Abstract
Background
The long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) is expressed in solid malignant tumors. The aim of this systematic review and meta-analysis was to determine whether expression of the lncRNA SNHG1 was associated with prognosis in patients with malignancy.
Material/Methods
A literature review from Jan 1970 to July 2018 identified publications in the English language. Databases searched included: PubMed, OVID, Web of Science, the Cochrane Database, Embase, EBSCO, Google Scholar. Systematic review and meta-analysis were performed according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. The Newcastle-Ottawa Scale (NOS) assessment tool for risk of bias was used.
Results
Eight publications (570 patients) and eight solid tumors were identified, including osteosarcoma, colorectal cancer, hepatocellular carcinoma, non-small cell lung cancer, esophageal cancer, ovarian cancer, glioma, and gastric cancer. Meta-analysis showed that expression of the lncRNA SNHG1 was significantly correlated with reduced overall survival (OS) (HR=1.917; 95% CI, 1.58–2.31) (P<0.001). Subgroup analysis showed that lncRNA SNHG1 expression was significantly correlated with TNM stage (OR=3.99; 95% CI, 2.48–6.43) and lymph node metastasis (OR=3.12; 95% CI, 1.95–4.98). There were no significant correlations between lncRNA SNHG1 expression and patient gender, tumor subtype, or tumor size.
Conclusions
Systematic literature review and meta-analysis identified eight publications that included 570 patients with eight types of solid malignant tumor, and showed that the expression of the lncRNA SNHG1 was significantly associated with worse clinical outcome.
MeSH Keywords: Meta-Analysis; Prognosis; RNA, Long Noncoding
Background
Worldwide, solid malignant tumors arising from epithelial cells, or cancers, are an increasing cause of morbidity and mortality as populations increase, live longer, environmental pollution increases, and other lifestyle factors exert carcinogenic effects [1]. In the United States (US) in 2017, there were 1,688,780 new cancer cases and 600,920 cancer deaths [2]. Currently, surgery, radiotherapy, and chemotherapy remain the first-line treatments for cancer, although new therapeutic approaches are being developed and for some types of cancer, including nasopharyngeal carcinoma, treatments have resulted in improved patient prognosis [3]. However, the overall survival (OS) rate for most types of cancer remains low, and the majority of patients with cancer have a poor prognostic [2]. Therefore, there remains a need to identify novel prognostic biomarkers and more effective therapeutic strategies for cancer.
Recently published studies have shown that long noncoding RNAs (lncRNAs), with a length of more than 200 nucleotides, and which are non-protein-encoding RNAs, have a role in transcriptional and posttranscriptional processing, and genomic imprinting in oncogenesis [4,5]. Studies have shown that increased expression of lncRNAs can be found in several human cancers, including breast cancer, ovarian cancer, gastric cancer, and lung cancer [6–9]. Also, lncRNAs have a regulatory role in cell proliferation, apoptosis, and cancer metastasis, which are associated with patient prognosis [10–13]. Therefore, lncRNAs have potential as a biomarker for cancer diagnosis, prognosis, or treatment [14]. However, these roles for lncRNAs in human cancer remain to be investigated.
The long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) (GenBank accession ID: 23642) is a newly identified non-protein coding RNA localized at 11q12.3, which is upregulated in several types of solid malignant tumors, and may have a role in tumorigenesis [15–17]. Recently published studies have shown that the lncRNA SNHG1 is expressed in the cellular processes involved in malignant neoplasia, including cell proliferation and migration, invasion and metastasis [18,19]. Also, lncRNA SNHG1 has been shown to be expressed during the initiation and progression of malignancy by affecting the expression of p53, regulating microRNA, and competing with the expression of endogenous RNA [20–22]. Lan et al. showed that the lncRNA SNHG1 functions as a competing endogenous RNA (ceRNA) to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cells [23]. Also, Lu et al. showed that the expression of the lncRNA SNHG1 reduced miR-145-5p and upregulated MTDH, the gene encoding metadherin, in non-small cell lung cancer (NSCLC) [24]. Based on these previously published findings, it is possible that the lncRNA SNHG1 might represent a diagnostic, prognostic, or therapeutic cancer biomarker.
Also, recently published studies have shown that the increased expression of the lncRNA SNHG1 was associated with reduced survival rates in patients with several types of malignant solid tumors, including osteosarcoma [25], colorectal cancer [18], hepatocellular carcinoma (HCC) [26], non-small cell lung cancer (NSCLC) [27], esophageal squamous cell cancer [28], epithelial ovarian cancer [29], glioma [30] and gastric cancer [31]. However, because most published studies have been limited by low study sample size, the prognostic value of expression of the lncRNA SNHG1 remains unclear. Therefore, the aim of this study was to undertake a systematic review of the literature and meta-analysis to determine whether expression of the lncRNA SNHG1 is associated with prognosis in human solid malignant tumors.
Material and Methods
Literature search
A search of the literature was performed to identify published studies on the expression of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) in human solid malignant tumors and patient outcome in the English language, published in the Peoples’ Republic of China. The following databases were searched: Pubmed, OVID, Web of Science, the Cochrane Database, Embase, EBSCO, and Google Scholar,
The following keywords were used in the database search: ‘snhg1,’ ‘SNHG1,’ ‘neoplasia,’ ‘neoplasm,’ ‘tumor,’ ‘tumors,’ ‘cancer,’ ‘cancers,’ ‘malignant neoplasm,’ ‘malignant neoplasms,’ ‘neoplasm, malignant,’ ‘malignancy,’ ‘malignancies,’ ‘benign neoplasms,’ ‘neoplasms, benign.’ All irrelevant articles were excluded by review of the publication title and abstract. Duplicated publications were excluded using the function in Endnote X8. The selected full text of the included publications that were identified our reviewers were read in full.
Design and implementation of the systematic literature review and meta-analysis
A review of the literature was undertaken from Jan 1970 to April 2018, with meta-analysis conducted according to the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (http://prisma-statement.org) (Figure 1).
Figure 1.
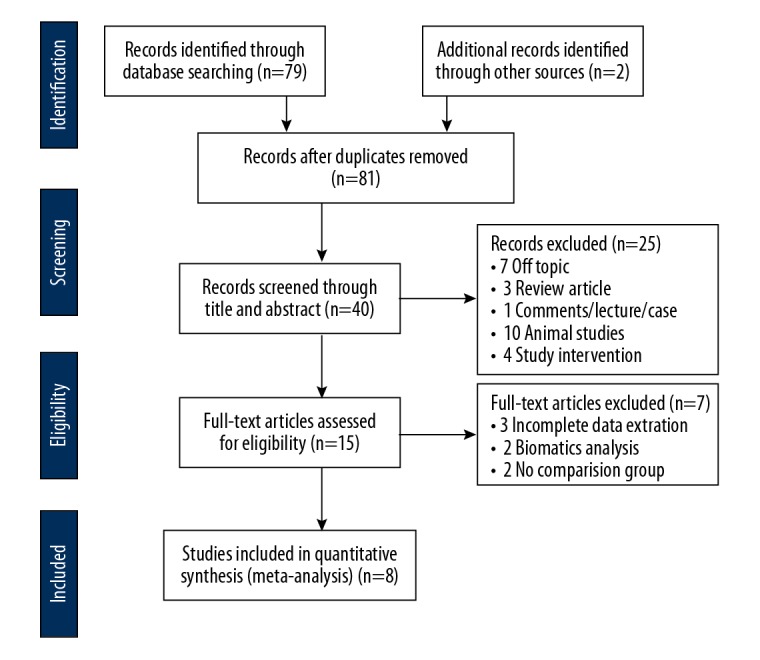
The flowchart shows the process of the literature search and study selection.
Publication inclusion criteria
All publications were initially screened and selected by two investigators, based on publication titles and abstracts, followed by a review of the published manuscript. The inclusion criteria for meta-analysis followed the population, intervention, control, and outcomes (PICO) criteria. (1) All articles were written and published in English with the availability of the full text of the publication. (2) Studies included those on human solid malignant neoplasms. (3) Expression of lncRNA SNHG1 in the tissue specimens of patients with malignant neoplasms were detected by established molecular methods, usually by quantitative reverse transcription polymerase chain reaction (qRT-PCR). (4) Studies were included that demonstrated the correlation between lncRNA SNHG1 expression, patient survival, and other clinical parameters of prognosis, including tumor stage and histological grade, and the presence of lymph node metastases. (5) Studies were included that provided survival information, including overall survival (OS), relapse-free survival (RFS), progression-free survival (PFS) and were correlated with lncRNA SNHG1 expression. (6) Sufficient statistical analysis was required, including hazard ratios (HRs) and the 95% confidence interval (CI) and hazard ratios (HRs) with prognostic endpoints, or data that could be used to estimate the HRs and 95% CIs, or other outcomes for OS such as Kaplan-Meier survival curves.
Publication exclusion criteria
(1) Preclinical in vitro or in vivo experimental studies were excluded. (2) Articles were not published in English were excluded. (3) Studies without access to the full text of the publication were excluded. (4) Case reports, letters, expert opinions, meeting records, review articles, commentaries, and clinical guidelines were excluded. (5) Studies without available clinical parameters, such as TNM stage, histological grade, lymph node metastasis data, were excluded. (6) Studies without HRs or 95% CIs were excluded. (7) Studies with a patient sample size <30 were excluded.
Data extraction
The databases were searched and the publications were assessed independently by two reviewers (BFX and ZHH). The included studies were chosen by consensus. The following data were obtained from each eligible study: author and year, ethnic background, country, sample size, cancer type, patient number, the method of detection used for lncRNA SNHG1 expression, the analysis type, the cut-off value, clinicopathological features, and the follow-up period. The prognostic endpoints included overall survival (OS), progression-free survival (PFS), recurrence-free survival (RFS), and disease-free survival (DFS). Hazard ratios (HRs) and 95% confidence intervals (CIs) were directly extracted from the univariate or multivariate analysis, or with the use of Engauge Digitizer4.1, a digitizing program, converting Kaplan-Meier survival curves [32].
Quality assessment
The quality of each study was assessed using the Newcastle-Ottawa Scale (NOS) and was quantitatively evaluated by two independent reviewers (BFX and ZHH) [33]. Any disagreements between the two reviewers were resolved by discussion and consensus. The scores for quality assessment ranged from 0 (minimum) to 9 (maximum); studies with a NOS score >6 were considered to be high quality.
Statistical analysis
Primary outcome data, including OS and HRs, were calculated using STATA 12 (Stata, College Station, TX, USA) and Engauge Digitizer version 4.1. Statistical heterogeneity was assessed using the I2 test as well as the chi-based Q-test, to determine heterogeneity between several studies. Heterogeneity was considered as statistically significant with I2 <50%. When the I2 was >50%, a random effects model was used. Otherwise, a fixed effects model was used to analyze the pooled results. According to the results from the STATA 12, there was the same outcome data in the fixed effects model and the random effect model. Also, a sensitivity analysis was used to check the stability of the combined results and to determine the source of any heterogeneity. The publication bias was evaluated using Begg’s test and Egger’s test. The trim and fill procedure was included in the meta-analysis to further evaluate any possible publication bias. Using a two-tailed statistical test, a P-value of <0.05 was considered as statistically significant.
Results
Study characteristics
A systematic literature review of published studies on the expression of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) and prognosis in patients with solid malignant tumors included publications from the Peoples’ Republic of China. As shown in the flow diagram of the literature search process (Figure 1), a total of 81 publications in the English language were identified from PubMed, OVID, Web of Science, the Cochrane Database, Embase, EBSCO, and Google Scholar from Jan 1970 to July 2018 (Figure 1). A total of 41 duplicate studies were excluded, and 25 publications were excluded following screening the abstracts. Seven full-text publications were excluded, including three without extractable data, two bioinformatics studies, as well as two uncontrolled studies. Based on the inclusion and exclusion criteria, there were eight different types of cancer evaluated, including osteosarcoma [25], colorectal cancer [18], hepatocellular carcinoma [26], non-small cell lung cancer [27], esophageal squamous cell cancer [28], epithelial ovarian cancer [29], glioma [30] and gastric cancer [31].
Of the eight publications identified, 570 patients were included in the meta-analysis. The characteristics of the seven published studies are summarized in Table 1. All patients with solid malignant tumors were diagnosed based on histology, with tissue specimens collected from tumor tissues and adjacent normal tissues, to determining the expression level of lncRNA SNHG1 by qRT-PCR. The Newcastle-Ottawa Scale (NOS) scores of all the included published studies were ≥7 [34].
Table 1.
Characteristics of studies included in the meta-analysis of the systematic literature review of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression in patients with solid malignant tumors.
| First author | Year | Ethnicity | Sample size | Cancer | AT | Specimen | Follow-up (mounths) | Outcome measures | HR | 95%CI |
|---|---|---|---|---|---|---|---|---|---|---|
| Cui Y [27] | 2017 | Asia | 68 | NSCLC | None | Tissue | 60 | OS | 1.83 | (1.14, 2.95) |
| Hu Y [31] | 2017 | Asia | 50 | GC | None | Tissue | 60 | OS | 2.40 | (1.12, 5.14) |
| Wang Q [17] | 2017 | Asia | 78 | Golima | N/A | Tissue | 45 | OS | 1.64 | (0.85, 3.17) |
| Zhu Y [18] | 2017 | Asia | 108 | CRC | None | Tissue | 60 | OS/PFS | 2.20 | (1.37, 3.54) |
| Zhang M [26] | 2016 | Asia | 82 | HCC | None | Tissue | 60 | OS/RFS | 2.13 | (1.20, 3.77) |
| Wang J [25] | 2018 | Asia | 45 | OS | None | Tissue | 60 | OS | 1.56 | (0.79, 3.10) |
| Wang S [29] | 2017 | Asia | 67 | EOC | None | Tissue | 60 | OS | 1.99 | (1.20, 3.30) |
| Zhang Y [28] | 2017 | Asia | 72 | ESCC | None | Tissue | 70 | OS | 1.78 | (1.19, 2.67) |
Quality assessment using the Newcastle-Ottawa Scale (NOS)
The quality scores of the NOS [34], which ranged from 6–8, indicating that all of the studies were eligible for inclusion and were of high quality. Of the eight identified publications, NOS assessment included two publications with a NOS score of 6, five publications with a NOS score of 7, and one publication with a NOS score of 8, with a median score of 7. Therefore, all eight eligible studies underwent meta-analysis (Table 2).
Table 2.
Quality assessment based on the Newcastle-Ottawa Scale (NOS).
| Author | Year | Selection | Comparability | Outcome | Total score |
|---|---|---|---|---|---|
| Cui Y | 2017 | 4 | 2 | 1 | 7 |
| Hu Y | 2017 | 3 | 1* | 2 | 6* |
| Wang Q | 2017 | 4 | 2 | 2 | 8* |
| Zhu Y | 2017 | 4* | 1 | 2 | 7 |
| Zhang M | 2016 | 4 | 2* | 1 | 7 |
| Wang J | 2018 | 3 | 2* | 1 | 6* |
| Wang S | 2017 | 4 | 1 | 2 | 7 |
| Zhang Y | 2017 | 4 | 2 | 1* | 7 |
The association between lncRNA SNHG1 expression and overall survival (OS)
The eight published studies that included 570 patients reported HRs for the OS according to the levels of lncRNA SNHG1 expression. As shown in Figure 2 and Table 3, due to the lack of statistically significant heterogeneity (P=0.981; I2=0.0%), the fixed-effects model was used for the pooled HR with corresponding 95% CI. The aggregated data showed that high expression levels of lncRNA SNHG1 expression were significantly correlated with poor OS (HR=1.917, 95% CI: 1.58,2.31, P<0.001), which means the lower lncRNA SNHG1 expression in patients with solid malignant tumors may result in a better clinical outcome (Figure 2).
Figure 2.
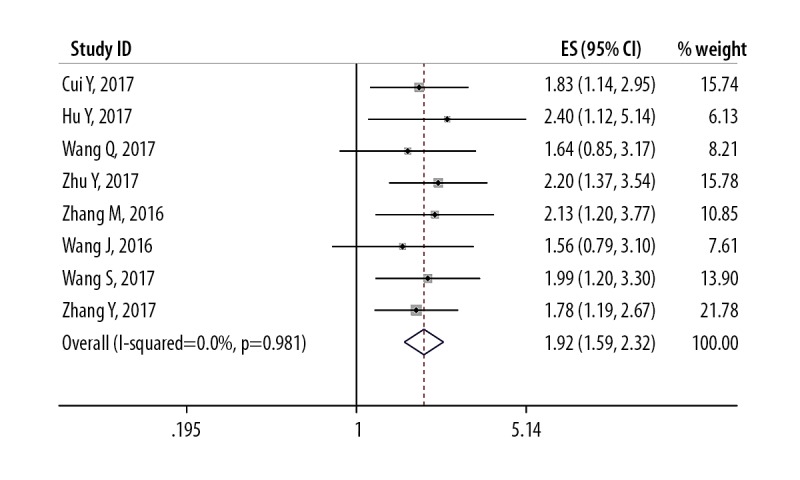
Forest plot shows the relationship between the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression and overall survival (OS).
Table 3.
Main results of the pooled hazard ratios (HRs) in the meta-analysis.
| Comparisons | Heterogeneity test | HR (95%CI) | Hypothesis test | Studies | ||||
|---|---|---|---|---|---|---|---|---|
| Q | P | I2 (%) | H | Z | P | |||
| Total | ||||||||
| OS | 1.54 | 0.981 | <25% | 1.0 | 1.917 (1.58,2.31) | 7.77 | <0.01 | 8 |
| PRS/RFS | 0.5 | 0.478 | <25% | 1.0 | 2.171 (1.50,3.12) | 3.50 | <0.01 | 2 |
| Cancer types | ||||||||
| Digestive | 0.69 | 0.707 | <25% | 1.0 | 2.004 (1.50,2.66) | 4.78 | <0.01 | 3 |
| Other cancers | 0.68 | 0.953 | <25% | 1.0 | 1.853 (1.44,2.38) | 4.81 | <0.01 | 5 |
| Tumor size | ||||||||
| <72 | 0.74 | 0.864 | <25% | 1.0 | 1.899 (1.42,2.52) | 4.39 | <0.01 | 4 |
| >72 | 0.80 | 0.851 | <25% | 1.0 | 1.931 (1.42,2.52) | 5.15 | <0.01 | 4 |
Digestive – Digestive cancer, including colorectal cancer, gastric Cancer, esophageal squamous cell carcinoma (ESCC).
Subgroup analysis
Although the study heterogeneity was low (I2 <25%; P=0.981), several subgroup analyses were performed. The subgroup analysis data is presented in Table 3. By stratifying the combined data according to tumor stage (OS vs. RFS/PFS), cancer type (gastrointestinal cancers vs. ‘other cancers’) (Figure 3), and the tumor size (>72 mm vs. <72 mm) (Figure 4). The correlations between SNHG1 lncRNA expression with clinicopathological features in the eight included studies are summarized in Table 4. The clinical outcomes showed that low expression of lncRNA SNHG1 was correlated with gender (OR=1.29; 95% CI, 0.81–2.05), TNM stage (OR=3.99; 95% CI, 2.48–6.43), and lymph node metastasis (OR=3.12; 95% CI, 1.95–4.98) (Figure 5). Owing to limited data for analysis further associations between other clinicopathological characteristics and lncRNA SNHG1 expression could not be analyzed.
Figure 3.
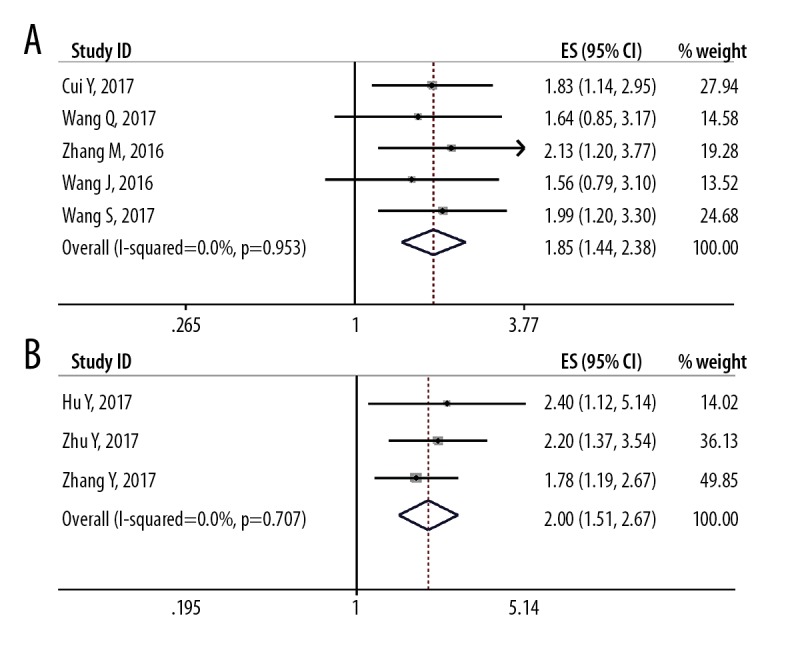
Forest plot shows the relationship between the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression and gastrointestinal cancers and ‘other cancers’. (A) Gastrointestinal cancers. (B) ‘Other cancers.’
Figure 4.
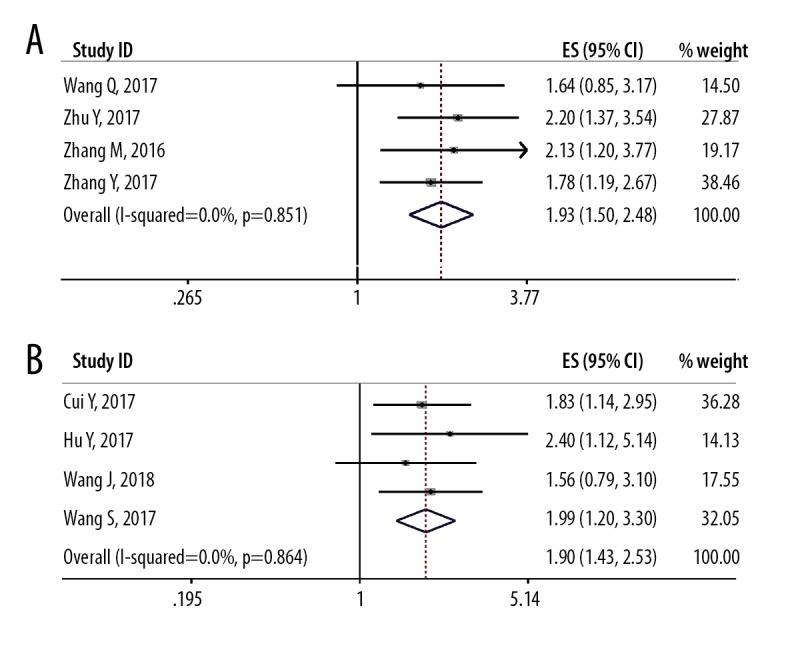
Forest plot shows the relationship between the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression and tumor size. (A) Tumor size >72 mm. (B) Tumor size <72 mm.
Table 4.
The association between low expression levels of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) and clinicopathological characteristics
| Characteristics | Pooled OR (95% CI) | Heterogeneity assessment | ||
|---|---|---|---|---|
| Chi2 | I2 | P value | ||
| Gender Male vs. Female |
1.292 (0.811, 2.057) | 1.43 | <20% | 0.700 |
| TNM stage I/II vs. III/IV |
3.994 (2.479, 6.433) | 0.81 | <20% | 0.848 |
| LNM N vs. P |
3.115 (1.948, 4.980) | 0.45 | <20% | 0.930 |
LNM – lymph node metastasis; N – negative; P – positive.
Figure 5.
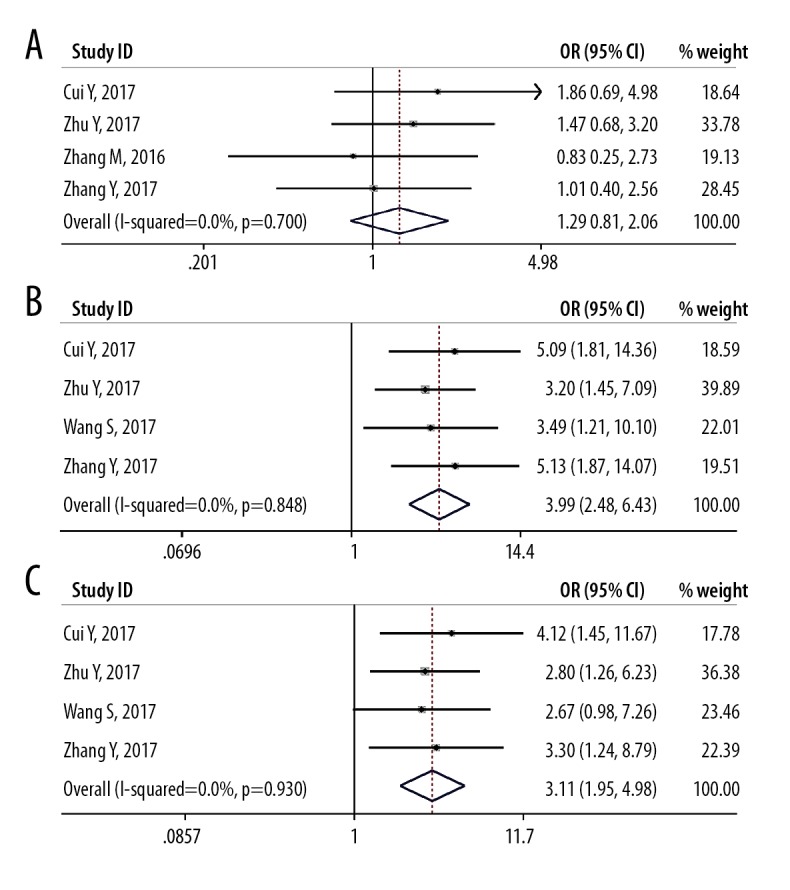
Forest plot shows the relationship between the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression and clinicopathological characteristics of patients with solid malignant tumors. (A) Gender. (B) Tumor TNM stage. (C) The presence of lymph node metastasis.
Analysis of sensitivity
A sensitivity analysis was performed using STATA 12 software to assess whether any individual study affected the overall results. The pooled results were not affected by the removal of individual studies, and the corresponding combined HRs were not significantly changed (Figure 6). Galbraith’s radial plot for heterogeneity between studies (Figure 7) confirmed the sensitivity and reliability of the meta-analysis.
Figure 6.
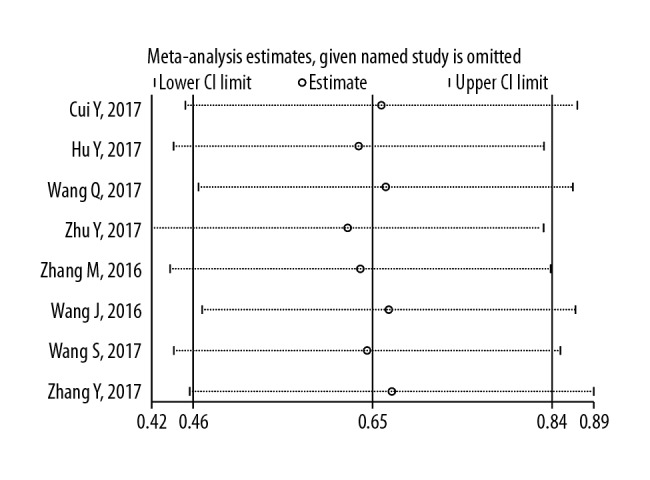
The sensitivity of the meta-analysis for overall survival (OS) in patients with solid malignant tumors.
Figure 7.
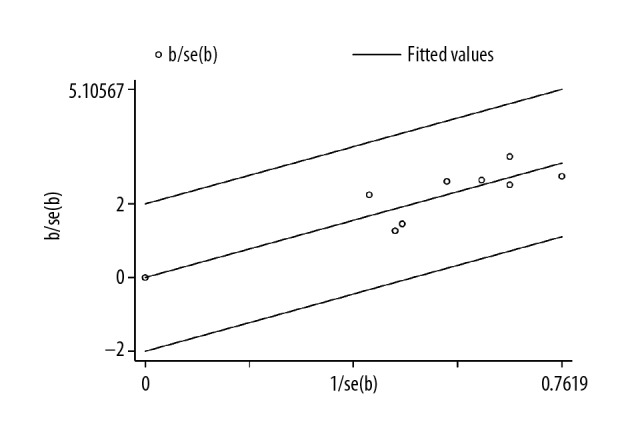
Galbraith’s radial plot for heterogeneity between studies on the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) expression.
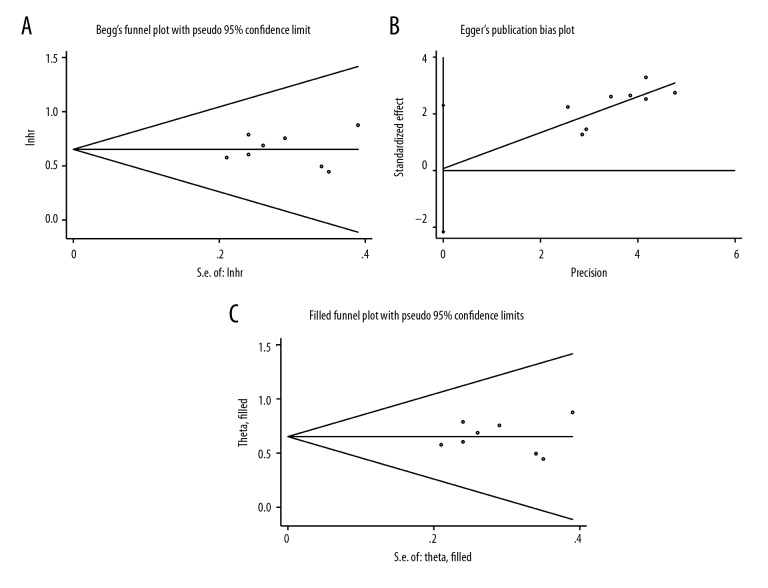
Analysis of publication bias
Begg’s funnel plot and Egger’s test were performed to detect publication bias (Figure 8). The findings showed that there was no significant publication bias in the evaluation of lncRNA SNHG1 expression and OS in the gastrointestinal cancer group and the ‘other cancers’ group (Figure 3, Table 4). And the data in detail of publication bias of SNHG1 for Begg’s test and Egger’s test were showed in Table 5.
Figure 8.
Publication bias analysis of overall survival (OS) data. (A) Begg’s funnel plot of overall survival (OS) in the published studies. (B) Egger’s publication bias plot of OS in the published studies. (C) Trim and fill publication bias plot to identify and correct for funnel plot asymmetry arising from publication bias of OS in the published studies.
Table 5.
Publication bias evaluation using Begg’s test and Egger’s test.
| Comparison | Begg’s test | Egger’s test | |||
|---|---|---|---|---|---|
| z | p | t | p | 95% CI | |
| OS | 0.12 | 1.000 | 0.06 | 0.941 | −2.158–2.298 |
| Digestive | 1.57 | 0.296 | 1.19 | 1.19 | −15.053–18.152 |
| Other cancers | 0.98 | 0.462 | 1.26 | 1.26 | −5.269–2.271 |
| <72 | 0.68 | 0.734 | 0.30 | 0.793 | −6.093–7.00 |
| >72 | 0.68 | 0.734 | 0.13 | 0.906 | −7.802–7.332 |
Discussion
Systematic literature review and meta-analysis identified eight publications that included 570 patients with eight types of solid malignant tumor and showed that increased expression of the long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) was significantly associated with worse clinical outcome.
Worldwide, cancer continues to be an increasing public health problem that results in patient morbidity with impaired quality of life, and the mortality rates for many solid malignant tumors remains high. Increasing numbers of studies have shown a role for long non-coding RNAs (lncRNAs) in tumorigenesis [35]. The lncRNAs can act either as an oncogene or a tumor suppressor gene to regulate cancer-related biological processes. For example, lnc-IGFBP4-1 has been shown to be upregulated in lung cancer, and the degree of expression is significantly associated with increased tumor stage, and lncRNA SNHG1 has been shown to be an oncogenic lncRNA [15–17]. However, there has previously been some controversy regarding the relationship between the expression of lncRNA SNHG1 and patient prognosis in solid malignant tumors, which is why this systematic literature review and meta-analysis was undertaken.
The findings of this systematic review of the literature identified eight eligible studies that underwent meta-analysis. The survival data analyzed included overall survival (OS), relapse-free survival (RFS), and progression-free survival (PFS). A fixed effects model was used to analyze the data, based on the findings of heterogeneity analysis. To our knowledge, at this time, this was the first meta-analysis of the prognostic value of lncRNA SNHG1 in solid malignant tumors. A high level of expression of lncRNA SNHG1 was significantly correlated with poor OS, based on meta-analysis. Cox multivariate analysis of combined hazard ratios (HRs), showed that there was a significant difference in OS between high and low lncRNA SNHG1 expression level groups (HR=1.917; 95% CI, 1.58–2.31) (P<0.001). However, because of lack of data, it was not possible to perform meta-analysis to verify whether lower expression of lncRNA SNHG1 in tumor tissues had an impact on PFS and RFS. Also, subgroup analysis showed that there was no significant difference between gastrointestinal cancers and ‘other cancers,’ or with tumor specimen size >72 mm and <72 mm, which further supported the pooled results. There was no significant correlation between the expression of lncRNA SNHG1 and other clinicopathological parameters, including gender, TNM stage, and lymph node metastasis.
However, this study had several limitations. Only eight studies were included in the meta-analysis. All patients in the analysis were Asian and from China, which means the data applied only to one ethnic group, which is a form of study bias. Although the outcomes the publication bias plots that included Begg’s, Egger’s and trim and fill were convincing, there is still the possibility of publication bias. Also, the meta-analysis was a retrospective analysis, and selection bias may have been a limitation. The cut-off values of the expression of lncRNA SNHG1 in tumor issues were not consistent in the published studies and were not reported in some of the studies. The values of HRs and 95% CIs were estimated from Kaplan-Meier survival curves, which might have overestimated the prognostic value of lncRNA SNHG1 expression. Finally, in studies that include different types of cancer, there are likely to be different oncogenic mechanisms involved and the same gene may play different roles in different cancers, which could affect the prognostic role of expression of lncRNA SNHG1 in different types of malignant solid tumor.
Although prognostic biomarkers in human cancer are used routinely and can assist treatment decisions, including CA19-9 [36], AFP [37], CEA [38], and prostate-specific antigen (PSA) the sensitivity and specificity of these clinical biomarkers require improvement [39]. Recent developments in tumor diagnosis with technological improvements have included the detection of circulating tumor cells (CTC) [40], and circulating tumor DNA (ctDNA) [41] in blood samples from patients with cancer. According to our previous review [42], the capability of exosomes to transfer functionally active components highlights their importance as promising biomarkers as well as diagnostic molecules. The findings from the present systematic review of the literature and meta-analysis have provided encouraging support for further studies to evaluate the expression levels of lncRNA SNHG1 in cancer patients, with the potential for practical clinical application in treatment decisions or in the detection of early-stage malignancy.
Conclusions
Previously, the prognostic value of detecting the expression levels of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) in patients in solid malignant tumors has been controversial. The findings of this systematic review of the literature and meta-analysis showed that increased expression of lncRNA SNHG1 was significantly correlated with poor prognosis in patients with solid malignant tumors. Because the number of studies evaluated was limited, further high-quality studies are required to provide more data on the role of lncRNA SNHG1 in different types of malignant tumor.
Acknowledgments
We gratefully acknowledge The Cancer Genome Atlas pilot project for providing data for this research. Also, Bufan Xiao would like to thank Liangke Liu for her support during the past year.
Footnotes
Conflicts of interest
None.
Source of support: This study was supported by the National Natural Science Foundation of China (Grant No. 81660391) and the Natural Science Foundation of Jiangxi, China (Grant No. 201562001) and the Major Project of Jiangxi, China (Grant No. 20141BBG70000) and Scientific Research Foundation of Scientific department of Jiangxi, China (Grant No. 201BBG70010)
References
- 1.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends – an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: Current practice and future perspective. J Clin Oncol. 2015;33:3356–64. doi: 10.1200/JCO.2015.60.9347. [DOI] [PubMed] [Google Scholar]
- 4.Lau E. Non-coding RNA: Zooming in on lncRNA functions. Nat Rev Genet. 2014;15:574–75. doi: 10.1038/nrg3795. [DOI] [PubMed] [Google Scholar]
- 5.Barra J, Leucci E. Probing long non-coding RNA-protein interactions. Front Mol Biosci. 2017;4:45. doi: 10.3389/fmolb.2017.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu X, Cao Y, Tang L, et al. Baicalein inhibits breast cancer growth via activating a novel isoform of the long noncoding RNA PAX8-AS1-N. J Cell Biochem. 2018;119(8):6842–56. doi: 10.1002/jcb.26881. [DOI] [PubMed] [Google Scholar]
- 7.Yang B, Zhang L, Cao Y, et al. Overexpression of lncRNA IGFBP4-1 reprograms energy metabolism to promote lung cancer progression. Mol Cancer. 2017;16:154. doi: 10.1186/s12943-017-0722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripathi MK, Doxtater K, Keramatnia F, et al. Role of lncRNAs in ovarian cancer: Defining new biomarkers for therapeutic purposes. Drug Discov Today. :2018. doi: 10.1016/j.drudis.2018.04.010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li L, Wang YY, Mou XZ, et al. Upregulation of long non-coding RNA M26317 correlates with tumor progression and poor prognosis in gastric cancer. Hum Pathol. 2018 doi: 10.1016/j.humpath.2018.04.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Yue B, Liu C, Sun H, et al. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26(5):1287–98. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Y, He Y, Zhang P, et al. LncRNAs regulate the cytoskeleton and related Rho/ROCK signaling in cancer metastasis. Mol Cancer. 2018;17:77. doi: 10.1186/s12943-018-0825-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang L, Xu J, Wang M, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9:372. doi: 10.1038/s41419-018-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Liu X, Zhang L, Li X. LncRNA BANCR facilitates vascular smooth muscle cell proliferation and migration through JNK pathway. Oncotarget. 2017;8:114568–75. doi: 10.18632/oncotarget.21603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu Y, Li B, Liu Z, et al. Up-regulation of lncRNA SNHG1 indicates poor prognosis and promotes cell proliferation and metastasis of colorectal cancer by activation of the Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:111715–27. doi: 10.18632/oncotarget.22903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Li Q, Zhou P, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. 2017;91:906–11. doi: 10.1016/j.biopha.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Yang YL, Li L, et al. LncRNA &ITSNHG1&IT enhances cell proliferation, migration, and invasion in cervical cancer. Biochem Cell Biol. 2018;96:38–43. doi: 10.1139/bcb-2017-0188. [DOI] [PubMed] [Google Scholar]
- 19.Tian T, Qiu R, Qiu X. SNHG1 promotes cell proliferation by acting as a sponge of miR-145 in colorectal cancer. Oncotarget. 2018;9:2128–39. doi: 10.18632/oncotarget.23255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Y, Qin ZS, Feng Y, et al. Long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) promote cell proliferation in colorectal cancer by affecting P53. Eur Rev Med Pharmacol Sci. 2018;22:976–84. doi: 10.26355/eurrev_201802_14379. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Luo X, Chen F, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem. 2018;119(7):5460–72. doi: 10.1002/jcb.26705. [DOI] [PubMed] [Google Scholar]
- 22.Tan H, Zhao L, Song R, et al. The long noncoding RNA SNHG1 promotes nucleus pulposus cell proliferation through regulating miR-326/CCND1. Am J Physiol Cell Physiol. 2018;315(1):C21–27. doi: 10.1152/ajpcell.00220.2017. [DOI] [PubMed] [Google Scholar]
- 23.Lan X, Liu X. LncRNA SNHG1 functions as a ceRNA to antagonize the effect of miR-145a-5p on the down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13497. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Q, Shan S, Li Y, et al. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32(7):3957–67. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 25.Wang JD, Cao L, Wu JH, Wang QG. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52:77–88. doi: 10.3892/ijo.2017.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Wang W, Li TY, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi: 10.1016/j.biopha.2016.02.036. [DOI] [PubMed] [Google Scholar]
- 27.Cui Y, Zhang F, Zhu C, et al. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/beta-catenin signaling pathway. Oncotarget. 2017;8:17785–94. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Jin X, Wang Z, et al. Downregulation of SNHG1 suppresses cell proliferation and invasion by regulating Notch signaling pathway in esophageal squamous cell cancer. Cancer Biomark. 2017;21:89–96. doi: 10.3233/CBM-170286. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Jiang JY, Wang ZY, et al. Long non-coding RNA SNHG1 is an unfavorable prognostic factor and promotes cell proliferation and migration by Wnt/beta-catenin pathway in epithelial ovarian cancer. International Journal of Clin Exp Pathol. 2017;10:9284–92. [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Li Q, Zhou P, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. 2017;91:906–11. doi: 10.1016/j.biopha.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 31.Hu YB, Ma Z, He YM, et al. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–31. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
- 32.Tierney JF, Stewart LA, Ghersi D, et al. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong WC, Cheung CS, Hart GJ. Development of a quality assessment tool for systematic reviews of observational studies (QATSO) of HIV prevalence in men having sex with men and associated risk behaviours. Emerg Themes Epidemiol. 2008;5:23. doi: 10.1186/1742-7622-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 35.Yuan J, Yue H, Zhang M, et al. Transcriptional profiling analysis and functional prediction of long noncoding RNAs in cancer. Oncotarget. 2016;7:8131–42. doi: 10.18632/oncotarget.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong W, Yu Z, Zhan J, et al. Association of serum levels of CEA, CA199, CA125, CYFRA21-1 and CA72-4 and disease characteristics in colorectal cancer. Pathol Oncol Res. 2015;21:83–95. doi: 10.1007/s12253-014-9791-9. [DOI] [PubMed] [Google Scholar]
- 37.Toyoda H, Kumada T, Tada T, et al. Tumor markers for hepatocellular carcinoma: Simple and significant predictors of outcome in patients with HCC. Liver Cancer. 2015;4:126–36. doi: 10.1159/000367735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai PL, Su WJ, Leung WH, et al. Neutrophil-lymphocyte ratio and CEA level as prognostic and predictive factors in colorectal cancer: A systematic review and meta-analysis. J Cancer Res Ther. 2016;12:582–89. doi: 10.4103/0973-1482.144356. [DOI] [PubMed] [Google Scholar]
- 39.Pezaro C, Woo HH, Davis ID. Prostate cancer: Measuring PSA. Intern Med J. 2014;44:433–40. doi: 10.1111/imj.12407. [DOI] [PubMed] [Google Scholar]
- 40.Tjensvoll K, Nordgard O, Smaaland R. Circulating tumor cells in pancreatic cancer patients: Methods of detection and clinical implications. Int J Cancer. 2014;134:1–8. doi: 10.1002/ijc.28134. [DOI] [PubMed] [Google Scholar]
- 41.Chaudhuri AA, Binkley MS, Osmundson EC, et al. Predicting radiotherapy responses and treatment outcomes through analysis of circulating tumor DNA. Semin Radiat Oncol. 2015;25:305–12. doi: 10.1016/j.semradonc.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou R, Chen KK, Zhang J, et al. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol Cancer. 2018;17:75. doi: 10.1186/s12943-018-0823-z. [DOI] [PMC free article] [PubMed] [Google Scholar]