SUMMARY
To discover microRNAs that regulate sleep, we performed a genetic screen using a library of miRNA sponge-expressing flies. We identified 25 miRNAs that regulate baseline sleep; 17 were sleep-promoting and 8 promoted wake. We identified one miRNA that is required for recovery sleep after deprivation and 8 miRNAs that limit the extent of recovery sleep. 65% of the hits belong to human-conserved families. Interestingly, the majority (75%), but not all, of the baseline sleep-regulating miRNAs are required in neurons. Sponges that target miRNAs in the same family, including the miR-92a/92b/310 family and the miR-263a/263b family, have similar effects. Finally, mutation of one of the screen’s strongest hits, let-7, using CRISPR/Cas-9, phenocopies sponge-mediated let-7 inhibition. Cell-type-specific and temporally restricted let-7 sponge expression experiments suggest that let-7 is required in the mushroom body both during development and in adulthood. This screen sets the stage for understanding the role of miRNAs in sleep.
In Brief
To examine the role of microRNAs in sleep, Goodwin et al. screened a Drosophila microRNA sponge library, identifying 25 microRNAs that regulate sleep. The majority of these were from families well conserved in vertebrates. Let-7, a strong hit from the screen, has both adult and developmental roles.
Graphical Abstract
INTRODUCTION
Changes in sleep/wake status are accompanied by changes in gene expression. Sleep not only changes mRNA expression, it also alters levels of non-coding RNAs called microRNAs (miRNAs). miRNAs are small, 20- to 22-nt RNA transcripts that bind to mRNAs in conjunction with the RISC complex. miRNA binding to mRNA targets results in decreased protein synthesis, either via mRNA degradation or decreased translation (Hunt-zinger and Izaurralde, 2011). In mammals, miRNAs are differentially expressed during sleep and wake (Davis et al., 2007; Davis et al., 2012). Sleep fragmentation can change miRNA expression, and miRNA levels are altered in patients with sleep disorders, including narcolepsy and idiopathic hypersomnia (Holm et al., 2014). Many miRNAs also regulate the function of the circadian clock, which has a role in determining when sleep occurs (Chen et al., 2014a; Vodala et al., 2012). Furthermore, miRNAs are essential regulators of neuronal differentiation, development, and morphology and could therefore be required for the formation of sleep-regulating neuronal circuits (McNeill and Van Vactor, 2012). Thus, miRNAs are promising candidate regulators of sleep, but the role of miRNAs in sleep has not been comprehensively investigated.
We used Drosophila to perform a genetic screen to identify miRNAs that regulate sleep and sleep homeostasis. To inhibit miRNA function in Drosophila, we employed a library of transgenic miRNA inhibitors called miRNA sponges. We aimed to identify miRNAs that regulate sleep and identify miRNAs that regulate the homeostatic response to sleep loss. We then determined whether these sleep-regulating miRNAs are required in neurons and compared the effects of sponge-mediated miRNA inhibition with the effects of genetic ablation of the cognate miRNA genes. Finally, we characterized the role of one sleep-regulating miRNA, let-7, in greater depth and found that let-7 is required during both development and adulthood for its effects on sleep and that it is required in the mushroom body.
RESULTS
Screen Design
To investigate the role of miRNAs in sleep, we screened a library of 143 transgenic miRNA sponge (miRSP) fly lines containing tandem repeats of miRNA response elements (MREs) that bind to the cognate miRNA and prevent it from interacting with its endogenous mRNA target (Fulga et al., 2015). Sponges have multiple advantages over null mutations for screening: they partially inhibit miRNA function, allowing the study of miRNAs that are required for development or survival; miRNAs with similar seed sequences can be inhibited with a single sponge, preventing compensation; and sponge expression can be controlled using the GAL4/UAS expression system. Because sleep is regulated by many cell types (Artiushin and Sehgal, 2017), we initially inhibited miRNA activity ubiquitously by expressing sponges with tubulin-GAL4. We screened females because their daytime sleep levels are lower than those of males, making it easier to find miRSPs that increase sleep.
We measured sleep in 12 hr light:12 hr dark (LD) conditions using an operational definition of sleep as more than 5 min of inactivity (Hendricks et al., 2000; Shaw et al., 2000). Flies from each tubulin>miRSP line were divided into two subgroups: a control subgroup, which had uninterrupted sleep for 6 days, and a sleep deprivation (SD) subgroup, which had 3 days of baseline sleep followed by 12 hr of mechanical SD during the dark period (night) and then 2 days of recovery sleep (Figure 1A). For each round of screening, miRSP-expressing flies were compared with concurrently assayed UAS-scramble control sponge flies (Figure 1B).
Figure 1. miR Screen for Sleep.
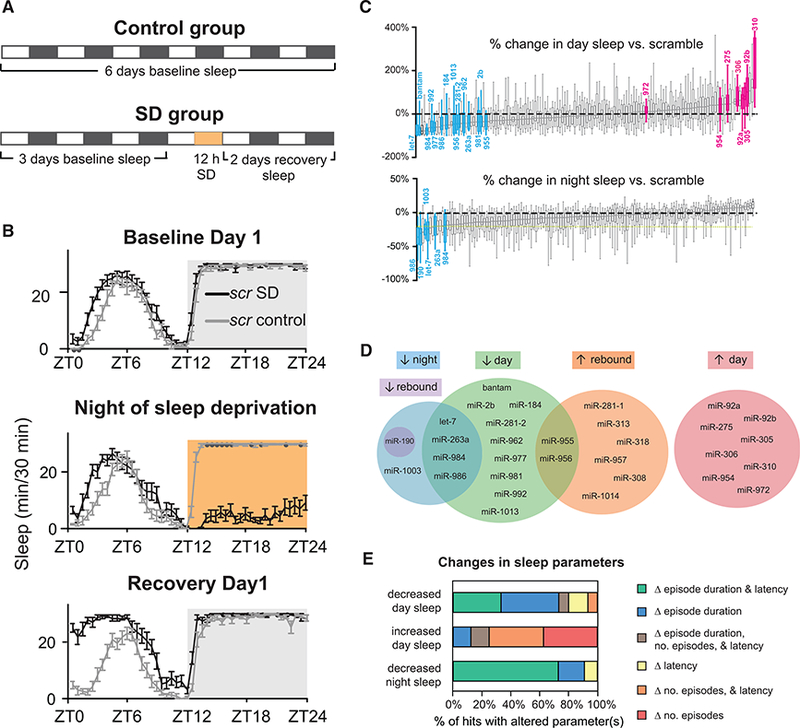
(A) Screen schematic. Sleep data were collected for 6 days under conditions of 12 hr light (white bars):12 hr dark (black bars). On day 4, an SD subgroup of each genotype was sleep-deprived for 12 hr at night (orange bar).
(B) Sleep of tubulin>scramble flies (control and SD subgroups) during a representative round of screening. Shown are minutes of sleep per 30 min across 24 hr for the first day of baseline sleep (top), the night of SD (center), and the first day of recovery sleep (bottom).
(C)Percent change in day (top) and night sleep (bottom) relative to scramble for all lines. miRSPs were considered hits when they varied from scramble by 20% or more. Blue, decreased sleep hit; pink, increased sleep hit; gray, non-hit. Values are shown in Table 1.
(D) Venn diagram illustrating the phenotypes hits.
(E) MiRNAs regulate sleep architecture. For baseline hits, changes in sleep architecture (relative to scramble) were identified.
Error bars represent SEM; whiskers represent maxima and minima.
An miRSP line was considered a ‘‘baseline sleep hit’’ when it met these criteria: (1) total day, night, or 24 hr sleep was significantly different (p < 0.05) from scramble flies; (2) the percent change in sleep was more than 20%, which represents two SDs of the scramble groups across multiple rounds of screening; and (3) both the control subgroup and the SD subgroup’s base-line sleep had to meet criteria (1) and (2). Because impaired locomotion could be misinterpreted as increased sleep, we confirmed that hits that decreased sleep had normal locomotion. For each hit, we performed follow-up assays that included UAS-miRSP/+ and tubulin-GAL4/+ controls.
To discover miRNAs that regulate sleep homeostasis, we examined the amount of sleep lost and recovered after SD. To adjust for differing levels of baseline sleep, we calculated the cumulative change in sleep versus each fly’s own baseline. This was compared with concurrently run scramble. To be considered a hit, recovery had to be either lower than scramble (under-recovery) or greater than scramble (hyper-recovery). We confirmed that changes in sleep after SD were not due to age or physical damage during shaking.
miRNAs Regulate Baseline Sleep
We identified 25 miRNAs that regulate baseline sleep. Figure 1C and Table 1 show results for light period (day) and dark period (night) sleep. MiRSPs affect sleep in both directions; 17 miRSPs decreased baseline sleep, whereas eight miRSPs increased baseline sleep. We did not detect any sponges that increased night sleep, likely because control flies slept near the maximum possible at night (Figure 1B). Four of the miRSPs that decreased sleep (let-7, 263a, miR-984, and miR-986) had effects on both day and night sleep (Figures 1C and 1D). Two (miR-190 and miR-1003) affected night sleep only. Conversely, nine lines caused specific decreases in day sleep. The finding that some miRNAs affect day sleep without also affecting night sleep (and vice versa) concurs with work showing that day and night sleep are regulated by different mechanisms (Ishimoto et al., 2012). Fifteen of the 25 miRNAs we found to regulate baseline sleep (60%) are conserved in humans (Table 1; Ibáñez-Ventoso et al., 2008).
Table 1.
miRNAs that Regulate Baseline Sleep
| Human Conserv. |
Control Group Change in Sleep versus Scramble |
SD Group Change in Sleep versus Scramble |
Control Group Sleep Architecture Changes |
Nsyb>miRSP Change in Sleep versus Scramble |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Screen Hit | 5’ Sequence | 24 hr | Day | Night | 24 hr | Day | Night | Day | Night | 24 hr | Day | Night |
| let-7.5p |
let-7, miR-98, miR-196a/b |
−42%* | −72%* | −27%* | −32%* | −49%* | −24%* | ↓ep dur., ↑ lat. |
↑freq., ↓ep. dur., ↑ lat. |
−73%* | −99%* | −65%* |
| miR-984.5p | − | −37%* | −77%* | −22%* | −35%* | −63%* | −23%* | ↓freq., ↑ep. dur., ↓ lat. |
↑freq., ↓ep. dur. | −23%* | −67%* | −13%* |
| miR-986.5p | miR-513c | −37%* | −41%* | −36%* | −36%* | −38%* | −35%* | ↓ep. dur. | ↑freq., ↓ep. dur., ↑ lat. |
−28%* | −57%* | −24%* |
| miR-977.3p |
let-7, miR-98, miR-202 |
−31%* | −51%* | −21%* | −24%* | −49%* | −12%* | ↓ep. dur., ↑ lat. |
↑freq., ↓ep. dur., ↑ lat. |
−20%* | 0% | −25%* |
| bantam.3p | miR-450b-3p | −29%* | −72%* | −16%* | −31%* | −62%* | −18%* | −43%* | −96%* | −31%* | ||
| miR-1003.3p | miR-342-3p | −24%* | −23%* | −25%* | −32%* | −30% | −33%* | ↑ lat. | ↑freq., ↓ep. dur., ↑ lat. |
−17%* | −5% | −20%* |
| miR-263a.5p | miR-569 | −24%* | −32%* | −21%* | −29%* | −46%* | −21%* | ↓ep. dur. | ↑freq., ↓ep. dur. | 7% | 20% | 2% |
| miR-190.5p | miR-190 | −23%* | 6% | −36%* | −19%* | 4% | −30%* | ↑freq., ↓ep. dur., ↓ lat. |
↑freq., ↓ep. dur., ↑ lat. |
−5% | 160%* | −32%* |
| miR-184.3p | miR-184 | −22%* | −35%* | −18%* | −26%* | −30%* | −25%* | ↓ep. dur. | ↑ lat. | −22%* | −11% | −25%* |
| miR-955.5p | - | −22%* | −37%* | −16%* | −23%* | −46%* | −12%* | ↓ep. dur., ↑ lat. |
↑freq., ↓ep. dur., ↑ lat. |
−15%* | −33% | −10%* |
| miR-956.3p | - | −21%* | −42%* | −14%* | −23%* | −58%* | −8% | ↓ep. dur. | − | −5% | −14% | −3% |
| miR-2b.3p | miR-499-3p | −20%* | −44%* | −11%* | −20%* | −42%* | −9%* | ↓ep. dur., ↑ lat. |
↓ep. dur., ↑ lat. |
−24%* | −38%* | −16%* |
| miR-981.3p | - | −19%* | −30%* | −16%* | −18%* | −26%* | −14%* | ↓freq., ↑ lat. | ↓ep. dur., ↑ lat. |
−21%* | −71%* | −8% |
| miR-1013.3p | - | −18%* | −36%* | −13% | −22%* | −47%* | −12% | ↓ep. dur. | - | −10% | −3% | −12%* |
| miR-992.3p | - | −18%* | −56%* | −7% | −12% | −32%* | −6% | - | −5% | −16% | −1% | |
| miR-281-2.5p |
miR-146a, miR-146b-5p, miR-9, miR-320 |
−13%* | −37%* | −3% | −14%* | −36%* | −4% | ↓Yep. dur., ↑ lat. |
||||
| miR-962.5p | - | −11% | −30%* | −5% | −14%* | −34%* | −7% | ↑ lat. | - | −13% | −23% | −11% |
| miR-972.3p | - | 1% | 35%* | −13%* | 4% | 27%* | −7% | ↑freq. | - | −24%* | −20% | −25%* |
| miR-954.5p | - | 8% | 39%* | −5% | 9% | 36%* | −4% | ↑freq. | - | 4% | 64%* | −15%* |
| miR-275.3p | - | 10% | 60%* | −6% | 10% | 47%* | −4% | ↑freq. | - | −1% | 37%* | −11%* |
| miR-92a.3p |
miR-25, miR-32, miR-92a/b, miR-363, miR-367, miR-866-5p |
15% | 56%* | −5% | 11%* | 46%* | −5% | ↑freq., ↓ep. dur., ↓ lat. |
−36% | −60%* | −25%* | |
| miR-306.5p | miR-450b-3p | 15%* | 82%* | 2% | 20%* | 87%* | 5% | ↑ep dur. | - | 2% | 17% | −2% |
| miR-92b.3p |
miR-25, miR-32, miR-92a/b, miR-363, miR-367, miR-866-5p |
20% | 101%* | −5% | 4%* | 32%* | −8% | ↑freq., ↓ lat. | - | −5% | 12%* | −10%* |
| miR-305.3p | - | 30%* | 79%* | 13%* | 17%* | 65%* | 1% | ↑freq., ↓ lat. | - | −1% | 34%* | −9%* |
| miR-310.3p |
miR-25, miR-32, miR-92a/b, miR-363, miR-367, miR-866-5p |
45%* | 244%* | 7% | 10%* | 46%* | −6% | ↑freq., ↑ep. dur., ↓ lat. |
- | −12%* | −21%* | −10%* |
Tub-Gal4-driven UAS-miRSP female animals were assayed for sleep in LD, and total sleep and sleep structure parameters were compared with the Tub-Gal4>scramble control assayed on the same day to identify lines that affect sleep. Only lines in which sleep (day, night, or 24 hr) changed by more than 20% (italic typeface) were considered hits. The control subgroup was analyzed for significant (p < 0.05) changes in sleep parameters, including episode duration (ep. dur.), frequency/number of episodes (freq.), and latency to sleep (lat.). Arrows indicate the direction of change for each sleep parameter. Secondary screening with nsyb-Gal4 was carried out to determine whether the miRNA acts in neurons.
p < 0.05, Mann-Whitney test with comparison with the same-day scramble control. Human homologs are listed as presented by Ibáñez-Ventoso et al. (2008).
miRNAs Regulate Sleep Structure
Total sleep in a given time period depends on the number of sleep episodes, episode duration, and the delay before sleep initiation (sleep latency). For miRSPs that affected baseline sleep, we observed changes in all structural parameters (Figure 1E; Table 1). Table 1 reports all changes in parameters, whereas Figure 1E reports only changes that could be causative of the observed change in total sleep.
Increased day sleep was more often associated with an increase in episode number (occurring in 88% of increased day sleep hits) rather than an increase in episode length (25% of increased day sleep hits). Conversely, decreased day sleep was more often associated with shorter episode length (80% of decreased day sleep hits) than with fewer sleep episodes (13% of decreased day sleep hits). miRSPs that affected night sleep, on the other hand, tended to primarily affect latency and episode duration. For all of the miRSPs that reduced night sleep, except for miR-184, sleep episode length was reduced. In no case was the amount of night sleep reduced because there were fewer sleep episodes.
miRNAs Regulate Sleep Homeostasis
We discovered nine miRNAs that affect sleep homeostasis. Only one (miR-190) was required for rebound sleep (Figure 2A). Most of the hits (8 of 9) appear to function as negative regulators. Figure 2B shows data for miR-956SP, which is representative. After SD, these flies recover significantly more sleep than scramble. Figure 2C shows the difference in cumulative change in sleep relative to scramble at 36 hr post-SD for all lines that alter recovery. Most of the homeostasis hits did not have significant effects on baseline sleep. There were three exceptions, miR-956SP and miR-955SP, which reduced baseline day sleep but increased rebound sleep, and miR-190SP, which reduced baseline for both night and rebound sleep (Figure 2B). Of the nine miRNAs that were found to regulate homeostasis, six (67%) are conserved in humans (Ibáñez-Ventoso et al., 2008).
Figure 2. MiRNAs Regulate Rebound Sleep.
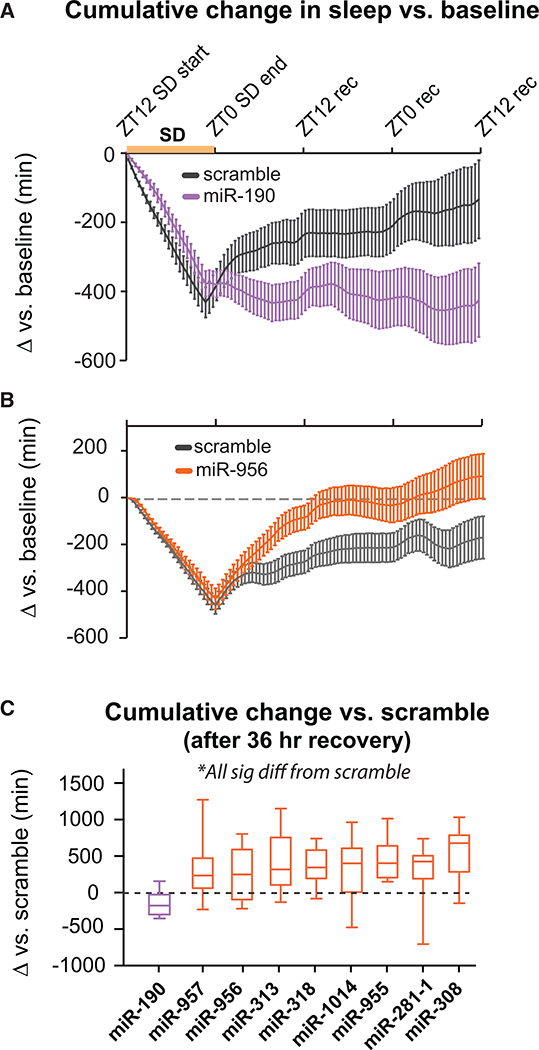
(A)Cumulative change in sleep for miR-190SP flies versus scramble.
(B)Cumulative change in sleep for miR-956SP flies versus scramble.
(C)Difference in cumulative change in sleep between miRSP flies and scramble (in minutes) 36 hr after SD ended.
Purple, decreased rebound; orange, increased rebound. All lines were significantly different from scramble.
miRNAs Act Predominantly in Neurons to Regulate Sleep
To determine where sleep-regulating miRNAs are acting, we inhibited them specifically in neurons using a panneuronal GAL4 (nsyb-GAL4). Of the 24 hits we assayed (Table 1), three (miR-263a, miR-92b, and miR-956) failed to show a sleep phenotype. In 11 cases (46%), nsyb>miRSP was able to recapitulate the tub>miRSP phenotype with a similar magnitude effect. Seven miRSPs (29%) produced weak effects in the same direction as tubulin-Gal4 when expressed in neurons. The remaining three hits had sleep phenotypes that were qualitatively different when expressed with nsyb-GAL4 compared with tubulin-Gal4. Ubiquitous expression of miR-972SP increased daytime sleep; however, expression of this sponge in neurons decreased both day and night sleep. Inhibition of miR-92a and miR-310, which strongly increased daytime sleep in the initial screen, inhibited sleep when limited to neurons. These differences suggest that these three miRNAs may have multiple sites of action with different effects on sleep.
Validation of Hits Using Endogenous Mutants
We compared phenotypes generated by sponge-mediated miRNA inhibition with mutation of the cognate miR gene. For sponges that inhibit miRNAs with unique seed sequences, we would expect sponge to produce similar effects as mutation. Conversely, if a miRNA has multiple homologs that can compensate for its loss, then mutation may have weaker or different effects than sponge. Many miRNA genes are located in clusters with other miRNAs; therefore, some mutants were not selective for the miRNA we wanted to assess. In addition, a number of the hits from our screen have functions essential for viability. In total, we were able to find specific, viable null mutants for only 9 of our 25 hits, highlighting one aspect of the utility of miRSPs as an approach.
Endogenous mutations in mir-184, mir-956, mir-986, let-7, and mir-1003 caused significant or borderline (p = 0.06) changes in total sleep in the same direction as ubiquitous expression of the sponge (Figures 3A–3F). This suggests that the sponges are specific and that our screen correctly identified a role for these miRNAs in sleep. For two mutants, mir-2b-1 and mir-955, we observed no significant change in sleep. It is possible that the miR-2b sponge inhibits both miR-2b-1 and miR-2b-2 and that ablation of mir-2b-1 alone is insufficient to reduce sleep or that differences between miRNA sponges and endogenous mutants are due to different effects on the major versus the minor miR strand (Discussion).
Figure 3. Knockout of miRNA Genes Phenocopies miRSP.
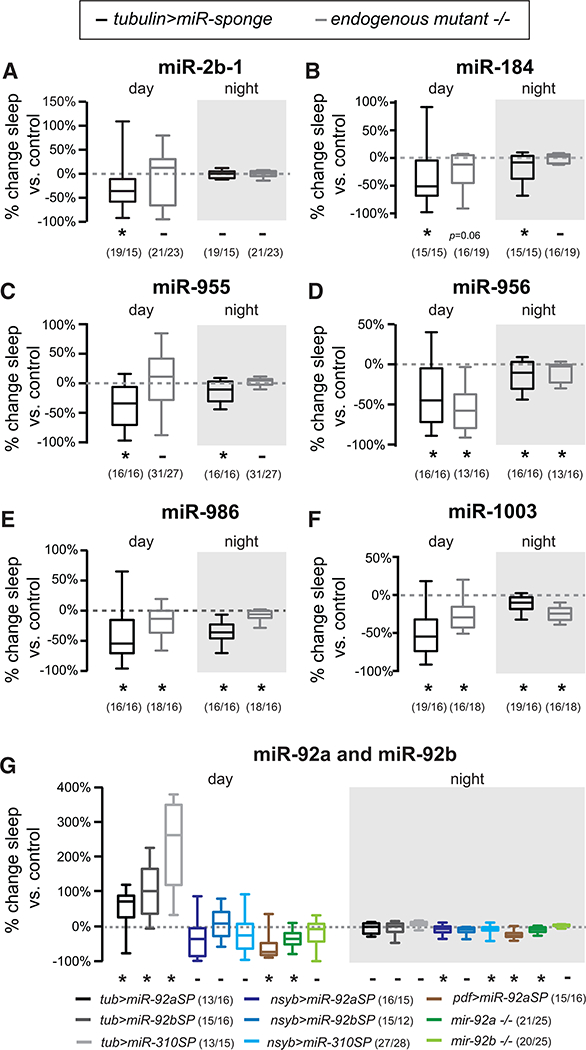
(A-F) Change in total day and night sleep for six miRNAs: miR-2b-1 (A), miR-184 (B), mir-955 (C), miR-956 (D), miR-986 (E), and miR-1003 (F). Change in tubulin>miRSP sleep relative to scramble is plotted in black. Change in knockout flies’ sleep relative to genetic control is plotted in gray.
(G)Percent change in day and night sleep relative to scramble or genetic background control after inhibition of miR-92 family members. miRNAs were either inhibited by expression of sponges via tubulinGal4 (gray plots), nsybGal4 (blue plots), or PDFGal4 (brown) or through knockout of the miR gene (green plots).
Whiskers represent maxima and minima. *p < 0.05, -p > 0.06 using Mann-Whitney test. n values are reported next to the genotype. See also Figure S1.
miRNA Family Members Have Similar Effects on Sleep
In our screen, we found that miRNA sponges designed to target miRNAs in the same family produce similar effects (Figure 3G, miR-92 family; Figure S1, miR-263 family). The miR-92a and miR-92b sponges’ MREs differ only by four nucleotides; miR-310 is also in this family. These sponges all increased day sleep with tubulinGal4 (Table 1). However, none of these sponges caused increased day sleep when expressed in neurons (Figure 3G). In fact, miR-92aSP and miR-310SP reduced 24-hr and night sleep when expressed in all neurons, consistent with published reports of miR-92a actions in PDF+ circadian neurons (Chen and Rosbash, 2017). Because these similar sponges produced similar effects on sleep in our cell specificity experiments, it suggests that our methodology can reliably interrogate the effects of miRNA families on sleep.
To determine which of the miRNAs in this family could be responsible for these effects, we measured sleep in homozygous nulls for both mir-92a and mir-92b. Because mir-310 is located in a genomic locus with multiple miRNAs, we were not able to investigate its role using mutants. Similar to inhibition of miR-92a in neurons, knockout of mir-92a reduced sleep (Figure 3G). In contrast, mir-92b nulls had no significant change in sleep. These results suggest that miR-92a is likely acting in PDF+ neurons (Figure 3G) to promote 24-hr per night sleep.
Let-7 Promotes Sleep in LD and DD
Let-7SP had some of the strongest effects on sleep. Let-7 has well-characterized roles in neuronal development in mammals and Drosophila. Let-7’s role in sleep regulation in Drosophila could be related to its previously described roles in circadian rhythms (Chen et al., 2014a) and/or development of the mushroom body (Kucherenko et al., 2012; Wu et al., 2012), a structure known to regulate sleep. The Drosophila let-7 gene is located in a cluster (the let-7 complex) with two other miRNAs, mir-100 and mir-125. Knockout of all three miRNAs is lethal at the pupal stage (Sokol et al., 2008), and the functions of the three encoded miRNAs are intimately intertwined (Caygill and Johnston, 2008; Chawla et al., 2016); hence, the effects of let-7 complex deletion mutations are difficult to properly interpret. To avoid these problems, we generated let-7-specifc mutations using CRISPR/Cas9 technology. We chose to examine two mutant alleles, let-7Δ12−15 and let-7Δ13−43, which both disrupt the let-7.5p seed sequence but leave the let-7.3p, mir-100, and mir-125 sequences intact.
To ensure that changes in sleep are due to let-7 mutation, we tested these alleles in trans with one another or a previously characterized deletion that removes the entire let-7 complex (let-7CKO; Sokol et al., 2008). As shown in Figure 4A, all three let-7 trans-heterozygote groups (let-7CKO/let-7Δ13−43, let-7CKO/let-7Δ12−15, and let-7Δ13−43/let-7Δ12−15) had reduced day sleep relative to controls in LD. In DD, both subjective day and subjective night sleep were reduced in all three trans-heterozygotes (Figure 4B) These effects are not limited to female flies (Figure S2). Additionally, flies with only one mutated copy of let-7.5p (jet-7Δ13−43/+ and let-7Δ12−15/+) also showed reduced sleep. Attempts to rescue mutants by expression of UAS-let7 under the control of nsyb-GAL4 were unsuccessful because of lethality of let-7 overexpression (data not shown).
Figure 4. Let-7 Has Developmental and Adult-Specific Roles.
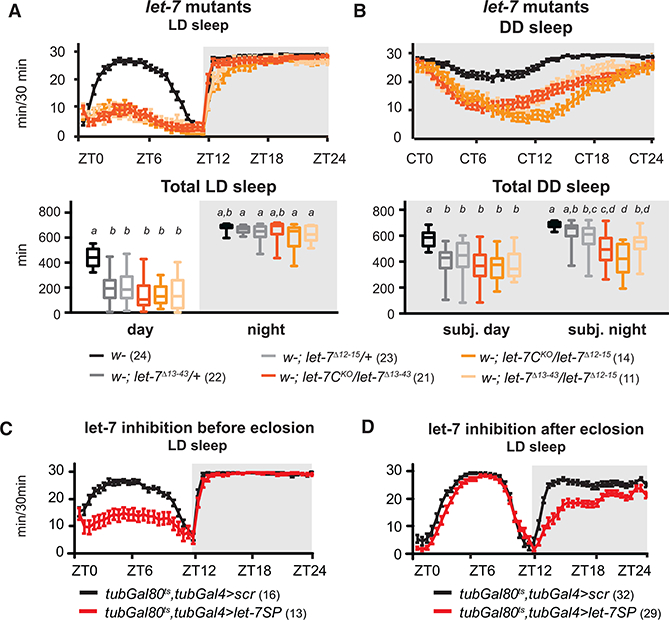
(A and B) Deletions of the entire let-7 complex (let-7C), most of the let-7.5p sequence (let-7Δ13−43), or the let-7.5p seed sequence (let-7Δ12−15), assayed as heterozygotes (gray plots) or trans-heterozygotes (orange plots), decreased day sleep in LD and DD.
(A)Sleep per 30 min (top) and total day and night sleep (bottom) in let-7 mutants in LD. let-7 trans-heterozygotes had reduced day sleep (Kruskal-Wallis test, p < 0.05).
(B)Sleep per 30 min (top) and total sleep (bottom) in let-7 mutants in DD. All trans-heterozygous let-7 mutants had reduced sleep (Kruskal-Wallis test, p < 0.05).
(C)Let-7SP was restricted to the larval and pupal stages by shifting tubGal4, tubGal80ts > let-7SP flies from 29°C to 18°C after eclosion. Day sleep was significantly decreased (Mann-Whitney test, p < 0.05).
(D)Let-7SP expression was restricted to adulthood by shifting tubGal4, tubGal80ts > let-7SP from 18°C to 29°C after eclosion. Night sleep was significantly decreased (Mann-Whitney test, p < 0.05).
Error bars represent SEM; whiskers represent maxima and minima. Numbers are reported next to the genotype. See also Figures S2 and S3.
Both sponge-mediated let-7 inhibition in neurons (Table 1) and mutation of the let-7 gene reduced sleep. To confirm that these manipulations act in the same genetic pathway, we expressed let-7SP neuronally in a let-7 mutant background. If these two manipulations affect the same pathway, then we should not see a greater reduction in sleep when they are combined. Indeed, sponge-mediated inhibition of let-7 on a let-7 mutant background does not further sleep relative to either manipulation alone (Figure S3). These results suggest that the let-7 sponge and CRISPR mutations specifically target let-7.
Let-7 Has Developmental and Adult Roles in Sleep
To determine whether let-7’s role in sleep was due to a developmental function or whether it had an ongoing role in the adult nervous system, we employed a temperature-sensitive variant of Gal80 (McGuire et al., 2004) to control the timing of let-7SP expression. Gal80ts blocks Gal4 activity at low temperature, but at high temperature, Gal80ts repression is lost. Tubulin-Gal80ts; tubulin-Gal4 > let-7SP flies were grown at 29°C and shifted to 18°C after eclosion to inhibit let-7 activity only during the larval and pupal stages. This eliminated the effect of let-7 inhibition on night sleep but did not eliminate the effect of let-7 inhibition on day sleep (Figure 4C). Conversely, inhibition of let-7 only during adulthood reduced night sleep without affecting day sleep (Figure 4D). This suggests that let-7 acts in development to ultimately affect adult day sleep but may also be required during adulthood to promote nighttime sleep.
Let-7’s Role in Sleep Is Independent of the Clock
One potential neuronal locus of let-7 action was the clock. Chen et al. (2014a) reported that overexpression of the let-7 complex in PDF+ neurons lengthens the period of circadian locomotor rhythms; overexpression of let-7 alone had a weaker effect. Surprisingly, knockout of the let-7 complex did not alter the circadian period (Chen et al., 2014a). In agreement with this study, we found that nsyb>let-7SP flies had a lower rhythmicity index (RI; Levine et al., 2002) compared with nsyb>scramble (Table 2). In contrast to this study, however, we found that let-7 inhibition (not gain of function) lengthened the circadian period. Importantly, when arrhythmic flies (with RI < 0.2) were excluded from sleep analysis, nsyb>let-7SP flies still displayed less sleep than nsyb>scramble, indicating that the changes in sleep were not due to loss of rhythmicity (Figure S4A). Similarly, when let-7SP was expressed in PDF+ neurons, we observed no change in sleep in LD and no change in period or RI (Figure S4B; Table 2). Importantly, not all let-7 trans-heterozygous mutant fly lines had a change in RI or period (Table 2), but all let-7 trans-heterozygous mutants did have reduced sleep (Figure 4A). Together, these results suggest that let-7’s role in sleep is separable from its role in circadian rhythms. The differences between our results and those of Chen et al. (2014a) likely lie in their utilization of mutants and overexpression constructs that affect expression of both the 5p and 3p forms of let-7 in addition to mir-100 and mir-125.
Table 2.
Circadian Parameters for let-7miRSP and let-7 Mutants
| Period | RI | % Rhythmic | |
|---|---|---|---|
| nsyb>scramble | 24.29 ± 0.09 | 0.44 ± 0.02 | 100% |
| nsyb>let-7SP | 25.21 ± 0.24* | 0.31 ± 0.04* | 64% |
| Pdf>scramble | 24.12 ± 0.06 | 0.39 ± 0.02 | 94% |
| Pdf>let-7SP | 24.15 ± 0.13 | 0.35 ± 0.02 | 85% |
| w | 24.42 ± 0.28 | 0.28 ± 0.02 | 75% |
| let-7C/let-7Δ13−43 | 24.76 ± 0.25 | 0.25 ± 0.03 | 62% |
| let-7C/let-7Δ12-15 | 24.96 ± 0.28 | 0.29 ± 0.02 | 93% |
| let-7Δ13−43/let-7Δ12−15 | 24.23 ± 0.32 | 0.19 ± 0.05* | 45% |
Animals of the indicated genotypes were entrained in LD and released into DD to assay clock function. Period and rhythmicity index (RI; Levine et al., 2002) were calculated. Flies with RI > 0.2 were considered rhythmic. nsyb>let-7SP was significantly different from its genetic control (nsyb>scramble) for period and RI. let-7Δ13−43/let-7Δ12−15 mutants were significantly different from the w genetic control for RI.
p < 0.05, t test (period) or Mann-Whitney test (RI). See also Figure S4.
Let-7 Is Required in Mushroom Body Neurons for Normal Sleep
Although inhibition of let-7 in PDF+ neurons did not change sleep, our data (Table 1) suggest that it acts in neurons. To determine whether the MBs were the neuronal locus of let-7 action, we used VT030559Gal4 (a strong pan-MB driver) to express let-7SP. Both LD and DD sleep were significantly reduced (Figures 5A and 5B), although the reduction during the day was not as great as that seen for let-7 nulls. This could be due to residual activity of let-7 in the mushroom body (MB) because sponges are not knockouts, or it could suggest that there are other sleep-regulating brain regions that require let-7.
Figure 5. Let-7 Is Required in MB Neurons to Promote Sleep.
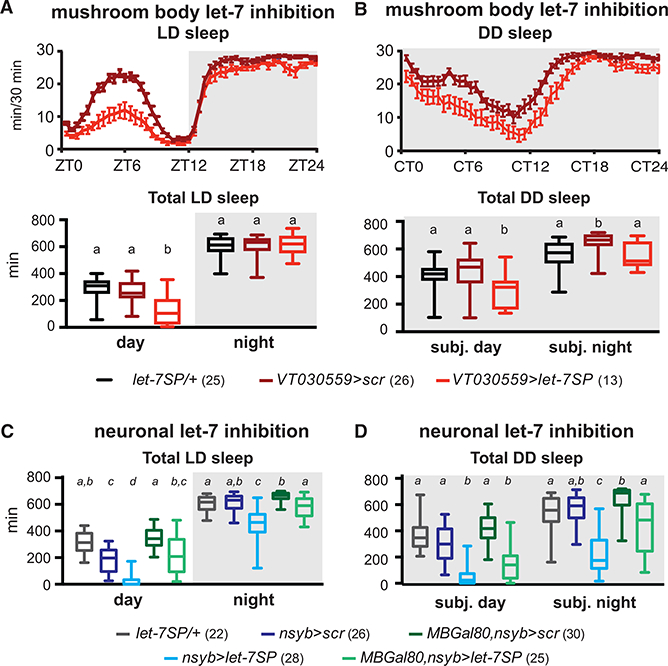
(A) Let-7SP expressed in Kenyon cells (VT030559Gal4) reduces day sleep in LD.
(B) MB-specific expression of let-7SP reduces both day and night sleep in DD.
(C) Let-7SP was expressed in all neurons (nsybGal4) or in all neurons except MB neurons (nsybGal4, MBGal80). The effect of panneuronal inhibition (blue) was partially blocked by restoring let-7 in the MB(green, Kruskal Wallis test, p < 0.05).
(D) Panneuronal let-7 inhibition decreased subjective day and night sleep in DD (blue). Restoring let-7 to the MB reversed the effect of neuronal inhibition (green, Kruskal-Wallis test, p < 0.05).
Error bars represent SEM; whiskers represent maxima and minima.
To test the contribution of the MB to the panneuronal let-7 phenotype (Table 1), we combined nsybGal4-driven expression of let-7SP with MB-specific repression of Gal4 activity using MB-Gal80. When let-7 is inhibited panneuronally, day and night sleep are reduced to levels comparable with let-7 nulls in both LD and DD (Figure 5C). Restoring let-7 activity to the MB partially reversed the effects of panneuronal inhibition on day and night sleep in LD and subjective night sleep in DD (Figure 5D). These data suggest that, although MBs are an important locus of let-7 action, there are likely other neurons that contribute. In support of this, experiments driving let-7SP with ChaTGal4, a driver that expresses both in MBs and other neurons, produced a phenotype that was very similar to nsybGal4 (data not shown).
DISCUSSION
We identified 25 miRNAs that regulate aspects of sleep. Although little is known in general about the role of miRNAs in human sleep, increases in the mammalian homologs of four of the sleep-promoting hits from our screen are associated with low-sleep states (Davis et al., 2007, 2012; Holm et al., 2014; Matos et al., 2014). These studies suggest that there may be conservation of function for certain miRNAs in regulation of sleep across phylogeny.
Unexpectedly, our screen identified eight miRNAs that negatively regulate rebound sleep but only one miRNA (miR-190) that is required. This raises a question: why might it be advantageous to limit rebound? Rebound sleep after SD exists within a hierarchy of multiple competing behaviors (e.g., foraging, mating, egg laying), and sleep is modulated by multiple physiological and sensory inputs (e.g., hunger, circadian rhythms, light, mating status, and temperature). These miRNAs may help prioritize some other behavior over rebound sleep. Alternatively, these miRNAs might be sensors of recovery processes that occur during rebound and function to prolong sleep when recovery is ineffective. Future studies are needed to determine the role these miRNAs play in sleep homeostasis.
There are many important differences between sponge-mediated inhibition and mutation of miRNA genes, but these strategies provide complementary insights into miRNA function. miRNA sponges have hypomorphic effects, whereas gene knockouts are nulls. Sponges, therefore, may fail to show effects when complete loss of the miRNA is required for a phenotype. On the other hand, sponges can be used to investigate the function of miRNAs that are essential for development or survival. Sponges inhibit multiple miRNAs in the same family, which can create ambiguity around which miRNA is causative; however, they eliminate the possibility that homologous miRNAs mask a phenotype. MiRNA sponges target only the 5p or 3p miR sequence, whereas knockouts typically eliminate both mature strands. Finally, miRNA sponges are transgenes, and coupling with the Gal4/UAS system allows interrogation of when and where miRNAs are required.
In the case of let-7, sponge technology allowed us to identify the MB as one of let-7’s sites of action and to discover that let-7 plays distinct roles in development and adulthood, whereas let-7 mutants allowed us to verify that let-7.5p itself (and not other homologous miRNAs inhibited by the sponge) increases sleep. Previously described let-7 complex knockout approaches also disrupt let-7’s neighboring miRNAs, miR-100 and miR-125; therefore, previous attempts to specifically study let-7 relied on transgenic rescue of miR-100 and miR-125. This approach can cause neomorphic effects because of overexpression, particularly because miR-125 and let-7 have some mRNA targets in common (Chawla et al., 2016). To avoid this issue, we generated mutations in let-7 that specifically affect the 5p sequence without affecting mir-100 or mir-125. These mutations, either in trans with one another or with a let-7 complex knockout, reduce sleep in both males and females under LD and DD conditions.
Although we found some evidence to support the idea that let-7 regulates circadian rhythms (Table 2), not all of our results were consistent with the let-7 complex deletion literature (Chen et al., 2014a). Most importantly, we found that let-7 is not required in PDF+ neurons for its role in sleep and that reductions in sleep caused by let-7 inhibition were not due to loss of circadian rhythmicity (Figure S4). Therefore, let-7 appears to play a role in sleep that is independent of any effects it may have on circadian rhythms.
Our evidence suggests that let-7’s role in sleep is partially mediated through the MB, a region known to be involved in regulation of sleep (Joiner et al., 2006; Pitman et al., 2006). Inhibition of let-7 in MB neurons reduced day and night sleep, and restoring let-7 activity in the MB in nsyb>let-7SP flies partially reversed the effects of panneuronal knockdown. This is consistent with let-7’s known role in regulation of MB lobe development (Kucherenko et al., 2012; Wu et al., 2012) and our finding that let-7 is required during development for its effects on day sleep. Recent work has implicated the α’, β’, and γ lobes of the MB in sleep regulation (Sitaraman et al., 2015). Our results suggest that let-7 may affect the function of these neuronal subsets. Comparison of the magnitude of inhibition of sleep using MB drivers with that of let-7 mutants and panneuronal or ChaTGal4-driven inhibition, however, argues that there are also other neurons that are important.
There may also be an acute function of let-7 in adults. let-7 levels change with time of day in adult Drosophila (Chen et al., 2014a) and are modulated by SD in adult mammals (Davis et al., 2007), suggesting a potential role for let-7 in the adult brain. We found that let-7SP expression in adulthood reduced night sleep in females, although night sleep did not significantly change in males or let-7 mutants. Let-7 may therefore have a weak female-specific role in night sleep in the fly.
Sleep is regulated by many cell types, including neurons, glia, and endocrine cells. Consistent with this idea, roughly a quarter of our hits were not required in neurons for their effects. We identified one miRNA family, miR-263a/b, that appears to be required in glia rather than neurons for its effects on sleep. This is of particular interest because a complementary miRSP-based genetic screen has identified multiple miRNAs that function in adult glial astrocytes to regulate circadian rhythmicity (You et al., 2018). However, determining where a miR is required for its effects on sleep is only a starting point for understanding the specific genetic pathways. Because miRNAs regulate many cellular functions and aspects of development, the miRNAs identified here could regulate sleep through widely divergent mechanisms. Future studies will be required to dissect those mechanisms and, in particular, to identify the relevant sleep-related mRNA targets, which could shed light on the mechanisms of sleep and sleep homeostasis.
EXPERIMENTAL PROCEDURES
Drosophila Husbandry
Drosophila were raised on cornmeal-dextrose-yeast agar medium at 25°C in a 12-hr LD cycle with two exceptions. To enhance viability, after eclosion, let-7 mutants were housed and assayed at 18°C. To restrict let-7 sponge expression to development or adulthood with tubulinGal80ts, flies were reared and assayed at either 18°C or 29°C as described. For the screen and for all other experiments (except for Figure S2), we assayed 3- to 5-day-old mated females. In experiments involving let-7 mutants, we expanded the age range to 3–7 days old using similarly aged controls.
Drosophila Lines
UAS-miRNA sponge lines (w; UAS-mCherry-sponge/CyO; UAS-mCherry sponge/TM6b) have been described previously (Fulga et al., 2015). Gal4 strains included w;;pTub-Gal4DCR1014/TM3,w;; pnsyb-Gal4/TM6b, w; pTub-Gal80ts, w; MB-Gal80 (Thum et al., 2007), w; pPDF-Gal4, repo-Gal, and w;; VT030559Gal4 (VDRC ID# v206077). miRNA knockout alleles included w; {wMw.hs}mir-2b-1KO (Bloomington #58915), w; {wmW.hs)mir-184KO (BL#58896), w; mir-263bΔ (BL#58903), w; {wmW.hsGal4}mir-955KO (BL#58940), w; {wmW.hsGal4}mir-956/TM3,Ser (BL#58941), w;{TI}mir-986KO (BL#58959), and w; {TI}mir-1003KO (BL#58883), described by Chen et al. (2014b), and let-7CKO1, described by Sokol et al. (2008). Mutants for miR-92a and miR-92a (Yuva-Aydemir et al., 2015) were obtained from Fen-Biao Gao (University of Massachusetts [UMASS] Medical School).
Circadian and Sleep Assays
Sleep was assayed as described previously (Do-nelson et al., 2012) using the Drosophila activity monitoring (DAM) system (Trikinetics, Waltham, MA). For flies in the SD subgroup, 3 days of base-line sleep were recorded, followed by 12 hr SD. During SD, flies were shaken for 2 s every 10 s, beginning at Zeitgeber time (ZT)12 and ending at ZT0. Recovery sleep was measured for 2 days. The strength of shaking was adjusted so that scramble flies had pronounced rebound sleep on day 1 after SD but returned to baseline sleep levels by day 2. To measure circadian rhythms, mated female flies were entrained for 3 days in LD and then switched to dark:dark (DD) for 6 to 7 days.
Generation of let-7 Mutants
To generate specific let-7 mutants, a 20-nt guide RNA (gRNA) (5՛-CTC TGGCAAATTGAGGTAGT-3՛), designed to target the AGG PAM sequence located in the +20 position of pre let-7, was ligated into the dU6-J28gRNAvector (Port et al., 2014). The vector was injected into w;;P{CaryP}attP2 (Rainbow Transgenic Flies).Transgenic flies were crossed with yw, {pActin5c-Cas9, w+} (BL#54590), and male flies with putative mutations in let-7 were housed singly with w; Sp/CyO virgin females. Progeny were crossed and homozygotes and screened by PCR and sequencing of let-7 locus.
Multiple viable let-7 alleles were isolated. let-7Δ12−15, a4-bp (GAGG) deletion spanning the +2–5 position of let-7.5p removes half of the genomic region encoding let-7.5p’s seed. let-7Δ13−43, a larger 31-bp deletion with a 2-bp (TA) insertion that spans positions 13–41 of the let-7 hairpin, removes 20 of 22 bp of the mature let-7.5p sequence but does not disrupt the genomic region encoding the let-7.3p strand. Both alleles were outcrossed at least 4 times to a Cantonized w line (wCS), and mutations were verified with PCR and sequencing. Let-7 mutant homozygotes had reduced viability and fecundity but were viable for up to a week after eclosion as adults.
Analysis Software
Sleep analysis was performed using an in-house MATLAB program called Sleep and Circadian Analysis MATLAB Program (SCAMP) (Donelson et al., 2012). Sleep recovery after SD was measured as the cumulative change in sleep relative to each fly’s own baseline. Circadian rhythmicity patterns were analyzed using SCAMP’s autocorrelation function (Levine et al., 2002). The height of the third peak is designated as the RI. We considered RI < 0.2 to be ‘‘arrhythmic.’’
Statistical Testing
For each sponge, the percent change in day, night, and 24-hr sleep relative to concurrently tested scramble flies was calculated. For other sleep parameters, data were tested for normality using a Kolmogorov-Smirnov test and then compared with scramble data using either t test or Mann-Whitney test. For experiments in which more than 2 groups were compared, Dunn’s multiple comparison tests were used. To compare circadian periods, we used ANOVA with Tukey post hoc testing. To compare RI values, we performed Kruskal-Wallis ANOVA with Dunn’s multiple comparison tests.
Controls for Locomotion- and Age-Related Artifacts
For hits that increased sleep, we calculated activity during waking epochs to rule out decreased locomotion. To rule out age as a factor in homeostasis hits, we examined the cumulative change in sleep in the non-SD subgroup across the last 60 hr of the experiment to confirm that sponge effects remained stable.
Supplementary Material
Highlights.
A genetic screen identifies sleep-regulating microRNAs in Drosophila
microRNAs regulate amount, structure and homeostatic regulation of sleep
Let-7 regulates sleep through developmental and adult-specific pathways
ACKNOWLEDGMENTS
This work was supported by NIH grants P01 NS090994 (to D.V.V. and L.C.G.) and F32 GM109531 and T32 NS7292–28 (to P.R.G.). We thank Stephen Alkins and Rob Jackson for comments on the manuscript.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.05.078.
AUTHOR CONTRIBUTIONS
Conceptualization, D.V.V., L.C.G., and P.R.G.; Methodology, D.V.V., T.A.F., and P.R.G.; Investigation, P.R.G., A.M., J.M., and M.H.; Writing - Original Draft, P.R.G.; Writing - Review and Editing, L.C.G. and D.V.V.; Supervision and Funding Acquisition, L.C.G. and D.V.V.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Artiushin G, and Sehgal A (2017). The Drosophila circuitry of sleep-wake regulation. Curr. Opin. Neurobiol 44, 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caygill EE, and Johnston LA (2008). Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr. Biol 18, 943–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla G, Deosthale P, Childress S, Wu YC, and Sokol NS (2016). A let-7-to-miR-125 MicroRNA Switch Regulates Neuronal Integrity and Lifespan in Drosophila. PLoS Genet. 12, e1006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, and Rosbash M (2017). MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat. Commun 8, 14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Liu Z, Li T, Zhang R, Xue Y, Zhong Y, Bai W, Zhou D, and Zhao Z (2014a). Regulation of Drosophila circadian rhythms by miRNA let-7 is mediated by a regulatory cycle. Nat. Commun 5, 5549. [DOI] [PubMed] [Google Scholar]
- Chen YW, Song S, Weng R, Verma P, Kugler JM, Buescher M, Rouam S, and Cohen SM (2014b). Systematic study of Drosophila micro-RNA functions using a collection of targeted knockout mutations. Dev. Cell 31, 784–800. [DOI] [PubMed] [Google Scholar]
- Davis CJ, Bohnet SG, Meyerson JM, and Krueger JM (2007). Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci. Lett 422, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CJ, Clinton JM, and Krueger JM (2012). MicroRNA 138, let-7b, and 125a inhibitors differentially alter sleep and EEG delta-wave activity in rats. J. Appl. Physiol (1985) 113, 1756–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson NC, Kim EZ, Slawson JB, Vecsey CG, Huber R, and Griffith LC (2012). High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the ‘‘tracker’’ program. PLoS ONE 7, e37250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, McNeill EM, Binari R, Yelick J, Blanche A, Booker M, Steinkraus BR, Schnall-Levin M, Zhao Y, DeLuca T, et al. (2015). A trans-genic resource for conditional competitive inhibition of conserved Drosophila microRNAs. Nat. Commun 6, 7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, and Pack AI (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. [DOI] [PubMed] [Google Scholar]
- Holm A, Bang-Berthelsen CH, Knudsen S, Kornum BR, Modvig S, Jen-num P, and Gammeltoft S (2014). miRNA profiles in plasma from patients with sleep disorders reveal dysregulation of miRNAs in narcolepsy and other central hypersomnias. Sleep (Basel) 37, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, and Izaurralde E (2011). Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet 12, 99–110. [DOI] [PubMed] [Google Scholar]
- Ibáñez-Ventoso C, Vora M, and Driscoll M (2008). Sequence relationships among C. elegans, D. melanogaster and human microRNAs highlight the extensive conservation of microRNAs in biology. PLoS ONE 3, e2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H, Lark A, and Kitamoto T (2012). Factors that Differentially Affect Daytime and Nighttime Sleep in Drosophila melanogaster. Front. Neurol 3, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joiner WJ, Crocker A, White BH, and Sehgal A (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. [DOI] [PubMed] [Google Scholar]
- Kucherenko MM, Barth J, Fiala A, and Shcherbata HR (2012). Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J. 31, 4511–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, and Hall JC (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci. 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos G, Scorza FA, Mazzotti DR, Guindalini C, Cavalheiro EA, Tufik S, and Andersen ML (2014). The effects of sleep deprivation on microRNA expression in rats submitted to pilocarpine-induced status epilepticus. Prog. Neuropsychopharmacol. Biol. Psychiatry 51, 159–165. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, and Davis RL (2004). Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci. STKE 2004, pl6. [DOI] [PubMed] [Google Scholar]
- McNeill E, and Van Vactor D (2012). MicroRNAs shape the neuronal landscape. Neuron 75, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitman JL, McGill JJ, Keegan KP, and Allada R (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, and Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, and Tononi G (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. [DOI] [PubMed] [Google Scholar]
- Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, and Nitabach MN (2015). Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr. Biol 25, 2915–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol NS, Xu P, Jan YN, and Ambros V (2008). Drosophila let-7 micro-RNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22, 1591–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum AS, Jenett A, Ito K, Heisenberg M, and Tanimoto H (2007). Multiple memory traces for olfactory reward learning in Drosophila. J. Neurosci 27, 11132–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodala S, Pescatore S, Rodriguez J, Buescher M, Chen YW, Weng R, Cohen SM, and Rosbash M (2012). The oscillating miRNA 959–964 cluster impacts Drosophila feeding time and other circadian outputs. Cell Metab. 16, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YC, Chen CH, Mercer A, and Sokol NS (2012). Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev. Cell 23, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Fulga TA, Van Vactor D, and Jackson FR (2018). Regulation of Circadian Behavior by Astroglial MicroRNAs in Drosophila. Genetics 208, 1195–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuva-Aydemir Y, Xu XL, Aydemir O, Gascon E, Sayin S, Zhou W, Hong Y, and Gao FB (2015). Downregulation of the Host Gene jigr1 by miR-92 Is Essential for Neuroblast Self-Renewal in Drosophila. PLoS Genet. 11, e1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


