Abstract
Although the clinical use of recombinant adeno-associated virus (rAAV) vectors is constantly increasing, the development of suitable quality control methods is still needed for accurate vector characterization. Among the quality criteria, the titration of infectious particles is critical to determine vector efficacy. Different methods have been developed for the measurement of rAAV infectivity in vitro, based on detection of vector genome replication in trans-complementing cells infected with adenovirus, detection of transgene expression in permissive cells, or simply detection of intracellular vector genomes following the infection of indicator cells. In the present study, we have compared these methods for the titration of infectious rAAV8 vector particles, and, to assess their ability to discriminate infectious and non-infectious rAAV serotype 8 particles, we have generated a VP1-defective AAV8-GFP vector. Since VP1 is required to enter the cell nucleus, the lack of VP1 should drastically reduce the infectivity of rAAV particles. The AAV8 reference standard material was used as a positive control. Our results demonstrated that methods based on measurement of rAAV biological activity (i.e., vector genome replication or transgene expression) were able to accurately discriminate infectious versus non-infectious particles, whereas methods simply measuring intracellular vector genomes were not. Several cell fractionation protocols were tested in an attempt to specifically measure vector genomes that had reached the nucleus, but genomes from wild-type and VP1-defective AAV8 particles were equally detected in the nuclear fraction by qPCR. These data highlight the importance of using suitable controls, including a negative control, for the development of biological assays such as infectious unit titration.
Keywords: AAV vectors, gene therapy, quality control, titration, infectivity
Introduction
Adeno-associated viruses (AAVs) were discovered in electron micrographs as contamination in adenovirus preparations, and soon became the subject of interest of scientists around the world.1 Over 50 years later, the interest in AAV as a vector in gene therapy continues to grow. The AAV belongs to the parvovirus family, specifically the dependoparvovirus subfamily. The members of this subfamily require a helper virus, such as the adenovirus (Ad) or herpes simplex virus (HSV), to facilitate productive infection and replication. Although it is estimated that 90% of the human population is AAV seropositive,2, 3 these viruses do not cause any known disease in humans, being an important safety criterion for their use in gene therapy approaches.
Clinical trials using recombinant AAV (rAAV) vectors have shown impressive results for Leber congenital amaurosis,4 hemophilia B,5 spinal muscular atrophy (ClinicalTrials.gov: NCT02122952), and other diseases. The first commercial product based on rAAV was approved in 2012 by the European Medicine Agency for the treatment of lipoprotein lipase deficiency, and a second drug could be approved soon according to positive results of a phase III trial.4 Nonetheless, a major bottleneck to commercialize these products is the manufacturing of rAAV in accordance with current good manufacturing practices (cGMPs) on a large scale. Production of rAAV in human cells (HEK293) transiently transfected with plasmids is probably the most common approach,6 but the use of insect cells and baculoviruses is highly convenient for industrial manufacturing.7 Other viable approaches consist of using a recombinant HSV complementation system in suspension-cultured mammalian cells (BHK21 or HEK293)8, 9 or mammalian-derived producer cell lines containing the rep and cap genes and the AAV vector integrated into the genome. In the latter case, the amplification of the rAAVs is initiated upon infection by a helper virus, such as Ad.10, 11
Given that quality attributes of rAAV stocks could be different depending on the manufacturing platform, it is highly important to have accurate analytical methods for their characterization, as emphasized by the FDA.12 Among the quality attributes, the infectious titer is critical to ensure the efficacy of the product.
AAV infection does not result in cytopathic effect, and, therefore, plaque assays cannot be used to determine infectious titers; but, in the presence of a helper virus, it is possible to induce the replication of AAV genomes and measure infectious events. One of the most widely used methods to titer infectious units is the median tissue culture infective dose (TCID50); the assay utilizes an HeLa-derived AAV2 rep and cap-expressing cell line, grown in 96-well plates and infected with replicate 10-fold serial dilutions of AAV vector in the presence of Ad type 5. After infection, vector genome replication is determined by qPCR.13 Similarly, the infectious center assay (ICA) uses HeLa rep-cap cells and Ad, but, after incubation, cells are transferred to a membrane and infectious centers (representing individual infected cells) are detected by hybridization with a labeled probe complementary to a portion of the recombinant genome.14, 15, 16
In this study, we compared these titration methods using rAAV serotype 8 vectors. In particular, we produced and characterized a VP1-defective AAV8-GFP vector that was used to mimic a non-infectious rAAV vector.17, 18, 19 This non-infectious vector lot allowed us to assess the ability of the different methods to discriminate between infectious and non-infectious rAAV serotype 8 vectors.
In addition, another objective of our study was to develop a new protocol for the titration of infectious AAV vector particles using sensitive qPCR-based quantification of intracellular or intranuclear vector genomes following the transduction of a permissive cell line, without helper virus co-infection. Such a procedure could be very useful for the titration of any AAV serotype, including those that do not infect standard cell lines such as HeLa rep-cap cells. Ideally, the protocol could be adapted to any type of cultured cells, including differentiated cells mimicking a targeted tissue, and it could result in infectious titers more predictive of in vivo vector efficiency.
Our results demonstrated that ICA was the most selective method to discriminate between infectious AAV8 particles and AAV8ΔVP1 negative control and correlated with vector-encoded transgene expression. Moreover, all methods tested for cytoplasm and nuclei fractionation of infected cells and measure of AAV genomes failed to distinguish infectious AAV8 and VP1-deficient particles. These data highlight the need for using appropriate biological assays to accurately measure the infectivity of rAAV stocks and the importance of including relevant controls in testing protocols.
Results
Production and Characterization of a VP1-Defective AAV8 Vector
The aim of the present study was to evaluate the accuracy of different methods for the titration of rAAV infectious particles; thus, we decided to generate a non-infectious AAV vector for use as a negative control. To this end, the ATG initiation codon of VP1 was changed to a stop (TGA) codon in the pKO-R2C8 packaging plasmid encoding AAV2 Rep and AAV8 capsid proteins. This mutated (pKO-R2C8ΔVP1) plasmid was co-transfected in HEK293 cells with pAdΔF6 helper and pTR-UF11 vector plasmids to produce an AAV8-GFP vector lacking VP1. The AAV8-GFP control vector was produced in parallel using the original pKO-R2C8 plasmid to get an infectious vector produced by the same method (i.e., three-plasmid transfection). Preliminary testing of AAV8ΔVP1 production demonstrated not only that vector genome packaging actually occurred into VP2 and VP3 particles but also that vector genome (VG) titers were reduced compared to a vector with wild-type AAV8 capsid composed of VP1, VP2, and VP3 polypeptides (data not shown). Thus, AAV8ΔVP1-GFP vector stock was produced through transfection of three CellStack-5 chambers (CS5), whereas a single CS5 was used for the control AAV8-GFP vector with wild-type capsid, but both vectors were then processed identically. This resulted in an AAV8ΔVP1-GFP vector stock with a higher VG titer (3.3 × 1013 and 2.4 × 1013 VG/mL based on bGH and SV40 polyA sequences, respectively) than the AAV8-GFP control vector stock (8.8 × 1012 and 8.8 × 1012 VG/mL based on bGH and SV40 polyA sequences, respectively), following purification through CsCl gradients (Table 1).
Table 1.
Capsids and Vector Genome Titers of AAV8 Vectors Used in This Study
| Vector | Capsids/mL (ELISA) | Vector Genome/mL |
Total:Full Particle Ratioa | |
|---|---|---|---|---|
| qPCR bGHpA | qPCR SV40pA | |||
| AAV8 control | 9.7 × 1012 | 8.8 × 1012 | 8.9 × 1012 | 1.1 |
| AAV8ΔVP1 | 7.0 × 1013 | 3.3 × 1013 | 2.4 × 1013 | 3.2 |
| AAV8 IC1 | ND | 2.7 × 1013 | 3.0 × 1013 | NA |
| AAV8 IC2 | 1.7 × 1013 | 1.7 × 1013 | 1.6 × 1013 | 1.1 |
| AAV8RSM | 5.5 × 1011b | ND | 5.65 × 1011c | 1.0 |
Total AAV8 capsid titers were determined by ELISA for the calculation of total:full particle ratio from CsCl-purified preparations, indicative of vector quality (Table 1). For vectors with wild-type AAV8 capsid, ELISA (total particles) and qPCR (full, recombinant genome-containing particles) titers were basically the same, indicating that they essentially contained particles with encapsidated VG, similar to the rAAV8RSM.20 In contrast, the AAV8ΔVP1-GFP vector stock contained 3-fold more total particles than VG-containing particles, i.e., two-thirds of the particles with no (empty) or illegitimate (non-vector) DNA encapsidated. Thus, although rAAV-UF11 VGs were actually encapsidated into VP2 and VP3 particles, the absence of VP1 apparently resulted in an apparent lower packaging efficiency. However, we cannot exclude that this result was not due to an ELISA quantification bias. Indeed, the ADK8 antibody used in the ELISA has its conformational epitope localized in VP3,21 and it may be more accessible in VP1-deleted capsid than in the wild-type AAV8 capsid, thus resulting in a higher ELISA signal.
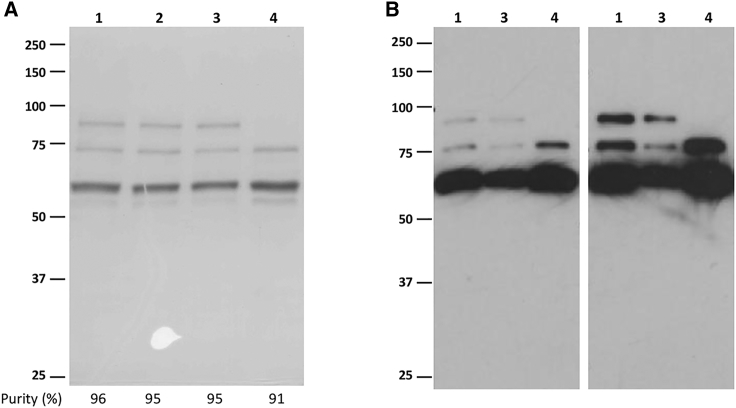
SDS-PAGE analysis of vector preparations showed that all AAV8-GFP vectors had similar purity, ranging from 91% for AAV8ΔVP1-GFP to 96% for internal control 1 (IC1), as determined by optical density scanning of Coomassie blue-stained gels (Figure 1A). It also showed the complete absence of VP1 in AAV8ΔVP1-GFP preparation, which was further confirmed by western blot analysis with anti-VP1/2/3 antibody B1 (Figure 1B, lane 4).22
Figure 1.
SDS-PAGE Analysis of AAV8-GFP Vector Preparations
(A) Coomassie blue staining performed after loading 1.5 × 1011 viral genomes per lane. Purity (% of VP1, VP2, and VP3 within total proteins) is indicated below gel image for each vector preparation. (B) Western blot analysis with anti-AAV capsid protein B1 antibody performed after loading 1.5 × 109 (left panel) or 3.0 × 109 (right panel) viral genomes per lane. Vector preparations were AAV8 IC1 (1), AAV8 IC2 (2), AAV8 control (3), and AAV8ΔVP1 (4).
As an additional quality control, particle size was measured in AAV8-GFP control and AAV8ΔVP1-GFP vector preparations by dynamic light scattering (DLS) (Figure S2). The results showed that both vector preparations had a very similar particle size distribution (24.57 ± 7.02 and 24.33 ± 7.38 nm, respectively), and they were quite homogeneous with no detectable particle aggregates (Table S2). AAV8-GFP control and AAV8ΔVP1-GFP vectors were also analyzed by differential scanning fluorimetry (DSF), and they were shown to have similar thermal stability with a melting temperature (Tm) of ∼70.5°C.23 These data indicated that the absence of VP1 had no major impact on AAV8 particle size and capsid stability in Dubelcco’s phosphate buffer saline (DPBS).
Analysis of AAV8 Vector Intracellular Trafficking
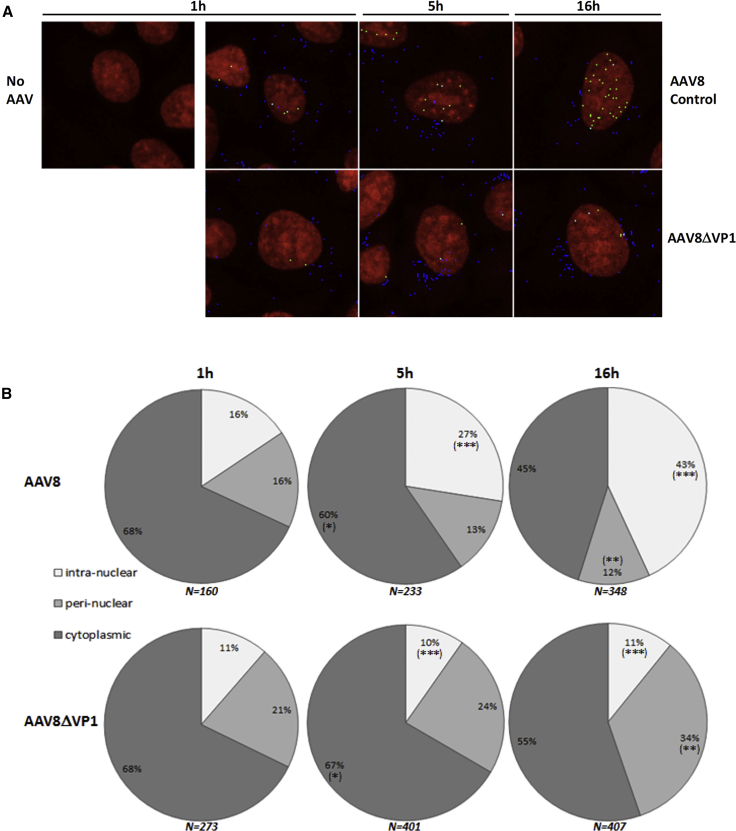
To further characterize the AAV8ΔVP1 vector, we analyzed AAV8 particle intracellular trafficking in HeLa cells infected with AAV8ΔVP1-GFP or AAV8-GFP control vector. To this end, HeLa cells were infected at an MOI of 20,000 VG/cell during 1, 5, or 16 hr, intracellular AAV8 particles were labeled with ADK8 antibody that specifically recognizes assembled AAV8 capsids24 and Alexa Fluor 555 secondary antibody, and nuclei were stained with DraQ5 fluorescent dye. Confocal microscopy images (Figure 2A) were then analyzed, using the compartmentalization task of Volocity software, for counting viral capsids according to their localization. Intranuclear particles were counted using the “inside” class (full colocalization with DraQ5 staining), perinuclear particles were calculated as the difference between “overlapping” (full or partial colocalization with DraQ5) and “inside” classes, and cytoplasmic particles were the difference between “nearest by edge” (no colocalization with DraQ5) and “overlapping” classes.
Figure 2.
Immunofluorescence Analysis of Intracellular Localization of AAV8 Particles
(A) Representative pictures of infected HeLa cells. Cells were non-infected (no AAV) or infected with AAV8 control or AAV8ΔVP1 at a multiplicity of 20,000 VG/cell, and then they were fixed after 1, 5, or 16 hr. Cell nuclei stained with DraQ5 appear in red, and assembled AAV8 particles stained with Alexa Fluor 555 appear in blue, green, or cyan, depending on their localization (cytoplasmic, intranuclear, or perinuclear, respectively). (B) Quantitative analysis of the immunofluorescence pictures. AAV8-assembled particles were quantified in the intranuclear, perinuclear, and cytoplasmic cellular compartments at 1, 5, and 16 hr post-infection with AAV8-GFP (upper panel) or AAV8ΔVP1-GFP (lower panel). Results obtained with AAV8ΔVP1 and AAV8 were compared by a two-tailed Mann-Whitney test for each cell compartment. *p < 0.05, **p < 0.005, ***p ≤ 0.0001; N = total number of AAV8 particles counted at each time point. Data are presented as mean ± SD.
The results indicated that assembled AAV8 particles entered the cytoplasm and then accumulated in the nucleus over time, the ratio of perinuclearly localized particles remaining constant (Figure 2B), as already described for AAV2.19, 25 In contrast, AAV8ΔVP1 particles were not actively entering the nucleus, as expected, and rather accumulated in the perinuclear region (Figure 2B). About 10% of intracellular AAV8ΔVP1 particles were detected within the nucleus, which may represent the background of the method, possibly due to a technical artifact, but not to non-specific antibody binding, since no signal was detected in non-infected HeLa cells. Since HeLa cells were dividing, we hypothesized that intracellular AAV8ΔVP1 particles were able to enter the nucleus during cell division, when the nuclear membrane was disrupted. When comparing AAV8 and AAV8ΔVP1 particle distribution by a two-tailed Mann-Whitney test, a significant difference was found at 5 hr post-infection (p = 0.0001) for intranuclear capsids, and at 16 hr post-infection for both intranuclear (p < 0.0001) and perinuclear (p = 0.0028) capsids. Overall, this analysis clearly confirmed that intracellular trafficking of AAV8 particles was altered in the absence of VP1, resulting in poor or absent nuclear translocation.
Titration of Infectious AAV8 Particles through the Detection of VG Replication
One method that is largely used to quantify infectious AAV vectors consists in infecting trans-complementing cells that have stably integrated AAV2 rep and cap genes,26 such as HeRC32 cells.27 When infected with Ad, these so-called packaging cells express both the AAV Rep and Ad helper proteins, allowing replication of the recombinant AAV genomes that have reached the nucleus, which correspond to infectious vector particles. Here we compared two methods based on this principle for the titration of infectious units (IUs) in AAV8 vector lots, which differ in particular by the way VG replication is detected. The TCID50 uses qPCR as the detection method, and VG replication is calculated by the Spearman-Kärber method.13, 28 In contrast, the ICA uses whole-cell DNA hybridization to detect cells in which VG replication happened.14 Another major difference is that infected cells are harvested 72 hr post-infection in the standard TCID50 assay but only 24 hr post-infection in the ICA currently performed in our laboratory.
The results obtained with the AAV8 control and AAV8RSM vectors indicated that both vectors have similar infectivity when comparing the VG:IU ratio obtained with each method (Table 2; Figure S3A). For both vectors, the VG:IU ratio calculated with the TCID50 titers (4.6 × 102 and 3.5 × 102) were 40- to 60-fold lower than those calculated with the ICA titers (2.5 × 104 and 1.5 × 104), indicating either that the TCID50 is more sensitive and can detect infectious particles not detected by the ICA or that it overestimates the infectious titer due to higher background. For the AAV8ΔVP1 vector, the VG:IU ratio obtained with both assays was higher than controls, confirming an altered infectivity for VP1-defective particles (Table 2; Figure S3A), but, surprisingly, it was not null. The detection of positive cells in the ICA method was not due to non-specific hybridization of the GFP probe, as shown by the absence of background on non-infected (Figure S3A) and AAV8-LacZ-infected cells (Figure S3B). We hypothesized that infectious AAV8ΔVP1 particles could be explained by the relatively high AAV multiplicity used and the presence of Ad, known to be an intracellular carrier for various biological molecules, especially through its endosomolytic activity.29, 30 Thus, the high Ad multiplicity (500 IUs/cell) used in the TCID50 and ICA may help non-infectious AAV particles to escape the endosomes and reach the cell nucleus. Importantly, in the case of AAV8ΔVP1, the difference in the VG:IU ratio calculated with the TCID50 (2.9 × 103) was about 4-log10 higher compared to that calculated with the ICA (3.1 × 107). In addition, the mean of TCID50 results from 3 independent assays indicated only a 6-fold difference in VG:IU ratio between AAV8 control and AAV8ΔVP1 vectors (p = 0.1), while the VG:IU ratio calculated by the ICA method resulted in a 1,240-fold difference between AAV8 control and AAV8ΔVP1, that difference being statistically significant (p = 0.0002).
Table 2.
Results Obtained with AAV8 Vectors Using the Different Infectious Titration Methods
| Method | AAV8RSM |
AAV8 Control |
AAV8ΔVP1 |
||||
|---|---|---|---|---|---|---|---|
| IU/mL | VG:IU Ratioa | IU/mL | VG:IU Ratioa | IU/mL | VG:IU Ratioa | ||
| TCID50 (n = 2–3) | 1.6 × 109b | 3.5 × 102 | 2.0 × 1010 | 4.6 × 102 | 8.3 × 109 | 2.9 × 103 | |
| Modified TCID50c (n = 2) | ND | NA | 3.0 × 109 | 3.0 × 103 | 7.5 × 108 | 3.2 × 104 | |
| ICA (n = 2) | 3.8 × 107 | 1.5 × 104 | 3.5 × 108 | 2.5 × 104 | 7.8 × 105 | 3.1 × 107 | |
| TU (n = 3) | −Ad | ND | NA | 6.7 × 107 | 1.3 × 105 | 1.8 × 105 | 1.3 × 108 |
| +Ad | ND | NA | 1.6 × 108 | 5.5 × 104 | 2.3 × 106 | 1.0 × 107 | |
| IG (n = 4) | pro. 1 | ND | NA | 1.4 × 1010 | 6.4 × 102 | 1.2 × 1011 | 2.0 × 102 |
| pro. 2 | ND | NA | 4.5 × 108 | 2.0 × 104 | 1.2 × 1010 | 2.0 × 103 | |
| pro. 3 | ND | NA | 4.5 × 1010 | 2.0 × 102 | 2.4 × 1011 | 1.0 × 102 | |
| pro. 4 | ND | NA | 2.0 × 109 | 4.5 × 103 | 1.9 × 1010 | 1.3 × 103 | |
IU, infectious unit; VG, vector genome (full particles); ND, not determined; NA, not applicable; pro. 1–4, procedures 1–4 described in the Materials and Methods.
Calculated using VG titer based on SV40pA qPCR (Table 1).
Note that infectious titer published by the AAVRSMWG was 1.26 × 109 IUs/mL.20
Modified assay analyzed at 24 hr post-infection.
One additional observation that confirmed the infectivity defect of AAV8ΔVP1 was the absence of AAV replication in the control ICA performed on HeLa cells co-infected with Ad5 (Figures S3A and S4A). In contrast, some AAV replication was detected with the AAV8 control vector, which was correlated with the presence of infectious rep-positive particles that were clearly detected by probing the ICA membranes with a rep probe (Figure S4B). This vector lot was found to contain 8.5 × 103 rep-positive infectious particles per milliliter (i.e., 1 rep-positive into 4 × 104 AAV8-GFP infectious particles). These rep-positive particles are known to be generated during vector production when using three-plasmid transfection with a rep-cap plasmid containing a full-length p5 promoter.31 Importantly, no replication was detected in HeLa cells with the AAV8RSM (Figure S3A) and the AAV8 internal control 2 vector (Figure S4), both produced by double transfection using the large helper plasmid pDP8 with the MMTV LTR replacing the p5 promoter.20, 32 The high background observed with the humanized green fluorescent protein (hGFP) probe on HeLa cells infected with AAV8-GFP IC2 and AAV8-GFP ΔVP1 vectors was likely due to the very high vector input (Figure S4A). The signal observed with the rep probe on HeRC32 cells was due to Ad-induced amplification of the integrated rep-cap sequences (Figure S4B).
The discrepancy between TCID50 and ICA results obtained with the AAV8ΔVP1 vector may indicate that the current TCID50 assay indeed leads to a higher background and overestimates vector infectivity compared to ICA. In addition to the sensitivity of the detection method (qPCR versus blotting), this difference could also be explained by the different infection duration used in the assays (i.e., 3 days for the TCID50 versus 1 day for the ICA), which may allow non-infectious AAV genomes to enter the nucleus during cell divisions. To assess the impact of the incubation time on TCID50 titers, additional assays were performed using 24-hr incubation, similar to the ICA. The infectious titers obtained with this modified TCID50 assay were reduced 7-fold and 11-fold for the AAV8 control and AAV8ΔVP1 vectors, respectively, and they resulted in an increased difference (11-fold versus 6-fold) in VG:IU ratio between both vectors (Table 2). Thus, by reducing the infection time of the protocol, it is possible to improve accuracy of the TCID50 assay. Indeed, the VG:IU ratio calculated for the AAV8 control vector using this modified TCID50 was closer (less than 10-fold higher) to that calculated with the ICA. However, this ratio was still 1,000-fold higher for the AAV8ΔVP1 vector.
Comparison of the TCID50 and ICA Methods Using AAV2 Vectors
To further investigate the differences between the TCID50 and ICA, additional experiments were conducted using a different AAV serotype, i.e., AAV2. When the TCID50 was performed with the AAVRSMWG28 vector and an internal control AAV2 vector (AAV2 IC) following the standard adeno-associated virus reference strain material working group (AAVRSMWG) method, i.e., using 72-hr incubation, the calculated VG:IU ratio was 1.5 and 1.3, respectively (Table S4), indicating almost 100% infectious particles in both vector lots, which seems rather unlikely and supports the assumption that the standard TCID50 may overestimate the infectious titer in some cases. Indeed, the VG:IU ratio published by the adeno-associated virus type 2 reference strain materials working group (AAVRSMWG) was 7.5.28
By reducing the incubation time to 24 hr, the TCID50 infectious titers were reduced 7.8- and 25-fold for the AAVRSMWG and AAV2 IC vectors, respectively, which resulted in VG:IU ratios of 11.7 and 33.1, respectively. The infectious titers determined by this modified TCID50 assay were closer to those obtained by the ICA method (Table S4), i.e., almost identical for the AAV2 IC vector and only 6.7-fold higher for the AAVRSMWG.
Based on these results and those obtained with the AAV8 control vector, it appears that 24-hr incubation is sufficient to determine consistent infectious titers using both TCID50 and ICA methods, and it decreases the discrepancy between both assays. In addition, reducing the incubation time is likely decreasing the variability of the TCID50, as already shown for the ICA. Indeed, in the original ICA protocol published by our laboratory,14 the infection time was fixed at 42–44 hr using an Ad5 multiplicity of 50 IUs/cell, but subsequent studies demonstrated that 24–26 hr of infection using a higher Ad5 multiplicity (500 IUs/cell) was sufficient to detect rAAV replication in infected cells. This shorter incubation also resulted in less background signal and more reproducible results, which was correlated with the appearance of Ad-induced cytopathic effect around 36 hr post-infection. Since high Ad5 multiplicity is also used in the TCID50 and the qPCR-based detection is highly sensitive, the use of 24-hr incubation was considered an improvement for this assay.
Titration of Infectious AAV8 Particles by Transgene Expression Assay
Another biological assay that is commonly used to test infectivity of AAV preparation consists of infection of permissive cells and measuring transgene expression to determine a titer in transducing units (TUs). Similar to the replication assays described above, transduction requires the entry of rAAV vectors into the cells, the translocation of VGs to the nucleus and their conversion in double-stranded DNA, and, in addition, transcription of the transgene. The readout is the detection of the transgene-encoded protein.
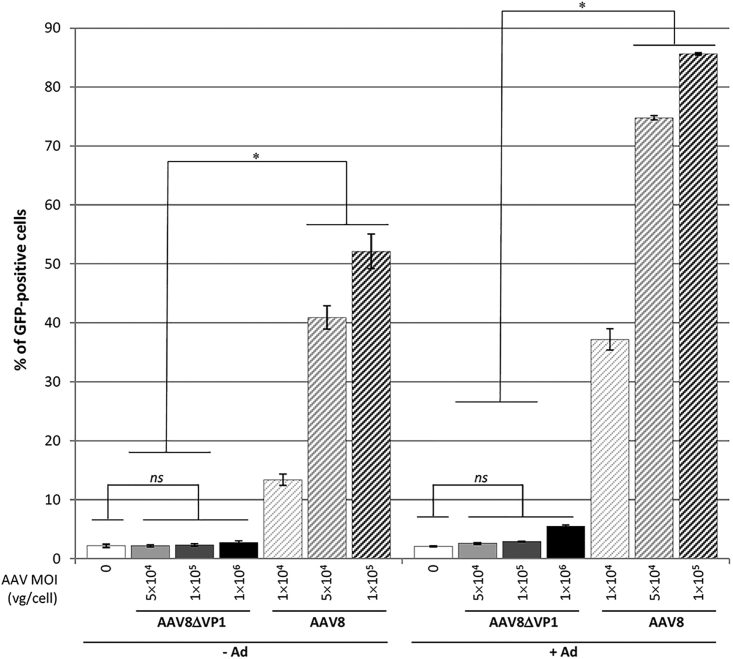
To analyze GFP transgene expression from AAV8 and AAV8ΔVP1 vectors, we used HeLa cells because they have similar permissiveness compared with HeRC32 cells, thus allowing comparison of infectious titers obtained by the TU, TCID50, and ICA methods. HeLa cells were infected with controlled AAV multiplicities in the presence or absence of Ad (Addl324) at 50 IUs/cell. Analysis of GFP expression by fluorescence-activated cell sorting (FACS) showed a defect in infectivity for the AAV8ΔVP1 vector (p = 0.0782), and the amount of GFP-positive cells was found above background only at the higher AAV multiplicity (Figure 3). In contrast, a significant amount of GFP-positive cells was found with AAV8 compared to AAV8ΔVP1, in particular at MOIs greater than or equal to 5 × 104 VG/cell. Transgene expression was enhanced by Ad transduction, by promoting most likely second-strand DNA synthesis33 and possibly endosomal escape of AAV as discussed above. According to infectious titers calculated by FACS analysis, the difference in VG:IU ratio between AAV8 and AAV8ΔVP1 vectors was 3-log without Ad and 2.3-log when Addl324 was added, which were consistent with the results obtained using the ICA method.
Figure 3.
Titration of AAV8-Transducing Units by FACS Analysis of GFP Expression
HeLa cells were infected or not with AAV8ΔVP1-GFP or AAV8-GFP vector at the indicated MOIs (in VG/cell), in the presence (+Ad) or absence (−Ad) of Addl324 (50 infectious particles/cell). Cells were harvested after 48 hr and GFP-positive cells were counted by FACS. Bars represent the mean percentage of GFP-positive cells (±SD) from 3 independent wells. Results of AAV8ΔVP1-infected cells were compared to non-infected cells by a two-tailed Mann-Whitney test, and to cells infected with AAV8 at the same MOI by a one-tailed Mann-Whitney test. ns, p > 0.05; *p ≤ 0.05. Data are presented as mean ± SD.
Evaluation of Sample Preparation Methods for the Isolation of Intracellular AAV VGs
In an attempt to develop a new, more sensitive and accurate assay, we evaluated the possibility to quantify infectious AAV particles through a qPCR-based detection method of intracellular or intranuclear VGs. Indeed, such a procedure would be of interest for vector serotypes that do not infect HeLa rep-cap cells, such as HeRC32, and it would be virtually adaptable to any type of permissive cells. In addition, the procedure would not require Ad co-infection to avoid possible bias caused by Ad-induced endosome disruption. To this end, we tested several protocols to isolate vector DNA from AAV8-infected cells.
For the development of the so-called infectious genome (IG) assay, we used HeLa cells to allow comparison of titers with the other methods. Cells were seeded in 12-well plates, infected (or not) for 16 hr with different MOIs (2,000 or 10,000 VG/cell) of AAV8 control or AAV8ΔVP1 vectors, and harvested by trypsin-EDTA treatment. Next, we tested four sample preparation procedures prior to DNA isolation. After washing with PBS, harvested cells were kept intact and washed again with PBS to remove extracellular AAV particles (procedure 1), or they were submitted to cytoplasm/nucleus fractionation (procedures 2, 3, and 4). Fractionation was performed in order to remove cytoplasmic AAV particles and to isolate the VGs of infectious particles from the nucleus. To achieve this, three different protocols were tested and compared. The first one (procedure 2) used cell lysis and nuclei-stabilizing solutions, allowing the preparation of a single-cell nuclei suspension for cell counting and viability analysis (http://shop.chemometec.com/product/reagent-a100-500-ml/). The second one (procedure 3) is based on a commercial cell fractionation kit (NE-PER Nuclear and Cytoplasmic Extraction, Thermo Scientific) that has been already used for the analysis of AAV2 intracellular trafficking in a recent study.34 The last one (procedure 4) was adapted from a protocol that was used for the analysis of the subcellular localization of factors of the RNAi pathway.35 All procedures are described in detail in the Materials and Methods.
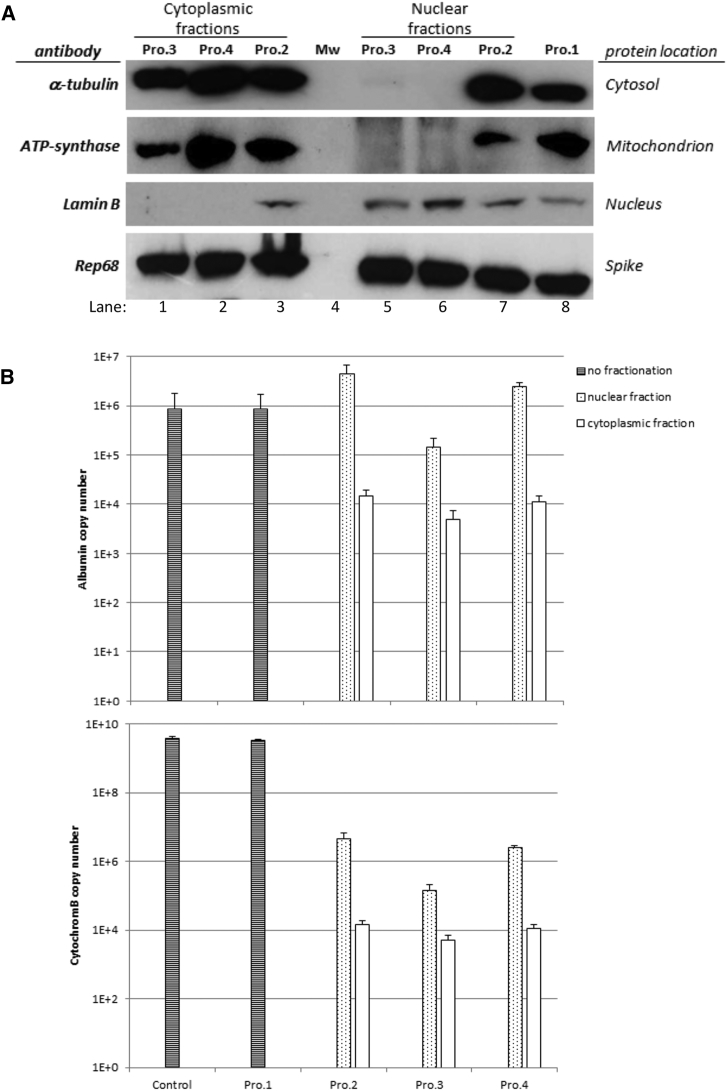
The purity of both cytoplasmic and nuclear fractions was controlled by western blot using antibodies against proteins localized exclusively in the cytosol (α-tubulin), the mitochondrion (ATP-synthase β subunit), or the nucleus (lamin B). Since protein samples were precipitated with acetone to get equal sample volumes prior to SDS-PAGE, purified His-tagged Rep68 protein was added as a loading control. The signal obtained with the spiked Rep68 protein was equivalent in all lanes, showing that protein precipitation by acetone was equally efficient for all samples (Figure 4A). Results with procedure 2 showed that fractionation was not efficient, since the cytoplasmic (lane 3) and nuclear (lane 7) fractions both contained the 3 indicator proteins, similar to whole-cell lysate prepared by procedure 1 (lane 8). Both procedures 3 and 4 resulted in apparent pure cytoplasmic fractions in which lamin B was not detected (lanes 1 and 2), but a pure nuclear fraction was obtained only with procedure 4 (lane 6), showing no α-tubulin signal and no detection of ATP-synthase. Indeed, a weak but clear α-tubulin signal was detected in the nuclear fraction obtained with procedure 3 (lane 5).
Figure 4.
Molecular Analysis of HeLa Cell Fractionation
(A) Western blot analysis of whole-cell lysate (procedure 1) or cytoplasmic and nuclear fractions prepared by the different procedures (2, 3, and 4; see Materials and Methods for details). Samples were acetone-precipitated, resuspended in equal volumes of Laemmli buffer, and separated by SDS-PAGE for western blot analysis using antibodies to protein markers of different cell compartments. An equal amount of purified Rep68 protein was spiked in each sample to control protein recovery during acetone precipitation. (B) qPCR analysis of genomic albumin gene (upper panel) and mitochondrial cytochrome B gene (lower panel) in DNA samples prepared by the different procedures compared to control (untreated) cells. Bars represent the total copy number ± SD in each sample.
Since detection of intracellular VG is based on a DNA method and not a protein method, we decided also to control cell fractionating by qPCR. To this end, cytoplasmic and nuclear fractions as well as whole-cell samples were analyzed with albumin and cytochrome B primers, as markers of nuclear genomic DNA (gDNA) and cytoplasmic mitochondrial DNA (mtDNA), respectively. The qPCR analysis showed similar results for procedures 2 and 4 (Figure 4B), for which nuclear fractions were enriched in gDNA but still contained mtDNA. Although this contaminating mtDNA was less than 0.5% of the total mtDNA detected in control cells, it still represented more than 106 copies per sample. Similarly, more than 104 copies of gDNA were detected in the cytoplasmic fractions from both procedures, representing around 1.5% of total gDNA detected in control cells. Regarding procedure 3, the distribution of gDNA and mtDNA in the nuclear and cytoplasmic fractions was very similar, but the copy numbers in both fractions were lower for both albumin and cytochrome B compared to the other methods. In particular, albumin copy number in the nuclear fraction represented only 16.5% of that detected in control cells, which may reflect a problem of DNA recovery when using the NE-PER kit that is primarily intended to extract proteins. Strikingly, the amount of mtDNA was higher in the nuclear fractions than in the cytoplasmic fractions for all three fractionation procedures, demonstrating that nuclei are not efficiently separated from cytoplasmic organelles. The results obtained by qPCR were in contradiction with western blot analyses that showed efficient nuclei isolation when using procedure 4, maybe due to the higher sensitivity of the qPCR compared to immunoblotting. Although the method published by Gagnon et al.35 (procedure 4) appeared as the most efficient in terms of purity of the subcellular fractions, our results showed that none of the cell fractionation methods tested can completely separate nuclei from cytoplasmic components.
Titration of AAV8 Particles in Cellular Fractions by qPCR
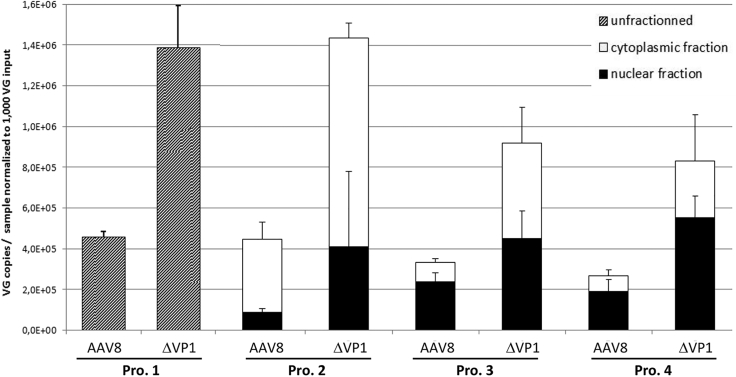
DNA samples prepared by the four different procedures were also analyzed using a qPCR assay targeting the SV40 poly(A) sequence for quantification of intracellular, cytoplasmic, and intranuclear AAV VGs. The qPCR results obtained with different MOIs were normalized to an input of 1,000 VG/cell (Figure 5) to facilitate comparison. Interestingly, the amount of intracellular VGs was higher with AAV8ΔVP1 compared to AAV8 (p = 0.015), showing that washing the cells with PBS does not remove defective particles lacking VP1. Nonetheless, it is known that AAV interaction with cellular receptors and cell entry do not depend on the presence of VP1.19 A more striking observation was the higher amounts of VGs detected in the nuclear fractions with AAV8ΔVP1 compared to AAV8 (p = 0.011), which is consistent with the confocal microscopy results showing perinuclear accumulation of VP1-defective particles (Figure 2) and the inefficient separation of nuclei (Figure 4B). Finally, similar results were observed in the cytoplasmic fractions (p = 0.047), suggesting that VP1 defect resulted in an overall increase of intracellular VGs at 16 hr post-infection. One possible explanation could be that AAV8 particles undergo a pH-induced conformational change during trafficking through the late endosome,36, 37 leading in particular to the externalization of VP1 N-terminal domain and capsid destabilization,38 which may increase their sensitivity to proteolytic degradation. In the absence of VP1, the effect of low pH on capsid stability may be less significant, leading to an increased intracellular persistence of VP1-defective particles.
Figure 5.
Distribution of Vector Genomes in Samples Prepared by Different Cell Fractionation Procedures
DNA samples from infected HeLa cells were prepared from whole cells (procedure 1) or subcellular fractions (procedures 2, 3, and 4), and vector genomes were quantified in each sample by qPCR targeting the SV40 poly(A) sequence. Results were normalized to an input of 1,000 vector genomes per cell at the time of infection, in order to compile data obtained with the different MOIs (2,000 and 10,000 VG/cell). Bars represent the amount of vector genomes per sample ± SD.
Infectious titers of AAV were calculated using the values obtained for intracellular (procedure 1) or intranuclear (procedures 2–4) VGs (Table 2). The formula used for titer calculation considered both the number of cells at infection and the amount of gDNA (based on the albumin gene copy number) in order to normalize the VG copy values to the efficiency of nuclear DNA recovery. The infectious titer for the AAV8 control was in the same range as those obtained with the standard methods, TCID50 and ICA (Table 2). However, lower VG:IU ratios were consistently obtained with the AAV8ΔVP1 vector with all four sample preparation procedures, wrongly indicating a higher infectivity for AAV8ΔVP1. This result is in conflict with the VG:IU ratio calculated using the TCID50, ICA, and TU methods, and it does not reflect the current knowledge of VP1 function, recognized as critical for AAV transduction. In summary, none of these methods was able to discriminate between a fully functional (i.e., infectious) AAV8 vector and a non-infectious counterpart lacking the VP1 capsid protein.
Discussion
The aim of the present study was to compare different methods for the titration of infectious AAV vector particles and to evaluate their ability to accurately discriminate infectious and non-infectious vectors. The study was focused on AAV serotype 8 vectors, a capsid serotype that allows highly efficient gene transfer in vivo in different tissues and animal models39 and has been used successfully in clinical trials.5 In addition, a fully characterized reference standard material (AAV8RSM) is available to the community for this serotype.20
To challenge the titration methods, we have generated an AAV8 vector lacking the large capsid protein VP1, known to be essential for AAV endosomal escape and nuclear translocation following cell entry.17, 19 In particular, virus infectivity is largely dependent on the phospholipase A217 and basic domains18 located in the VP1 N terminus sequence. To our knowledge, this is the first time that such a VP1 negative control is used in infectious titer assays.
To discriminate the infectivity of the AAV particles depending on the VP1 content is currently of high importance, because it has been shown that the baculovirus and insect cell system in particular could generate rAAV with variable VP1 content, depending on the cap sequence design.40, 41, 42 Moreover, during gene therapy drug development phases, a change in the upstream or downstream manufacturing process may be required to be more amenable to commercial scale. In this case, it will be mandatory to conduct equivalence studies, and then accurate characterization of the infectivity of the rAAV products will be critical for regulatory approval.
Here we show that AAV8 nuclear targeting following cell entry is clearly altered in the absence of VP1, as shown by immunofluorescence analysis of intracellular trafficking of AAV8ΔVP1 particles. In the same line, the ICA titration method showed that AAV8ΔVP1 was 1,000-fold less infectious than AAV8 and was consistent with the almost absence of GFP transgene expression. In contrast, by using the TCID50 titration method, the difference in infectivity (VG:IU ratio) between AAV8 and AAV8ΔVP1 was only 6-fold, suggesting that partial loss of infectivity of an AAV8 lot might be more difficult to detect by TCID50 compared to the ICA method. Thus, the ICA appears as a more discriminating method to distinguish between an infectious and a non-infectious AAV8 vector, and it is applicable independently of the transgene cassette, in contrast to transgene expression assays (not suitable for, e.g., coding sequences controlled by tissue-specific promoters and non-coding sequences such as small hairpin RNA [shRNA]). We acknowledge that there could be differences between serotypes, and, thus, it would be interesting to implement a similar method evaluation using VP1-defective control vectors for other AAV serotypes.
It is worth mentioning that the TCID50 method was originally developed with AAV2, a serotype that is much more infectious than AAV8 on cultured cell lines.43 Indeed, standard TCID50 assays performed using the AAVRSMWG and an internal AAV2 vector resulted in a VG:IU ratio close to 1, whereas this ratio was between 350 and 460 for AAV8RSM and AAV8 control vectors. Hence, some adjustments of the TCID50 method could be implemented to improve the accuracy of the method for AAV8 or other serotypes with low infectivity in vitro. We investigated, among the possible adjustments, a reduction of the infection time to 24 hr (instead of 72 hr in the standard method). This modification resulted in an almost 2-fold increased difference in infectivity (VG:IU ratio) between AAV8 and AAV8ΔVP1 (i.e., 11-fold versus 6-fold), and, thus, it could be considered as an improvement of the method. Reducing the incubation time in the TCID50 assay also resulted in about 10-fold lower infectious titers for both AAV8 and AAV2 vectors, thus mitigating the discrepancy between the TCID50 and ICA methods. However, the infection-defective AAV8ΔVP1 vector was still found to be 1,000-fold more infectious with the modified TCID50 compared to the ICA.
On the other hand, we found that none of the four physicochemical methods tested to fraction cytoplasm versus nuclei was selective enough to specifically detect AAV8 infectious genomes by qPCR. Nonetheless, cell fractionation could be suitable for AAV serotypes with high infectivity in vitro, as shown by Salganik et al.34 using AAV2 on HeLa cells. Indeed, the authors were able to distinguish VG distribution of wild-type and different capsid mutants using the NE-PER kit for cell fractionation (similar to procedure 3 in this study) and qPCR. Since none of the cell fractionation methods tested here was suitable for AAV8, further evaluation of these methods using AAV2 was not considered. Moreover, we did not include cell fractionation methods based on ultracentrifugation in sucrose gradient to separate cell compartments,18, 19 since they were considered very difficult to implement in quality control laboratories following good laboratory practices (GLPs), in particular because (1) they are laborious, (2) they need ultracentrifugation equipment, (3) they require large amounts of cells (and thus large amounts of vector preparation), and (4) they would be difficult to standardize.
In conclusion, our data demonstrate that transgene expression and ICA were the most sensitive methods to detect changes in infectivity of AAV8 stocks. Moreover, lessons learned during the development of cell-fractioning protocols were key to understanding the limitations of current tools (e.g., cell lines) and the need for developing more accurate protocols. This study also highlights the importance of including suitable positive and negative controls for the evaluation of analytical methods, and it suggests implementing VP1-defective reference vectors as negative controls for the validation of AAV infectivity assays.
Materials and Methods
Cell Lines
HEK293, HeLa, and HeRC32 (ATCC CRL-2972) cells were maintained in DMEM (Sigma-Aldrich, Saint-Quentin Fallavier, France) supplemented with 10% fetal bovine serum (FBS) (HyClone-GE Healthcare, Logan, UT, USA) and 1% pencillin-streptomycin (Thermo Scientific, Illkirch, France). Cell infections were performed in DMEM with 2% FBS.
Plasmids
pTR-UF11 (ATCC MBA-331) contains a recombinant AAV2 genome of 4.3 kb, with both CBA-GFP-SV40p(A) and PYF441/HSVtk-Neomycin-bGHp(A) expression cassettes (Figure S1).44
pAAV-cytomegalovirus (CMV)-nlsLacZ contains a recombinant AAV2 genome of 4.7 kb, with a 0.3-kb human CMV immediate early promoter, an alcohol dehydrogenase and beta-galactosidase fusion protein-coding sequence, and a bGH polyA signal cloned between the XbaI sites of AAV2 plasmid pSub201.45
Plasmid pKO-R2C8 contains the AAV2 rep gene from pSub201 and the AAV8 cap gene from p5E18-VD2/8.46
Plasmid pKO-R2C8ΔVP1 was generated by a QuickChange Site-Directed Mutagenesis modified protocol. Briefly, pKO-R2C8 DNA was amplified using KOD DNA polymerase (Merck Millipore, Molsheim, France) with two complementary primers, which introduce a TGA stop codon in place of the ATG initiation codon of VP1. Amplification reactions were digested with DpnI (New England Biolabs, Evry, France) and transformed into E. coli DH5α-competent cells (Life Technologies, Illkirch, France). Primers used for mutagenesis were DELVP1FOR (5′-GAACAATAAATGATTTAAATCAGGTTGACCTGCCGATGGTTATCTTCCAGATTGG-3′) and DELVP1REV (5′-CCAATCTGGAAGATAACCATCGGCAGGTCAACCTGATTTAAATCATTTATTGTTC-3′), with the 4 mutated bases shown in bold (underlined is a HincII restriction site created by the mutation). Rep and cap genes in pKO-R2C8ΔVP1 were verified by sequencing.
pAdΔF6 helper plasmid, which contains E2A, VA RNA, and E4 Ad helper functions, was obtained from the Penn Vector Core (Philadelphia, PA, USA).
pDP8-KanR plasmid contains the same elements as pDP8 used for manufacturing of the rAAV8RSM (i.e., E2A, VA RNA, and E4 Ad helper functions, as well as AAV2 rep and AAV8 cap sequences), except ampicillin resistance that was replaced by kanamycin for amplification in E. coli.
pCQAAV1 plasmid is a derivative of pSub201 containing all qPCR target sequences used in this study, i.e., human albumin gene, human cytochrome B gene, SV40, and bGH polyA signals.
Antibodies for Western Blot and Confocal Microscopy
Mouse monoclonal antibodies that recognized assembled AAV8 capsid (ADK8), AAV capsid proteins (B1), and AAV Rep proteins (303.9) were kindly provided by Dr. J.A. Kleinschmidt (DKFZ, Heidelberg, Germany). Goat polyclonal antibody detecting lamin B (C-20) was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Mouse monoclonal antibody to ATP-Synthase β subunit (3D5AB1) and goat anti-mouse Alexa Fluor 555 secondary antibody were from Life Technologies. Mouse monoclonal antibody to α-tubulin (B-5-1-2) was from Sigma-Aldrich. Goat anti-mouse-horseradish peroxidase (HRP) and rabbit anti-goat-HRP secondary antibodies were from Dako (Agilent Technologies, Santa Clara, CA, USA).
AAV8 Vector Preparations
AAV8 vectors were manufactured as described for the rAAV8RSM.20 Briefly, HEK293 cells in CS5 were transfected with two or three plasmids by the calcium phosphate precipitation method, and AAV8 particles in the culture supernatant were polyethylene glycol (PEG)-precipitated, then purified by double CsCl density gradient ultracentrifugation, and finally formulated in 1× DPBS containing Ca2+ and Mg2+ through dialysis in Slide-A-Lyzer 10K cassettes (Thermo Scientific, Illkirch, France). AAV8 internal control vectors (IC1 and IC2), used as quality control standards in our laboratory, were produced by co-transfection of pTR-UF11 vector plasmid and pDP8-KanR helper plasmid. AAV8 control and AAV8ΔVP1 vectors were produced by co-transfection of pTR-UF11, pAdΔF6 helper plasmid, and either pKO-R2C8 or pKO-R2C8ΔVP1 plasmid. AAV8 reference standard material (rAAV8RSM, ATCC VR-1816) was used as an additional control.
VG Titration by qPCR
Purified AAV vectors (3 μL) were treated with 4 U DNase I (Sigma-Aldrich) in DNase buffer (13 mM Tris [pH 7.5], 0.12 mM CaCl2, and 5 mM MgCl2) for 45 min at 37°C. Then, DNase I-resistant nucleic acids were purified by the NucleoSpin RNA Virus kit (Macherey-Nagel, Hoerdt, France), and VGs were quantified by TaqMan qPCR in Premix Ex Taq probe qPCR master mix (TaKaRa Bio, Saint-Germain-en-Laye, France). Primers were targeted to either the SV40 or the bGH polyA signal (Table S1). Standard curves were obtained using 102–108 copies pTR-UF11 plasmid linearized with ScaI (New England Biolabs, Evry, France). VG titers were calculated from at least 3 independent titrations, and mean values are reported in Table 1.
AAV8 Capsid Titration by ELISA
Titration of AAV8 assembled caspids by ELISA was performed as described for the rAAV8RSM using the Progen AAV8 Capsid ELISA Kit (PROGEN Biotechnik, Heidelberg, Germany), following the manufacturer’s instructions, also available on the ATCC website (https://www.lgcstandards-atcc.org/∼/media/AAV8_Information/AAV8%20RSM%20Infectious%20titer%20assay.ashx).
SDS-PAGE and Western Blot Analysis of AAV Vector Preparations
For SDS-PAGE analysis of rAAV preparations, vectors were denatured for 5 min at 95°C in Laemmli buffer and loaded on 10% Tris-Glycine polyacrylamide gels (Life Technologies). Precision Plus Protein All Blue Standards (Bio-Rad, Marnes-la-Coquette, France) was used as a molecular weight marker. Following electrophoresis, gels were either submitted to Coomassie blue staining (Imperial Protein staining, Thermo Fisher Scientific) or transferred on nitrocellulose membranes for western blot analysis. Membranes were probed with monoclonal antibody B1, which recognizes all three AAV capsid proteins. The amounts of VGs loaded on the gel were 1.5 × 1011 and 1.5–3.0 × 109 for Coomassie blue staining and western blotting, respectively.
For the determination of vector purity, Coomassie blue-stained gels images were analyzed with Gene Tools software (Syngene, Cambridge, UK) to determine signal intensity of each protein band. Purity was then calculated as the relative amount of VP1, VP2, and VP3 proteins over the total amount of proteins present in the sample, all extrabands being integrated as non-expected contaminant proteins.
Fluorescence Microscopy Analysis
HeLa cells were seeded in 8-well μ-Slide ibiTreat chambers (Biovalley, Nanterre, France) at 2 × 105 (for 1- or 5-hr infections) or 8 × 104 (for 16-hr infection) cells/well. The day after, cells were infected or not with the rAAV vectors at a multiplicity of 20,000 VG/cell in duplicate. After 1, 5, or 16 hr, cells were washed in PBS, fixed with 4% paraformaldehyde in PBS, and permeabilized with 0.2% Triton X-100 in PBS. Permeabilized cells were blocked with PBS containing 10% goat serum (Sigma-Aldrich), incubated with ADK8 antibody (undiluted hybridoma culture supernatant) recognizing AAV8 intact capsid,21, 47 and washed with PBS. Cells were then incubated with anti-mouse Alexa Fluor 555 secondary antibody (1:200 in PBS), washed with PBS, and incubated with DraQ5 (1:1,000 in PBS) for nuclei staining (Interchim, Montluçon, France). Slides were finally mounted with Prolong Gold antifade reagent (Life Technologies). Images were captured on a Nikon A1 confocal microscope (Nikon Instruments, Amsterdam, Netherlands), and quantification of AAV8 particles according to their localization was then performed using the compartmentalization task of Volocity software (PerkinElmer, Courtaboeuf, France). Images were processed with ImageJ software (https://imagej.nih.gov/ij/) for picture presentation.
Infectious AAV8 Particle Titration by the TCID50 Assay
Titration of infectious AAV8 particles by qPCR was performed following the procedure for the rAAV8RSM infectious titer of the Adeno-Associated Virus Reference Standard Working Group, available on the ATCC website (https://www.lgcstandards-atcc.org/∼/media/AAV8_Information/AAV8%20RSM%20Infectious%20titer%20assay.ashx). A modified assay was also tested where analysis was performed at 24 hr post-infection (instead of 72 hr in the standard procedure). For titration of AAV8 control and AAV8ΔVP1, the vector preparations were diluted in 1× DPBS to get a VG titer approaching that of the AAV8RSM, in order to perform serial dilutions exactly the same way. TCID50 titers were calculated from two independent assays.
Infectious AAV Particle Titration by the ICA
ICAs were performed as previously described,14 except incubation time and Ad concentration were modified. Briefly, HeRC32 cells were seeded in 48-well plates at 7.0 × 104 cells/well, and they were infected the next day with duplicate 10-fold dilutions of the AAV vector preparations, in the presence of wild-type Ad5 at a multiplicity of 500 IUs per cell. Negative controls were HeRC32 cells without Ad5 and HeLa cells with Ad5, both infected with the AAV vectors. Cells were harvested 24–26 hr post-infection and filtered through Zeta-Probe nylon membranes (Bio-Rad) using a vacuum device. Membrane filters were hybridized overnight at 65°C with vector-specific probes generated with the PCR Fluorescein Labeling Mix (Sigma-Aldrich), and detection was performed using the CDP-Star ready-to-use labeling kit (Sigma-Aldrich). Titers were determined by counting dots (i.e., AAV-infected cells) on membrane autoradiography, which was done blind by two independent operators. ICA titers were calculated from two independent assays.
GFP Expression by Flow Cytometry
HeLa cells were seeded in 24-well plates at 1.0 × 105 cells/well, and they were infected the next day with AAV vectors at different MOIs in triplicates, in the presence or absence of Addl324 virus48 at a multiplicity of 50 infectious particles per cell. Cells were harvested 48 hr post-infection, fixed with 4% paraformaldehyde in PBS, and resuspended in FACS buffer (1× PBS, 0.5% BSA, and 2 mM EDTA). Analysis of GFP expression was performed on 30,000–31,000 cells from each well in a LSRII cell analyzer instrument (Becton Dickinson, Le Pont de Claix, France).
The TU titer was calculated using the following formula:
where NGFP is the mean number of GFP-positive cells (% GFP-positive cells × total number of cells) per well minus background (% GFP-positive cells × total number of cells in non-infected wells), and V is the volume of vector used to infect cells in microliters.
Cell Processing for the Quantification of Intracellular AAV Genomes
HeLa cells were seeded in 12-well plates at 3.5 × 105 cells/well. The day after, cells (4–7 × 105 per well) were infected or not with the rAAV vectors at a multiplicity of 2,000 or 10,000 VG/cell in duplicate. At 16 hr post-infection, cells were washed with PBS, detached with Trypsin-EDTA (Sigma-Aldrich), and pelleted at 2,000 × g at 4°C, then washed again with 500 μL PBS and pelleted at 2,000 × g at 4°C. Cells were then either left unfractionated (procedure 1) or fractionated into nuclear and cytoplasmic fractions (procedures 2, 3, and 4) as described below, all centrifugation steps being performed at 4°C.
Procedure 1
Cell pellets were resuspended by repeated pipetting in 800 μL PBS, then cells were pelleted at 200 × g for 5 min and 700 μL supernatant was discarded.
Procedure 2
Cell pellets were resuspended by repeated pipetting in 400 μL Lysis Buffer (Reagent A100, Chemometec, Villeneuve-Loubet, France) before the addition of 400 μL Stabilizing Buffer (Reagent B, Chemometec). After centrifugation at 200 × g for 5 min, 700 μL supernatant was collected as the cytoplasmic fractions and the remaining 100 μL was kept as the nuclear fractions.
Procedure 3
This protocol makes use of the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific). Cells were resuspended in 100 μL ice-cold CER I solution by vortexing and incubated on ice for 10 min, then 5.5 μL CER II solution was added and samples were mixed by vortexing and left on ice for 1 min. The nuclear fractions were pelleted at 16,000 × g for 5 min and resuspended in 100 μL PBS, and the supernatants were collected as the cytoplasmic fractions.
Procedure 4
This protocol was described by Gagnon et al.35 Briefly, cell pellets were resuspended by repeated pipetting in 380 μL ice-cold hypotonic lysis buffer (HLB), containing 10 mM Tris (pH 7.5), 10 mM NaCl, 3 mM MgCl2, 0.3% (v:v) NP-40, and 10% (v:v) glycerol, and incubated 10 min on ice. After brief vortexing, nuclear fractions were pelleted at 1,000 × g for 3 min, and supernatants were collected as the cytoplasmic fractions. Nuclear pellets were then washed 3 times using 1 mL ice-cold HLB and centrifugation at 200 × g for 2 min, and nuclei were kept in 100 μL HLB after the last wash.
Analysis of Cellular Fractions and Quantification of Intracellular AAV Genomes by qPCR
For whole-cell (procedure 1) and nuclear fraction samples (procedures 2, 3, and 4), DNA was extracted and purified using the Gentra Puregene Blood kit (QIAGEN, Courtaboeuf, France), following the protocol for body fluids with proteinase K digestion, adapted to the different sample volumes. DNA pellets were resuspended in a final volume of 50 μL. DNA concentrations were measured by OD260 and adjusted to 20 ng/μL. For cytoplasmic fraction samples, DNA was extracted using the High Pure Viral Nucleic Acid kit (Roche, Meylan, France). TaqMan qPCR was performed using 5 μL sample DNA, in Premix Ex Taq 2X RT-PCR reagent (TaKaRa Bio) with primers targeted to the following: (1) the SV40 polyA signal for quantification of rAAV genomes, (2) the human albumin gene for quantification of cellular genomic (nuclear) DNA, and (3) the human cytochrome B gene for quantification of cellular mitochondrion (cytoplasmic) DNA (Table S1). Standard curves were obtained using 102–108 copies of ClaI-linearized pCQAAV1 plasmid.
IG titers were calculated from whole cell (procedure 1) and nuclear fraction samples (procedures 2, 3, and 4) using the following formula:
where Nvector is the SV40 copy number per sample, NAlb is the albumin gene copy number per sample, V is the volume of vector used to infect cells in microliters, and n is the number of cells at the time of infection. The first multiplication factor of 2 is because there are two albumin gene copies per cell genome. The second multiplication factor of 2 is because AAV genome is single stranded and plasmid used for the standard curve is double stranded.
Analysis of Cellular Fractions by Western Blot
For western blot analysis, we used a procedure allowing efficient recovery of proteins from all samples, i.e., whole cells or subcellular fractions. To this end, for all samples prepared by procedures 1–4, protease inhibitors (Complete Protease Inhibitor Cocktail, Roche) were added and samples underwent 3 freeze/thaw cycles to disrupt membranes. Purified His-tagged Rep68 protein (500 ng)30 was then spiked in each sample, before the addition of 4 sample volumes of ice-cold acetone and overnight precipitation at −20°C. Proteins were then pelleted at 16,000 × g for 15 min at 4°C and resuspended in 150 μL of 1× Laemmli buffer. SDS-PAGE was conducted using 25 μL of each protein sample, which was denatured at 95°C for 5 min before loading on 10% Tris-glycine gels (Life Technologies). Gels were transferred on nitrocellulose membranes and probed with antibody against lamin B, α-tubulin, ATP-synthase, or Rep proteins. Membranes were then incubated with HRP-conjugated secondary antibodies followed by chemiluminescence detection with ECL substrate (Thermo Scientific).
Statistical Analysis
Data are presented as mean ± SD. Statistical analyses were performed using PRISM 5 software (GraphPad). Differences between AAV8 control and AAV8ΔVP1 were assessed by Mann-Whitney tests. The p values lower than 0.05 were considered statistically significant.
Author Contributions
A.F., E.L., V.B., O.A., P.M., and E.A. conceived experiments and secured funding. M.B., E.L., and F.B. performed experiments. A.F., E.L., and M.P.-B. analyzed data. A.F. and E.A. wrote the manuscript.
Acknowledgments
We thank all the staff of the Vector Core at the University Hospital of Nantes (http://umr1089.univ-nantes.fr/facilities-cores/cpv/vector-core-2203910.kjsp?RH=1519296954157) for the production of the AAV reference vectors and for technical assistance. We are grateful to Nicolas Jaulin for performing the flow cytometry analysis, to Philippe Hulin and Steven Nedellec for their assistance in confocal imaging and analysis, and to Stéphanie Bucher and Sandy Douthe for their contribution to the intracellular AAV genome qPCR assay design. Flow cytometry and confocal microscopy analyses were conducted at the Cytocell and MicroPICell core facilities of Nantes University, respectively. This research was supported by the Fondation d’Entreprise Thérapie Génique en Pays de Loire, the Commissariat Général à l’Investissement (ANR Program, Investissements d’Avenir, Preindustrial Gene Therapy vector consortium), the Association Française contre les Myopathies (AFM), the Agence Nationale de Sécurité des Médicaments (ANSM), the Centre Hospitalier Universitaire (CHU) of Nantes, and the INSERM.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, five figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.07.004.
Contributor Information
Achille François, Email: achille.francois@univ-nantes.fr.
Eduard Ayuso, Email: eduard.ayuso@univ-nantes.fr.
Supplemental Information
References
- 1.Atchison R.W., Casto B.C., Hammon W.M. Adenovirus-Associated Defective Virus Particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 2.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 3.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nathwani A.C., Reiss U.M., Tuddenham E.G., Rosales C., Chowdary P., McIntosh J., Della Peruta M., Lheriteau E., Patel N., Raj D. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N. Engl. J. Med. 2014;371:1994–2004. doi: 10.1056/NEJMoa1407309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsushita T., Elliger S., Elliger C., Podsakoff G., Villarreal L., Kurtzman G.J., Iwaki Y., Colosi P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5:938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 7.Kotin R.M., Snyder R.O. Manufacturing Clinical Grade Recombinant Adeno-Associated Virus Using Invertebrate Cell Lines. Hum. Gene Ther. 2017;28:350–360. doi: 10.1089/hum.2017.042. [DOI] [PubMed] [Google Scholar]
- 8.Thomas D.L., Wang L., Niamke J., Liu J., Kang W., Scotti M.M., Ye G.J., Veres G., Knop D.R. Scalable recombinant adeno-associated virus production using recombinant herpes simplex virus type 1 coinfection of suspension-adapted mammalian cells. Hum. Gene Ther. 2009;20:861–870. doi: 10.1089/hum.2009.004. [DOI] [PubMed] [Google Scholar]
- 9.Adamson-Small L., Potter M., Byrne B.J., Clément N. Sodium Chloride Enhances Recombinant Adeno-Associated Virus Production in a Serum-Free Suspension Manufacturing Platform Using the Herpes Simplex Virus System. Hum. Gene Ther. Methods. 2017;28:1–14. doi: 10.1089/hgtb.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorne B.A., Takeya R.K., Peluso R.W. Manufacturing recombinant adeno-associated viral vectors from producer cell clones. Hum. Gene Ther. 2009;20:707–714. doi: 10.1089/hum.2009.070. [DOI] [PubMed] [Google Scholar]
- 11.Martin J., Frederick A., Luo Y., Jackson R., Joubert M., Sol B., Poulin F., Pastor E., Armentano D., Wadsworth S., Vincent K. Generation and characterization of adeno-associated virus producer cell lines for research and preclinical vector production. Hum. Gene Ther. Methods. 2013;24:253–269. doi: 10.1089/hgtb.2013.046. [DOI] [PubMed] [Google Scholar]
- 12.Gavin D.K. FDA statement regarding the use of adeno-associated virus reference standard materials. Hum. Gene Ther. Methods. 2015;26:3. doi: 10.1089/hgtb.2015.1501. [DOI] [PubMed] [Google Scholar]
- 13.Zen Z., Espinoza Y., Bleu T., Sommer J.M., Wright J.F. Infectious titer assay for adeno-associated virus vectors with sensitivity sufficient to detect single infectious events. Hum. Gene Ther. 2004;15:709–715. doi: 10.1089/1043034041361262. [DOI] [PubMed] [Google Scholar]
- 14.Salvetti A., Orève S., Chadeuf G., Favre D., Cherel Y., Champion-Arnaud P., David-Ameline J., Moullier P. Factors influencing recombinant adeno-associated virus production. Hum. Gene Ther. 1998;9:695–706. doi: 10.1089/hum.1998.9.5-695. [DOI] [PubMed] [Google Scholar]
- 15.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 16.Zeltner N., Kohlbrenner E., Clément N., Weber T., Linden R.M. Near-perfect infectivity of wild-type AAV as benchmark for infectivity of recombinant AAV vectors. Gene Ther. 2010;17:872–879. doi: 10.1038/gt.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girod A., Wobus C.E., Zádori Z., Ried M., Leike K., Tijssen P., Kleinschmidt J.A., Hallek M. The VP1 capsid protein of adeno-associated virus type 2 is carrying a phospholipase A2 domain required for virus infectivity. J. Gen. Virol. 2002;83:973–978. doi: 10.1099/0022-1317-83-5-973. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J.S., Li C., DiPrimio N., Weinberg M.S., McCown T.J., Samulski R.J. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 2010;84:8888–8902. doi: 10.1128/JVI.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Popa-Wagner R., Porwal M., Kann M., Reuss M., Weimer M., Florin L., Kleinschmidt J.A. Impact of VP1-specific protein sequence motifs on adeno-associated virus type 2 intracellular trafficking and nuclear entry. J. Virol. 2012;86:9163–9174. doi: 10.1128/JVI.00282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ayuso E., Blouin V., Lock M., McGorray S., Leon X., Alvira M.R., Auricchio A., Bucher S., Chtarto A., Clark K.R. Manufacturing and characterization of a recombinant adeno-associated virus type 8 reference standard material. Hum. Gene Ther. 2014;25:977–987. doi: 10.1089/hum.2014.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gurda B.L., Raupp C., Popa-Wagner R., Naumer M., Olson N.H., Ng R., McKenna R., Baker T.S., Kleinschmidt J.A., Agbandje-McKenna M. Mapping a neutralizing epitope onto the capsid of adeno-associated virus serotype 8. J. Virol. 2012;86:7739–7751. doi: 10.1128/JVI.00218-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wistuba A., Weger S., Kern A., Kleinschmidt J.A. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 1995;69:5311–5319. doi: 10.1128/jvi.69.9.5311-5319.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacouret S., Bouzelha M., Shelke R., Andres-Mateos E., Xiao R., Maurer A., Mevel M., Turunen H., Barungi T., Penaud-Budloo M. AAV-ID: A Rapid and Robust Assay for Batch-to-Batch Consistency Evaluation of AAV Preparations. Mol. Ther. 2017;25:1375–1386. doi: 10.1016/j.ymthe.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuck D., Kern A., Kleinschmidt J.A. Development of AAV serotype-specific ELISAs using novel monoclonal antibodies. J. Virol. Methods. 2007;140:17–24. doi: 10.1016/j.jviromet.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 25.Xiao P.J., Samulski R.J. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J. Virol. 2012;86:10462–10473. doi: 10.1128/JVI.00935-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark K.R., Voulgaropoulou F., Fraley D.M., Johnson P.R. Cell lines for the production of recombinant adeno-associated virus. Hum. Gene Ther. 1995;6:1329–1341. doi: 10.1089/hum.1995.6.10-1329. [DOI] [PubMed] [Google Scholar]
- 27.Chadeuf G., Favre D., Tessier J., Provost N., Nony P., Kleinschmidt J., Moullier P., Salvetti A. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J. Gene Med. 2000;2:260–268. doi: 10.1002/1521-2254(200007/08)2:4<260::AID-JGM111>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Lock M., McGorray S., Auricchio A., Ayuso E., Beecham E.J., Blouin-Tavel V., Bosch F., Bose M., Byrne B.J., Caton T. Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum. Gene Ther. 2010;21:1273–1285. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seth P., Rosenfeld M., Higginbotham J., Crystal R.G. Mechanism of enhancement of DNA expression consequent to cointernalization of a replication-deficient adenovirus and unmodified plasmid DNA. J. Virol. 1994;68:933–940. doi: 10.1128/jvi.68.2.933-940.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Awedikian R., François A., Guilbaud M., Moullier P., Salvetti A. Intracellular route and biological activity of exogenously delivered Rep proteins from the adeno-associated virus type 2. Virology. 2005;335:252–263. doi: 10.1016/j.virol.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 31.Nony P., Chadeuf G., Tessier J., Moullier P., Salvetti A. Evidence for packaging of rep-cap sequences into adeno-associated virus (AAV) type 2 capsids in the absence of inverted terminal repeats: a model for generation of rep-positive AAV particles. J. Virol. 2003;77:776–781. doi: 10.1128/JVI.77.1.776-781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimm D., Kern A., Rittner K., Kleinschmidt J.A. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 1998;9:2745–2760. doi: 10.1089/hum.1998.9.18-2745. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari F.K., Samulski T., Shenk T., Samulski R.J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salganik M., Aydemir F., Nam H.J., McKenna R., Agbandje-McKenna M., Muzyczka N. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J. Virol. 2014;88:1071–1079. doi: 10.1128/JVI.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gagnon K.T., Li L., Janowski B.A., Corey D.R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 2014;9:2045–2060. doi: 10.1038/nprot.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kronenberg S., Böttcher B., von der Lieth C.W., Bleker S., Kleinschmidt J.A. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 2005;79:5296–5303. doi: 10.1128/JVI.79.9.5296-5303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonntag F., Bleker S., Leuchs B., Fischer R., Kleinschmidt J.A. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 2006;80:11040–11054. doi: 10.1128/JVI.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nam H.J., Gurda B.L., McKenna R., Potter M., Byrne B., Salganik M., Muzyczka N., Agbandje-McKenna M. Structural studies of adeno-associated virus serotype 8 capsid transitions associated with endosomal trafficking. J. Virol. 2011;85:11791–11799. doi: 10.1128/JVI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 40.Mietzsch M., Grasse S., Zurawski C., Weger S., Bennett A., Agbandje-McKenna M., Muzyczka N., Zolotukhin S., Heilbronn R. OneBac: platform for scalable and high-titer production of adeno-associated virus serotype 1-12 vectors for gene therapy. Hum. Gene Ther. 2014;25:212–222. doi: 10.1089/hum.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mietzsch M., Casteleyn V., Weger S., Zolotukhin S., Heilbronn R. OneBac 2.0: Sf9 Cell Lines for Production of AAV5 Vectors with Enhanced Infectivity and Minimal Encapsidation of Foreign DNA. Hum. Gene Ther. 2015;26:688–697. doi: 10.1089/hum.2015.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urabe M., Nakakura T., Xin K.Q., Obara Y., Mizukami H., Kume A., Kotin R.M., Ozawa K. Scalable generation of high-titer recombinant adeno-associated virus type 5 in insect cells. J. Virol. 2006;80:1874–1885. doi: 10.1128/JVI.80.4.1874-1885.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis B.L., Hirsch M.L., Barker J.C., Connelly J.P., Steininger R.J., 3rd, Porteus M.H. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol. J. 2013;10:74. doi: 10.1186/1743-422X-10-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burger C., Gorbatyuk O.S., Velardo M.J., Peden C.S., Williams P., Zolotukhin S., Reier P.J., Mandel R.J., Muzyczka N. Recombinant AAV viral vectors pseudotyped with viral capsids from serotypes 1, 2, and 5 display differential efficiency and cell tropism after delivery to different regions of the central nervous system. Mol. Ther. 2004;10:302–317. doi: 10.1016/j.ymthe.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 45.Samulski R.J., Chang L.S., Shenk T. A recombinant plasmid from which an infectious adeno-associated virus genome can be excised in vitro and its use to study viral replication. J. Virol. 1987;61:3096–3101. doi: 10.1128/jvi.61.10.3096-3101.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonntag F., Köther K., Schmidt K., Weghofer M., Raupp C., Nieto K., Kuck A., Gerlach B., Böttcher B., Müller O.J. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011;85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thimmappaya B., Weinberger C., Schneider R.J., Shenk T. Adenovirus VAI RNA is required for efficient translation of viral mRNAs at late times after infection. Cell. 1982;31:543–551. doi: 10.1016/0092-8674(82)90310-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.