Abstract
Background
Lung adenocarcinoma with EGFR activating mutations will inevitably acquire resistance to first generation TKIs. Acquired EGFR T790M mutation causes about 50% of these resistance cases. Droplet digital PCR (ddPCR) and cobas enables quantification of T790M mutation. Whether these methods can predict clinical response of osimertinib treatment is unknown.
Methods
Tumor and blood samples from 69 stage IIIB-IV NSCLC patients acquired resistance to EGFR-TKI were collected. Cobas® Mutation Test v2 kit was used to detect EGFR mutations in FFPE or plasma samples. Plasma T790M mutation of both osimertinib naïve and treated patients were quantified by Droplet digital PCR (ddPCR).
Results
T790M mutation rate detected by FFPE tissue cobas, plasma ctDNA cobas and plasma ctDNA ddPCR test were 54.5, 21.3 and 30.4% respectively. The T790M positive rate was 52.2% considering all testing methods. The objective response rate (ORR) was 60.9% in 23 patients received osimertinib treatment. Quantification of T790M after treatment decreased to very low level, but no association was observed between clinical response and T790M mutation level decrease.
Conclusion
ddPCR is more sensitive in plama ctDNA testing and should be performed even in tumor tissue T790M test negative cases. EGFR T790M mutation level is not associated with clinical response after osimertinib treatment.
Keywords: NSCLC, ctDNA, Epidermal growth factor receptor, T790M
ᅟ
Part result of this study was published as abstract and poster presentation in IASLC 18th World Conference on Lung Cancer [1].
Background
Lung cancer remains the leading cause of cancer-related death worldwide [2]. Though epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) have significantly improved outcome of EGFR mutation positive advanced non-small cell lung cancer (NSCLC), most of these patients develop resistance to first generation EGFR TKIs after a median of 10–12 months, in which about 50–60% of these tumors harbor EGFR T790M resistance mutation [3–5]. Osimertinib is an oral, irreversible EGFR-TKI that is selective for both EGFR activating and T790M resistance mutations [6]. In AURA3 study, osimertinib showed significant advantage against platinum therapy plus pemetrexed in both progression-free survival and objective response rate [7].
To obtain enough tumor tissue in patients experienced disease progression is challenging, especially in patients receiving first generation EGFR-TKI treatment. In other cases, tumor biopsy tissue may not reflect the overall tumor mutational features due to high tumor burden and heterogeneity. The use of circulating tumor DNA (ctDNA) to detect genetic alterations by various platforms known as liquid biopsy has provided a non-invasive option [8]. The current study focuses on the efficacy of T790M detection by different platforms, as well as T790M level response after receiving osimertinib treatment.
Methods
Patients
Plasma and tumor tissue samples were collected from patients screened or enrolled in the ongoing ASTRIS study. ASTRIS (D5160C00022) is open label, multinational, multicenter, real world treatment study of single agent osimertinib for patients with advanced/metastatic EGFR T790M mutation positive NSCLC who have received prior therapy with an first generation EGFR-TKI. Patients were required to have the diagnosis of EGFR activating mutation positive NSCLC. Drug resistance was defined as disease progression after first generation EGFR-TKI (gefetinib, elortinib or ecotinib) treatment, which was evaluated by RECIST 1.1 criteria. EGFR T790M positive patients received osimertinib 80 mg Qd until disease progression evaluated by investigators.
Tumor tissue and plasma sample collection and mutational analysis
Tumor tissue or plasma samples were collected from patients screened or enrolled in ASTRIS study, following progression on a previous EGFR-TKI, but prior to dosing with osimertinib. CT-guided needle lung biopsy or Supraclavicular lymph node biopsy samples were snap frozen and stored at − 80 °C. A total of 8 ml of Plasma sample from each patient were assigned for cobas and ddPCR testing, and 4 ml of plasma from the time point of first evaluation (6 weeks) after osimertinib treatment were evaluated by ddPCR. ctDNA isolation followed standard protocols. Cobas® Mutation Test v2 kit was used to detect both EGFR activating mutation and T790M in FFPE or plasma samples. The Cobas is an allele-specific polymerase chain reaction assay to detect known common EGFR mutations. Analysis was confirmed by negative and positive controls. The PCR reactions were run on the cobas®z 480 analyzer with EGFR Blood Analysis Package Software. A semiquantitative index (SQI) was generated to reflect the proportion of mutated and wild-type copies of the EGFR gene. The SQI was derived from a dilution series with known copy numbers of mutated EGFR and a fixed amount of wild-type EGFR, with the wildtype DNA copy as internal control. Bio-rad PrimePCR™ ddPCR™ Mutation Assay kit were used to detect T790M in plsma ctDNA. The RNase P Copy Number Reference Assay (Life Technologies, Carlsbad, California) was used to determine the total amount of DNA in plasma samples. A total of 3000–3500 genome equivalents were analyzed per reaction with a sensitivity between 0.1–0.5%.
Statistical analysis
Associations between mutations and clinical and biological characteristics were analyzed by χ2 or Fisher’s exact test. All data were analyzed using the Statistical Package for the Social Sciences Version 16.0 Software (SPSS Inc., Chicago, IL). The two-sided significance level was set at p < 0.05.
Results
Study design and patient characteristics
The flowchart of study design was shown in Fig. 1. Among the 69 screened NSCLC patients, there were 23 men and 46 women. There were 22 smokers and 47 non-smokers, accounting for 31.9 and 68.1%, respectively. There were 67 adenocarcinoma and 2 Adenosquamous carcinoma. The number of patients in stages IIIB, IVA and IVB was 20, 21, and 28, respectively. The numbers of M1a, M1b and M1c patients were 19, 4 and 26. The detailed information is listed in Table 1.
Fig. 1.
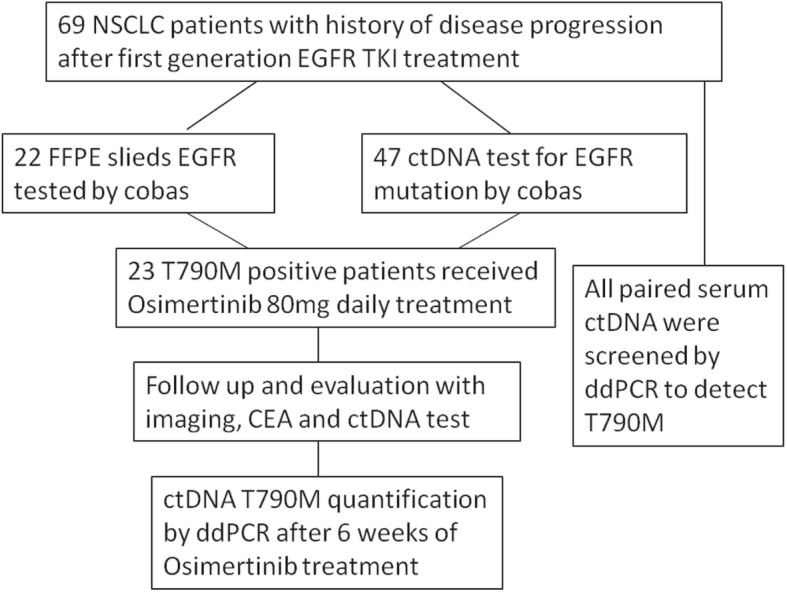
Flowchart of the study design
Table 1.
Demographics and clinicopathologic features of 69 NSCLC patients screened for T790 M mutation
| Variables | No. of patients (%) |
|---|---|
| Gender | |
| Male | 23(33.3) |
| Female | 46(66.7) |
| Age | |
| ≤ 40 | 5 (7.2) |
| > 40 | 64(92.8) |
| ECOG performance status | |
| 0–1 | 58(84.1) |
| 2 | 11(15.9) |
| Histologic subtype | |
| Adenocarcinoma | 67(97.1) |
| Adenosquamous carcinoma | 2 (2.9) |
| Smoking status | |
| Smoker | 22(31.9) |
| Non-smoker | 47(68.1) |
| TNM Stage | |
| IIIB | 20(29.0) |
| IVA | 21(30.4) |
| IVB | 28(40.6) |
| M Stage | |
| M1a | 19(38.8) |
| M1b | 4 (8.2) |
| M1c | 26(53.0) |
| First generation EGFR TKIs | |
| Gefitinib | 22(31.8) |
| Elortinib | 15(21.7) |
| Ecotinib | 31(44.9) |
| Two or more TKI drugs | 1 (1.4) |
| EGFR mutation detection | |
| FFPE slieds by cobas | 22 |
| cfDNA by cobas | 47 |
| T790 M ddPCR assay | 69 |
| Total | 69 |
Detection of T790M mutation by cobas® and ddPCR platform
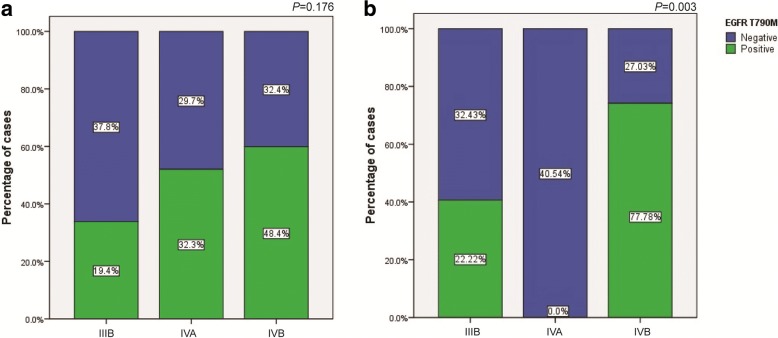
The T790M mutation rate of FFPE tissue cobas, plasma cobas and plasma ddPCR were 54.5, 21.3 and 30.4% respectively. The T790M positive rate was 52.2% considering all testing methods. T790M mutation was detected by ddPCR in all of the plasma cobas positive cases. However, in 10 tumor tissue biopsy cobas negative cases, 3 were positive by plasma ctDNA ddPCR. The proportion of T790M positive cases detected by ddPCR in stage IIIB, IVA and IVB patients were 30, 47.6 and 57.1%, respectively (P = 0.176) (Fig. 2a). However, T790M positive defined by cobas plasma ctDNA test were significantly different in stage IIIB, IVA and IVB (Fig. 2b) or M1a, M2b and M1c patients (P = 0.003, 0.004), suggesting the advantages of ddPCR test in any stages of disease. The T790M quantification by ddPCR significantly rises in stage IIIB, IVA and IVB cases (P = 0.033), while no such trend was observed in different M1a, M2b and M1c cases (P = 0.178). No association was observed between T790M status and duration of first generation EGFR TKI treatment (Table 2).
Fig. 2.
Distribution of T790 M positive and negative cases in stage IIIB-IVB NSCLC patients with acquired resistance to first generation EGFR TKIs. a.ctDNA T790 M mutation detected by ddPCR; b.ctDNA T790 M mutation detected by Cobas
Table 2.
Association between EGFR T790 M mutation and clinical features in patients with disease progression after receiving 1st generation EGFR TKI treatment
| EGFR T790M detected by any method | EGFR T790M detected by plama ctDNA ddPCR | T790M detected by cobas plama ctDNA test | Cobas Activating Mutation SQI | Cobas T790M SQI | ddPCR T790M quantitative | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristics | Total | Mutant | Wild type | P | Mutant | Wild type | P | Mutant | Wild type | P | Mean | P | Mean | P | Mean | P |
| Gender | ||||||||||||||||
| Male | 23 | 12 | 11 | 1 | 11 | 12 | 0.864 | 2 | 12 | 0.7 | 10.24 | 0.859 | 9.81 | 0.101 | 0.52 | 0.307 |
| Female | 46 | 24 | 22 | 21 | 25 | 8 | 25 | 10.55 | 8.17 | 1.4 | ||||||
| Smoking status | ||||||||||||||||
| Smoker | 22 | 11 | 11 | 0.805 | 10 | 12 | 0.916 | 3 | 11 | 1 | 10.59 | 0.897 | 9.8 | 0.472 | 1.63 | 0.384 |
| Non-smoker | 47 | 25 | 22 | 22 | 25 | 7 | 26 | 10.37 | 7.94 | 0.86 | ||||||
| Clinical Stage | ||||||||||||||||
| IIIB | 20 | 9 | 11 | 0.495 | 6 | 14 | 0.176 | 2 | 12 | 0.003 | 9.24 | 0.444 | 7.8 | 0.77 | 0.29 | 0.033 |
| IVA | 21 | 10 | 11 | 10 | 11 | 0 | 15 | 10.03 | NA | 0.2 | ||||||
| IVB | 28 | 17 | 11 | 16 | 12 | 8 | 10 | 11.51 | 8.68 | 2.37 | ||||||
| M Stage | ||||||||||||||||
| M1a | 19 | 9 | 10 | 0.627 | 9 | 10 | 0.827 | 0 | 15 | 0.004 | 10.03 | 0.541 | NA | NA | 0.21 | 0.178 |
| M1b | 4 | 2 | 2 | 2 | 2 | 0 | 1 | NA | NA | 1.07 | ||||||
| M1c | 26 | 16 | 10 | 15 | 11 | 8 | 9 | 11.51 | 8.5 | 2.4 | ||||||
| Time of EGFR-TKI treatment | ||||||||||||||||
| ≤ 6 months | 14 | 6 | 8 | 0.733 | 6 | 8 | 0.952 | 1 | 6 | 0.621 | 10.5 | 0.082 | 8.99 | 0.203 | 0.83 | 0.472 |
| 6–12 months | 26 | 14 | 12 | 12 | 14 | 5 | 12 | 12.71 | 10.32 | 1.75 | ||||||
| >12 months | 29 | 16 | 13 | 14 | 15 | 4 | 19 | 8.86 | 6.11 | 0.67 | ||||||
ddPCR Droplet Digital PCR, SQI Semi-Quantitative Index, Bold numbers showing statisticaly significant results
Evaluation of plasma T790M level during osimertinib treatment by ddPCR
In 23 patients received osimertinib treatment, the OOR was 60.9%. There were 14 patients evaluated as partial response (PR) and 8 were stable disease (SD), 1 patient experienced PR of liver metastasis tumor but progression of primary lung lesion. Quantification of T790M after 6 weeks of treatment decreased to very low level, while no association was observed between response status and T790M mutation level decrease (Fig. 3).
Fig. 3.
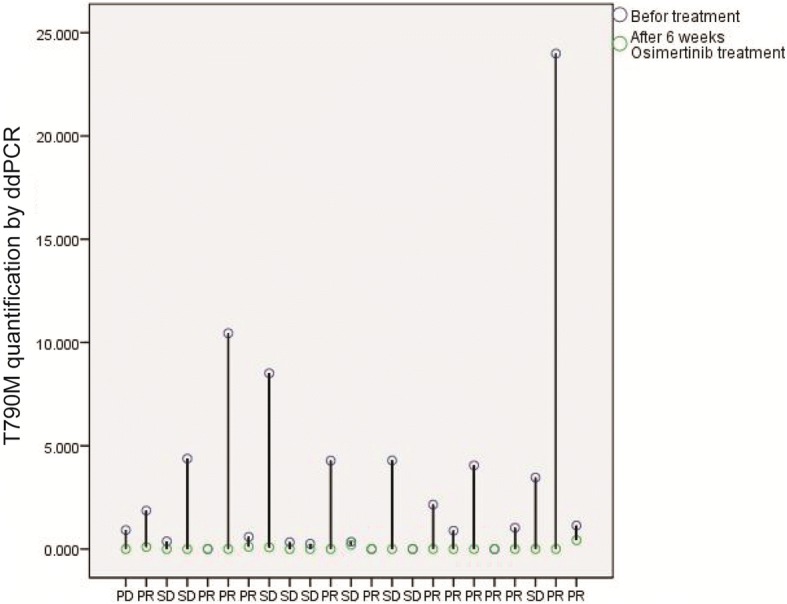
ctDNA T790 M quantification by ddPCR before and after osimertinib treatment. PD, Progressive Disease; PR, Partial Response; SD, Stable Disease
Discussion
The aim of this study was to evaluate different T790M detecting methods in advanced NSCLC patients who experienced disease progression after receiving EGFR TKI treatment, as well as T790M quantification after osimertinib treatment. Two quantification methods were tested on a cohort of 69 patients enrolled in this single center as part of the multicenter real-world ASTRIS study. These patients represent outline features of Chinese patients who experienced disease progression after gefetinib, elortinib or ecotinib treatment. Plasma samples were collected at screening and 6 weeks after receiving osimertinib treatment. The overall T790M positive rate was 52.2% considering all testing methods, the ORR of T790M positive patients receiving osimertinib treatment was 60.9%. These data were similar compared with published data [7, 9–11].
Our analysis revealed a rising trend of T790M positive rates detected by ddPCR in stage IIIB, IVA and IVB patients. In plasma ctDNA samples tested by cobas, T790M positive rate was significantly higher in stage IVB than stage IIIB and IVA, M1c than M1a and M1b patients. On one side, more advanced stage represents significantly higher tumor burden, in which case tumor shed more ctDNA to the bloodstream [12, 13]. On the other side, these results indicate that the cobas test is less capable of detecting relatively earlier stage cases.
In all of the plasma ctDNA cobas test T790M positive samples, ddPCR test also yielded positive results. Even in 10 tumor tissue test negative cases, 3 were positive defined by plasma ctDNA ddPCR test. These results suggest that plasma ctDNA ddPCR test is more sensitive and should be used as primary choice in managing patients with resistance to first line EGFR TKIs. The reason of inconsistency between tumor tissue test and ddPCR test is probably due to tumor heterogeneity in primary and metastatic tumors, as well as intratumor heterogeneity. These facts suggests co-existing of multiple resistant clones or single clone harboring multiple resistance mechanism [14, 15]. Plasma ctDNA ddPCR test should be routinely performed in such cases considering its noninvasive and low cost feature.
Most of patients showed a PR or SD status after the evaluation of 6 weeks after receiving osimertinib treatment, generating an ORR of 60.9%. We also compared the ctDNA T790M level in pre and post osimertinib treatment plasma samples. Though all plasma ctDNA T790M decreased to very low level, no association was observed with radiographic response. Previous studies dynamically monitored EGFR mutation status using plasma samples by ddPCR to evaluate response to first generation EGFR TKIs [16]. Another study quantified plasma T790M level in two cases of patients who received osimertinib treatment [17]. In the present study, we have quantified T790M level in relatively large number of samples. Though quantification of plasma ctDNA T790M didn’t predict response in short term, dynamic monitoring may indicate disease progression in the long run.
Conclusion
In conclusion, our data suggest that ddPCR is more sensitive in plama ctDNA testing and should be performed even in tumor tissue T790M test negative cases. Osimertinib significantly decreased plasma T790M level, but no association was observed between plasma ctDNA T790M level decrease and clinical response.
Acknowledgements
We thank Yongsheng Sha, Hao He, Lu Ye, Mei Liu, Aiping Xu and Yan Yan for managing clinical samples.
Funding
This work was supported by the National Natural Science Foundation of China (81302001, 81772484), National Key Research and Development Program of China (2016YFC0905501, 2016YFC0905500), China Postdoctoral Science Foundation (2014 M550147, 2015 T80225), Cancer translational medicine seed fund of Tianjin medical university cancer institute and hospital (Fund No.1606).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
CG L, RJ, BZ and HL L analyzed patient and experiment data. RJ and CG L was the major contributor in writing the manuscript. CL W and CG L designed and supervised the research. All authors read and approved the final manuscript.
This study was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent before screening. The ASTRIS study protocol was reviewed and approved by review boards of Tianjin medical university cancer institute and hospital.
Not applicable.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chenguang Li, Email: cgli82@yeah.net.
Rui Jia, Email: jiarui156@163.com.
Hailin Liu, Email: xiaolin-happy@163.com.
Bin Zhang, Email: binzh1028@163.com.
Changli Wang, Email: wangchangli309@163.com.
References
- 1.Li C, Liu H, Zhang B, Zhang Z, Su Y, Wang C. EGFR T790M mutation detection and osimertinib treatment response evaluation by liquid biopsy in advanced NSCLC patients. J Thorac Oncol. 2017;12(11):S2142. doi: 10.1016/j.jtho.2017.09.1291. [DOI] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–2247. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oxnard GR, Arcila ME, Chmielecki J, Ladanyi M, Miller VA, Pao W. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJ, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046–1061. doi: 10.1158/2159-8290.CD-14-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or platinum-Pemetrexed in EGFR T790M-positive lung Cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murtaza M, Dawson SJ, Tsui DW, Gale D, Forshew T, Piskorz AM, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 9.Thress KS, Jacobs V, Angell HK, Yang JC, Sequist LV, Blackhall F, et al. Modulation of biomarker expression by Osimertinib: results of the paired tumor biopsy cohorts of the AURA phase I trial. J Thorac Oncol. 2017;12:1588–1594. doi: 10.1016/j.jtho.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Satouchi M, Tanaka H, Yoshioka H, Shimokawaji T, Mizuno K, Takeda K, et al. Detection of epidermal growth factor receptor gene T790M mutation in cytology samples using the cobas(R) EGFR mutation test. Lung Cancer. 2017;111:190–194. doi: 10.1016/j.lungcan.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 11.Jenkins S, Chih-Hsin Yang J, Janne PA, Thress KS, Yu K, Hodge R, et al. EGFR mutation analysis for prospective patient selection in two phase II registration studies of Osimertinib. J Thorac Oncol. 2017;12:1247–1256. doi: 10.1016/j.jtho.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Diaz LA, Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knol AC, Vallee A, Herbreteau G, Nguyen JM, Varey E, Gaultier A, et al. Clinical significance of BRAF mutation status in circulating tumor DNA of metastatic melanoma patients at baseline. Exp Dermatol. 2016;25:783–788. doi: 10.1111/exd.13065. [DOI] [PubMed] [Google Scholar]
- 14.Belchis DA, Tseng LH, Gniadek T, Haley L, Lokhandwala P, Illei P, et al. Heterogeneity of resistance mutations detectable by nextgeneration sequencing in TKI-treated lung adenocarcinoma. Oncotarget. 2016;7:45237–45248. doi: 10.18632/oncotarget.9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan R, Wang Z, Lee JJ, Wang S, Li Q, Tang F, et al. Comprehensive analysis of the discordance of EGFR mutation status between tumor tissues and matched circulating tumor DNA in advanced non-small cell lung Cancer. J Thorac Oncol. 2017;12:1376–1387. doi: 10.1016/j.jtho.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Xiong L, Cui S, Ding J, Sun Y, Zhang L, Zhao Y, et al. Dynamics of EGFR mutations in plasma recapitulates the clinical response to EGFR-TKIs in NSCLC patients. Oncotarget. 2017;8:63846–63856. doi: 10.18632/oncotarget.19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chia PL, Do H, Morey A, Mitchell P, Dobrovic A, John T. Temporal changes of EGFR mutations and T790M levels in tumour and plasma DNA following AZD9291 treatment. Lung Cancer. 2016;98:29–32. doi: 10.1016/j.lungcan.2016.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.