glucose, 3 KCl, 3 MgSO4, 1.25 NaH2PO4, and 1 CaCl2 that was saturated with 95% O2/5% CO2 for 1 hr before use. The recording aCSF solution contained (in mM) 130 NaCl, 24 NaHCO3, 10 glucose, 3 KCl, 0.2 MgSO4, 1.5 CaCl2, and 1.25 NaH2PO4 saturated with 95%O2/5% CO2 and filtered through 0.45 μm filter immediately before being used. Recording electrodes pulled from thin walled glass (TW150F-4, World Precision Instruments) had resistances of 7-9 M and were filled with internal solution containing (in mM) 120 Cs-methanesulfonate, 15 CsCl, 10 tetraethylammonium chloride, 10 HEPES, 8 NaCl, 3 Mg-ATP, 1.5 MgCl2, 1 QX-314, 0.3 Na-GTP, and 0.2 EGTA, pH 7.3 (Guzman et al., 2009). Internal solutions were diluted by 5% v/v with deionized water to an osmolality of 290 mOsm and then filtered using a 0.2 μm syringe fitted nylon filter. Recordings were made using an Axopatch 200B amplifier (Molecular Devices), filtered at 1-2 kHz using an eight-pole Bessel filter (−3 dB; Frequency Devices), and digitized at 20 kHz using Axon pClamp8 software. A Picospritzer II (Parker Hannifin) was used to pressure apply brief pulses (6-12 psi for up to 100 ms) of NMDA (0.5 mM) and glycine (0.5 mM) or NMDA (0.5 mM) alone through a micropipette (3-4 MΩ resistance measured in 0.9% saline). During picospritzer recordings, the external solution contained 1 μM TTX (Tocris, Bristol, UK, Cat. No. 1078/1), 20 μM bicuculline (Tocris, Bristol, UK, Cat. No. 2503/10), and 10 μM CNQX purchase from Tocris, Bristol, UK, Cat. No. 1045/1 (Lee et al., 2007). Voltage clamp recordings were performed at a VHOLD of −60 mV unless mentioned otherwise; slices were maintained at room temperature (23°C). NMDAR modulators PYD-106 and CIQ were made as 20 mM stock solutions in DMSO and added dropwise to the aCSF external solution while stirring, and bath applied while currents were evoked by pressure application of NMDA and glycine; solutions were made fresh before each recording. The final DMSO was 0.1-0.5% in modulator-containing solution and vehicle. NMDA or NMDA and glycine were applied at an interval of 20-30 secs. Recording duration for each sweep was 25.2 or 10 seconds. Whole cell capacitance (Cm 3-6 pF), series resistance (Rs 8-38 mΩ), and resting membrane resistance (0.5-8 GΩ) were monitored during the recordings, or from the transient currents produced by a 5 mV step. Following the application of test drug, the NMDAR competitive antagonist D,L-2-amino-5-phosphonovalerate (DL-APV, 200 μM, Tocris, Bristol, UK, Cat. No. 0105/10) was applied to confirm that the currents measured in response to pressure application of NMDA and glycine were mediated by NMDARs (Lee et al., 2007). Data were individually analyzed in a blinded fashion using Clampfit 10.2. All reagents, salts and drugs were purchased from Sigma Aldrich (St. Louis, MO) and Tocris (Bristol, UK) respectively unless mentioned otherwise.
Neuronal culture
Cortical neurons were harvested from Sprague-Dawley rat embryos of both sexes at embryonic day 18 (Charles River Labs) as described previously (Banker and Goslin, 1998). Dissociated neurons (3 × 106) were suspended in 100 μl Rat Neuron Nucleofection reagent (Lonza) with 0.5 μg pMax-GFP cDNA (Lonza) and 1.5 μg rat GluN2C cDNA, and transfected using Amaxa Nucleofector according to manufacturer instructions with preset program O-3. Neurons were recovered in Roswell Park Memorial Institute (RPMI, Gibco, Cat. No: 11-875-101) media containing 10% horse serum for 15 min at 37°C, and then plated on poly-D-lysine-coated (0.5 mg/ml) glass coverslips in DMEM supplemented with 10% FBS. After 1 hour, the media was changed to RPMI media supplemented with GlutaMax-I and B-27 (Invitrogen). Twenty-four hours later, the media was changed to NeuroBasal media supplemented with GlutaMax-I and B-27. Whole-cell voltage-clamp recordings were performed 2-5 days after transfection at a holding potential of −60 mV using an Axopatch 200B amplifier (Molecular Devices) at room temperature. Recording electrodes (3–4 MΩ) were made from thin wall glass micropipettes pulled using a vertical puller (Narishige) and filled with intracellular recording solution consisting of (in mM) 110 D-gluconate, 110 CsOH, 30 CsCl2, 5 HEPES, 4 NaCl, 0.5 CaCl2, 2 MgCl2, 5 BAPTA, 2 NaATP, 0.3 NaGTP, the pH was adjusted to 7.35 with CsOH. Extracellular recording solution contained (in mM) 150 NaCl, 10 HEPES, 3 KCl, 1 CaCl2, 20 D-mannitol, and 10 glucose (pH 7.4 with NaOH). A Picospritzer II was used to evoke NMDAR currents by pressure applying brief pulses (4–12 psi; 3–50 ms) of NMDA (1 mM) and glycine (0.5 mM) through a borosilicate glass tube (3.5 MΩ). NMDAR responses were continuously evoked by single pressure pulses at 30 s intervals while NMDAR modulators were bath-applied in the extracellular recording solution. Recordings were digitized at 20 kHz using Axon pClamp10 software and filtered at 2 kHz using an eight-pole Bessel filter (−3 dB; Frequency Devices). Series resistance (10–20 MΩ) was monitored throughout the experiment and corrected offline using ChanneLab (Traynelis et al., 1998). Cells were excluded if series resistance changed >20%.
DNA constructs
Rat cDNAs of GluN1-1a (GenBank accession number U08261; hereafter GluN1), GluN2A (D13211), GluN2B (U11419), and GluN2C (M91563) were generously provided by Drs. S. Heinemann (Salk Institute, La Jolla, CA) and S. Nakanishi (Osaka Bioscience Institute, Osaka, Japan). To selectively express the triheteromeric GluN1/GluN2A/GluN2C at the membrane surface of the cell, we adapted the GABAB receptor leucine zipper motifs and constructed two C-terminal peptide tags made up of leucine zipper motifs from GABAB1 and GABAB2 (LZ1 and LZ2, respectively) placed upstream of the C terminal dilysine KKXX ER retention/retrieval motifs (Hansen et al., 2014; Khatri et al., 2014). The zipper motifs of the GABAB1 and GABAB2 subunits produce a heterodimeric coiled-coil formation (Kammerer et al., 1999). The ER retention motif is effective unless it is masked by a coiled-coil interaction formed between LZ1 and LZ2 (Brock et al., 2005). A peptide linker (L4) was added to the N-terminal of the constructs to generate L4-LZ1-KKXX and L4-LZ2-KKXX (C1 and C2 respectively). The peptide linker constituted four repetitive amino acid sequences- EAAAK (Arai et al., 2004). We added the C1 and C2 tags to the intracellular C-terminal of GluN2A to produce GluN2AC1 and GluN2AC2, respectively, and then created a chimeric receptor in which we replaced the wild type C-terminal of GluN2C starting at residue 837 with the C-terminal domain of GluN2AC2 starting at GluN2A residue 837 to produce GluN2CC2. Co-expression of GluN1/GluN2AC1/GluN2CC2 in oocytes led to selective expression of functional NMDARs composed of two GluN1 subunits, one GluN2AC1, and one GluN2CC2. To assess the extent to which any diheteromeric receptors that contain two copies of GluN2AC1 or two copies of GluN2CC2 escaped ER retention, and potentially contributed to the current responses, we measured the fraction of escape current. We constructed GluN2AC1 and GluN2CC2 subunits with mutations in the agonist binding pocket that abolish glutamate binding; we introduced the mutations R518K + T690I into 2AC1(RK+TI), and R529K + T701I into 2CC2(RK+TI) (Hansen et al., 2014; Khatri et al., 2014). Simultaneous injection of cRNA for GluN1/GluN2AC1/GluN2CC2(RK+TI) in oocytes should express nonfunctional GluN2CC2-containing receptors, and thus any detectable NMDAR current must arise from diheteromeric GluN1/GluN2AC1 receptors that escape ER retention. This approach was used to determine the contribution of escaped receptors to the overall current response in cells injected with GluN1/GluN2AC1/GluN2CC2. Similarly, injection of GluN1/GluN2AC1(RK+TI)/GluN2CC2 allowed us to determine the proportion of escape current mediated by diheteromeric GluN1/GluN2CC2 receptors (Figure S3).
To further reduce the escape current in our experiments, we introduced the N624K mutation into GluN2CC2 (referred to as NK) and the corresponding GluN2C(RK+TI) constructs, which reduces Mg2+ sensitivity (Hatton and Paoletti, 2005), allowing Mg2+ to block GluN2C-lacking receptors that escape ER retention. When expressed as GluN1/GluN2AC1/GluN2CC2(NK,RK+TI), recordings in presence of 0.1-1.0 mM Mg2+ produced no detectable escape current, suggesting most of our escape current was mediated by diheteromeric GluN1/GluN2AC1 NMDARs that were highly sensitive to block by Mg2+. Hence, all experiments (except those that might be affected by the presence of Mg2+) were repeated with oocytes injected with 2AC1/2CC2(NK) to further minimize any possible escape currents. We also expressed GluN1/GluN2BC1/GluN2CC2 receptors and its corresponding NK variant in oocytes using GluN2CC2 described above in addition to GluN2BC1, as described in Hansen et al. (2014).
Two-electrode voltage-clamp recordings
Defolliculated stage V-VI Xenopus laevis oocytes were obtained from EcoCyte Bioscience (Austin, TX) or dissected from ovaries obtained from a commercial vendor (Xenopus One, Dexter, MI) as previously described (Hansen et al., 2013), and injected with varying amounts of cRNA in 50 nl of water (GluN1: GluN2AC1: GluN2AC2 was injected 1:1:1 after dilution from 0.2 μg/μl stock as 1:15, 1:5 and 1:5 respectively, GluN1: GluN2AC1: GluN2CC2 was injected 1:1:1 after dilution from 0.2 μg/μl stock as 1:15, 1:5 and 1:5 respectively, and GluN1: GluN2CC1: GluN2CC2 was injected 1:1:1 after dilution from 0.2 μg/μl stock as 1:10, 1:3 and 1:3 respectively). The ratios for injection for the RK+TI mutations matched that of the triheteromeric construct. Following injection, oocytes were maintained at 19 °C in Barth’s culture medium composed of (in mM) 88 NaCl, 2.4 NaHCO3, 1 KCl, 0.33 Ca(NO3)2, 0.41 CaCl2, 0.82 MgSO4, 5 Tris-HCl (pH 7.4 with NaOH), 1 U/mL penicillin, 0.1 mg/mL gentamicin sulfate, and 1 g/mL streptomycin. Recordings were performed on the 2nd day post-injection based on pilot escape current measurement studies performed on different batches of oocytes on consecutive days post injections. Escape currents were also measured on each day of recording for all data included in this study. Oocytes were then transferred to a recording chamber and continuously perfused with extracellular recording solution composed of (in mM) 90 NaCl, 1 KCl, 10 HEPES, 0.5-1 BaCl2, 0.01 EDTA (pH 7.4). For recordings in the presence of magnesium, the extracellular recording solution did not contain EDTA. Microelectrodes were made from thin walled borosilicate glass with resistance of 3-5 MΩ (World Precision Instruments TW150F-4) using a vertical puller (Narishige P-10) and filled with 0.3 or 3 M KCl, and current recordings were achieved with a two-electrode voltage clamp amplifier (Warner Instruments model OC-725C). Currents were low-pass filtered at 10 Hz and digitized at 20 Hz using custom acquisition software and National Instruments data acquisition boards. Oocytes were held at −40 mV unless otherwise stated and all experiments were performed at room temperature. Agonist concentration-effect curves were fitted by the Hill equation
| (1) |
where maximum is the response in saturating agonist, EC50 is the concentration of agonist that produces a half-maximal response, and nH the Hill slope. The positive allosteric modulator concentration-response relationship was fitted by
| (2) |
where maximum is the response in saturating modulator as a percent of control, EC50 is the concentration of modulator that produces a half-maximal response, and nH the Hill slope. The negative allosteric modulator concentration-response relationship was fitted by
where minimum is the response in saturating modulator as a percent of control, IC50 is the concentration of modulator that produces a half-maximal inhibiting response, and nH is the Hill slope. Concentration-response curves were determined to calculate the EC50 or IC50 values for each experiment. For statistical significance, EC50 values were determined for individual concentration-response curves for each oocyte, expressed as log10, which were then used to determine the 95% confidence intervals. The mean as well as lower and upper confidence limits were converted back to a linear value and reported as EC50 or IC50 (lower confidence limit, upper confidence limit) for each data set.
The probability of channel opening (Popen) was measured as previously described (Yuan et al., 2005) using MTSEA (Toronto Research Chemicals Inc., Toronto, Canada, Cat. No: A609100) to covalently modify NMDARs that contained GluN1(A652C). NMDARs were activated by 100 μM glutamate and 30 μM glycine, and subsequently treated with 0.2 mM MTSEA. The change in current recorded after the bath application of MTSEA was calculated relative to the current produced without MTSEA in the bath. Popen was calculated from
| (3) |
where Potentiation is the ratio of current in MTSEA to control and γMTSEA/γCONTROL is 0.67 (Yuan et al., 2005).
Single channel recordings
Forty-eight hours prior to recording, the HEK-293 Tet-On Advanced cells (Takara, RRID: CVCL_KU43) were plated on 50 μg/ml poly-D-lysine-coated 5 mm glass coverslips (Warner Instruments) placed in a 12 well plate. Calcium phosphate transfection method was used to transiently transfect these cells 24 h after plating with GluN1, GluN2AC1, GluN2CC2 at a 1:1:1 ratio for a total of 200 ng of DNA per well. GluN1 was under control of an inducible promotor in the pTRE-tight vector, with GFP inserted between the inducible promoter and open reading frame of GluN1 (GFP and GluN1 were not expressed as a fusion protein; Hansen 2014). To decrease the cytotoxic effect of NMDAR expression on the cultured cells, the antagonists DL-APV (200 μM,) and 7-chlorokynurenic acid (200 μM, Tocris, Cat. No. 0237/10) were added to the culture medium. Doxycycline (5 ng/ml) was added to induce low level expression of GluN1, which controls the overall surface expression of NMDARs. Single channel recordings were made from excised outside-out patches 18 to 24 hrs after transfection. All recordings were performed at room temperature (23°C).
Fire-polished thick-wall electrodes (outer diameter 1.5 mm, inner diameter 0.86 mm, Warner Instruments) with a resistance of 9 to 14 MΩ were coated with SYLGARD 184 (World Precision Instruments) and used to pull outside-out patches from low expressing cells determined by the level of EGFP intensity. The intracellular recording solution was the same as that used for neuronal recordings as described above. The extracellular recording solution consisted of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, 0.01 EDTA, and 30 D-mannitol (pH 7.4). 1 mM glutamate and 0.1 mM glycine were used to activate the NMDARs.
Excised outside-out patches were voltage clamped at a holding potential (VHOLD) of −80 mV, filtered at 8 kHz (8-pole Bessel filter, −3 dB), and digitized at 40 kHz. These recordings were digitally filtered at 4 kHz (−3 dB) and idealized by fitting a filtered step response function to each transition [SCAN, David Colquhoun, University College London; (Colquhoun, 1983)]. An open resolution of 40 μs and shut resolution of 40 μs was imposed on the data. The open and closed duration histograms were fitted with the sum of 3-6 exponential functions using maximum likelihood method (Channel Lab, Synaptosoft). The mean open time, mean shut time, and open probability (Popen) were calculated from the summed duration of the individual openings. Bursts were identified using a critical time (Tcrit) that optimally separates the two slowest shut time components by minimizing the total number of misclassified events according to
| (3) |
where A1 and A2 are the areas of the two components, and τ1 and τ2 are the time constants for the two components (Clapham and Neher, 1984; Magleby and Pallotta, 1983).
Estimation of unitary current amplitudes
To determine the amplitude of the channel conductance levels, only the amplitudes of events with a duration longer than 2.5 filter rise time were analyzed to ensure the unitary current reflects the fully open channel. Amplitudes were fitted by the sum of 2 or 3 Gaussian components using an expectation-maximization algorithm; the standard deviation for all components were constrained to be equal. All resolvable openings were used to estimate the parameters of the Gaussian mixture distribution. From the estimated parameters of the Gaussian mixture distribution, each opening greater than 2.5 filter rise times was classified to one of the components of the Gaussian mixture distribution. To minimize the total number of misclassified openings, a Bayes classifier was used (James et al., 2013), which assigned each opening to the most probable component given the amplitude. In the two components case, this is similar to the use of a critical amplitude by Howe et al. (Howe et al., 1991).
Direct transitions between openings were defined as an opening longer than 2.5 filter rise times (Colquhoun, 1983) that transitioned directly to another open state with a non-zero amplitude that was also longer than 2.5 filter rise times. We only evaluated the first two transitions of every sequence of direct transitions, and determined the percentage of events for every possible direct transition.
Macroscopic currents recorded from outside-out patches
Xenopus oocytes were injected as described above. Before patch excision, the fractional escape currents from GluN1/GluN2AC1/GluN2AC1 and GluN1/GluN2CC2/GluN2CC2 receptors were determined for oocytes co-expressing GluN1, GluN2AC1, and GluN2CC2. The oocytes were then incubated for 1-5 min in high osmolality solution containing (in mM) 300 NaCl, 20 HEPES, 6 KCl, 1 CaCl2 and 0.02 EDTA (pH 7.4), and the oocyte vitelline layer was removed with the aid of syringe needles. The oocytes were then transferred to a recording chamber and perfused with extracellular recording solution composed of (in mM) 150 NaCl, 10 HEPES, 3 KCl, 0.5 CaCl2, 0.01 EDTA (pH 7.4 with NaOH). Outside-out patch-clamp recordings were performed with an Axopatch 200B amplifier (Molecular Devices) at room temperature (20°C). Recording electrodes (2-4 Ω) were made from thin-wall glass micropipettes (TW150F-4; World Precision Instruments) pulled using a horizontal puller (P-1000, Sutter Instruments). The electrodes were filled with internal solution as described in neuronal culture experiments. Rapid solution exchange was achieved by placing the outside-out patch in the stream of a two-barrel theta-glass pipette controlled by a piezoelectric translator (Burleigh Instruments). 10-90% open-tip solution exchange times were 0.6–0.8 ms. In the continuous presence of glycine (100 μM), current responses were activated with a brief application of 1 mM glutamate (<5 ms). At least 4 recorded episodes from each outside-out patch recording were then averaged for further analysis. For outside-out patches pulled from oocytes expressing GluN1/GluN2A or GluN1/GluN2C receptors, the deactivation time course was fitted with a mono-exponential function. The deactivation time course for the current recorded from outside-out patches pulled from oocytes expressing GluN1/GluN2AC1/GluN2CC2 receptors was fitted with three exponentials:
| (4) |
where A is the amplitude of the response,τ2A, τ2AC, and τ2C are the deactivation time constants for GluN1/GluN2A, GluN1/GluN2AC1/GluN2CC2, and GluN1/GluN2C, respectively, F2A, F2AC, and F2C are the fraction of GluN1/GluN2AC1/GluN2AC1, GluN1/GluN2AC1/GluN2CC2, and GluN1/GluN2CC2/GluN2CC2 components to the total current amplitude (A) respectively. F2A and F2C were fixed to the values determined by assessing the fractional escape currents of GluN1/GluN2AC1/GluN2AC1 and GluN1/GluN2CC2/GluN2CC2 receptors. τ2A and τ2C were fixed to the values determined for outside-out patches pulled from oocytes expressing GluN1/GluN2A or GluN1/GluN2C receptors. Four batches of oocytes co-expressing GluN1, GluN2AC1, and GluN2CC2 were used to determine the deactivation time courses of GluN1/GluN2AC1/GluN2CC2 receptors. No significant difference was detected among the four batches (Figure 4C and D).
QUANTIFICATION AND STATISTICAL ANALYSIS
Sample sizes to detect an effect size of 1 were determined using G-Power software version 3.1.9.2 to be 0.85-0.99. Effect sizes were approximately determined using previously published data using similar methods. Statistical analyses were performed in GraphPad Prism 5.0 and OriginPro 9.0. Datasets were tested for normality using Kolmogorov-Smirnov test and homogeneity of variances using the Bartlett’s test. The specific statistical methods, n values and the number of model organisms used are reported in figure and table legend or in the relevant text section. Significance level was set as 0.05. EC50 values were determined to be significantly different if there was no overlap of the 95% confidence interval determined from the log EC50 transformed data. When multiple parameters were measured from the same current record, family wise error was corrected using the Holm-Bonferroni method. Specific software used to analyze data are mentioned in respective methods. Electrophysiological data from cerebellar granule cells were blindly analyzed by two individuals.
Supplementary Material
Highlights.
Functional GluN1/GluN2C NMDARs appear to be absent in cerebellar granule cells
GluN2C is preferentially inserted into triheteromeric GluN1/GluN2A/GluN2C NMDARs
GluN1/GluN2A/GluN2C receptors have distinct single channel properties
Acknowledgments
We thank Dr. T. J. Murphy for critical discussions, Dr. Kevin Ogden for development of single channel analysis methods, Phuong Le, Gil Shaulsky, and Dr. Jing Zhang for their outstanding technical support.
This work was supported by the NIH (NS036654 and NS065371 to S.F.T., GM103546 and NS097536 to K.B.H., NS086361 to S.A.S., and NS078873 to A.K).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contribution: SB performed experiments, analyzed data, wrote manuscript, AK performed experiments, analyzed data, wrote manuscript, SAS performed experiments, analyzed data, wrote manuscript, JOR performed experiments, analyzed data, FY performed experiments, analyzed data, wrote manuscript, KBH performed experiments, analyzed data, wrote manuscript, HY performed experiments, analyzed data, and SFT designed experiments, analyzed data, wrote manuscript.
Declaration of Interests: S.F.T. is a member of the Scientific Advisory Board for Sage Therapeutics, principle investigator on a research grant to Emory University from Janssen, and a co-founder of NeurOp Inc.
References
- Akazawa C, Shigemoto R, Bessho Y, Nakanishi S, Mizuno N. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in the cerebellum of developing and adult rats. Journal of Comparative Neurology. 1994;347:150–160. doi: 10.1002/cne.903470112. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Sucher N, Bradley D, Tafazzoli A, Trinh D, Hetrick W, Potkin S, Sandman C, Bunney W, Jones E. Selective alterations in gene expression for NMDA receptor subunits in prefrontal cortex of schizophrenics. Journal of Neuroscience. 1996;16:19–30. doi: 10.1523/JNEUROSCI.16-01-00019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghoul WM, Meeker RB, Greenwood RS. Differential expression of five N-methyl-D-aspartate receptor subunit mRNAs in vasopressin and oxytocin neuroendocrine cells. Molecular brain research. 1997;44:262–272. doi: 10.1016/s0169-328x(96)00205-7. [DOI] [PubMed] [Google Scholar]
- Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ. NMDA diheteromeric receptor populations and associated proteins in rat hippocampus. Journal of Neuroscience. 2007;27:8334–8343. doi: 10.1523/JNEUROSCI.2155-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allgaier C, Durmaz M, Müller D, Franke H, Poelchen W, Wirkner K, Illes P. Single-cell RT-PCR analysis of N-methyl-D-aspartate receptor subunit expression in rat locus coeruleus neurones. Naunyn-Schmiedeberg’s archives of pharmacology. 2001;363:120–123. doi: 10.1007/s002100000348. [DOI] [PubMed] [Google Scholar]
- Arai R, Wriggers W, Nishikawa Y, Nagamune T, Fujisawa T. Conformations of variably linked chimeric proteins evaluated by synchrotron X-ray small-angle scattering. PROTEINS: Structure, Function, and Bioinformatics. 2004;57:829–838. doi: 10.1002/prot.20244. [DOI] [PubMed] [Google Scholar]
- Banker G, Goslin K. Culturing nerve cells. MIT press; 1998. [Google Scholar]
- Benveniste M, Mayer ML. Kinetic analysis of antagonist action at N-methyl-D-aspartic acid receptors. Two binding sites each for glutamate and glycine. Biophysical journal. 1991;59:560–573. doi: 10.1016/S0006-3495(91)82272-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock C, Boudier L, Maurel D, Blahos J, Pin JP. Assembly-dependent surface targeting of the heterodimeric GABAB Receptor is controlled by COPI but not 14-3-3. Molecular biology of the cell. 2005;16:5572–5578. doi: 10.1091/mbc.E05-05-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller AL, Larson HC, Schneider BE, Beaton JA, Morrisett RA, Monaghan DT. The molecular basis of NMDA receptor subtypes: native receptor diversity is predicted by subunit composition. Journal of Neuroscience. 1994;14:5471–5484. doi: 10.1523/JNEUROSCI.14-09-05471.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Loo LS, Atlashkin V, Hanson BJ, Hong W. Deficiency of sorting nexin 27 (SNX27) leads to growth retardation and elevated levels of N-methyl-D-aspartate receptor 2C (NR2C) Molecular and cellular biology. 2011;31:1734–1747. doi: 10.1128/MCB.01044-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathala L, Misra C, Cull-Candy S. Developmental profile of the changing properties of NMDA receptors at cerebellar mossy fiber–granule cell synapses. Journal of Neuroscience. 2000;20:5899–5905. doi: 10.1523/JNEUROSCI.20-16-05899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazot PL, Coleman SK, Cik M, Stephenson FA. Molecular characterization of N-methyl-D-aspartate receptors expressed in mammalian cells yields evidence for the coexistence of three subunit types within a discrete receptor molecule. Journal of Biological Chemistry. 1994;269:24403–24409. [PubMed] [Google Scholar]
- Cheffings CM, Colquhoun D. Single channel analysis of a novel NMDA channel from Xenopus oocytes expressing recombinant NR1a, NR2A and NR2D subunits. JOURNAL OF PHYSIOLOGY-LONDON THEN CAMBRIDGE. 2000;526:481–492. [PubMed] [Google Scholar]
- Chen BS, Roche KW. Growth factor-dependent trafficking of cerebellar NMDA receptors via protein kinase B/Akt phosphorylation of NR2C. Neuron. 2009;62:471–478. doi: 10.1016/j.neuron.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YB, Lipton SA. Identification and mechanism of action of two histidine residues underlying high-affinity Zn 2+ inhibition of the NMDA receptor. Neuron. 1999;23:171–180. doi: 10.1016/s0896-6273(00)80763-1. [DOI] [PubMed] [Google Scholar]
- Chung C, Wu WH, Chen BS. Identification of novel 14-3-3 residues that are critical for isoform-specific interaction with GluN2C to regulate N-methyl-D-aspartate (NMDA) receptor trafficking. Journal of Biological Chemistry. 2015;290:23188–23200. doi: 10.1074/jbc.M115.648436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham D, Neher E. Substance P reduces acetylcholine-induced currents in isolated bovine chromaffin cells. The Journal of Physiology. 1984;347:255–277. doi: 10.1113/jphysiol.1984.sp015065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark JP, Kofuji P. Stoichiometry of N-methyl-D-aspartate receptors within the suprachiasmatic nucleus. Journal of neurophysiology. 2010;103:3448–3464. doi: 10.1152/jn.01069.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J, Westbrook G. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron. 1991;7:605–613. doi: 10.1016/0896-6273(91)90373-8. [DOI] [PubMed] [Google Scholar]
- Colquhoun D, Sigworth FJ. Fitting and statistical analysis of single-channel records. In Single Channel Recording. 1983:191–263. [Google Scholar]
- Colquhoun D, Hawkes A. Stochastic properties of ion channel openings and bursts in a membrane patch that contains two channels: evidence concerning the number of channels present when a record containing only single openings is observed. Proceedings of the Royal Society of London B: Biological Sciences. 1990;240:453–477. doi: 10.1098/rspb.1990.0048. [DOI] [PubMed] [Google Scholar]
- Cotman CW, Monaghan DT, Ottersen OP, Storm-Mathisen J. Anatomical organization of excitatory amino acid receptors and their pathways. Trends in Neurosciences. 1987;10:273–280. [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Current opinion in neurobiology. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Cull-Candy SG, Howe JR, Ogden DC. Noise and single channels activated by excitatory amino acids in rat cerebellar granule neurones. The Journal of Physiology. 1988;400:189–222. doi: 10.1113/jphysiol.1988.sp017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw N, Stein P, Fox K. The role of NMDA receptors in information processing. Annual review of neuroscience. 1993;16:207–222. doi: 10.1146/annurev.ne.16.030193.001231. [DOI] [PubMed] [Google Scholar]
- Dravid SM, Burger PB, Prakash A, Geballe MT, Yadav R, Le P, Vellano K, Snyder JP, Traynelis SF. Structural determinants of D-cycloserine efficacy at the NR1/NR2C NMDA receptors. Journal of Neuroscience. 2010;30:2741–2754. doi: 10.1523/JNEUROSCI.5390-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dravid SM, Prakash A, Traynelis SF. Activation of recombinant NR1/NR2C NMDA receptors. The Journal of Physiology. 2008;586:4425–4439. doi: 10.1113/jphysiol.2008.158634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erreger K, Traynelis SF. Zinc inhibition of rat NR1/NR2A N-methyl-d-aspartate receptors. The Journal of physiology. 2008;586:763–778. doi: 10.1113/jphysiol.2007.143941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant M, Feldmeyer D, Takahashi T, Cull-Candy SG. NMDA-receptor channel diversity in the developing cerebellum. Nature. 1994;368:335. doi: 10.1038/368335a0. [DOI] [PubMed] [Google Scholar]
- Gray JA, Shi Y, Usui H, During MJ, Sakimura K, Nicoll RA. Distinct modes of AMPA receptor suppression at developing synapses by GluN2A and GluN2B: single-cell NMDA receptor subunit deletion in vivo. Neuron. 2011;71:1085–1101. doi: 10.1016/j.neuron.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilarte TR, Miceli RC, Jett DA. Biochemical evidence of an interaction of lead at the zinc allosteric sites of the NMDA receptor complex: effects of neuronal development. Neurotoxicology. 1994;16:63–71. [PubMed] [Google Scholar]
- Guthmann A, Herbert H. Expression of N-methyl-D-aspartate receptor subunits in the rat parabrachial and Kölliker-Fuse nuclei and in selected pontomedullary brainstem nuclei. Journal of Comparative Neurology. 1999;415:501–517. [PubMed] [Google Scholar]
- Guzman JN, Sánchez-Padilla J, Chan CS, Surmeier DJ. Robust pacemaking in substantia nigra dopaminergic neurons. Journal of Neuroscience. 2009;29:11011–11019. doi: 10.1523/JNEUROSCI.2519-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Ogden KK, Yuan H, Traynelis SF. Distinct functional and pharmacological properties of Triheteromeric GluN1/GluN2A/GluN2B NMDA receptors. Neuron. 2014;81:1084–1096. doi: 10.1016/j.neuron.2014.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Tajima N, Risgaard R, Perszyk RE, Jørgensen L, Vance KM, Ogden KK, Clausen RP, Furukawa H, Traynelis SF. Structural determinants of agonist efficacy at the glutamate binding site of N-methyl-D-aspartate receptors. Molecular pharmacology. 2013;84:114–127. doi: 10.1124/mol.113.085803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton CJ, Paoletti P. Modulation of triheteromeric NMDA receptors by N-terminal domain ligands. Neuron. 2005;46:261–274. doi: 10.1016/j.neuron.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Hood WF, Compton RP, Monahan JB. D-cycloserine: a ligand for the N-methyl–aspartate coupled glycine receptor has partial agonist characteristics. Neuroscience letters. 1989;98:91–95. doi: 10.1016/0304-3940(89)90379-0. [DOI] [PubMed] [Google Scholar]
- Howe J, Cull-Candy S. On the kinetics of large-conductance glutamate-receptor ion channels in rat cerebellar granule neurons. Proc R Soc Lond B. 1988;233:407–422. doi: 10.1098/rspb.1988.0030. [DOI] [PubMed] [Google Scholar]
- Howe JR, Cull-Candy SG, Colquhoun D. Currents through single glutamate receptor channels in outside-out patches from rat cerebellar granule cells. The Journal of physiology. 1991;432:143–202. doi: 10.1113/jphysiol.1991.sp018381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima K, Abe H, Okazawa M, Moriyoshi K, Nakanishi S. Dual regulation of NR2B and NR2C expression by NMDA receptor activation in mouse cerebellar granule cell cultures. Proceedings of the National Academy of Sciences. 2008;105:12010–12015. doi: 10.1073/pnas.0805574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James G, Witten D, Hastie T, Tibshirani R. An introduction to statistical learning. Vol. 112. Springer; 2013. [Google Scholar]
- Javitt DC, Zukin SR. Recent advances in the phencyclidine model of schizophrenia. Am J Psychiatry. 1991;148:1301–1308. doi: 10.1176/ajp.148.10.1301. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nature neuroscience. 2003;6:803. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Kadotani H, Hirano T, Masugi M, Nakamura K, Nakao K, Katsuki M, Nakanishi S. Motor discoordination results from combined gene disruption of the NMDA receptor NR2A and NR2C subunits, but not from single disruption of the NR2A or NR2C subunit. Journal of Neuroscience. 1996;16:7859–7867. doi: 10.1523/JNEUROSCI.16-24-07859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TM, Kell SA, Kusumoto H, Shaulsky G, Bhattacharya S, Epplin MP, Strong KL, Miller EJ, Cox BD, Menaldino DS. The bioactive protein-ligand conformation of GluN2C-selective positive allosteric modulators bound to the NMDA receptor. Molecular pharmacology. 2018;93:141–156. doi: 10.1124/mol.117.110940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammerer RA, Frank S, Schulthess T, Landwehr R, Lustig A, Engel J. Heterodimerization of a functional GABAB receptor is mediated by parallel coiled-coil α-helices. Biochemistry. 1999;38:13263–13269. doi: 10.1021/bi991018t. [DOI] [PubMed] [Google Scholar]
- Karavanova I, Vasudevan K, Cheng J, Buonanno A. Novel regional and developmental NMDA receptor expression patterns uncovered in NR2C subunit-β-galactosidase knock-in mice. Molecular and Cellular Neuroscience. 2007;34:468–480. doi: 10.1016/j.mcn.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentros C, Hargreaves E, Hawkins RD, Kandel ER, Shapiro M, Muller RV. Abolition of long-term stability of new hippocampal place cell maps by NMDA receptor blockade. Science. 1998;280:2121–2126. doi: 10.1126/science.280.5372.2121. [DOI] [PubMed] [Google Scholar]
- Khatri A, Burger PB, Swanger SA, Hansen KB, Zimmerman S, Karakas E, Liotta DC, Furukawa H, Snyder JP, Traynelis SF. Structural determinants and mechanism of action of a GluN2C-selective NMDA receptor positive allosteric modulator. Molecular pharmacology. 2014;86:548–560. doi: 10.1124/mol.114.094516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagréze WA, Darstein M, Feuerstein TJ, Otto T, Landwehrmeyer GB. N-methyl-D-aspartate receptor subunit mRNA expression in human retinal ganglion cells. Graefe’s archive for clinical and experimental ophthalmology. 2000;238:486. doi: 10.1007/pl00007888. [DOI] [PubMed] [Google Scholar]
- Landwehrmeyer G, Standaert D, Testa C, Penney J, Young A. NMDA receptor subunit mRNA expression by projection neurons and interneurons in rat striatum. Journal of Neuroscience. 1995;15:5297–5307. doi: 10.1523/JNEUROSCI.15-07-05297.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Mannaioni G, Yuan H, Woo DH, Gingrich MB, Traynelis SF. Astrocytic control of synaptic NMDA receptors. The Journal of Physiology. 2007;581:1057–1081. doi: 10.1113/jphysiol.2007.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichnerova K, Kaniakova M, Skrenkova K, Vyklicky L, Horak M. Distinct regions within the GluN2C subunit regulate the surface delivery of NMDA receptors. Frontiers in cellular neuroscience. 2014;8:375. doi: 10.3389/fncel.2014.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J. Long-term potentiation: outstanding questions and attempted synthesis. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2003;358:829–842. doi: 10.1098/rstb.2002.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proceedings of the National Academy of Sciences. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Fu Z, Karavanov I, Yasuda RP, Wolfe BB, Buonanno A, Vicini S. NMDA receptor subtypes at autaptic synapses of cerebellar granule neurons. Journal of neurophysiology. 2006;96:2282–2294. doi: 10.1152/jn.00078.2006. [DOI] [PubMed] [Google Scholar]
- Luo J, Wang Y, Yasuda RP, Dunah AW, Wolfe BB. The majority of N-methyl-D-aspartate receptor complexes in adult rat cerebral cortex contain at least three different subunits (NR1/NR2A/NR2B) Molecular pharmacology. 1997;51:79–86. doi: 10.1124/mol.51.1.79. [DOI] [PubMed] [Google Scholar]
- Magleby K, Pallotta B. Burst kinetics of single calcium-activated potassium channels in cultured rat muscle. The Journal of Physiology. 1983;344:605–623. doi: 10.1113/jphysiol.1983.sp014958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta I, Katayama M, Watanabe M, Mishina M, Ishii K. Developmental expression of NMDA receptor subunits and the emergence of glutamate neurotoxicity in primary cultures of murine cerebral cortical neurons. Cellular and Molecular Life Sciences CMLS. 1998;54:721–725. doi: 10.1007/s000180050199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Mullasseril P, Hansen KB, Vance KM, Ogden KK, Yuan H, Kurtkaya NL, Santangelo R, Orr AG, Le P, Vellano KM. A subunit-selective potentiator of NR2C-and NR2D-containing NMDA receptors. Nature communications. 2010;1:90. doi: 10.1038/ncomms1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara BF, Andretic R, Heller HC, Carter DB, Kilduff TS. GABA A, GABA C, and NMDA receptor subunit expression in the suprachiasmatic nucleus and other brain regions. Molecular brain research. 1995;28:239–250. doi: 10.1016/0169-328x(94)00212-w. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1– NR2A receptors. Journal of Neuroscience. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P, Neyton J. NMDA receptor subunits: function and pharmacology. Current opinion in pharmacology. 2007;7:39–47. doi: 10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Perin-Dureau F, Fayyazuddin A, Le Goff A, Callebaut I, Neyton J. Molecular organization of a zinc binding N-terminal modulatory domain in a NMDA receptor subunit. Neuron. 2000;28:911–925. doi: 10.1016/s0896-6273(00)00163-x. [DOI] [PubMed] [Google Scholar]
- Rauner C, Köhr G. Triheteromeric NR1/NR2A/NR2B receptors constitute the major N-methyl-D-aspartate receptor population in adult hippocampal synapses. Journal of Biological Chemistry. 2011;286:7558–7566. doi: 10.1074/jbc.M110.182600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Hernandez A, Simorowski N, Karakas E, Furukawa H. Molecular basis for subtype specificity and high-affinity zinc inhibition in the GluN1-GluN2A NMDA receptor amino-terminal domain. Neuron. 2016;92:1324–1336. doi: 10.1016/j.neuron.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Sheinin A, Shavit S, Benveniste M. Subunit specificity and mechanism of action of NMDA partial agonist D-cycloserine. Neuropharmacology. 2001;41:151–158. doi: 10.1016/s0028-3908(01)00073-9. [DOI] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- Smart TG, Hosie AM, Miller PS. Zn2+ ions: modulators of excitatory and inhibitory synaptic activity. The Neuroscientist. 2004;10:432–442. doi: 10.1177/1073858404263463. [DOI] [PubMed] [Google Scholar]
- Soares C, Lee KF, Nassrallah W, Béïque JC. Differential subcellular targeting of glutamate receptor subtypes during homeostatic synaptic plasticity. Journal of Neuroscience. 2013;33:13547–13559. doi: 10.1523/JNEUROSCI.1873-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standaert DG, Friberg IK, Landwehrmeyer GB, Young AB, Penney J. Expression of NMDA glutamate receptor subunit mRNAs in neurochemically identified projection and interneurons in the striatum of the rat. Molecular brain research. 1999;64:11–23. doi: 10.1016/s0169-328x(98)00293-9. [DOI] [PubMed] [Google Scholar]
- Stern P, Béhé P, Schoepfer R, Colquhoun D. Single-channel conductances of NMDA receptors expressed from cloned cDNAs: comparison with native receptors. Proc R Soc Lond B. 1992;250:271–277. doi: 10.1098/rspb.1992.0159. [DOI] [PubMed] [Google Scholar]
- Stroebel D, Carvalho S, Grand T, Zhu S, Paoletti P. Controlling NMDA receptor subunit composition using ectopic retention signals. Journal of Neuroscience. 2014;34:16630–16636. doi: 10.1523/JNEUROSCI.2736-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Shipley MT, Lidow MS. Expression of NR1, NR2A-D, and NR3 subunits of the NMDA receptor in the cerebral cortex and olfactory bulb of adult rat. Synapse. 2000;35:212–221. doi: 10.1002/(SICI)1098-2396(20000301)35:3<212::AID-SYN6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Sun W, Hansen KB, Jahr CE. Allosteric interactions between NMDA receptor subunits shape the developmental shift in channel properties. Neuron. 2017;94:58–64.e53. doi: 10.1016/j.neuron.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström E, Whittemore S, Mo LL, Seiger Å. Analysis of NMDA receptors in the human spinal cord. Experimental neurology. 1997;148:407–413. doi: 10.1006/exnr.1997.6691. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Feldmeyer D, Suzuki N, Onodera K, Cull-Candy SG, Sakimura K, Mishina M. Functional correlation of NMDA receptor ε subunits expression with the properties of single-channel and synaptic currents in the developing cerebellum. Journal of Neuroscience. 1996;16:4376–4382. doi: 10.1523/JNEUROSCI.16-14-04376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, McGinley MJ, Westbrook GL. Triheteromeric NMDA receptors at hippocampal synapses. Journal of Neuroscience. 2013;33:9150–9160. doi: 10.1523/JNEUROSCI.0829-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. Journal of Neuroscience. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Burgess MF, Zheng F, Lyuboslavsky P, Powers JL. Control of voltage-independent zinc inhibition of NMDA receptors by the NR1 subunit. Journal of Neuroscience. 1998;18:6163–6175. doi: 10.1523/JNEUROSCI.18-16-06163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Huerta PT, Tonegawa S. The essential role of hippocampal CA1 NMDA receptor–dependent synaptic plasticity in spatial memory. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- Tu W, Xu X, Peng L, Zhong X, Zhang W, Soundarapandian MM, Belal C, Wang M, Jia N, Zhang W. DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell. 2010;140:222–234. doi: 10.1016/j.cell.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Tsukamoto M, Hirano T, Kikuchi K, Yamada MK, Nishiyama N, Nagano T, Matsuki N, Ikegaya Y. Mossy fiber Zn2+ spillover modulates heterosynaptic N-methyl-D-aspartate receptor activity in hippocampal CA3 circuits. The Journal of cell biology. 2002;158:215–220. doi: 10.1083/jcb.200204066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDongen AM. Biology of the NMDA Receptor. CRC Press; 2008. [Google Scholar]
- Vicini S, Wang JF, Li JH, Zhu WJ, Wang YH, Luo JH, Wolfe BB, Grayson DR. Functional and pharmacological differences between recombinant N-methyl-D-aspartate receptors. Journal of neurophysiology. 1998;79:555–566. doi: 10.1152/jn.1998.79.2.555. [DOI] [PubMed] [Google Scholar]
- Wafford KA, Bain CJ, Le Bourdelles B, Whiting PJ, Kemp JA. Preferential co-assembly of recombinant NMDA receptors composed of three different subunits. Neuroreport. 1993;4:1347–1349. doi: 10.1097/00001756-199309150-00015. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Inoue Y, Sakimura K, Mishina M. Developmental changes in distribution of NMDA receptor channel subunit mRNAs. Neuroreport. 1992;3:1138–1140. doi: 10.1097/00001756-199212000-00027. [DOI] [PubMed] [Google Scholar]
- Watson GB, Bolanowski MA, Baganoff MP, Deppeler CL, Lanthorn TH. d-Cycloserine acts as a partial agonist at the glycine modulatory site of the NMDA receptor expressed inXenopus oocytes. Brain research. 1990;510:158–160. doi: 10.1016/0006-8993(90)90745-w. [DOI] [PubMed] [Google Scholar]
- Wenzel A, Fritschy JM, Mohler H, Benke D. NMDA receptor heterogeneity during postnatal development of the rat brain: differential expression of the NR2A, NR2B, and NR2C subunit proteins. Journal of neurochemistry. 1997;68:469–478. doi: 10.1046/j.1471-4159.1997.68020469.x. [DOI] [PubMed] [Google Scholar]
- Westbrook GL, Mayer ML. Micromolar concentrations of Zn2+ antagonize NMDA and GABA responses of hippocampal neurons. Nature. 1987;328:640–643. doi: 10.1038/328640a0. [DOI] [PubMed] [Google Scholar]
- Williams K. Separating dual effects of zinc at recombinant N-methyl-D-aspartate receptors. Neuroscience letters. 1996;215:9–12. doi: 10.1016/s0304-3940(96)12924-4. [DOI] [PubMed] [Google Scholar]
- Wood MW, VanDongen H, VanDongen A. Structural conservation of ion conduction pathways in K channels and glutamate receptors. Proceedings of the National Academy of Sciences. 1995;92:4882–4886. doi: 10.1073/pnas.92.11.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Erreger K, Dravid SM, Traynelis SF. Conserved structural and functional control of N-methyl-D-aspartate receptor gating by transmembrane domain M3. Journal of Biological Chemistry. 2005;280:29708–29716. doi: 10.1074/jbc.M414215200. [DOI] [PubMed] [Google Scholar]
- Zhang L, Zheng X, Paupard MC, Wang AP, Santchi L, Friedman LK, Zukin RS, Bennett M. Spermine potentiation of recombinant N-methyl-D-aspartate receptors is affected by subunit composition. Proceedings of the National Academy of Sciences. 1994;91:10883–10887. doi: 10.1073/pnas.91.23.10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


