Abstract
Although plasmids provide additional functions for cellular adaptation to the environment, they also create a metabolic burden, which causes the host cells to be less competitive with their siblings. Low-copy-number plasmids have thus evolved several mechanisms for their long-term maintenance in host cells. pMF1, discovered in Myxococcus fulvus 124B02, is the only endogenous autonomously replicated plasmid yet found in myxobacteria. Here we report that a post-segregational killing system, encoded by a co-transcriptional gene pair of pMF1.19 and pMF1.20, is involved in maintaining the pMF1 plasmid in its host cells. We demonstrate that the protein encoded by pMF1.20 is a new kind of nuclease, which is able to cleave DNA in vitro. The nuclease activity can be neutralized by the protein encoded by pMF1.19 through protein–protein interaction, suggesting that the protein is an immune protein for nuclease cleavage. We propose that the post-segregational killing mechanism of the nuclease toxin and immune protein pair encoded by pMF1.20 and pMF1.19 is helpful for the stable maintenance of pMF1 in M. fulvus cells.
Keywords: pMF1 plasmid, nuclease, immune protein, co-transcriptional gene pair, post-segregational killing system, Myxococcus fulvus
Introduction
Plasmids are self-replicating extra-chromosomal genetic elements that are dispensable to their bacterial host cells. They usually contain some essential genes for independent replication and segregation (Dmowski and Jagura-Burdzy, 2013), and some accessory genes (Hulter et al., 2017). The accessory genes may bring specific characters to their bacterial hosts, such as virulence factors in pathogens (Elwell and Shipley, 1980; Sengupta and Austin, 2011) and antibiotic resistance (Martinez and Baquero, 2002; Tett et al., 2007). However, the presence of plasmids also leads to extra metabolic burdens, which cause the host cells to be less competitive with their siblings. To decrease metabolic burden, plasmids are normally maintained at a low copy number, but this raises the risk of losing the plasmid during cell division. Plasmids have developed several maintenance mechanisms, such as sufficient copy numbers for daughter cells (Tabata et al., 1989; del Solar et al., 1998; Paulsson, 2002), site-specific resolution systems to avoid plasmid multimerization (Austin et al., 1981; Summers, 1998), partition systems (par) to actively segregate plasmid copies to daughter cells prior to cell division (Thomas, 2000; Ebersbach and Gerdes, 2005) and post-segregational killing (PSK) systems to kill competitive plasmid-free cells (Gerdes et al., 1986; Engelberg-Kulka and Glaser, 1999; Sengupta and Austin, 2011).
Myxobacteria are gram-negative bacteria that possess complex multicellular social behavior and large genomes (Shimkets, 1990; Dworkin, 1996; Han et al., 2013; Chen et al., 2016; Sharma et al., 2016). We previously isolated pMF1, which is the only endogenous plasmid yet found in myxobacteria, from Myxococcus fulvus 124B02 (Zhao et al., 2008). M. xanthus–Escherichia coli shuttle plasmids have been constructed based on the original replication region of pMF1 (Feng et al., 2012), and improved by containing the partitioning system of pMF1 (Sun et al., 2011); they are still losable from M. xanthus cells in the environment with no antibiotics selection pressure. In this study, we found a new nuclease and immune protein pair encoded by the co-transcriptional plasmid genes pMF1.20 and pMF1.19. The presence of the gene pair made plasmids more stable in Myxococcus cells. We propose that together the two genes play a major role in the stable maintenance of plasmid pMF1 in M. fulvus cells using a post-segregational killing mechanism.
Results
pMF1.19 and pMF1.20 are co-transcribed, forming an operon in pMF1
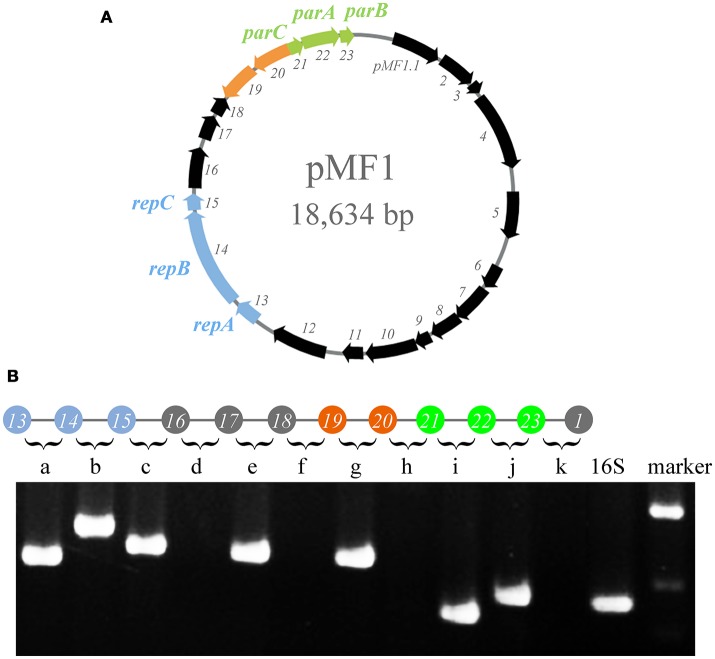
The genome sequence of pMF1 in M. fulvus 124B02 has been reported previously (Chen et al., 2016). The pMF1 plasmid is 18,634 bp in length and contains 23 predicted genes (pMF1.1 to pMF1.23, Figure 1A and Table S1). We previously determined that genes pMF1.13 to pMF1.15 form the replication origin region and genes pMF1.21 to pMF1.23 constitute the partitioning system (Sun et al., 2011; Feng et al., 2012). BLASTp searching against GenBank databases showed that the predicted amino acid sequences of many genes in the pMF1 plasmid had homologs in various myxobacterial genome sequences (Table S1), all of which were functionally undetermined. The pieced construction of pMF1 with various myxobacteria-sourced genes suggests that the plasmid had been widely present in different myxobacteria, and had exchanged with the host genomes.
Figure 1.
Operons in pMF1. (A) pMF1 consists of 23 genes. The blue arrows (pMF1.13-pMF1.15) represent the replication origin region. pMF1.21-pMF1.23 (green arrows) is the partitioning system of pMF1. The orange parts are the two genes with an opposite direction to the others. (B) Co-transcription of the pMF1.13-pMF1.23 genes by PCR mapping. The upper panel is a schematic drawing of the pMF1 genetic locus, and the lower panel is the amplification results of reverse transcription PCR for the fragments containing the 3′ end of the upstream gene, the intergenic sequence, and 5′ end of the downstream gene, corresponding to the sequence between two adjacent genes. The cDNA template was used to amplify the 16S rRNA gene as the positive control. The yield of RT-PCR product with the expected size indicated that the two neighboring genes were co-transcribed. The migration positions of molecular weight markers are indicated.
We performed transcriptomic analysis of the pMF1-harboring M. fulvus 124B02 to investigate the transcription of pMF1.19 and pMF1.20 genes (Figure S1). The results showed that the transcriptional level of pMF1.20 was similar to that of the replication origin and partition system genes, while the transcriptional level of pMF1.19 was much higher, indicating that pMF1.20 and pMF1.19 function in cells.
The transcriptomic results also indicated that pMF1.19 and pMF1.20 were in an operon, like pMF1.13 to pMF1.16 and pMF1.21 to pMF1.23. The reverse transcriptional PCR result showed that the intergenic region between pMF1.19 and pMF1.20 could be amplified, while the intergenic regions between pMF1.18 and pMF1.19 or pMF1.20 and pMF1.21 were not (Figure 1B). Co-transcription in an operon suggest that the pMF1.19 and pMF1.20 genes are functionally related.
The pMF1.19-pMF1.20 gene pair have homologues in the 124B02 genome
Among the 23 genes, 21 were situated on the leading strand, while pMF1.19 and pMF1.20 were on the lagging strand. The most similar homologous genes of pMF1.19 and pMF1.20 were from M. fulvus 124B02 (Table S1). The amino acid sequence identities of pMF1.19 and pMF1.20 were 75 and 88% similar to MFUL124B02_RS18140 and MFUL124B02_RS18145, respectively. MFUL124B02_RS18140 is downstream of MFUL124B02_RS18145 in the genome, which is the same as the order of pMF1.19 and pMF1.20. This result suggests that the pMF1.19 and pMF1.20 genes are probably repeats derived from the genome of M. fulvus 124B02, and vice versa.
Notably, the genome of M. fulvus 124B02 contains six homologs of pMF1.20, all of which are upstream of the homologs of pMF1.19 (Table S2). All of these homologous gene pairs, apart from MFUL124B02_RS18140 and MFUL124B02_RS18145, had low similarities with pMF1.19 and pMF1.20. Interestingly, the phylogenies of the pMF1.19 homologs were completely consistent with those of their pMF1.20 homolog partners in the gene pairs (Figure S2). These results suggest that the pMF1.19 and pMF1.20 genes, as well as their homologous gene pairs in the genome of 124B02, co-evolved, and the functions of the paired genes are probably related but divergent.
The pMF1.19-pMF1.20 operon plays a role in plasmid maintenance in myxococcus
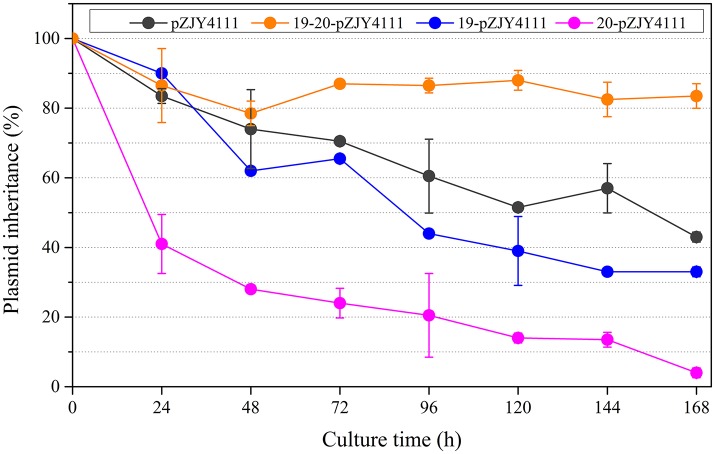
The shuttle plasmid pZJY4111 contains the replication origin genes and partitioning genes of pMF1 (Sun et al., 2011). To investigate the functions of pMF1.19 and/or pMF1.20 in the plasmid, we introduced them into the pZJY4111 plasmid, producing plasmids 19-pZJY4111, 20-pZJY4111, and 19-20-pZJY4111 (the construction is shown in Figure S3). These recombinant plasmids were further separately transferred into the M. xanthus DZ1 strain, which grows dispersively in liquids (Breton et al., 1985; Zhao et al., 2008). We assayed the maintenance abilities of the plasmids in these strains, using the DZ1 transformant with plasmid pZJY4111 as a control. Without kanamycin selection, the retention of pZJY4111 in DZ1 was 43% after 168 h. A similar retention curve was observed in the 19-pZJY4111 plasmid stability test, but with weakly lower retention ability (Figure 2). However, after construction with the pMF1.20 gene, the retention of 20-pZJY4111 plasmid was only 4% after 168 h, which suggests that pMF1.20 alone was harmful to the hosting cells. In contrast, with the complete pMF1.19-pMF1.20 operon, the retention of plasmid 19-20-pZJY4111 was 84% after 168 h, which was not only markedly higher than the retention of the 20-pZJY4111 plasmid, but also significantly higher than the retention of pZJY4111 or 19-pZJY4111 (t-test, p < 0.01). These results suggest that the co-presence of pMF1.19 and pMF1.20 had a stabilizing effect on the retention of the plasmid in Myxococcus cells. The pMF1.19-pMF1.20 operon seemed to function using a post-segregational killing mechanism, i.e., plasmids with the operon could apparently eliminate plasmid-free cells, thus guaranteeing the survival of plasmid-containing cells in competition with plasmid-free cells. The pMF1.20 gene might encode products that are toxic to the host cells whereas the product encoded by pMF1.19 counteracts the toxicity of pMF1.20.
Figure 2.
Effects of the presence of pMF1.19 and/or pMF1.20 on plasmid stabilities in M. xanthus DZ1. Plasmids 19-pZJY4111, 20-pZJY4111, and 19-20-pZJY4111 were constructed and electroporated into M. xanthus DZ1. The strains were cultivated in CTT medium without antibiotics for 168 h and plasmid retention was examined every 24 h.
The toxic effect of pMF1.20 on cellular growth is suppressed by pMF1.19
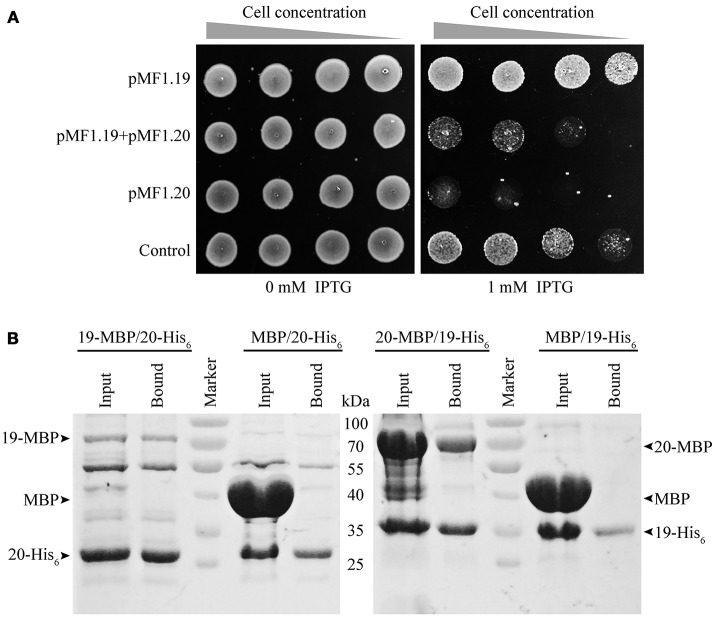
To determine the in vivo functions of pMF1.19 and pMF1.20, we expressed them in E. coli cells. We performed a growth inhibition assay with E. coli strains (Figure 3A). The presence of plasmids had almost no effect on the growth of the E. coli strains. When 1 mM IPTG was added to the medium, the growth abilities of the strains with 19-pACYC/empty pMAL-c5x (pMF1.19) and with pACYC-Duet/pMAL-c5x (control) were weakened slightly. However, when the expression of pMF1.20 was induced by IPTG, the growth ability of the strain containing 20-pMAL/pACYC-Duet (pMF1.20) was greatly decreased (Figure 3A). This result indicates that the presence of pMF1.20 is toxic to the cellular growth of E. coli. Compared with the single expression of pMF1.20, co-expression of pMF1.20 and pMF1.19 increased the growth of the strain. These results suggest that the pMF1.20 expression product is toxic to cellular growth, and the pMF1.19 expression product is capable of neutralizing the toxicity of pMF1.20.
Figure 3.
Results of in vivo and in vitro experiments with pMF1.19 and pMF1.20. (A) Growth abilities of the BL21 strains containing empty pMAL-c5x and pACYC-Duet plasmid (control), pMF1.20-harboring plasmid and empty pACYC-Duet, 19-pACYC and empty pMAL-c5x, and the two pMF1.19- and pMF1.20-harboring plasmids. The medium was with or without IPTG supplementation. Equal numbers of BL21 cells were five-fold serially diluted and spotted on the plates. Pictures were taken after overnight cultivation. (B) Pull-down experiments on the binding activities between pMF1.19 and pMF1.20. 19-MBP (71 kDa) and 20-MBP (69 kDa) were purified by amylose resin and 19-His6 (31 kDa) and 20-His6 (29 kDa) were purified by Ni2+ beads. 19-MBP (1.9 mg/ml, 300 μl) was incubated with 20-His6 (1.5 mg/ml, 300 μl). In a similar manner, 20-MBP (7 mg/ml, 150μl) was mixed with 19-His6 (1.5 mg/ml, 450 μl). MBP protein was incubated with 19-His6 or 20-His6 as negative controls. The input and bound samples were tested using SDS-PAGE. The PageRulerTM Prestained Protein Ladder was used.
We performed pull-down experiments to determine the in vitro binding interactions between the pMF1.20 and pMF1.19 proteins. 19-His6/20-MBP, 20-His6/19-MBP, 19-His6/MBP, and 20-His6/MBP mixtures were made. The mixtures were further extracted with Ni2+ beads, and the extracts were detected by SDS-PAGE to assay the binding abilities of the pMF1.20 and pMF1.19 proteins. If the two proteins were bound with each other, the 20-MBP and 19-MBP would be detectable in the extracts of 19-His6/20-MBP and 20-His6/19-MBP, respectively. As shown in the SDS-PAGE image (Figure 3B), when mixed with 19-His6 or 20-His6, the MBP protein did not appear in the extracts of the bound proteins. In contrast, 20-MBP and 19-MBP appeared in the extracts of the bound proteins when they were mixed with 19-His6 and 20-His6, respectively. These results demonstrate that pMF1.19 is able to bind to pMF1.20 in vitro.
pMF1.20 possesses nuclease activity
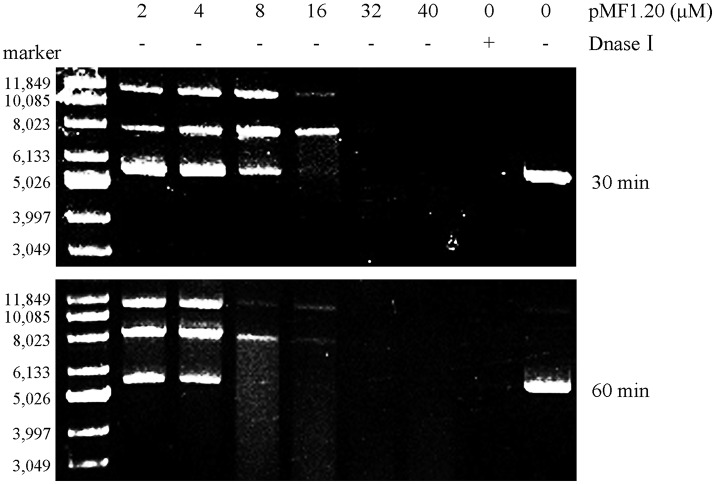
Bioinformatics analysis revealed that the pMF1.20 protein and its homologs are all single-domain proteins containing a DUF2380 domain (pfam09533), and they were predicted to be lipoproteins with a type II signal peptide. The alignment of the amino acid sequences of pMF1.20 and its homologs indicated many conserved histidine sites (Figure S4). Some proteins with highly conserved histidine sites are known to be nucleases, in which histidine residues are in the active center (Midon et al., 2012; Belieres et al., 2015; Sivagnanam et al., 2016). We speculated that pMF1.20 was probably a new kind of nuclease. We therefore mixed purified pMF1.20 with supercoiled pMAL-c5x plasmid DNA in a nuclease reaction solution at 37°C for 30 or 60 min, followed by the DNA agarose gel electrophoresis. Dnase I was used as the control. At low concentrations, pMF1.20 loosened and linearized the plasmid DNA; at high concentrations, the protein digested the plasmid DNA completely (Figure 4). These results indicate that pMF1.20 possesses in vitro nuclease activity.
Figure 4.
In vitro nuclease activity assay of the pMF1.20 proteins. Different concentrations of purified pMF1.20 were mixed with the supercoiled plasmid pMAL-c5x DNA for 30 and 60 min. DNase I was used for the positive control and the negative control contained no enzyme. The reactions were detected by gel and ethidium bromide staining.
We further mixed pMF1.20 with pMF1.19, and incubated the mixture with supercoiled pMAL-c5x plasmid DNA in a nuclease reaction solution at 37°C for 60 min. The results showed that the presence of 10 μM pMF1.19 alone did not lead to loosening, linearization, or digestion of the supercoiled DNA, whereas incubation with 10 μM pMF1.20 for 60 min caused the supercoiled DNA to disappear (Figure 5). Comparatively, the co-presence of 10 μM pMF1.19 and 10 μM pMF1.20 slowed the activities of pMF1.20 on the DNA substrate. These results suggest that the binding of pMF1.20 with pMF1.19 inhibited the enzymatic activity of the nuclease.
Figure 5.
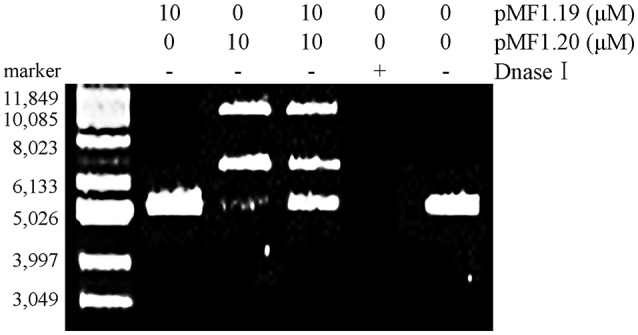
In vitro assay of pMF1.19 inhibition of pMF1.20 nuclease activity. Purified pMF1.19, pMF1.20 and equal parts pMF1.19 plus pMF1.20 were incubated with supercoiled pMAL-c5x DNA for 60 min. DNase I was used for the positive control and the negative control contained no enzyme.
Discussion
Lowering the copy number is a strategy of plasmids to reduce the metabolic burden on their cell host. However, a low copy number means a high risk of loss, and some mechanisms for retaining plasmids are necessary. Although the gene sequence and the replication origin region and partitioning system of pMF1 had been determined previously (Zhao et al., 2008; Sun et al., 2011; Feng et al., 2012), the known mechanisms were not sufficient to explain the long-term maintenance of pMF1 in M. fulvus. In this study, we revealed that the presence of the pMF1.19-pMF1.20 operon can improve plasmid maintenance. We determined that pMF1.20 encoded a novel nuclease protein, which was able to bind with pMF1.19 proteins. The in vivo and in vitro experiments showed that binding with pMF1.19 repressed the enzymatic activities of the pMF1.20 proteins. Notably, in M. fulvus 124B02, the expression of pMF1.19 was approximately eight-fold that of pMF1.20, which implies that neutralizing the toxicity of the pMF1.20 nuclease required high concentrations of pMF1.19 proteins. We propose that the pMF1.19-pMF1.20 operon probably functions like a PSK system for the maintenance of plasmids in M. fulvus cells.
The PSK system has been classified into two groups according to the type of the antidote. The antidote for the type I system is an antisense RNA complementary to the mRNA of the toxin gene, such as hok-sok from plasmid R1 (Gerdes et al., 1997). In the type II system, the antidote, as well as the toxin, is protein. In plasmid carrying cells, the two proteins form an inactive complex, which prevent the toxin from killing the cell. Plasmids F (ccdA-ccdB), P1 (phd-doc), and R1 (kid-kis) are well-understood type II PSK systems (Hayes and Van Melderen, 2011). For example, CcdB from the F plasmid and ParE from the RK2 plasmid influence the DNA gyrase actions (Jiang et al., 2002; Van Melderen, 2002). The Kid toxin of plasmid R1 is an RNase cleaving at the 5′-UUACU-3′ site to inhibit the expression of host gene so that stops cell growth, which is inactivated after binding with the Kis protein (Pimentel et al., 2005; de La Cueva-Mendez and Pimentel, 2007). The pMF1.20 toxin from pMF1 acts as nuclease effector, which may be delivered into neighboring cells and kill the co-existing plasmid-free cells that lack the corresponding immune proteins.
pMF1 is the only endogenous autonomously replicated plasmid that has been found in myxobacteria. The replication and partitioning systems are essential for the stable maintenance of pMF1 in Myxococcus cells. The nuclease-effector and immunity system consisting of pMF1.20-pMF1.19 is also significant for the stable maintenance of pMF1. pMF1.20 has a lipoprotein signal peptide at the N-terminus of the protein, implying that the protein is probably located in the cellular membrane. Vassallo et al. reported that in the outer-membrane exchange system of Myxococcus, the TraA proteins are able to transfer nuclease proteins to adjacent cells and kill cells with no immune proteins (Vassallo et al., 2017). We speculate that pMF1.20 might be secreted to the target cells by a system similar to outer-membrane exchange. Further evidence of such a system is needed.
In the pMF1 plasmid, the direction of pMF1.20 and pMF1.19 is different from the other pMF1 genes. In addition, the most similar homologs of pMF1.20 and pMF1.19 are from M. fulvus 124B02, while the most similar homologs of other genes in pMF1 are not. There are at least six pairs of homologs of pMF1.20-pMF1.19 in M. fulvus 124B02. Thus, we speculate that the pMF1.20-pMF1.19 operon is the latest member of pMF1 and is derived from the genome of M. fulvus 124B02. The new member pMF1.20-pMF1.19 constitutes a post-segregational killing system of pMF1. The stable maintenance of pMF1 relies on cooperation between the replication, partitioning and post-segregational killing systems in M. fulvus 124B02.
Experimental procedures
Plasmids, strains, and culture conditions
The plasmids and bacteria used in this study are listed in Table S3. The M. xanthus strains were cultivated at 30°C in casitone-based rich-nutrient CTT agar or CTT broth (Hodgkin and Kaiser, 1977). E. coli cells were cultivated at 37°C on Luria-Bertani (LB) agar or in LB broth. When required, final concentrations of 40 μg/ml kanamycin, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol were added to the medium.
RNA extraction and reverse transcriptional PCR
Total RNA of pMF1-harboring M. fulvus 124B02 was extracted using an SV Total RNA Purification Kit (Promega USA). Residual genomic DNA (gDNA) was removed and cDNA was synthesized using a Prime ScriptTM RT Reagent Kit with gDNA Eraser (Takara, China).The 16S rRNA gene sequence was amplified from the cDNA template to evaluate the quality of cDNA sample. The cDNA products were further amplified using specific primer pairs to determinate the co-transcription.
Construction of plasmids for introduction into myxococcus
The pMF1.19, pMF1.20, and pMF.19-pMF1.20 genes were cloned from pMF1, digested with BamH I and Kpn I and ligated into M. xanthus-E. coli shuttle plasmid pZJY4111 (which was also digested with BamH I and Kpn I) to obtain 19-pZJY4111, 20-pZJY4111, and 19-20-pZJY4111. The primers used in this study are listed in Table S4. The recombinant plasmids were electroporated into M. xanthus DZ1 according to the protocol described previously (Kashefi and Hartzell, 1995).
Plasmid stability assay
To test stability, M. xanthus DZ1 strains harboring the plasmids were grown to the late exponential phase in liquid CTT medium supplemented with 40 μg/ml kanamycin. Then we diluted the cultures by 1:25 in fresh CTT liquid medium with no antibiotics and grew them at 30°C and 200 rpm. After 24 h of incubation, the cultures were serially diluted and plated on CTT agar without antibiotics. The dilutions and plating were routinely repeated every 24 h until 168 h of incubation. In each round, 100 single colonies were patched onto CTT agar with and without kanamycin, and plasmid stability was measured as the percentage of antibiotic-resistant clones (Sun et al., 2011; Shen et al., 2016). The data presented are the averages of three independent experiments.
Pull-down assay for protein–protein interaction
We inserted pMF1.19 in pACYC-Duet and pMAL-c5x, following the His6 and MBP fragments, respectively, to produce His6-tagged pMF1.19 (19-His6) and MBP-tagged pMF1.19 (19-MBP). The pMF1.20 gene was similarly inserted into pET28a and pMAL-c5X to produce His6-tagged pMF1.20 (20-His6) and MBP-tagged pMF1.20 (20-MBP), respectively. We purified 19-His6 and 20-His6 using Ni beads, separately. 19-MBP and 20-MBP were purified by amylose resin, separately. The 19-His6 and 20-MBP mixture was incubated with Ni beads at 4°C for 2 h. We also incubated the MBP and 19-His6 mixture with Ni beads as the control. In turn the 19-MBP and 20-His6 mixture was incubated with Ni beads at 4°C for 2 h and MBP with 20-His6 as the control. After incubation, the proteins extracted by the beads were tested by SDS-PAGE.s.
Protein expression and purification
The proteins were expressed in E. coli BL21 (DE3), induced by the addition of 0.1 mM of IPTG when the OD600 value of the culture reached 1.0. The BL21 cells were grown at 37°C in LB broth with antibiotics. After the addition of IPTG, the cultures were grown at 16°C for 20 h. The cells were then collected and resuspended in lysis buffer (25 mM Tris-HCl, pH 8.0, 200 mM NaCl and 5% glycerol, pH 8.0) and lysed via ultrasonication. The mixtures were centrifuged at 4°C for 30 min at 12,000 rpm. The soluble proteins were mixed with Ni beads (GE Healthcare, Sweden) or amylose resin (New England Biolabs) according to the manufacturer's protocols.
Nuclease cleavage assay
19-MBP was purified using amylose resin (New England Biolabs) and 20-His6 was purified using Ni beads (GE Healthcare, Sweden). The substrate was supercoiled pMAL-c5x plasmid DNA, which was extracted using a TIANprep Mini Plamid Kit (Tiangen, China). Ten micrometers of purified pMF1.20 protein and 0.5 μg DNA were incubated for 30 and 60 min at 37°C in a reaction buffer (20 mM Tris-HCl, 200 mM NaCl and 5% glycerol, pH 8.0). The reaction was stopped by the addition of phenol/chloroform/isoamyl-alcohol, and the solutions were examined using 0.8% agarose gel electrophoresis. A supercoiled DNA ladder marker (Takara, China) was used.
Growth inhibition assay in E. coli
pMF1.19 was inserted into pACYC-Duet (19-pACYC), and the promoter was replaced with a constructive promoter. pMF1.20 was inserted in pMAL-c5x (20-pMAL) following an IPTG-inducible promoter. The two compatible plasmids were introduced into E. coli BL21 simultaneously. In a similar manner, 20-pMAL with empty pACYC-Duet, 19-pACYC with empty pMAL-c5x and pACYC-Duet with pMAL-c5x were also separately introduced into the BL21 strain simultaneously as the control. Overnight LB cultures of the E. coli BL21 strains were five-fold serially diluted and spotted onto the LB agar with and without 1 mM IPTG. Photographs were taken after overnight incubation.
Bioinformatics analyses
The gene information of plasmid pMF1 was obtained from the NCBI genome database (NC_010372.1). Using the amino acid sequences of pMF1.19 and pMF1.20 as query sequences, we searched for homologs in the M. fulvus 124B02 genome with BLASTp (Boratyn et al., 2012). Multiple sequence alignments of the full-length protein sequences of the pMF1.19 homologs and pMF1.20 homologs were implemented using MAFFT (Katoh and Standley, 2013). Phylogenetic analysis was conducted using the bootstrap neighbor joining method in MEGA version 7 (Kumar et al., 2016). A sequence logo was generated as a graphical representation of multiple sequence alignment (Crooks et al., 2004). The conserved domain information was obtained from the CDD protein family (Marchler-Bauer et al., 2017).
Author contributions
YJL and YZL designed researches. YJL, YL, XC, ZZ, and YG performed researches. YJL, YL, XC, ZZ, and YG analyzed data. YJL and YZL wrote the paper. YZL provided funding for the project.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (NSFC) (No. 31670076 and 31471183) and the Key Program of Shandong Natural Science Foundation (No. ZR2016QZ002) to YzL.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00274/full#supplementary-material
Transcriptional analyses of the pMF1.13-pMF1.23 genes in pMF1. (A) The transcriptional levels revealed by transcriptomic analysis. (B) The transcriptional levels of pMF1.19 and pMF1.20 by qPCR.
Comparison of the phylogenies of the amino acid sequences of pMF1.19 and pMF1.20 and their homologues in Myxococcus fulvus 124B02.
Construction of the 19-pZJY4111, 20-pZJY4111 and 19-20-pZJY4111 plasmids.
Consensus sequence of pMF1.20 and its homologues in Myxococcus fulvus 124B02. The sequence diversity at each position was assessed by multiple sequence alignment and displayed as a WebLogo.
Protein Details for pMF1.
Homologues in Myxococcus fulvus 124B02 of pMF1.19 and pMF1.20.
Strains and plasmid used in this study.
Primers used in this study.
References
- Austin S., Ziese M., Sternberg N. (1981). A novel role for site-specific recombination in maintenance of bacterial replicons. Cell 25, 729–736. 10.1016/0092-8674(81)90180-X [DOI] [PubMed] [Google Scholar]
- Belieres M., Dejugnat C., Chouini-Lalanne N. (2015). Histidine-based lipopeptides enhance cleavage of nucleic acids: interactions with DNA and hydrolytic properties. Bioconjug. Chem. 26, 2520–2529. 10.1021/acs.bioconjchem.5b00542 [DOI] [PubMed] [Google Scholar]
- Boratyn G. M., Schaffer A. A., Agarwala R., Altschul S. F., Lipman D. J., Madden T. L. (2012). Domain enhanced lookup time accelerated BLAST. Biol. Direct 7:12. 10.1186/1745-6150-7-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton A. M., Jaoua S., Guespin-Michel J. (1985). Transfer of plasmid RP4 to Myxococcus xanthus and evidence for its integration into the chromosome. J. Bacteriol. 161, 523–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. J., Han K., Feng J., Zhuo L., Li Y. J., Li Y. Z. (2016). The complete genome sequence and analysis of a plasmid-bearing myxobacterial strain Myxococcus fulvus 124B02 (M 206081). Stand. Genomic Sci. 11:1. 10.1186/s40793-015-0121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Cueva-Mendez G., Pimentel B. (2007). Gene and cell survival: lessons from prokaryotic plasmid R1. EMBO Rep. 8, 458–464. 10.1038/sj.embor.7400957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M., Diaz-Orejas R. (1998). Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62, 434–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmowski M., Jagura-Burdzy G. (2013). Active stable maintenance functions in low copy-number plasmids of Gram-positive bacteria I. Partition systems. Pol. J. Microbiol. 62, 3–16. Available online at: http://www.pjm.microbiology.pl/archive/vol6212013003.pdf [PubMed] [Google Scholar]
- Dworkin M. (1996). Recent advances in the social and developmental biology of the myxobacteria. Microbiol. Rev. 60, 70–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G., Gerdes K. (2005). Plasmid segregation mechanisms. Annu. Rev. Genet. 39, 453–479. 10.1146/annurev.genet.38.072902.091252 [DOI] [PubMed] [Google Scholar]
- Elwell L. P., Shipley P. L. (1980). Plasmid-mediated factors associated with virulence of bacteria to animals. Annu. Rev. Microbiol. 34, 465–496. 10.1146/annurev.mi.34.100180.002341 [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H., Glaser G. (1999). Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53, 43–70. 10.1146/annurev.micro.53.1.43 [DOI] [PubMed] [Google Scholar]
- Feng J., Chen X. J., Sun X., Wang N., Li Y. Z. (2012). Characterization of the replication origin of the myxobacterial self-replicative plasmid pMF1. Plasmid 68, 105–112. 10.1016/j.plasmid.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Gerdes K., Gultyaev A. P., Franch T., Pedersen K., Mikkelsen N. D. (1997). Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31, 1–31. 10.1146/annurev.genet.31.1.1 [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. (1986). Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. U.S.A. 83, 3116–3120. 10.1073/pnas.83.10.3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Li Z. F., Peng R., Zhu L. P., Zhou T., Wang L. G., et al. (2013). Extraordinary expansion of a Sorangium cellulosum genome from an alkaline milieu. Sci. Rep. 3:2101. 10.1038/srep02101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F., Van Melderen L. (2011). Toxins-antitoxins: diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 46, 386–408. 10.3109/10409238.2011.600437 [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. (1977). Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc. Natl. Acad. Sci. U.S.A. 74, 2938–2942. 10.1073/pnas.74.7.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulter N., Ilhan J., Wein T., Kadibalban A. S., Hammerschmidt K., Dagan T. (2017). An evolutionary perspective on plasmid lifestyle modes. Curr. Opin. Microbiol. 38, 74–80. 10.1016/j.mib.2017.05.001 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Pogliano J., Helinski D. R., Konieczny I. (2002). ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979. 10.1046/j.1365-2958.2002.02921.x [DOI] [PubMed] [Google Scholar]
- Kashefi K., Hartzell P. L. (1995). Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF- defect. Mol. Microbiol. 15, 483–494. 10.1111/j.1365-2958.1995.tb02262.x [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J. L., Baquero F. (2002). Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15, 647–679. 10.1128/CMR.15.4.647-679.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midon M., Gimadutdinow O., Meiss G., Friedhoff P., Pingoud A. (2012). Chemical rescue of active site mutants of S. pneumoniae surface endonuclease EndA and other nucleases of the HNH family by imidazole. Chembiochem 13, 713–721. 10.1002/cbic.201100775 [DOI] [PubMed] [Google Scholar]
- Paulsson J. (2002). Multileveled selection on plasmid replication. Genetics 161, 1373–1384. Available online at: http://www.genetics.org/content/genetics/161/4/1373.full.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel B., Madine M. A., De La Cueva-Mendez G. (2005). Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J. 24, 3459–3469. 10.1038/sj.emboj.7600815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M., Austin S. (2011). Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79, 2502–2509. 10.1128/IAI.00127-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma G., Khatri I., Subramanian S. (2016). Complete genome of the starch-degrading Myxobacteria Sandaracinus amylolyticus DSM 53668T. Genome Biol. Evol. 8, 2520–2529. 10.1093/gbe/evw151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Patil R. D., Sahin O., Wu Z., Pu X. Y., Dai L., et al. (2016). Identification and functional analysis of two toxin-antitoxin systems in Campylobacter jejuni. Mol. Microbiol. 101, 909–923. 10.1111/mmi.13431 [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. (1990). Social and developmental biology of the myxobacteria. Microbiol. Rev. 54, 473–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam U., Narayana Murthy S., Gummadi S. N. (2016). Identification and characterization of the novel nuclease activity of human phospholipid scramblase 1. BMC Biochem. 17:10. 10.1186/s12858-016-0067-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. (1998). Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol. 29, 1137–1145. 10.1046/j.1365-2958.1998.01012.x [DOI] [PubMed] [Google Scholar]
- Sun X., Chen X. J., Feng J., Zhao J. Y., Li Y. Z. (2011). Characterization of the partitioning system of Myxococcus plasmid pMF1. PLoS ONE 6:e28122. 10.1371/journal.pone.0028122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata S., Hooykaas P. J., Oka A. (1989). Sequence determination and characterization of the replicator region in the tumor-inducing plasmid pTiB6S3. J. Bacteriol. 171, 1665–1672. 10.1128/jb.171.3.1665-1672.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tett A., Spiers A. J., Crossman L. C., Ager D., Ciric L., Dow J. M., et al. (2007). Sequence-based analysis of pQBR103; a representative of a unique, transfer-proficient mega plasmid resident in the microbial community of sugar beet. ISME J. 1, 331–340. 10.1038/ismej.2007.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. (2000). Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491. 10.1046/j.1365-2958.2000.02006.x [DOI] [PubMed] [Google Scholar]
- Van Melderen L. (2002). Molecular interactions of the CcdB poison with its bacterial target, the DNA gyrase. Int. J. Med. Microbiol. 291, 537–544. 10.1078/1438-4221-00164 [DOI] [PubMed] [Google Scholar]
- Vassallo C. N., Cao P., Conklin A., Finkelstein H., Hayes C. S., Wall D. (2017). Infectious polymorphic toxins delivered by outer membrane exchange discriminate kin in myxobacteria. Elife 6:e29397. 10.7554/eLife.29397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. Y., Zhong L., Shen M. J., Xia Z. J., Cheng Q. X., Sun X., et al. (2008). Discovery of the autonomously replicating plasmid pMF1 from Myxococcus fulvus and development of a gene cloning system in Myxococcus xanthus. Appl. Environ. Microbiol. 74, 1980–1987. 10.1128/AEM.02143-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptional analyses of the pMF1.13-pMF1.23 genes in pMF1. (A) The transcriptional levels revealed by transcriptomic analysis. (B) The transcriptional levels of pMF1.19 and pMF1.20 by qPCR.
Comparison of the phylogenies of the amino acid sequences of pMF1.19 and pMF1.20 and their homologues in Myxococcus fulvus 124B02.
Construction of the 19-pZJY4111, 20-pZJY4111 and 19-20-pZJY4111 plasmids.
Consensus sequence of pMF1.20 and its homologues in Myxococcus fulvus 124B02. The sequence diversity at each position was assessed by multiple sequence alignment and displayed as a WebLogo.
Protein Details for pMF1.
Homologues in Myxococcus fulvus 124B02 of pMF1.19 and pMF1.20.
Strains and plasmid used in this study.
Primers used in this study.