Abstract
Purpose
The objectives of this study were to investigate the expression of MSX1 in cervical cells and tissues, the methylation status of the MSX1 promoter, the influence of overexpression of gene MSX1 on the proliferation, migration, and invasion of HeLa and SiHa cells, and finally the possible molecular mechanisms responsible for the suppressive effects of MSX1 upon cervical cancer cells.
Patients and methods
Semi-quantitative and quantitative reverse transcription-polymerase chain reactions were used to investigate the expression levels of MSX1, and methylation-specific polymerase chain reaction (MSP) was performed to investigate promoter methylation status in cervical cancer cell lines, primary cervical tissues, and normal cervical tissues. Clone formation, Cell Counting Kit-8 (CCK-8), cell wound scratch, and transwell assays were performed to verify whether MSX1 could inhibit the proliferation and migration of cervical cancer cells. Western blot was used to analyze the effect of MSX1 upon Notch1, Jagged1, c-Myc, cleaved PARP, cleaved caspse-3, and cyclin D1 (CCND1).
Results
MSX1 was frequently downregulated or silenced in 60.0% (3/5) of cervical cancer cell lines. The promoter methylation of MSX1 was detected in 42.0% (42/100) of primary tumor tissues, while no methylation was observed in normal cervical tissues. Pharmacological demethylation reduced MSX1 promoter methylation levels and restored the expression of MSX1. The overexpression of MSX1 in cervical cancer cells thus inhibited the proliferation and migration of cervical cancer cells. The overexpression of MSX1 in cervical cancer cells downregulated the expression levels of Notch1, Jagged1, and c-Myc but upregulated the expression levels of CCND1, cleaved PARP, and cleaved caspase-3.
Conclusion
MSX1 appears to be a functional tumor suppressor that regulates tumorigenesis in cervical cancer by antagonizing Notch signaling.
Keywords: MSX1, cervical cancer, tumor suppressor, methylation, Notch1
Introduction
Cervical cancer is the fourth most common cancer to affect women worldwide, next only to lung, breast, and colorectal cancers.1 Therefore, early diagnosis and the exploration of new treatment methods are of great significance to improve the survival and quality of life of patients with cervical cancer. The occurrence of malignant tumors involves the aberrant expression of oncogenes and tumor suppressor genes (TSGs). Recent studies have found that promoter methylation is a common mechanism underlying the aberrant expression of genes associated with the development of malignant tumors.2 Moreover, a study has detected the methylation in the promoters of a number of TSGs in cervical cancer.3
The Notch signaling pathway plays an important role in maintaining the balance between cell proliferation, differentiation, and apoptosis, and it is closely related to the development of malignant tumors in humans. Indeed, the Notch signaling pathway has been shown to be involved in the development of multiple types of tumors.4–12 A previous study has detected Notch1 in cervical cancer, suggesting that this gene may play a key role in the development of cervical cancer.12 Notch receptors and their ligand protein levels rise from cervical precancerous lesions to invasive cancers.12
The MSX1 gene is located on chromosome 4p16.2; it belongs to the homeobox family and encodes for a transcriptional repressor that can interact with a core protein of the transcription complex or other homeodomain protein. Consequently, gene MSX1 plays an important role in the process of embryo development.13 Previous studies have linked the aberrant methylation of MSX1 promoter DNA with lung cancer, gastric cancer, ovarian cancer, childhood acute T lymphoblastic leukemia, Wilms tumor, and breast cancer.14–20
A search using the Oncomine bioinformatic resource21,22 showed that the expression of MSX1 was significantly reduced in cervical cancer tissues, although its specific expression and the specific role of its expression in the development of cervical cancer are still unknown.
In the present study, we investigated the expression of gene MSX1 in cervical cancer and the methylation status of the gene MSX1 promoter and its specific function in vitro and analyzed the mechanisms underlying tumor suppressor function in cervical cancer. Collectively, our findings suggest that gene MSX1 acts as a TSG in cervical cancer and exerts influence via the Notch signaling pathway.
Patients and methods
Cell lines, tumor samples, and normal tissues
Human cervical cancer cell lines (SIHA, CASKI, HELA, C4-1, and C33A) and normal cervical epithelial cell line Ect1/E6E7 were obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in the Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific) in a humidified atmosphere (37°C) with 5% CO2 and 1 × penicillin/streptomycin, depending upon the medium being used. RNA from normal human cervical tissues was purchased from Stratagene (Santa Clara, CA, USA), BioChain (Newark, CA, USA), or Chemicon (Billerica, MA, USA). Primary tumor tissues of cervical cancer and normal cervical tissues were obtained from patients undergoing primary surgery at the Surgery Department of the First Affiliated Hospital of Chongqing Medical University, China. The status of all samples was defined and confirmed pathologically by physicians at Chongqing Medical University. We collected a range of clinical and pathological data from all patients with cervical tumors, including age, International Federation of Gynecology and Obstetrics (FIGO) stage, histological grade, tumor size, lymph node metastasis, and lymph vascular space invasion. All patients provided written informed consent for the research during the initial clinical investigation. This study was approved by the ethics committee of the First Affiliated Hospital of Chongqing Medical University, approval notice: 2012/2013(23).
Reverse transcriptase-polymerase chain reaction (RT-PCR) analysis and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from tissue and infected cells with TRIzol reagent in accordance with the manufacturer’s specifications. RT-PCR was performed as described previously23 using GAPDH as an internal control. The four primer sequences used for polymerase chain reaction (PCR) amplification in this study are given in Table 1. RT-PCR was carried out with 23 cycles for GAPDH and 32 cycles for gene MSX1 with Go-Taq DNA polymerase. The PCR program began with an initial denaturation at 95°C for 2 min, followed by amplification reaction cycles (95°C for 30 s, 55°C for 30 s, and 72°C for 30 s) with a final extension at 72°C for 3 min. Quantitative PCR was performed using a SYBR® Green PCR Master Mix kit (Thermo Fisher Scientific) and an Applied Biosystem 7500 Real-time PCR System (Thermo Fisher Scientific). β-actin served as a control. The relative expression of MSX1 was evaluated using the method. All assays were performed three times independently. The primer sequences are shown in Table 1.
Table 1.
List of primers used in this study
| PCR | Primer | Sequence (5′–3′) | Product size (bp) | PCR cycles | Annealing temperature (°C) |
|---|---|---|---|---|---|
| RT-PCR | MSX1-F | CATTCGAATACCGGGGCCGACGA | 176 | 32 | 55 |
| MSX1-R | CGCCTAGGGCTCAGTCCACCATGT | ||||
| GAPDH-F | CCTCAGTTGCCTAAACCA | 202 | 23 | 55 | |
| GAPDH-R | CACTACCCTAAAGGTAACTA | ||||
| MSP | MSX1m3 | GCGCCTCATCACATCAGCGC | 116 | 41 | 60 |
| MSX1m4 | GCGATTTCTGATGCTGGCGC | ||||
| MSX1u3 | CAAGGCTAGTCATCATCAACCA | 121 | 41 | 58 | |
| MSX1u4 | CGCCTAGGGCTCAGTCCACCATGT |
Abbreviations: PCR, polymerase chain reaction; RT-PCR, reverse transcriptase-polymerase chain reaction; MSP, methylation-specific polymerase chain reaction.
DNA bisulfite treatment and methylation-specific polymerase chain reaction (MSP)
Genomic DNA was extracted from tumors and cell pellets using the DNA Mini Kit (Qiagen NV, Venlo, the Netherlands). DNA bisulfite treatment and MSP were conducted as described in our previous report.24 Bisulfite DNA was amplified by MSP with gene MSX1 methylation-specific primer sets or non-methylation-specific primer sets (Table 2). MSP was used to amplify methylated gene alleles for 35 cycles and non-methylated gene alleles for 40 cycles using AmpliTaq Gold Polymerase with annealing temperatures of 60°C or 58°C. Methylated and non-methylated human DNAs were used as positive and negative controls, respectively. MSP products were visualized and identified on a 2% agarose gel containing 100 bp DNA markers.
Table 2.
Promoter methylation status of MSX1 in primary cervical tumors
| Tissues | Samples (number) |
MSX1 promoter
|
Frequency of methylation | |
|---|---|---|---|---|
| Methylation | Unmethylation | |||
| Cervical cancer tissues | 100 | 42 | 58 | 42/100 (42%) |
| Normal cervical tissues | 10 | 0 | 10 | 0/10 (0%) |
5-Aza-2′-deoxycytidine (Aza) and trichostatin A (TSA) treatment
The HeLa and SiHa cell lines were used for pharmacological demethylation. Briefly 1 × 106 cells were treated with 10 mmol/L Aza for 72 h and then with 100 nmol/L TSA (both from Sigma-Aldrich Co., St Louis, MO, USA and EMD Millipore, Billerica, MA, USA) for 24 h at 37°C. The cells were then harvested for DNA and RNA extraction and further analysis.
Cell-proliferation assays
HeLa cells were infected by MSX1 lentivirus or control virus, trypsinized, and then resuspended. Then, these cells were seeded, in triplicate, into a 96-well plate at a density of 1 × 104 cells/well in each well and then incubated overnight at 37°C in an environment with 5% CO2. Cell proliferation was evaluated by the Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan), which only stains living cells. At 24, 48, and 72 h, the culture medium was removed, and α-MEM (100 µL), with 10 µL CCK-8, was added to each well, followed by incubation for 2 h at 37°C. The absorbance was then measured by a microplate reader scanning at 450 nm. All assays were performed in triplicate.
Colony-formation assays
HeLa cells were infected with MSX1 lentivirus or negative control virus and seeded at a density of 300 cells/well into six-well plates and grown for 2 weeks in the regular culture medium. These cells were subsequently washed twice with phosphate-buffered saline (PBS), and colonies were fixed with 10% formaldehyde, dried and then stained with 2% crystal violet. The number of colonies formed (≥50 cells/colony) were manually counted in four different microscopic fields, and the mean value was calculated. Each experiment was performed in triplicate; three wells were measured for each treatment group, and the experiments were repeated at least three times.
Wound-healing assays
Cells that had been infected with MSX1 lentivirus or the negative control were evenly seeded in six-well plates and grown to 100% confluence in DMEM containing 10% FBS. A straight wound was induced by scratching the cells with a 200 µL plastic pipette tip. Cells were then incubated and allowed to migrate in the medium and were then washed twice with PBS to remove dead cells. At 0, 12, 24, and 36 h post-induction of injury, photographs were acquired using a TE2000 inverted phase contrast microscope (Nikon Corporation, Tokyo, Japan) in four random fields at ×100 magnification. The proportion (%) of wound closure was quantified according to the space of migration tumor cells with Image-Pro Plus software (Media Cybernetics, Inc., Rockville, MD, USA). Each experiment was performed in triplicate.
Transwell cell migration and invasion assay
Cell migration and invasion assays were carried out using Transwell chambers (8 µm pore size; Corning Incorporated, Corning, NY, USA). Cells were placed in the upper chamber at a density of 2.5 × 105 and cultured in the serum-free culture medium. Then, 300 µL of DMEM supplemented with 10% FBS as a chemoattractant was added into the lower chamber. After 48 h of incubation at 37°C in 5% CO2 atmosphere, non-migrating and non-invading cells on the upper surface of the filter were removed with a cotton-tipped swab. The cells that had migrated and invaded were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet staining solution. Ten photographs were then taken from each sample at 10 random points on five random microscopic fields. Cell numbers were counted at ×100 magnification, and the experiment was carried out in triplicate.
Flow cytometry analysis of the cell cycle
HeLa cells infected with puma lentivirus (PLV)-MSX1 or PLV-empty were seeded (1 × 106 cells/well) into six-well plates and incubated overnight at 37°C in 5% CO2. Then, cells were digested by trypsinization using 0.1% trypsin 48 h after infection and centrifuged at 1,000 rpm for 5 min. Cells were washed twice with PBS, fixed in 70% ethanol at 4°C for 2 h, and then treated with 100 µL of 50 mg/L propidium iodide for 30 min at room temperature (RT) in the dark. The cell-cycle data were analyzed by CellQuest software (BD Biosciences, San Jose, CA, USA). All experiments were performed in triplicate.
Western blot analysis
Infected cells were harvested and lysed in lysis buffer. Protein was then extracted from infected cells using Protein Extraction Reagent (Pierce, Thermo Fisher Scientific) containing a protease inhibitor cocktail (Sigma-Aldrich Co.). The same concentration of proteins was then separated by 12% sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer to polyvinylidenedifluoride (PVDF) membrane (Bio-Rad Laboratories Inc., Hercules, CA, USA) for antibody incubation. After blocking the membrane with 5% skimmed milk in Tris-buffered saline with Tween-20 for 1 h at RT, the membranes were then incubated with the primary antibodies at the manufacturer’s recommended dilutions with gentle shaking at 4°C overnight in a shaker. The details of the primary antibodies used are as follows: MSX1 (#5378; Cell Signaling Technology, Boston, MA, USA), Notch1 (#3608; Cell Signaling Technology), Jagged1 (#70109; Cell Signaling Technology), c-Myc (#1472-1; Epitomics, Cambridge, MA, USA), cyclin D1 (CCND1; #1677-1; Epitomics), cleaved PARP (#5625; Cell Signaling Technology), cleaved caspase-3 (#9661; Cell Signaling Technology), and GAPDH (#AE082046; Beijing Biosynthesis Biotechnology, Beijing, China) as a control. Samples were subsequently washed and then incubated with secondary antibodies at RT for 1 h. Finally, separated proteins were detected and photographed using an enhanced chemiluminescence detection system (Amersham Pharmacia Biotech, Piscataway, NJ, USA) according to the manufacturer’s instructions. The final MSX1 films were then used for densitometric analysis using ImageJ open source software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Statistical analyses were performed using the Student’s t-test, χ2 test, and Fisher’s exact test to determine P-values. For all tests, P < 0.05 was considered to be statistically significant. Results are shown as mean ± standard deviation (SD).
Results
Expression of MSX1 in cervical cancer cell lines was reduced but was broadly expressed in normal tissues
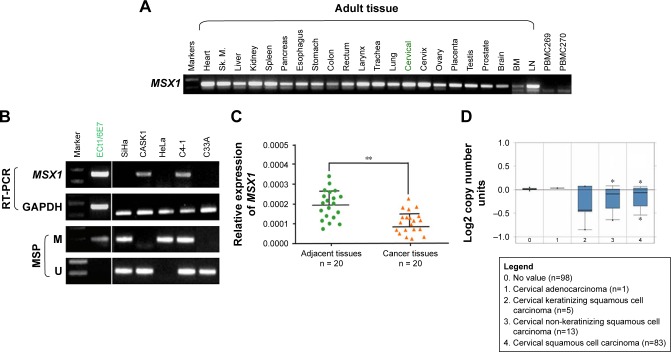
First, we examined the expression of gene MSX1 in a series of normal human adult tissues including cervical tissues and cervical cancer cell lines by RT-PCR. As shown in Figure 1A, MSX1 was broadly expressed in normal adult tissues, including normal cervical tissues, with variable expression levels, but significant reduction or silencing of MSX1 expression was frequently observed in cervical cancer cell lines (Figure 1B). MSX1 expression was then examined at the mRNA level in primary cervical tumors. qPCR demonstrated that MSX1 mRNA was downregulated in the cervical cancer tissues compared with that in the normal cervical tissues (P < 0.05; Figure 1C). Similarly, analysis using Oncomine23,24 showed that the expression of MSX1 in cervical cancer tissues was significantly reduced (Figure 1D). Collectively, these data suggest that MSX1 may represent a candidate TSG.
Figure 1.
MSX1 downregulation/silencing in cervical cancer cell lines.
Notes: (A) Detection of MSX1 expression in normal human adult tissues with semi-quantitative RT-PCR using GAPDH as the control. (B) MSX1 is frequently silenced in cervical cancer cell lines by promoter methylation, demonstrated with RT-PCR and MSP. (C) Expression of MSX1 in normal cervical tissues (n = 20) and cervical cancer tissues (n = 20) detected by real-time PCR, with β-actin as a control. Mean ± SD, P = 0.0241. **P<0.01. (D) MSX1 expression is reduced in cervical carcinoma. Oncomine platform provides all data (https://www.oncomine.org/). *P<0.05.
Abbreviations: RT-PCR, reverse transcriptase-polymerase chain reaction; MSP, methylation-specific polymerase chain reaction; PCR, polymerase chain reaction; SD, standard deviation; M, methylated; U, unmethylated; Sk.m, Sk. muscle; BM, bone marrow; LN, lymph node.
Promoter methylation mediates the downregulation of MSX1 expression in cervical cancer
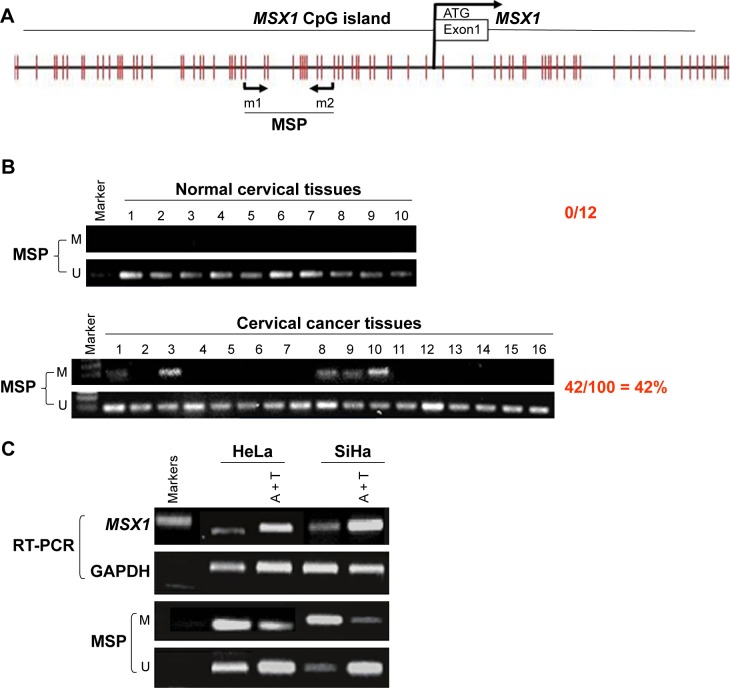
Next, we analyzed the promoter of gene MSX1 to evaluate whether MSX1 repression was due to promoter methylation. Using bioinformatic analysis, we found that gene MSX1 contained typical 5′-C-phosphate-G-3′ (CpG) islands (CGIs) spanning the proximal promoter and exon regions (Figure 2A), which indicated the potential role of promoter CpG methylation in MSX1 silencing. We further detected MSX1 methylation by MSP assay in normal cervical tissues and cervical cancer tissues. Promoter methylation of MSX1 was detected in 42% (42/100) of cervical cancer tissues but was not found in 10 normal cervical tissues. Pharmacological demethylation was conducted to test whether promoter methylation directly mediates the reduction of MSX1 levels in cervical cancer cells. Two (HeLa, SiHa) lacking MSX1 expression were treated with Aza and the histone deacetylase inhibitor TSA. Following treatment, the expression of MSX1 in these cell lines was significantly increased compared with that prior to treatment, accompanied by decreased methylated alleles of MSX1 (Figure 2C). These results indicate that promoter methylation is responsible for MSX1 silencing in cervical cancer cells. These results indicate that promoter methylation is responsible for MSX1 silencing in cervical cancer cells.
Figure 2.
MSX1 was methylated in primary cervical tumors.
Notes: (A) Schematic structure of the MSX1 promoter CGI. Exon 1 (gray rectangle), CpG sites (short vertical lines), and the MSP sites analyzed are shown. The transcription start site is indicated by a curved arrow. (B) Representative MSP analysis of MSX1 methylation in primary cervical tumors, surgical margin tissues, and normal cervical tissues. (C) The expression and methylation status of MSX1 were measured in HeLa and SiHa cells treated with Aza with or without TSA by RT-PCR and MSP.
Abbreviations: CpG, 5′-C-phosphate-G-3′; CGI, CpG island; MSP, methylation-specific polymerase chain reaction; Aza, 5-Aza-2′-deoxycytidine; TSA, trichostatin A; RT-PCR, reverse transcriptase-polymerase chain reaction; M, methylated; U, unmethylated; A+T, 5-aza-2′-deoxycytidine plus trichostatin A.
MSX1 promoter methylation and its correlation with the clinicopathological features of cervical cancer patients
We next used MSP analysis to investigate methylation in the promoter of gene MSX1 in 100 cervical primary tumor tissues and 10 normal cervical tissues. As expected, we observed MSX1 promoter methylation in 42 out of 100 (42%) cervical primary cancer tissues but not in normal cervical tissues (Figure 2B and Table 2). This suggests a pattern of tumor-specific methylation of gene MSX1 in cervical cancer. We further analyzed the correlation between MSX1 methylation and the clinicopathological features of cervical cancer patients, including age, FIGO stage, tumor size, histological grade, lymph node metastasis, and lymph vascular space invasion. However, there was no significant correlation between MSX1 promoter methylation and the clinicopathological features of these patients (Table 3).
Table 3.
Clinicopathological features of cervical cancer patients according to MSX1 methylation status
| Clinicopathological features | Number (n = 100) |
MSX1 promoter methylated status
|
P-value | |
|---|---|---|---|---|
| Methylated | Unmethylated | |||
| Age (years) | 0.37 | |||
| ≥45 | 34 | 18 | 16 | |
| <45 | 66 | 27 | 39 | |
| FIGO stage | 0.14 | |||
| I | 29 | 19 | 10 | |
| II | 38 | 20 | 18 | |
| III | 33 | 12 | 21 | |
| Size (cm) | 0.58 | |||
| <4 | 32 | 19 | 13 | |
| ≥4 | 68 | 31 | 37 | |
| Lymph node metastasis | 0.31 | |||
| Positive | 51 | 22 | 29 | |
| Negative | 49 | 24 | 25 | |
| Lymph vascular space invasion | 0.82 | |||
| Positive | 60 | 28 | 32 | |
| Negative | 40 | 21 | 19 | |
| Histological grade | 0.94 | |||
| G1 | 14 | 10 | 4 | |
| G2–3 | 86 | 46 | 40 | |
Abbreviation: FIGO, International Federation of Gynecology and Obstetrics.
Ectopic expression of gene MSX1 inhibited clonogenicity, proliferation, and migration of cervical cancer cells
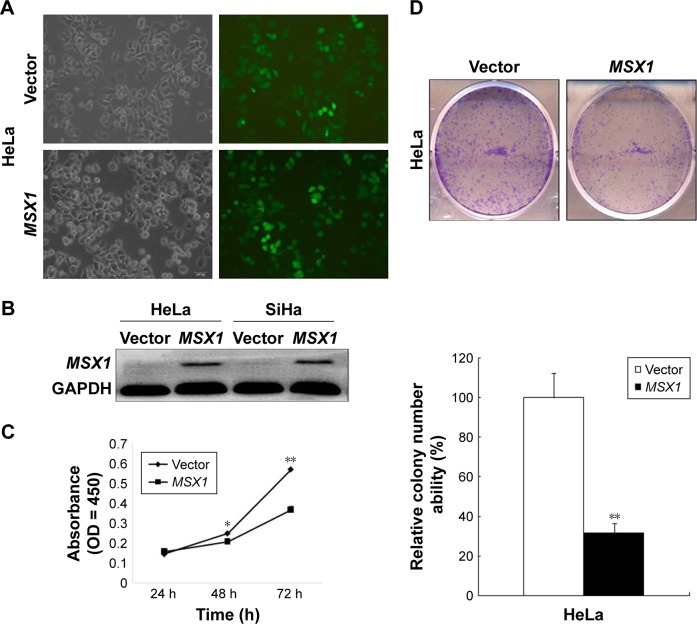
MSX1 repression by promoter methylation in cervical cancer cell lines, as well as primary tumors, indicated that gene MSX1 may be a functional tumor suppressor in cervical tumorigenesis. In our next experiment, we infected HeLa and SiHa cells with either MSX1-expressing lentivirus or empty lentivirus (Figure 3A). Using this method, we successfully obtained stable overexpression of MSX1 in both HeLa and SiHa cells, as indicated by Western blot (Figure 3B). Colony-formation assays and CCK-8 cell-proliferation assays were then used to evaluate the suppressive effect of MSX1 on cervical cancer cell proliferation. Colony-formation assays showed 40%–80% reductions in the clonogenicity of MSX1-infected HeLa cancer cells compared to controls (P < 0.01; Figure 3D). Cell viability was significantly reduced at 24, 48, and 72 h after infection by MSX1 in both HeLa and SiHa cells (P < 0.01, P < 0.05; Figure 3C). Western blot also showed that the expression of CCND1 was clearly downregulated in MSX1-expressing HeLa cells (Figure 4B). Collectively, these results showed that MSX1 possesses the ability to inhibit tumor cell clonogenicity and migration and thus functions as a TSG in cervical cancer.
Figure 3.
MSX1 is a functional TSG that inhibits cervical cancer tumor cell growth, clonogenicity.
Notes: (A) Lentivirus-infected cervical cancer cells. (B) MSX1 expression was measured by Western blot in vector- and MSX1-infected HeLa and SiHa cells. (C) Proliferative capacities of cervical cancer cell lines were lower than those of the control group 24, 48, and 72 h after infection with MSX1, detected with a CCK-8. *P < 0.05 and **P < 0.01. (D) Effect of MSX1 overexpression on colony formation in HeLa and SiHa cells (right panel, upper). Quantitative analyses of colony numbers in three replicates are shown as mean ± SD (right panel, lower). **P < 0.01.
Abbreviations: TSG, tumor suppressor gene; CCK-8, Cell Counting Kit-8; SD, standard deviation.
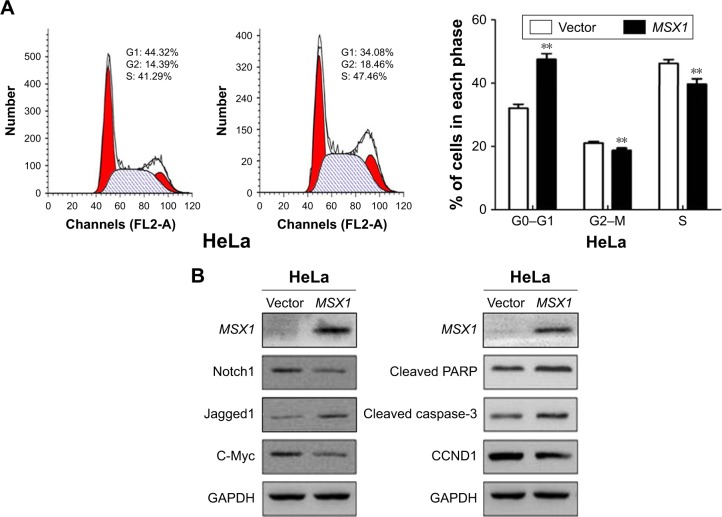
Figure 4.
Ectopic expression of MSX1 induced cell-cycle G1/S arrest and apoptosis, suppressed Notch signal pathway activity, and upregulated apoptotic markers in cervical cancer cells.
Notes: (A) Flow-cytometric analysis of MSX1 expression in HeLa cells showed an increased percentage of G1/S-phase cells compared with the control. Quantitative representation of the results is shown on the right. **P < 0.01. (B) Western blot analysis of Notch signal pathway components, CCND1, and apoptosis markers.
Abbreviation: CCND1, cyclin D1.
MSX1 suppressed the migration/invasion of cervical tumor cells
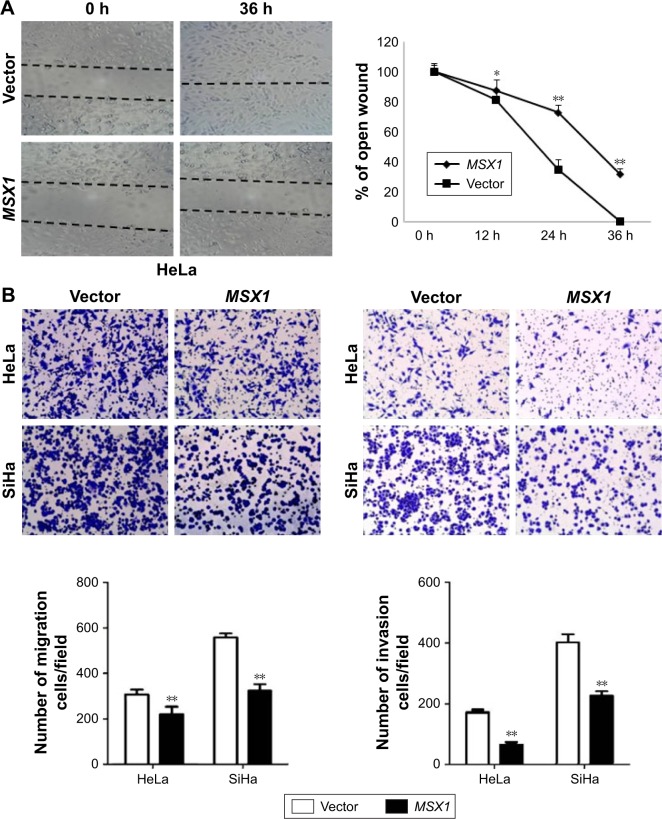
In order to understand the contribution of MSX1 to cellular motility, we next used wound-healing and transwell assays. Scratch wound-healing assays showed that MSX1-expressing cells exhibited reduced ability in closing an artificial wound than the vector-infected cells compared on a confluent monolayer (P < 0.05, P < 0.01; Figure 5A). Moreover, in a Matrigel invasion assay, MSX1-overexpressing cells had reduced ability to migrate and invade across the Matrigel (by up to 80%) compared to controls (P < 0.01; Figure 5B). Collectively, these results suggest that the overexpression of MSX1 could inhibit the metastasis and migration of cervical tumor cells.
Figure 5.
Ectopic expression of MSX1 inhibits cervical cancer cell migration and invasion.
Notes: (A) Effect of MSX1 on cell migration was assessed by wound-healing motility assay. Left: Representative images of wound healing at 0 and 36 h after wound scratch. Right: The percentage of wound sealing compared with that of controls at each time point as indicated. *P < 0.05 and **P < 0.01. (B) Transwell assay indicated that the ectopic expression of MSX1 inhibited the invasion potential of HeLa and SiHa cells. Photographs show cells that have traveled through the Matrigel, and the histogram represents the percentage of invasive cells. **P < 0.01.
MSX1 induced cell-cycle G0/G1 arrest and apoptosis in cervical cancer cells
In order to gain further insight into the possible mechanisms underlying the growth-inhibitory effect of MSX1 on human cervical cancer cells, we next examined cell-cycle distribution using flow cytometry. Our analysis showed a significant increase in the number of MSX1-expressing cells in the G0/G1 phase, accompanied by a reduction of cells in the S and G2/M phases compared with controls (P < 0.01; Figure 4A), indicating that MSX1 causes cell-cycle G0/G1 arrest in cervical cancer cells.
We further found that the expression of CCND1 was clearly downregulated using Western blot. These results suggest that the inhibitory effect of cell proliferation by MSX1 is likely to be mediated by G0/G1 arrest in the cell cycle. At the same time, we found that the expression of cleaved PARP and cleaved caspase-3 was clearly upregulated in MSX1-expressing HeLa cells. MSX1 may induce apoptosis in cervical cancer cells through upregulating the expression of cleaved PARP and cleaved caspase-3.
MSX1 negatively modulates the Notch signaling pathway and the activity of its downstream-target gene c-Myc
To elucidate whether the molecular mechanisms underlie the tumor suppression by MSX1, we next investigated whether MSX1 counteracts the Notch signaling pathway in order to exhibit its tumor-suppression function in tumor cells. The expression levels of Notch1, Jagged1, and its downstream-target gene c-Myc were reduced in MSX1-expressing HeLa cells (Figure 4B). This indicates that MSX1 causes a reduction in Notch1 levels, which antagonizes the Notch signaling pathway in cervical cancer.
Discussion
The pathogenesis of cervical cancer is complex. A number of oncogenes and TSGs have been identified in cervical cancer, but only few of these can be applied for the early diagnosis of cervical cancer and gene target therapy.25–30 Consequently, it is of great significance to continue to find and identify additional oncogenes and TSGs associated with the occurrence and development of cervical cancer.
The main function of the homeobox gene family is to encode transcription regulators, which play an important role in normal human embryonic development.31 MSX1-deficient mice died early in their development because of their craniofacial structural defects.32 MSX1 point mutations and deletions can also lead to limb hypoplasia in humans and also selective tooth hypoplasia and Wolf-Hirschhorn syndrome.33,34 The occurrence and development of tumors also involve the abnormal regulation of cell-proliferation and differentiation processes, which are similar to those involved in developmental defects. Gene MSX1 may play an important role in the occurrence and development of tumors. The TSGs are often missing in chromosome loci, thus leading to tumorigenesis.35
In the present study, we investigated the MSX1 gene, located on chromosome 4p16.2.36 Previous studies have shown that this site is absent in a range of tumors,15,16 and we therefore hypothesized that MSX1 may be a TSG and that its downregulation or deletion may lead to the occurrence of cervical cancer. By investigating gene MSX1 at cellular and tissue levels, we were able to fully demonstrate that the expression of MSX1 in cervical cancer cells was downregulated. MSX1 may therefore be a new TSG associated with cervical cancer, as its downregulation or deletion may be closely related to the occurrence of cervical cancer.
DNA methylation is a form of DNA sequence modification and represents one of the most important mechanisms in genetics as it can regulate genome function without changing the molecular structure of DNA.37 DNA methylation is the process of transferring methyl from S-adenosyl methionine (SAM) onto the corresponding bases of DNA molecules under the catalysis of DNA methyltransferase (DNMT).37 The common process of DNA methylation involves the covalent binding of cytosine to the fifth carbon atom of cytosine on the DNA chain and modifies cytosine to 5-methylcytosine.38 DNA methylation can lead to the inactivation of some genes, whereas demethylation can reactivate silenced genes.38
In carcinogenesis, the CGI of a TSG can be highly methylated, thus leading to inactivation.37 DNA methylation of TSGs has been detected in many different types of tumor and usually occurs in precancerous stages;37 thus, DNA methylation represents an ideal marker for the early diagnosis of tumors. In this study, we used RT-PCR and MSP to demonstrate the hypermethylation of the MSX1 gene promoter in a cervical cancer cell line, and the expression of MSX1 was downregulated or deleted. These results demonstrate that the main reason for the downregulation of MSX1 expression in cervical cancer cells was promoter methylation. However, the expression of MSX1 in cervical cancer tissues and cells and its methylation status are not consistent, which suggests that there may be other mechanisms (such as histone modification or microRNAs) involved in the downregulation of MSX1. Figure 1B shows that the expression of MSX1 in a C33A cell line is silent; however, in the MSP, methylation of the MSX1 gene promoter did not occur in this particular cervical cancer cell line. Given that the methylation of TSG promoters occurs in the early stages of tumorigenesis and the effect of methylated drugs can restore gene expression, it follows that intensive research may lead to use of TSG methylation in the early diagnosis of cervical cancer and therefore become a new therapeutic target.
The process of cell signal transduction includes a wide range of different signals, including molecular signals and exogenous stimuli. These signals can induce changes in intracellular signaling, which are mediated by messenger molecules in cells or on cell membranes, and ultimately leads to changes in the expression of target genes in the nucleus. Normal molecular signal transduction plays an important role in maintaining cell growth, differentiation, and apoptosis. Because of the stimulation of external or internal factors, cell signal transduction process can become abnormal, which will inevitably lead to a series of abnormal biological changes and, ultimately, the malignant proliferation of cells.39 In recent years, our understanding of the molecular biology of tumors has developed rapidly. Previous studies have shown that dysfunction of the Notch signaling pathway is an important reason for the occurrence and development of multiple tumors.40 Notch receptors are activated by direct contact with ligands expressed in adjacent cells and subsequently regulate cell proliferation, differentiation, and apoptosis.40 A previous study has shown that MSX1 is involved in regulating the expression of the Notch signaling pathway.41 In this study, we detected the expression of Notch1, Jagged1, and c-Myc in MSX1-infected HeLa cells by Western blot and found that the expression of these genes was downregulated in stable-infected tumor cells. Exogenous infection of MSX1 was able to effectively inhibit the growth, migration, and invasion of cervical cancer cells. These results suggest that MSX1 may play the role of a tumor suppressor by inhibiting activity of the Notch signaling pathway.
In other studies, MSX1 has been shown to inhibit the growth of tumor cells by upregulating the expression of CCND1, thus exerting effect upon the cell cycle and apoptosis.42 CCND1 is also a downstream-target gene of the Notch signaling pathway.43 Some studies have shown that the expression of CCND1 and the Notch signaling pathway is closely related to the occurrence and development of tumors and can readily affect the growth and metastasis of tumor cells.43 By studying the stable expression of MSX1 transgenic mice, Hu et al42 showed that breast tissue differentiation was blocked, accompanied by increased expression levels of CCND1 during the course of pregnancy in female rats. In ovarian cancer cells, the exogenous expression of MSX1 can arrest the ovarian cancer cell cycle in the G1 phase and thus inhibit tumor cell growth.44 CCND1 plays an important role in transition from G1 to S phase of the cell cycle. In this study, we analyzed the effect of MSX1 on cervical cancer cells by flow cytometry and showed that the exogenous expression of MSX1 could affect the cell cycle of cervical cancer, resulting in G1 phase arrest in cervical cancer cells. On the other hand, we also found that the exogenous expression of MSX1 can upregulate the expression of CCND1, thus providing further evidence that MSX1 plays a role in cell-cycle arrest by upregulating CCND1 in cervical cancer.
The high-risk human papillomavirus (HPV), which is implicated in the pathogenesis of >90% of cervical cancer cases, leads to the production of E6/E7 protein. Previous research has shown that the overexpression of MSX1 stabilizes wild-type p53 and restores the apoptotic function of p53 even in the presence of HPV-E6 oncoprotein, which abrogates the apoptotic function of wild-type endogenous p53 expression. Studies45,46 have also revealed crosstalk between the Notch signaling pathway and p53. The interaction between MSX1, the Notch signaling pathway, and P53 thus requires further investigation.
Conclusion
Our findings suggest that the low expression of MSX1 in cervical cancer affects the proliferation, apoptosis, and metastasis of tumor cells due to epigenetic silencing. These results suggest that MSX1 plays the role of a TSG in cervical cancer by inhibiting the Notch signaling pathway. Therefore, MSX1 may be used as an early diagnostic marker for cervical cancer or as a new therapeutic method for cervical carcinoma.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol Oncol. 2012;6(6):620–636. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Q, Balasubramanian A, Hawes SE, et al. Detection of hypermethylated genes in women with and without cervical neoplasia. J Natl Cancer Inst. 2005;97(4):273–282. doi: 10.1093/jnci/dji041. [DOI] [PubMed] [Google Scholar]
- 4.Kannan S, Sutphin RM, Hall MG, et al. Notch activation inhibits AML growth and survival: a potential therapeutic approach. J Exp Med. 2013;210(2):321–337. doi: 10.1084/jem.20121527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Razumilava N, Gores GJ. Notch-driven carcinogenesis: the merging of hepatocellular cancer and cholangiocarcinoma into a common molecular liver cancer subtype. J Hepatol. 2013;58(6):1244–1245. doi: 10.1016/j.jhep.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu S, Evans H, Buckle C, et al. Impaired osteogenic differentiation of mesenchymal stem cells derived from multiple myeloma patients is associated with a blockade in the deactivation of the Notch signaling pathway. Leukemia. 2012;26(12):2546–2549. doi: 10.1038/leu.2012.126. [DOI] [PubMed] [Google Scholar]
- 7.Li GG, Li L, Li C, et al. Influence of up-regulation of Notch ligand DLL4 on biological behaviors of human gastric cancer cells. World J Gastroenterol. 2013;19(28):4486–4494. doi: 10.3748/wjg.v19.i28.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guijarro MV, Dahiya S, Danielson LS, et al. Dual Pten/Tp53 suppression promotes sarcoma progression by activating Notch signaling. Am J Pathol. 2013;182(6):2015–2027. doi: 10.1016/j.ajpath.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Dong Y, Zhu N, et al. MicroRNA-139-5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014;13:124. doi: 10.1186/1476-4598-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Ahn YH, Gibbons DL, et al. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121(4):1373–1385. doi: 10.1172/JCI42579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY, Chan DW. Epigenetic silencing of microRNA-199b-5p is associated with acquired chemoresistance via activation of JAG1-Notch1 signaling in ovarian cancer. Oncotarget. 2014;5(4):944–958. doi: 10.18632/oncotarget.1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16(17):2252–2263. doi: 10.1101/gad.988902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Kumar RM, Biggs VJ, et al. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev Cell. 2011;21(3):575–588. doi: 10.1016/j.devcel.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shames DS, Girard L, Gao B, et al. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. Tumour Biol. 2012;33:287–296. doi: 10.1371/journal.pmed.0030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rauch TA, Wang Z, Wu X, Kernstine KH, Riggs AD, Pfeifer GP. DNA methylation biomarkers for lung cancer. Tumour Biol. 2012;33(2):287–296. doi: 10.1007/s13277-011-0282-2. [DOI] [PubMed] [Google Scholar]
- 16.Bonito NA, Borley J, Wilhelm-Benartzi CS, Ghaem-Maghami S, Brown R. Epigenetic regulation of the homeobox gene MSX1 associates with platinum-resistant disease in high-grade serous epithelial ovarian cancer. Clin Cancer Res. 2016;22(12):3097–3104. doi: 10.1158/1078-0432.CCR-15-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang T, Xu Y, Hou P. Identifying novel biomarkers of gastric cancer through integration analysis of single nucleotide polymorphisms and gene expression profile. Int J Biol Markers. 2015;30(3):e321–e326. doi: 10.5301/jbm.5000145. [DOI] [PubMed] [Google Scholar]
- 18.Chetcuti A, Aktas S, Mackie N, et al. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr Blood Cancer. 2011;57(6):950–957. doi: 10.1002/pbc.23003. [DOI] [PubMed] [Google Scholar]
- 19.Nagel S, Ehrentraut S, Meyer C, Kaufmann M, Drexler HG, MacLeod RA. Oncogenic deregulation of NKL homeobox gene MSX1 in mantle cell lymphoma. Leuk Lymphoma. 2013;55(8):1893–1903. doi: 10.3109/10428194.2013.864762. [DOI] [PubMed] [Google Scholar]
- 20.Sliwinski T, Synowiec E, Czarny P, et al. The c.469+46_56del mutation in the homeobox MSX1 gene – a novel risk factor in breast cancer? Cancer Epidemiol. 2010;34(5):652–655. doi: 10.1016/j.canep.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Kalyana S, Shanker, et al. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37:579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- 22.Zhou ZC, Ji ZZ, Wang Y, et al. TRIM59 is up-regulated in gastric tumors, promoting ubiquitination and degradation of p53. Gastroenterology. 2014;147:1043–1054. doi: 10.1053/j.gastro.2014.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Xiang T, Li L, Yin X, et al. The ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and apoptosis through stabilizing p53 and is frequently silenced in breast cancer. PLoS One. 2012;7(1):e29783. doi: 10.1371/journal.pone.0029783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Q, Huang H, Geiman TM, et al. Defective de novo methylation of viral and cellular DNA sequences in ICF syndrome cells. Hum Mol Genet. 2002;11(18):2091–2102. doi: 10.1093/hmg/11.18.2091. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Xu C, Long J, et al. E6 and E7 gene silencing results in decreased methylation of tumor suppressor genes and induces phenotype transformation of human cervical carcinoma cell lines. Oncotarget. 2015;6(27):23930–23943. doi: 10.18632/oncotarget.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senchenko VN, Kisseljova NP, Ivanova TA, et al. Novel tumor suppressor candidates on chromosome 3 revealed by NotI-microarrays in cervical cancer. Epigenetics. 2013;8(4):409–420. doi: 10.4161/epi.24233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huisman C, Wisman GB, Kazemier HG, et al. Functional validation of putative tumor suppressor gene C13ORF18 in cervical cancer by Artificial Transcription Factors. Mol Oncol. 2013;7(3):669–679. doi: 10.1016/j.molonc.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huisman C, van der Wijst MG, Schokker M, et al. Re-expression of selected epigenetically silenced candidate tumor suppressor genes in cervical cancer by TET2-directed demethylation. Mol Ther. 2015;24(3):536–547. doi: 10.1038/mt.2015.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen SY, Lin Y, Yu YQ, et al. MiR-506 acts as a tumor suppressor by directly targeting the hedgehog pathway transcription factor Gli3 in human cervical cancer. Oncogene. 2014;34(6):717–725. doi: 10.1038/onc.2014.9. [DOI] [PubMed] [Google Scholar]
- 30.Fan D, Wang Y, Qi P, et al. MicroRNA-183 functions as the tumor suppressor via inhibiting cellular invasion and metastasis by targeting MMP-9 in cervical cancer. Gynecol Oncol. 2016;141(1):166–174. doi: 10.1016/j.ygyno.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Fernàndez J. The genesis and evolution of homeobox gene clusters. Nat Rev Genet. 2005;6(12):881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- 32.Nassif A, Senussi I, Meary F, et al. Msx1 role in craniofacial bone morphogenesis. Bone. 2014;66:96–104. doi: 10.1016/j.bone.2014.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Lallemand Y, Nicola MA, Ramos C, Bach A, Cloment CS, Robert B. Analysis of Msx1; Msx2 double mutants reveals multiple roles for Msx genes in limb development. Development. 2005;132(13):3003–3014. doi: 10.1242/dev.01877. [DOI] [PubMed] [Google Scholar]
- 34.Saadi I, Das P, Zhao M, et al. Msx1 and Tbx2 antagonistically regulate Bmp4 expression during the bud-to-cap stage transition in tooth development. Development. 2013;140(13):2697–2702. doi: 10.1242/dev.088393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elder PA, Bell SM, Knowles MA. Deletion of two regions on chromosome 4 in bladder carcinoma: definition of a critical 750 kB region at 4p16.3. Oncogene. 1994;9(12):3433–3436. [PubMed] [Google Scholar]
- 37.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60(6):376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 38.Gosden RG, Feinberg AP. Genetics and epigenetics nature’s pen-and-pencil set. N Engl J Med. 2007;356(7):731–733. doi: 10.1056/NEJMe068284. [DOI] [PubMed] [Google Scholar]
- 39.Rowinsky EK. Signal events: cell signal transduction and its inhibition in cancer. Oncologist. 2003;8(suppl 3):5–17. doi: 10.1634/theoncologist.8-suppl_3-5. [DOI] [PubMed] [Google Scholar]
- 40.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol Rev. 2017;97(4):1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 41.Revet I, Huizenga G, Chan A, et al. The MSX1 homeobox transcription factor is a downstream target of PHOX2B and activates the delta-Notch pathway in neuroblastoma. Exp Cell Res. 2008;314(4):707–719. doi: 10.1016/j.yexcr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 42.Hu G, Lee H, Price SM, Shen MM, Abate-Shen C. Msx homeobox genes inhibit differentiation through upregulation of cyclin D1. Development. 2001;128(12):2373–2384. doi: 10.1242/dev.128.12.2373. [DOI] [PubMed] [Google Scholar]
- 43.Cohen B, Shimizu M, Izrailit J, et al. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res Treat. 2009;123(1):113–124. doi: 10.1007/s10549-009-0621-9. [DOI] [PubMed] [Google Scholar]
- 44.Park J, Park K, Kim S, Lee JH. Msx1 gene overexpression induces G1 phase cell arrest in human ovarian cancer cell line OVCAR3. Biochem Biophys Res Commun. 2001;281(5):1234–1240. doi: 10.1006/bbrc.2001.4474. [DOI] [PubMed] [Google Scholar]
- 45.Yun J, Pannuti A, et al. p53 Modulates Notch Signaling in MCF-7 Breast Cancer Cells by Associating With the Notch Transcriptional Complex Via MAML1. J Cell Physiol. 2015;230:3115–3127. doi: 10.1002/jcp.25052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ban J, Bennani-Baiti IM, Kauer M, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 2008;68:7100–7109. doi: 10.1158/0008-5472.CAN-07-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]