Abstract
Background
The grass family (Poaceae), ca. 12,075 species, is a focal point of many recent studies that aim to use complete plastomes to reveal and strengthen relationships within the family. The use of Next Generation Sequencing technology has revealed intricate details in many Poaceae plastomes; specifically the trnI - trnL intergenic spacer region. This study investigates this region and the putative mitochondrial inserts within it in complete plastomes of Paspalum and other Poaceae.
Results
Nine newly sequenced plastomes, seven of which contain an insert within the trnI - trnL intergenic spacer, were combined into plastome phylogenomic and divergence date analyses with 52 other species. A robust Paspalum topology was recovered, originating at 10.6 Ma, with the insert arising at 8.7 Ma. The alignment of the insert across Paspalum reveals 21 subregions with pairwise homology in 19. In an analysis of emergent self-organizing maps of tetranucleotide frequencies, the Paspalum insert grouped with mitochondrial DNA.
Conclusions
A hypothetical ancestral insert, 17,685 bp in size, was found in the trnI - trnL intergenic spacer for the Paspalum lineage. A different insert, 2808 bp, was found in the same region for Paraneurachne muelleri. Seven different intrastrand deletion events were found within the Paspalum lineage, suggesting selective pressures to remove large portions of noncoding DNA. Finally, a tetranucleotide frequency analysis was used to determine that the origin of the insert in the Paspalum lineage is mitochondrial DNA.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1379-1) contains supplementary material, which is available to authorized users.
Keywords: Grasses, Inserts, mtDNA, Next generation sequencing, Paspalum, Poaceae, ptDNA
Background
The grass family (Poaceae) comprises 12 subfamilies containing ca. 12,075 species [1]. Recent studies have delved into the phylogenomic framework of Poaceae using complete plastid genomes (plastomes) as a genomic marker; many of which have obtained plastomes by means of Next Generation Sequencing (NGS) methods. NGS technology has allowed for more extensive and deeper sampling within the grass subfamilies [2–15], tribes [16–19] and even genera (Alloteropsis: [20, 21]; Zea: [22]). This in-depth sampling strategy has allowed researchers to discover unique features of poaceous plastomes.
One such feature is the transfer of mitochondrial DNA (mtDNA) to plastome DNA (ptDNA). This type of mutational event was originally thought to be nonexistent or extremely rare [23–25]. The first reported case of mtDNA transfer to ptDNA was discovered in Daucus carota [26] and then a second discovery in Asclepias syriaca [27]. With both discoveries occurring within eudicots, it was not until Wysocki et al. [14], that mtDNA to ptDNA transfers were found in monocots.
The first discovery in monocots implicated transfers in two bamboos, Eremitis sp. and Pariana radiciflora, from the same subtribe, Parianinae. The inserts within Eremitis sp. and Pariana radiciflora were roughly 5 kilobase pairs (kbp) and 2.7 kbp, respectively, and with highly similar sequence for a 2.7 kbp section [14]. This was later expanded on with the inclusion of two more Parianinae, Pariana sp. and P. campestris [9], which also contained homologous inserts in the same region, the trnI - trnL intergenic spacer (IGS). While there have been other discoveries of mtDNA in ptDNA inserts found in grasses [Triticum monococcum: 11] and other eudicots [Lavandula angustifolia, Orobanche californica, Scutellaria baicalensis: 28], the placement of the insert within the trnI - trnL IGS has been of special interest.
The authors of a recent study [3] investigated this region, and found putative mtDNA inserts within the trnI - trnL IGS in two species of Paspalum. These inserts contained no similarity to each other, nor any similarity to the species of Parianinae in the same region. Their conclusions were either that multiple inserts within Paspalum arose independently of each other or that the two events were portions of a larger insert. If a mechanism for such intercompartmental DNA transfers were known, the taxonomic distribution of such events might be more evident. However, no such mechanism has been found [25]. This study will build on the previous study of Burke et al. [3], by increased sampling of species in Paspalum, to determine the origin, mechanism, and timing of the insertions found in plastomes of Paspalum.
Methods
Sampling
Species of Paspalum from another study [29] that were found to be closely related to those with and without the mtDNA insert [3] were selected for further analysis in this paper. Based on these criteria, eight new Paspalum plastomes (Table 1) were selected to be sequenced, adding to the three that were previously published. Taxa outside of Paspalum were selected to accommodate the available fossils within Poaceae [4]. Thus the analysis consists of the nine new plastomes, eight Paspalum species and Paraneurachne muelleri, and 45 other Poaceae plastomes from all subfamilies, which includes taxa for the divergence date analysis. The sampling also includes taxa in which mtDNA was found in the plastome, whether it is located in the trnI–trnL region [Parianinae: 9, 14] or in other regions [Triticum: 11].
Table 1.
Insert size, IR size, and plastome size of Paspalum species and Paraneurachne muelleri with their vouchers and Genbank accession numbers. Newly sequenced plastomes are in bold
| Taxa | Insert Size (bp) | IR Size (bp) | Plastome Size (bp) | Vouchera | Accession |
|---|---|---|---|---|---|
| Paraneurachne muelleri | 2808 | 22,774 | 140,886 | [19] | MG524000 |
| Paspalum dilatatum | 976 | 21,022 | 135,950 | [3] | NC_030614 |
| P. fimbriatum | 2914 | 23,711 | 140,804 | [3] | NC_030495 |
| P. glaziovii | 0 | 22,769 | 139,255 | [3] | NC_030496 |
| P. inaequivalve | 0 | 22,783 | 140,312 | PI 508769 | MG524001 |
| P. ionanthum | 11,393 | 34,039 | 162,086 | PI 404449 | MG524002 |
| P. juergensii | 1012 | 21,968 | 137,513 | PI 508779 | MG523994 |
| P. minus | 6500 | 29,160 | 152,184 | PI 404465 | MG523995 |
| P. pubiflorum | 10,619 | 31,803 | 157,521 | PI 304147 | MG523996 |
| P. simplex | 3678 | 24,848 | 143,254 | [19] | MG523997 |
| P. vaginatum | 0 | 22,777 | 140,451 | [19] | MG523998 |
| P. virgatum | 3204 | 24,396 | 142,796 | PI 364978 | MG523999 |
aPI = Plant Introduction number, U.S. National Plant Germplasm System (https://www.ars-grin.gov/npgs/)
Extraction and library preparation
DNA extractions from species of Paspalum were performed on young green leaf material dried in silica or herbarium specimens using the DNeasy Plant Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions after a liquid nitrogen homogenization step. Samples were then prepared for NGS using the Illumina Nextera protocol. DNA samples were diluted to 2.5 ng/ul (50 ng total) and paired-end libraries were prepared using the Nextera DNA Sample Preparation kit. All libraries were sequenced at the core DNA facility at Iowa State University (Ames, Iowa, USA) on an Illumina HiSEq 3000. The sequence reads, from Washburn et al. [19], for Paspalum simplex (SRR2162764), Paspalum vaginatum (SRR2163016), and Paraneurachne muelleri (SRR2163452) were retrieved from the SRA archive at NCBI (https://www.ncbi.nlm.nih.gov/sra) for whole plastome assembly.
NGS Plastome assembly, verification and annotation
Illumina and SRA reads were filtered and assembled following the methods of Burke et al. [3]. Reads of low quality were removed at default settings (DynamicTrim, SolexaQA++; [30]), excising adapters that were still attached to the reads (CutAdapt; [31]) and then the discarding of any read less than 25 base pairs (bp) long [LengthSort, SolexaQA++; 30]. SPAdes v3.6.1 [32] was used for de novo assembly with k-mers set from 19 to 73 bp with intervals of six. CD-Hit v4.6 [33] removed redundant sequences from the contig file. ACRE [34] was used to scaffold contigs together.
ACRE scaffolds and clean reads were imported into Geneious Pro v9.1.6 [35]. For each new accession, a closely related reference plastome, banked at NCBI, was chosen. The scaffolded contigs were then aligned to the reference plastome using the MAFFT v7.222 [36] plugin in Geneious. The gaps between the contigs were closed by using the map to reference function in Geneious to in silico genome walk [3]. A final verification of each plastome was performed by mapping the reads to the finalized plastome and manual adjustments were made when incongruences between the reads and the plastome occurred.
Verified plastome accessions were then pairwise aligned to their reference plastome, and annotations were applied using the transfer annotation feature in Geneious Pro. Coding sequence boundaries were inspected and manually adjusted to preserve reading frames. The inverted repeats (IRs) were located using the methods of Burke et al. [37]. BLASTn [38] was used to locate IR boundaries by aligning the new plastome to itself, and looking for segments in which the orientation of the reads transition from plus/plus to plus/minus. These boundaries were then flagged using the motif feature in Geneious Pro, and annotations for the IR were made.
Phylogenomic analyses and divergence date estimation
A 61 taxa matrix of Poaceae plastid DNA (ptDNA) was assembled containing nine new plastomes and 11 Paspalum species overall. An alignment of these 61 complete plastomes, excluding one IR copy, was created using the MAFFT v7.222 [36] plugin in Geneious Pro, with default settings.
The trnI–trnL regions from species of Paspalum were extracted from the MAFFT alignment and manually aligned. This alignment was manually examined for shared or unique rare genomic changes and sequences with unexpected homologies were identified by BLASTn searches. Manual searches were also performed in this region to find sequence evidence for molecular mechanisms indicative of insertions, deletions, or site-specific recombinations. Such searches were also conducted on all pairwise alignments (55, total) of the trnI - trnL insert region of Paspalum spp. In particular, the presence of tandem repeats, dispersed repeats, and inverted repeats was investigated. Intermediate results suggested the presence of significantly placed dispersed repeats in the trnI – trnL insert region. Dispersed repeats were localized to endpoints of subregions of the trnI - trnL insert region via the motif search option in the Annotate and Predict menu in the sequence viewer of Geneious Pro. Searches were conducted from a minimum sequence length of five bp for dispersed repeats that mediate intrastrand deletions as suggested by Graur et al. [39].
Prior to plastome phylogenomic analyses of the 61 taxon matrix, any gaps that were produced by the alignment were then stripped from the matrix. A model for the stripped nucleotide alignment was selected using jModelTest v2.1.10 [40], and the GTR + I + G model was selected under the Akaike information criterion [41]. Maximum Likelihood (ML) and Bayesian MC3 Inference (BI) analyses were conducted on the alignment. The ML analysis was done via RAxML-HPC2 on XSEDE v8.2.9 [42] on the CIPRES Science Gateway [43]. The number of bootstrap replicates was set to 1000, and all other parameters were set to default. The BI analysis used MrBayes on XSEDE v3.2.3 [44] at the CIPRES Science Gateway. Settings for two independent analyses with four chains of twenty million generations each were specified, with a default burn-in value of 25%. The model was set to “invgamma” and “nst = 6”, with all other parameters at default.
An estimation of divergence times was performed on the 61 taxa stripped alignment with one IR in BEAST v2.4.3 [45]. Parameters included an uncorrelated relaxed clock model [46], a GTR substitution model with a gamma category count of six, an estimated shape parameter of 0.92 and an initial value of 0.52 for estimated proportion of invariant sites. Initial values for the shape parameter and proportion of invariant sites were obtained from the ML analysis.
The seven fossils that were chosen as calibration points are based on the recent use in divergence date analyses [4] and are reliably dated and confidently associated extant homologous species [47–49]. The specifics of these fossils are given (Additional file 1: Table S1). The fossil calibrations were placed on the assigned nodes (Additional file 1: Table S1) with the minimum node age set to the stratigraphic or radiometric date of the fossil and the maximum age set to the oldest known fossil in Poaceae [110 Ma; 49], generally following the methods of Christin et al. [46].
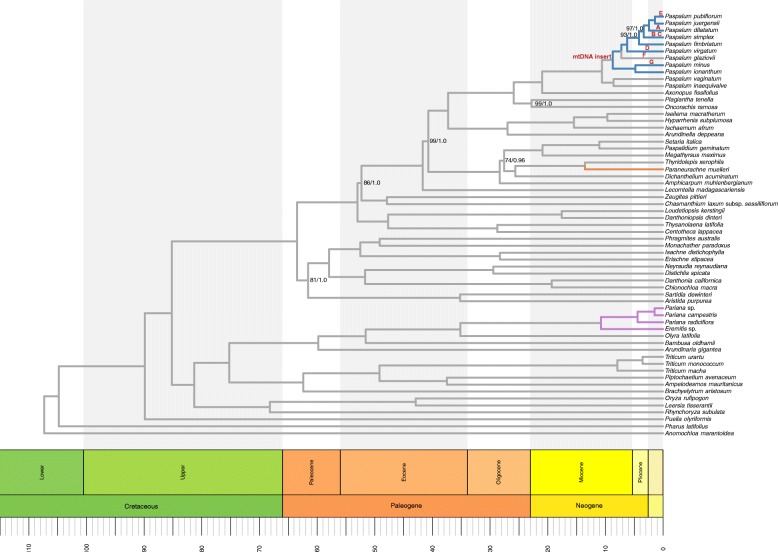
The BEAST analysis was conducted on the CIPRES Science Gateway for 40 million generations, logging at every 10,000 trees. Tracer v1.6 [50] was used to assess convergence. TreeAnnotator [51] was used to summarize the trees with a burn-in value of 25%. The plastome chronogram, with support values, was then visualized with the packages “strap” [52] and “ips” [53] in R [54]; (Fig. 1).
Fig. 1.
Chronogram of Poaceae species with support values (ML / BI) for nodes that did not recover maximum support. All other nodes are maximally supported. Highlighted branches depict different taxonomic groups with similar inserts in the trnI - trnL IGS. Letters in the Paspalum lineage designate IDEs (Table 2)
Tetramer identity analysis
The contigs assembled in SPAdes for Paspalum were filtered in BLASTn at default parameters with a database containing the 11 published complete mitochondrial sequences from Poaceae (NC_007579.1, NC_008362.1, NC_011033.1, NC_007886.1, NC_007982.1, NC_029816.1, NC_013816.1, NC_008360.1, NC_008333.1, NC_008332.1, and NC_008331.1). The contigs, which showed high mitochondrial identity, were extracted using the BBmap “filterbyname.sh” executable script [55]. Five partitions were binned using Binning-Master [56] to determine tetramer identity with a sliding window of four nucleotides, advancing by one, for the Large Single Copy (LSC), Small Single Copy (SSC), IR without the insert, mitochondrial contigs retrieved from BLASTn filtering, and the Paspalum inserts in the IR. Files were then imported into Emergent Self-Organizing Maps (ESOM) software [57] to visualize the tetramers for each partition. Training was performed with the K-Batch algorithm of 20 epochs, with a starting radius of 50 and the dimensions of a 150 × 150 plot. The other parameters were set to default.
In contrast to pairwise comparisons, the ESOM method provides a more thorough and precise estimate of insert origin. The ESOM method statistically groups tetrameric sequence regions and determines if they have similar identities. Our hypothesis is as follows. If the Paspalum inserts were of plastid origin, then the tetramers would display strong elevation boundaries around other plastid regions: LSC, SSC and IR; otherwise they would be distributed among the mitochondrial sequences.
Results
Plastome features
GenBank accession numbers were obtained for the nine new complete plastomes in this study (Table 1). Not including the trnI-trnL IGS insert regions of the 11 Paspalum species, the overall pairwise identity was 98.3%, while the manually aligned trnI-trnL IGS insert region was 28.6% identical, due to the large number of gapped positions. The inserts in Paspalum ranged from 976 bp in P. dilatatum to 11,393 bp in P. ionanthum. Due to the length of this insert, P. ionanthum is now the longest known Poaceae plastome at 162086 bp. Other notable indels in the species of Paspalum were a 776 bp deletion in the psbE-petL IGS in all Paspalum except for P. inaequivalve and P. vaginatum. All Paspaleae shared a 222 bp deletion in the petA-psbJ IGS.
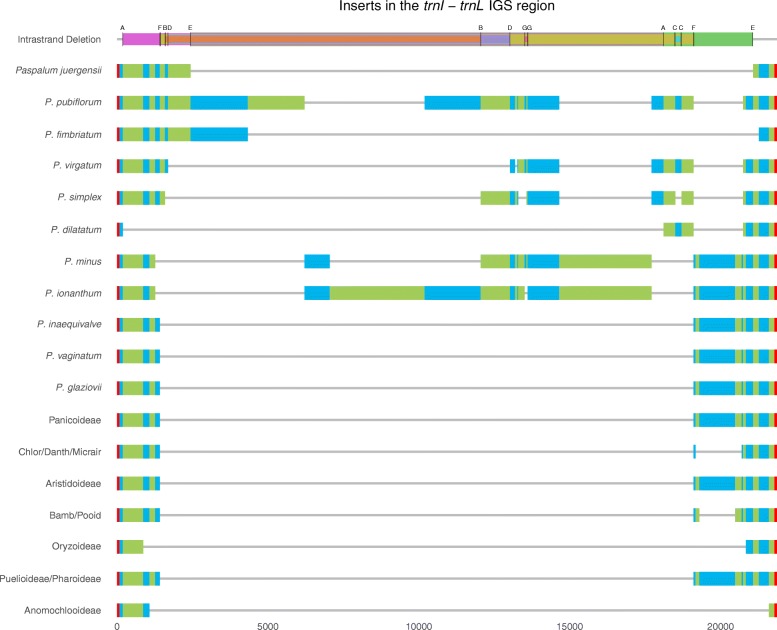
Based on the manual alignment, 21 separate subregions in the trnI-trnL IGS insert were identified. Of these, 19 shared homology between two or more Paspalum species, and the other two, found in P. publiforum at 1878 bp and P. ionanthum at 3146 bp in length respectively, were unique to only those species. This was visualized (Fig. 2) using the function “geom_linerange” in ggplot2 [58] with the use of “select” and “gather” functions in tidyverse [59] to coerce the data, in the R statistical suite. Based on extensive searches of annotated regions, seven pairs of dispersed repeats were identified that immediately flanked presumed intrastrand deletion events (IDE). The size of the dispersed repeats ranged from 8 to 17 bp, and the deleted regions based on the alignment ranged from 112 bp, “IDE G,” to 18,645 bp, “IDE E” (Table 2). Note that since excisions via intrastrand deletion, which are mediated by recombination between repeats, eliminates one copy of the repeat in any given species, the existence of dispersed repeats was only identified when comparing homologous plastome regions between Paspalum sp. Finally, a BLASTn search of the concatenated Paspalum insert sequence representing the progenitor insert, 17,685 bp in length, returned the top hit of the complete mitochondrial genome of Tripsacum dactyloides (DQ984517) at 99% identity for 33% of the sequence.
Fig. 2.
The inserts compared among Paspalum species and across generalized subfamilies. Aligned subregions are differentiated with alternating blue and green bars. These subregions are flanked by red bars representing trnI (left) and trnL (right). IDEs are represented with overlapping colored bars and their designated letter (Table 2)
Table 2.
A list of intrastrand deletion events (IDEs) in the Paspalum lineage
| IDE | Sequence | Deletion Length (bp) | Divergence Time (Ma) | Paspalum with Deletion | Both repeats present at each end of the deletion | One repeat present at one end of the deletion |
|---|---|---|---|---|---|---|
| A | CAAATATTCGGA | 17,929 | 2.4 | P. dilatatum | P. pubiflorum, P. virgatum, P. simplex | P. juergensii, P. fimbriatum, P. minus, P. ionanthum, P. glaziovii, P. inaequivalve, P. vaginatum |
| B | ATATAACTTCCC | 10,466 | 3.3 | P. simplex | P. pubiflorum | P. juergensii, P. fimbriatum, P. virgatum, P. minus, P. ionanthum |
| C | TCCTGAAACGTAAAAAA | 220 | 3.3 | P. simplex | P. dilatatum, P. pubiflorum, P. virgatum | |
| D | ACTTGCTC | 11,340 | 6.2 | P. virgatum | P. pubiflorum | P. juergensii, P. fimbriatum, P. simplex, P. minus, P. ionanthum |
| E | ATGATTTGG | 18,645 | 1.4 | P. juergensii | P. pubiflorum | P. fimbriatum, P. dilatatum, P. virgatum, P. simplex, P. minus, P. ionanthum, P. glaziovii, P. inaequivalve, P. vaginatum |
| F | GGAATCCGTTAGAAAT | 17,700 | 2.0 | P. glaziovii, P. inaequivalve, P. vaginatum | P. dilatatum, P. juergensii, P. fimbriatum, P. pubiflorum, P. virgatum, P. simplex, P. minus, P. ionanthum, | |
| G | GGCTCGGCGAC | 112 | 4.8 | P. ionanthum | P. pubiflorum, P. virgatum, P. minus | P. simplex |
The trnI-trnL IGS insert in Paraneurachne muelleri was 2808 bp long, and contained no similarity to the subregion found in Paspalum or Parianinae. BLASTn results returned a 92% shared identity to Sorghum bicolor mitochondrial genome (DQ984518.1) for 68% of the sequence.
Plastome Phylogenomic and divergence date analyses
The same topologies were retrieved for the ML and BI analyses. Most nodes were supported maximally except for seven nodes in the ML analysis and one node in the Panicinae in the BI analysis (Fig. 1). In both analyses Panicoideae were sister to the rest of the PACMAD clade with a bootstrap value of 81% and a posterior probability (PP) of 1.0. The Paspalum species were retrieved as monophyletic, with only two bootstrap values being less than maximum and all had a PP of 1.0.
The same topology was retrieved in the divergence date analysis performed by BEAST v2.4.3. The crown node of the PACMAD clade was dated at 63.5 Ma with crown nodes of Panicoideae diverging at 53.0 Ma, Paspaleae at 25.9 Ma and Paspalinae at 21.0 Ma. The crown node of Paspalum diverged at 10.6 Ma, with the most recent diversification event at 1.4 Ma with the divergence of P. pubiflorum and P. juergensii from their common ancestor. The shortest time between species divergences is 0.9 Ma, from which P. simplex diverges from the sister clade comprised of: P. dilatatum, P. pubiflorum, and P. juergensii. The longest time between divergence events is 8.6 Ma with the bifurcation of P. vaginatum and P. inaequivalve, the earliest lineage in Paspalum sampled here.
Tetramer identity analysis
The tetramer matrices were generated for each of the five partitions. These files were made into the U-Matrix, which is a visualization of distance in the tetramer frequency between data points and is represented as map elevation. These distances between clusters of points are further visualized with elevation barriers acting as cutoffs, thus representing large differences between data sets [56, 57]. This was used to interpret the origin of Paspalum inserts, depending on where they occurred and how they were separated in the map.
Inspection of the ESOM results shows that most of the plastome clusters together. The LSC and SSC cluster with high ‘elevation’ (white and light grey areas) around most of the points, and with lower ‘elevation’ (dark grey and black areas) compartmentalizing clusters of LSC and SSC within that barrier. The IR is close to the LSC/SSC regions along an area where the elevation is not as high. There is one cluster of IRs that is not near LSC/SSC area, and is located among mtDNA with no high ‘elevation’ barriers nearby. The Paspalum inserts were also located within the mtDNA region, also in an area with very few high ‘elevation’ barriers. Some of the insert data points clustered, with two groups of two and one group of four. The mtDNA points were located throughout the ESOM, but with fewer data points located within the LSC/SSC area (Fig. 3).
Fig. 3.
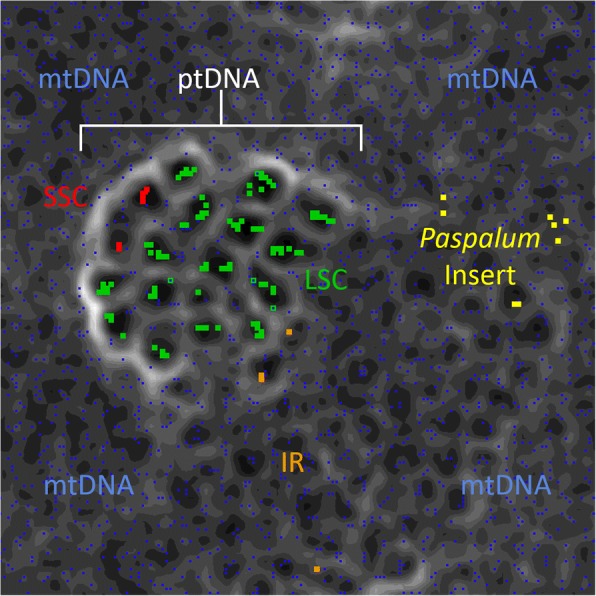
ESOM of mtDNA and partitions of ptDNA sequences based on tetranucleotide frequency. The mtDNA is in blue with the ptDNA divided into LSC (green), SSC (red), IR (orange), and the Paspalum inserts (yellow). High ‘elevations’ are in white visualizing major distances in tetranucleotide frequency, while ‘low elevations’ are in darker grays showing similarity in tetranucleotide frequency
Discussion
Plastome features
In the eight newly assembled Paspalum species, six contained an insert in the trnI-trnL IGS. We investigated whether the origin of the inserts was one or multiple events. Inserts from different species had little overall similarity and only one species, P. fimbriatum, had a secondary insert, which had homology to a smut parasite of grasses [3]. However, while none of the sequences are exactly the same for each species, these six new inserts contain multiple subregions of high identities that are shared between sections of newly and previously discovered Paspalum inserts [3]. Thus, 19 out of the 21 subregions (Fig. 2) contain shared sequence similarity across the sampled Paspalum, which suggests a single insertion event at some point in the Paspalum lineage (Fig. 1). With the current sampling, the hypothetical ancestral insert size is estimated to be around 17,685 bp long, and would have only occurred once. This is in agreement with previous characterizations of mitochondrial inserts in plastomes, which suggest that while these insert events happen, they are rare [25].
The homologous IGS also contained an insert for the newly sequenced Paraneurachne muelleri. While this 2808 bp insert did not match any subregion of the Paspalum inserts or the Parianinae inserts, it does illustrate that this IGS in the plastome is a hotspot for indel mutations. On average the lengths of plastid IGSs for all Poaceae in this sampling, removing species with probable mitochondrial inserts, is 422 bp, yet the length of the trnI-trnL IGS is on average 3071 bp. The greater length of this IGS is likely due to the disintegration of the large ycf2 CDS, which is one of the defining features for poaceous plastomes [60]. The larger size of the plastome IGS potentially offers less steric hindrance during recombination with the larger mitochondrial insert. Future discoveries of more grass genera with insertions in this trnI - trnL IGS, would support the idea of this spacer as a mutational hotspot for recombination mutations.
The most likely mutational mechanism that can explain the varying lengths of these inserts is gene conversion followed by repeated rounds of intrastrand deletions. Gene conversion, or nonreciprocal recombination, occurs when homologous regions have uneven replacement of sequence, causing the loss of one variant sequence [39]. To help put this mechanism in context, an understanding of plant mitochondrial genomes is warranted.
Mitochondria are known to actively take up and incorporate foreign DNA, which includes ptDNA [61]. One study [62], found that 22,593 bp of ptDNA was located in a mitochondrial genome (AB076665–6). That only accounts for 6.3% of the mitochondrial DNA, but it is nearly 20% of the donor plastome (NC_001320). Another study [63], found two major ptDNA inserts, of 12.6 kb and 4.1 kb that were primarily IR sequence, and with other inserts totaled 23.9 kb. This only accounts for 4.2% of the mitochondrial genome (AY506529), but is 17.0% of the respective plastome (X86563). Within these plastome inserts in mitochondrial genomes, complete genes can be found like trnI (NC_007982 at 326704–327726) and trnL (NC_007982 at 336020–336100). Since mitochondrial genomes are known for rearranging to the extent that gene order is not conserved between genera or even congeneric species [64–67], it is likely that the intergenic distance of genes in the mitochondrial genome with homology to those in the plastome would differ in length and sequence between grass taxa.
Thus, sequences in the mitochondrial genome that contain mtDNA flanked by conserved regions of ptDNA (trnI and trnL) would have the potential for gene conversions. The conserved ptDNA would act as recombination points between the plastome loci and homologous loci in the mitochondrial genome to create mtDNA insertions. The varying lengths seen among different genera are also explained by rearrangements within the mitochondrial genome that are specific to a taxonomic level, such as within the Paspalum or Parianinae lineages. This creates varying insert lengths and with sequence content unique to the separate insertion events at different taxonomic levels (Paspalum progenitor: 17685 bp, Paraneurachne: 2808 bp, Parianinae: 4920 bp).
Once the inserts are established in the plastome, the mechanism for the differential degradation that is seen within Paspalum can easily be explained. The identification of dispersed repeats localized to the endpoints of these areas clearly indicate IDE as the causal mechanism. There are seven examples of IDEs (Table 2) that have removed sequences of varying lengths, ranging from 112 bp to 18,645 bp, throughout the insert subregions in Paspalum and flanking ptDNA, which precisely terminate in short dispersed repeats. These IDEs, with other probable ones that are undetectable due to the taxonomic subset that was sampled, create sequences that are seemingly unique, but are just the result of various ways to excise sequence from the original insert.
An IDE may also explain a singular situation for one species, P. glaziovii, which does not contain mtDNA inserts. There is evidence for an IDE within Paspalum that can remove the entire insert. An upstream repeat (16 bp in length) can be found early in the sequence, near trnI, for P. juergensii, P. fimbriatum, P. pubiflorum, P. virgatum, and P. simplex, while a downstream exact repeat is found closer to the trnL gene in P. minus, P. ionanthum, P. glaziovii, P. inaequivalve, and P. vaginatum. At this point there is no Paspalum species that has both copies of this repeat. In those species that contain one repeat copy and insert sequence, it is likely that a smaller IDE removed one of the repeats. Doing so would preserve a larger portion of the original insert, and potentially create contingencies on what other IDEs can occur based on whether the first or second repeat was removed.
The hypothesized smaller IDEs that preserved the insert by removing one of the repeats appears to have also removed the possibilities of other IDEs. An example of this is at the beginning of P. minus, and P. ionanthum, which are missing one subregion prior to “IDE F,” likely due to a smaller IDE. This unidentified IDE most likely removed not only the first repeat of “IDE F,” but also the first of the dispersed repeats for “B”, “D”, and “E”. Thus, the contingency of future IDEs based on a previous IDE and understanding priority of IDEs will be better determined with further sampling within the Pasplaum lineage (for example Piot et al. [10], and future studies).
Phylogenomic and divergence date analyses
The topology of Paspalum species for this study was well supported with all but two nodes being maximally supported in the ML analysis, and all nodes maximally supported in the BI analysis. With little doubt as to the divergence of the represented species, we find that species with no evidence of an insert are non-monophyletic. P. inaequivalve and P. vaginatum, which both lack the insert, are in the earliest diverging clade in this study, but P. glaziovii, which also lacks the insert, is more recently diverged within the Paspalum lineage. This demonstrates that the lack of an insert is not an ancestral trait. Based on the current sampling, it is likely the insert arose at the clade comprising all Paspalum except P. inaequivalve and P. vaginatum. This might explain why P. ionanthum has the longest insert (11,393 bp), as it is near the origin of the insertion event. With the likely point of origin for the insertion, determining the time from foreign DNA insertions to IDEs will further the understanding of how plastomes interact with inserted non-ptDNA.
In the chronogram phylogeny, selected divergence dates for speciation effectively represent the hypothetical time of the insertion event and the time it takes for IDEs to become fixed in the plastome. If the insert arose around 8.7 Ma, and the subsequent loss of the insert in P. glaziovii occurred at 7.2 Ma, then the fixation of “IDE F” occurred within a time period of roughly 1.5 Ma. Other IDEs (A, B, D, E, F), became fixed in relatively short time periods as well, ranging from as little as 1.4 Ma (“IDE E”) to 3.3 Ma, except for “IDE D” at 6.2 Ma. These larger time estimates could be due to a lack of sampling, and not being able to break up longer branches with a species rich genus like Paspalum (~ 350 spp.) [29]. Thus, IDEs, especially larger ones, becoming fixed in a relatively short time could suggest that the original insert is being selected against.
The progenitor’s hypothesized insert size is over 17,000 bp, and contains no recognizable coding sequence. The longer the insert, the more time, energy and materials will be needed in the replication process of the plastome. This might explain for the variety of different IDEs seen throughout the phylogeny. While some species of Paspalum are missing one or both of the repeats that can remove the insert, “IDE F,” they contain other IDEs that either shorten the insert or remove most of the insert with the ptDNA that flanks the insert (P. juergensii, P. fimbriatum, and P. dilatatum). Thus, a smaller plastome, with little or no insert, could be more beneficial overall to the plastid compared to larger plastomes.
Tetramer identity analysis
It is also important to determine the origin of the insert as it could have other implications if the source is not from mtDNA. For this, the ESOM analysis (see visual matrix output, Fig. 3) was conducted to compare tetranucleotide frequencies to evaluate support for our hypotheses of origin. The Paspalum insert within the ESOM matrix did not cluster with other ptDNA, but rather clustered into smaller groups among the mtDNA. From the ESOM matrix (Fig. 3), there is a distinct high ‘elevation’ around the ptDNA. This circumscription of ptDNA is more apparent near the top and left side portion of the ptDNA cluster, with the loss of ‘elevation’ as it gets closer to the sections with IR sequences. This can be explained as there are large portions of IR that are located in the mtDNA of many Poaceae species [62, 63]. Thus, there is some similarity creating a lower ‘elevation’ around IR points. The incorporation of ptDNA into mtDNA is also why there are points from the mtDNA within the ptDNA cluster [62, 63]. There is also one IR point that is noticeably distant from the rest of the ptDNA cluster. This is most likely the rRNAs that are found within the inverted repeats. These regions are very GC rich (Paspalum and other Poaceae species in this study: GC% = 54.7%) and thus would look more like mtDNA, which is overall also more GC rich (AY506529: GC% = 43.9%, AB076665–6: GC% = 43.9%) than the overall AT rich ptDNA (Paspalum: GC% = 37.8%, Poaceae species in this study: GC% = 37.6%).
Our interpretation of these results is that the insert is mtDNA in origin, which was then transferred into ptDNA by mechanisms suggested previously. This possibility, which was once thought to be impossible or highly unlikely [23, 24], is only now becoming evident as more NGS work is done [3, 9, 10, 14, 28].
Conclusion
In conclusion, the Paspalum insert in the trnI - trnL IGS was observed in eight out of the eleven banked plastomes. The original insert was determined to have similarities to mtDNA suggesting mtDNA origins. This is supported by the high pairwise identity compared to other mitochondrial plastomes, and the placement of the insert in the tetranucleotide analysis. It is likely that this mtDNA insert arose at the clade comprising all Paspalum except possibly P. inaequivalve and P. vaginatum around 8.7 Ma, and mtDNA sequence was subsequently lost in certain species. The deletion events in the insert were not always complete removals, but there is a general trend of removing large subregions of sequence. This is seen throughout the Paspalum lineage, with seven different IDEs identified. These IDEs remove large portions of sequence in a given species lineage within a relatively small amount of time, the shortest of which was noted at 1.4 Ma. Thus, this suggests that there might be evolutionary pressure to remove this excess mtDNA from the plastome.
Additional file
Table S1 Information of fossils calibrations used for divergence date analysis [49, 68–73]. (DOCX 14 kb)
Acknowledgements
We would like to thank L. Attigala for submitting orders to the ISU DNA facility. Special thanks to J. Craine who provided samples for use in this study. We also thank the students in the Molecular Evolution class of 2017 for preliminary PCR based screening of Paspalum inserts.
Funding
This work was supported in part by the Plant Molecular Biology Center, the Department of Biological Sciences at Northern Illinois University and the National Science Foundation under Grant Numbers DEB-1120856 to MCU and MRD, and DEB-1120761 to MRD. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Availability of data and materials
The data supporting our conclusions are available from the corresponding author on reasonable request. All nucleotide sequences were deposited in the NCBI Genbank repository, and accession numbers can be found in Table 1.
Abbreviations
- ACRE
Anchored conserved region extension
- BEAST
Bayesian Evolutionary Analysis Sampling Trees
- BI
Bayesian MC3 Inference
- BLASTn
Nucleotide Basic Local Alignment Search Tool
- bp
Base pair
- CIPRES
Cyber infrastructure for phylogenetic research
- contigs
Contiguous sequences
- ESOM
Emergent Self-Organizing Maps
- GTR + G + I
General time reversible plus gamma distribution plus proportion of invariant sites
- IDE
Intrastrand deletion event
- IGS
Intergenic spacer
- IR
Inverted repeat
- Kbp
Kilobase pairs
- LSC
Large single copy
- Ma
Megaannus or million years
- MAFFT
Multiple alignment using fast fourier transform
- ML
Maximum likelihood
- mtDNA
Mitochondrial DNA
- NCBI
National Center for Biotechnology Information
- NGS
Next generation sequencing
- PACMAD
Panicoideae Arundinoideae Chloridoideae Micrairoideae Arundinoideae Danthonioideae
- PP
Posterior probability
- ptDNA
Plastid DNA
- SSC
Small single copy
Authors’ contributions
SVB performed data acquisition, Nextera library preparations, plastome assembly, alignment, phylogenomic analyses and drafted the manuscript. MCU provided plant material from USDA National Germplasm Bank for sequencing. MRD assisted in data acquisition, Nextera library preparations, contributed to the design of the study and facilitated interactions between co-authors. All authors read and assisted in the drafting of the manuscript in an editorial fashion. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing interests.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sean V. Burke, Email: sburke5@niu.edu
Mark C. Ungerer, Email: mcungere@ksu.edu
Melvin R. Duvall, Email: mel-duvall@niu.edu
References
- 1.Judd WS, Campbell CS, Kellogg EA, Stevens PF, Donoghue MJ. Plant systematics: a phylogenetic approach. Rhodora. 2016;118(976):418–420. doi: 10.3119/0035-4902-118.976.418. [DOI] [Google Scholar]
- 2.Burke SV, Clark LG, Triplett JK, Grennan CP, Duvall MR. Biogeography and phylogenomics of new world Bambusoideae (Poaceae), revisited. Am J Bot. 2014;101(5):886–891. doi: 10.3732/ajb.1400063. [DOI] [PubMed] [Google Scholar]
- 3.Burke SV, Wysocki WP, Zuloaga FO, Craine JM, Pires JC, Edger PP, et al. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae) BMC Plant Biol. 2016;16(1):140. doi: 10.1186/s12870-016-0823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke SV, Lin CS, Wysocki WP, Clark LG, Duvall MR. Phylogenomics and plastome evolution of tropical forest grasses (Leptaspis, Streptochaeta: Poaceae). Front Plant Sci. 2016;7:1-12. [DOI] [PMC free article] [PubMed]
- 5.Cotton JL, Wysocki WP, Clark LG, Kelchner SA, Pires JC, Edger PP, et al. Resolving deep relationships of PACMAD grasses: a phylogenomic approach. BMC Plant Biol. 2015;15(1):178. doi: 10.1186/s12870-015-0563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duvall MR, Fisher AE, Columbus JT, Ingram AL, Wysocki WP, Burke SV, et al. Phylogenomics and plastome evolution of the chloridoid grasses (Chloridoideae: Poaceae) Int J Plant Sci. 2016;177(3):235–246. doi: 10.1086/684526. [DOI] [Google Scholar]
- 7.Duvall MR, Yadav SR, Burke SV, Wysocki WP. Grass plastomes reveal unexpected paraphyly with endemic species of Micrairoideae from India and new haplotype markers in Arundinoideae. Am J Bot. 2017;104(2):286–295. doi: 10.3732/ajb.1600285. [DOI] [PubMed] [Google Scholar]
- 8.Jones SS, Burke SV, Duvall MR. Phylogenomics, molecular evolution, and estimated ages of lineages from the deep phylogeny of Poaceae. Plant Syst Evol. 2014;300(6):1421–1436. doi: 10.1007/s00606-013-0971-y. [DOI] [Google Scholar]
- 9.Ma PF, Zhang YX, Guo ZH, Li DZ. Evidence for horizontal transfer of mitochondrial DNA to the plastid genome in a bamboo genus. Sci Rep. 2015;5:1-9. [DOI] [PMC free article] [PubMed]
- 10.Piot A, Hackel J, Christin PA, Besnard G. One-third of the plastid genes evolved under positive selection in PACMAD grasses. Planta. 2018;247(1):255-66. [DOI] [PubMed]
- 11.Saarela JM, Wysocki WP, Barrett CF, Soreng RJ, Davis JI, Clark LG, Duvall MR. Plastid phylogenomics of the cool-season grass subfamily: clarification of relationships among early-diverging tribes. AoB Plants. 2015;7:1-27. [DOI] [PMC free article] [PubMed]
- 12.Teisher JK, McKain MR, Schaal BA, Kellogg EA. Polyphyly of Arundinoideae (Poaceae) and evolution of the twisted geniculate lemma awn. Annals Botany. 2017;120(5):725-38. [DOI] [PMC free article] [PubMed]
- 13.Wu ZQ, Ge S. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Mol Phylogenet Evol. 2012;62(1):573–578. doi: 10.1016/j.ympev.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 14.Wysocki WP, Clark LG, Attigala L, Ruiz-Sanchez E, Duvall MR. Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis. BMC Evol Biol. 2015;15(1):50. doi: 10.1186/s12862-015-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang YJ, Ma PF, Li DZ. High-throughput sequencing of six bamboo chloroplast genomes: phylogenetic implications for temperate woody bamboos (Poaceae: Bambusoideae) PLoS One. 2011;6(5):e20596. doi: 10.1371/journal.pone.0020596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arthan, W., M. R. McKain, P. Traiperm, C. A. D. Welker, J. K. Teisher, and E. A. Kellogg. (2017). Relationships of southeast Asian Andropogoneae (Poaceae). Syst Bot: in press.
- 17.Attigala L, Wysocki WP, Duvall MR, Clark LG. Phylogenetic estimation and morphological evolution of Arundinarieae (Bambusoideae: Poaceae) based on plastome phylogenomic analysis. Mol Phylogenet Evol. 2016;101:111–121. doi: 10.1016/j.ympev.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Besnard G, Christin PA, Malé PJG, Coissac E, Ralimanana H, Vorontsova MS. Phylogenomics and taxonomy of Lecomtelleae (Poaceae), an isolated panicoid lineage from Madagascar. Ann Bot. 2013;112(6):1057–1066. doi: 10.1093/aob/mct174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washburn JD, Schnable JC, Davidse G, Pires JC. Phylogeny and photosynthesis of the grass tribe Paniceae. Am J Bot. 2015;102(9):1493–1505. doi: 10.3732/ajb.1500222. [DOI] [PubMed] [Google Scholar]
- 20.Lundgren MR, Besnard G, Ripley BS, Lehmann CE, Chatelet DS, Kynast RG, et al. Photosynthetic innovation broadens the niche within a single species. Ecol Lett. 2015;18(10):1021–1029. doi: 10.1111/ele.12484. [DOI] [PubMed] [Google Scholar]
- 21.Olofsson JK, Bianconi M, Besnard G, Dunning LT, Lundgren MR, Holota H, et al. Genome biogeography reveals the intraspecific spread of adaptive mutations for a complex trait. Mol Ecol. 2016;25(24):6107–6123. doi: 10.1111/mec.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orton LM, Burke SV, Wysocki WP, Duvall MR. Plastid phylogenomic study of species within the genus Zea: rates and patterns of three classes of microstructural changes. Curr Genet. 2017;63(2):311–323. doi: 10.1007/s00294-016-0637-8. [DOI] [PubMed] [Google Scholar]
- 23.Rice DW, Palmer JD. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 2006;4(1):31. doi: 10.1186/1741-7007-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2006;58(1):1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 25.Smith DR. Mitochondrion-to-plastid DNA transfer: it happens. New Phytol. 2014;202(3):736–738. doi: 10.1111/nph.12704. [DOI] [PubMed] [Google Scholar]
- 26.Goremykin VV, Salamini F, Velasco R, Viola R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol Biol Evol. 2009;26(1):99–110. doi: 10.1093/molbev/msn226. [DOI] [PubMed] [Google Scholar]
- 27.Straub SC, Cronn RC, Edwards C, Fishbein M, Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (Apocynaceae) Genome Biol Evol. 2013;5(10):1872–1885. doi: 10.1093/gbe/evt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandini CL, Sanchez-Puerta MV. Foreign plastid sequences in plant mitochondria are frequently acquired via Mitochondrion-to-Mitochondrion horizontal transfer. Sci Rep. 2017;7:1-8. [DOI] [PMC free article] [PubMed]
- 29.Scataglini MA, Zuloaga FO, Giussani LM, Denham SS, Morrone O. Phylogeny of New World Paspalum (Poaceae, Panicoideae, Paspaleae) based on plastid and nuclear markers. Plant Syst Evol. 2014;300(5):1051–1070. doi: 10.1007/s00606-013-0944-1. [DOI] [Google Scholar]
- 30.Cox MP, Peterson DA, Biggs PJ. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC bioinformatics. 2010;11(1):485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 34.Wysocki WP, Clark LG, Kelchner SA, Burke SV, Pires JC, Edger PP, Duvall MR. A multi-step comparison of short-read full plastome sequence assembly methods in grasses. Taxon. 2014;63(4):899–910. doi: 10.12705/634.5. [DOI] [Google Scholar]
- 35.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke SV, Grennan CP, Duvall MR. Plastome sequences of two New World bamboos—Arundinaria gigantea and Cryptochloa strictiflora (Poaceae)—extend phylogenomic understanding of Bambusoideae. Am J Bot. 2012;99(12):1951–1961. doi: 10.3732/ajb.1200365. [DOI] [PubMed] [Google Scholar]
- 38.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graur D, Sater AK, Cooper TF. Molecular and genome evolution. Massachusetts: Sinauer Associates, Incorporated; 2016.
- 40.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9(8):772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19(6):716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 42.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller MA, Pfeiffer W, Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- 44.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014;10(4):e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christin PA, Spriggs E, Osborne CP, Strömberg CA, Salamin N, Edwards EJ. Molecular dating, evolutionary rates, and the age of the grasses. Syst Biol. 2014;63(2):153–165. doi: 10.1093/sysbio/syt072. [DOI] [PubMed] [Google Scholar]
- 47.Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. The age of the grasses and clusters of origins of C4 photosynthesis. Glob Chang Biol. 2008;14(12):2963–2977. doi: 10.1111/j.1365-2486.2008.01688.x. [DOI] [Google Scholar]
- 48.Iles WJ, Smith SY, Gandolfo MA, Graham SW. Monocot fossils suitable for molecular dating analyses. Bot J Linn Soc. 2015;178(3):346–374. doi: 10.1111/boj.12233. [DOI] [Google Scholar]
- 49.Poinar GJ, Alderman STEPHEN, Wunderlich JOERG. One hundred million year old ergot: psychotropic compounds in the cretaceous. Palaeodiversity. 2015;8:13–19. [Google Scholar]
- 50.Rambaut, A., Suchard, M. A., Xie, D., & Drummond, A. J. (2015). Tracer v1. 6. 2014.
- 51.Rambaut A, Drummond AJ. TreeAnnotator v2. 1.2. Edinburgh: University of Edinburgh, Institute of Evolutionary Biology; 2014. [Google Scholar]
- 52.Bell Mark A, Lloyd GT. Strap: stratigraphic tree analysis for Palaeontology. R package version 1.4. 2014. [Google Scholar]
- 53.Heibl C. Onwards. PHYLOCH: R language tree plotting tools and interfaces to diverse phylogenetic software packages. 2008. [Google Scholar]
- 54.R Core Team (2017). R: a language and environment for statistical computing. R Foundation for statistical computing, Vienna, Austria. URL https://www.R-project.org/.
- 55.Bushnell B. BBMap short read aligner. California: University of California, Berkeley; 2016. [Google Scholar]
- 56.Dick GJ, Andersson A, Baker BJ, Simmons SS, Thomas BC, Yelton AP, Banfield JF. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ultsch A, Mörchen F. ESOM-maps: tools for clustering, visualization, and classification with emergent SOM. 2005. [Google Scholar]
- 58.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York: Springer-Verlag; 2009.
- 59.Wickham H. Tidyverse: Easily Install and Load 'Tidyverse' Packages: R package version 1.1. 0. 2017. [Google Scholar]
- 60.Maier RM, Neckermann K, Igloi GL, Kössel H. Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Biol. 1995;251(5):614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 61.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22(6):1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol Gen Genomics. 2002;268(4):434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 63.Clifton SW, Minx P, Fauron CMR, Gibson M, Allen JO, Sun H, et al. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 2004;136(3):3486–3503. doi: 10.1104/pp.104.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr Genet. 2004;46(3):123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 65.Palmer JD, Herbon LA. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J Mol Evol. 1988;28(1–2):87–97. doi: 10.1007/BF02143500. [DOI] [PubMed] [Google Scholar]
- 66.Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11(1):29. doi: 10.1186/1741-7007-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Satoh M, Kubo T, Nishizawa S, Estiati A, Itchoda N, Mikami T. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol Gen Genomics. 2004;272(3):247–256. doi: 10.1007/s00438-004-1058-9. [DOI] [PubMed] [Google Scholar]
- 68.Prasad V, Strömberg CAE, Leaché AD, Samant B, Patnaik R, Tang L, et al. Late cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nat Commun. 2011;2:480. doi: 10.1038/ncomms1482. [DOI] [PubMed] [Google Scholar]
- 69.MacGinitie HD. Fossil plants of the Florissant beds, Colorado. Carnegie Inst. Washington Publ. 559. 198 p. 1962. The Kilgore Flora, a Late Miocene flora from northern Nebraska. Univ Calif Publ Geol Sci. 1953;3(5):67-158.
- 70.Walther H. Ergänzungen zur Flora von Seifhennersdorf (Sachsen), I. Teil.–Abh. Staatl. Mus. Mineral. Geol. 1967;12:259–277. [Google Scholar]
- 71.Dugas DP, Retallack GJ. Middle miocene fossil grasses from fort Ternan, Kenya. J Paleontol. 1993;67(1):113–128. doi: 10.1017/S0022336000021223. [DOI] [Google Scholar]
- 72.Thomasson JR. Observations on the characteristics of the lemma and Palea of the late Cenozoic grass Panicum elegans. Am J Bot. 1978:34–9.
- 73.Elias MK. Tertiary prairie grasses and other herbs from the high plains. Geol Soc Spec Pap. 1942;41:7–171. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Information of fossils calibrations used for divergence date analysis [49, 68–73]. (DOCX 14 kb)
Data Availability Statement
The data supporting our conclusions are available from the corresponding author on reasonable request. All nucleotide sequences were deposited in the NCBI Genbank repository, and accession numbers can be found in Table 1.