Abstract
Dopamine (DA) is a central monoamine neurotransmitter involved in many physiological and pathological processes. A longstanding yet largely unmet goal is to measure DA changes reliably and specifically with high spatiotemporal precision, particularly in animals executing complex behaviors. Here we report the development of genetically-encoded GPCR-Activation-Based-DA (GRABDA) sensors that enable these measurements. In response to extracellular DA, GRABDA sensors exhibit large fluorescence increases (ΔF/F0 ~90%) with subcellular resolution, sub-second kinetics, nanomolar to sub-micromolar affinities, and excellent molecular specificity. GRABDA sensors can resolve a-single-electrical-stimulus evoked DA release in mouse brain slices, and detect endogenous DA release in living flies, fish, and mice. In freely-behaving mice, GRABDA sensors readily report optogenetically elicited nigrostriatal DA release and depict dynamic mesoaccumbens DA signaling during Pavlovian conditioning or during sexual behaviors. Thus, GRABDA sensors enable spatiotemporally precise measurements of DA dynamics in a variety of model organisms while exhibiting complex behaviors.
ToC
The development of GPCR-Activation-Based-DA (GRABDA) sensors enables measurements of dopamine dynamics in freely-behaving animals with high spatiotemporal precision
Introduction
Dopamine is a crucial monoamine neurotransmitter across many species. In the vertebrate central nervous system, DA regulates a wide range of complex processes, including reward signaling (Schultz, 2016; Wise, 2004), reinforcement learning (Holroyd and Coles, 2002), attention (Nieoullon, 2002) and motor control (Graybiel et al., 1994). In the human brain, impaired DA transmission is associated with neurological diseases, including ADHD (Cook Jr et al., 1995), schizophrenia (Howes and Kapur, 2009) and Parkinson’s disease (Lotharius and Brundin, 2002). In addition, psychostimulants like cocaine act by altering extracellular DA levels in the brain to exert addictive effects (Di Chiara and Imperato, 1988).
Despite these important roles for DA, precise measurements of the spatial and temporal patterns of DA release during complex behaviors are lacking, due in large part to the limitations of existing methods for DA detection. Intracerebral microdialysis has long been the gold standard for quantitative measurements of extracellular DA concentration. However, its relatively slow sampling rate (> 1 min between sampling, typically ~10 mins) (Chefer et al., 2009) is not well suited to detect DA dynamic changes during complex and rapidly evolving behaviors (Tidey and Miczek, 1996). Fast-scan cyclic voltammetry (FSCV) is an electrochemical method that can measure changes in extracellular DA concentrations with 10 ms temporal resolution and ~1 nM sensitivity (Robinson et al., 2008). However, FSCV requires substrate oxidization for signal detection, therefore it is not readily able to distinguish DA from other structurally similar neurotransmitters, such as norepinephrine (NE) (Robinson et al., 2003). Moreover, both microdialysis and FSCV require implantation of a relatively large probe (approximately 70–300 μm in diameter) into brain tissue, which limits the ability to achieve spatially precise measurements of endogenous DA release (Jaquins-Gerstl and Michael, 2015).
In lieu of direct measurements, indirect methods such as measuring the activation of downstream targets of DA receptors have been used to approximate the dynamics of DA. Cell-based DA reporters, such as CNiFERs (Muller et al., 2014), use transplanted HEK293 cells constitutively expressing DA receptors together with an intracellular Ca2+ indicator to couple extracellular DA signals to fluorescence changes. Despite its high sensitivity, this approach requires cell transplantation, and only reports the volume transmission of DA. The TANGO assay, and next-generation versions (Barnea et al., 2008; Inagaki et al., 2012; Kim et al., 2017; Lee et al., 2017), have been used to measure endogenous DA release by coupling the β-arrestin signaling pathway to the expression of reporter genes. Although this approach enables cell-type specific expression of DA reporters and is suitable for in vivo measurements, the long signal amplification time (on the order of hours) precludes the ability to monitor rapid, physiologically relevant dynamics of DA signaling.
Here, we report the development of genetically-encoded fluorescent sensors for direct, rapid, sensitive, and cell-type specific detection of extracellular DA. These sensors, which we call GRABDA sensors, were engineered by coupling a conformationally sensitive circular-permutated EGFP (cpEGFP) to a selected human DA receptor. Through iterative engineering and optimization, we arrived at two GRABDA sensors: GRABDA1m (abbreviated to DA1m), with medium EC50 to DA (~130 nM); and GRABDA1h (abbreviated to DA1h), with high EC50 to DA (~10 nM). These two sensors enable real-time detection of endogenous DA in acute brain slices of mice and in the intact brains of versatile animal models including flies, fish, and mice.
Results
Development and characterization of GRABDA sensors in cultured HEK293T cells and culture neurons
To develop a genetically-encoded sensor for DA, we started with natural DA receptors as the sensing module and coupled a cpEGFP as a fluorescent light output module. We hypothesized that upon DA binding, the conformational changes in the receptor could alter the arrangement of the associated cpEGFP, resulting in a DA-dependent change in fluorescence. Indeed, a similar strategy was recently applied in creating the genetically-encoded acetylcholine sensor GACh (Miao Jing, 2018).
We used a three-step approach to engineer GRABDA sensors (Fig. 1A): First, a cpEGFP was inserted into the third intracellular loop (ICL3) of each human dopamine receptor subtype (DR). Based on preliminary results, we subsequently focused on the D2R-cpEGFP chimera due to its superior membrane trafficking and high affinity for DA (Beaulieu and Gainetdinov, 2011; Missale et al., 1998) (Fig. S1A). Second, the position of the cpEGFP insertion and the linker residues were systematically screened (Fig. 1A, B). Finally, mutations were introduced to expand the response range (Fig. 1C). After screening, we chose two variants, DA1m and DA1h for further characterization. Both sensors have ~90% maximal ΔF/F0 responses to DA with ~70% brightness of EGFP (Fig. 1E, F and S1B), but differ by an order of magnitude with respect to apparent affinity for DA (EC50 130 nM for DA1m, 10 nM for DA1h) (Fig. 1C). We also generated DA-insensitive control sensors containing the mutations C118A and S193N in the DA-binding pocket (Fig. 1C, E) that prevent the sensor from DA binding.
Figure 1. Design, optimization and characterization of GRABDA sensors in cultured HEK293T cells.
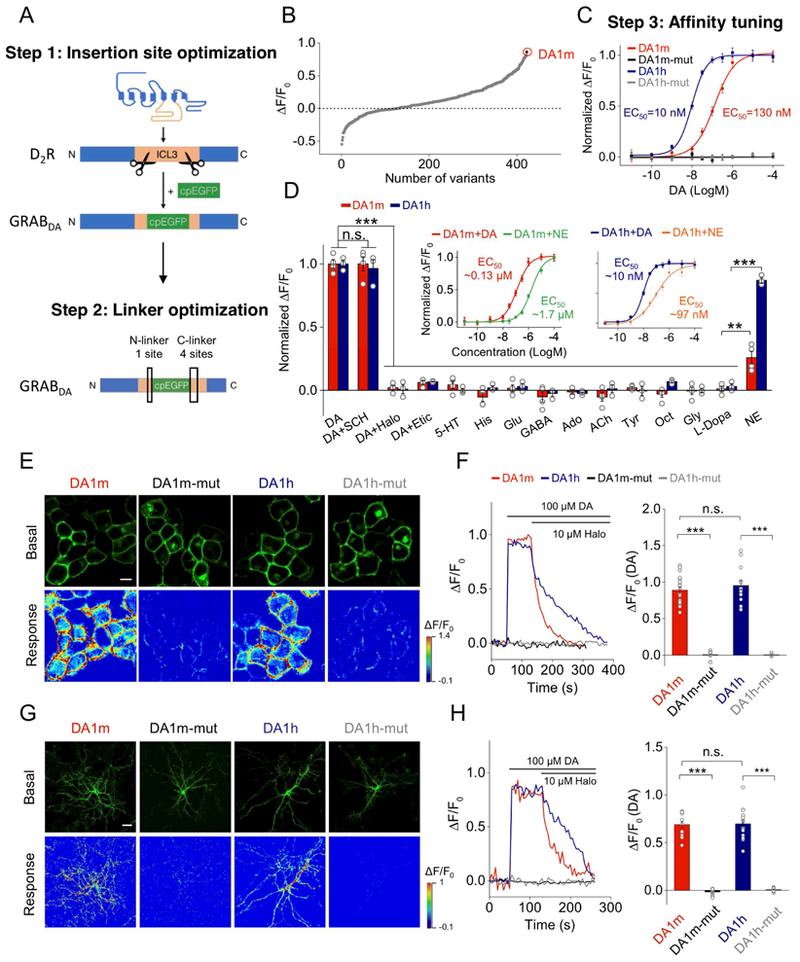
(A) Schematic diagrams showing the strategy of insertion site and linker optimization.
(B) Optimization of the cpEGFP insertion site within the third intracellular loop (ICL3) of D2R and the linkers between D2R and cpEGFP. The fluorescence responses of variant-expressing cells in response to 100 μM DA application. DA1m, with the highest ΔF/F0, was selected for further optimization. Each point represents the average of at least 3–5 cells.
(C) Affinity tuning. Either the T205M single mutation (generating DA1h), or the C118A/S193N double mutations (generating DA1m/h-mut), were introduced into DA1m. The normalized dose-dependent fluorescence responses of various GRABDA-expressing cells in response to DA application are plotted. Each point represents average of 6 wells containing 100–400 cells/well.
(D) Normalized fluorescence changes in DA1m (red), DA1h (blue) expressing cells in response to the application of indicated compounds at 1 μM: DA, DA + SCH, DA + Halo, DA + Etic, 5-HT, histamine (His), glutamate (Glu), gamma-aminobutyric acid (GABA), adenosine (Ado), acetylcholine (ACh), tyramine (Tyr), octopamine (Oct), glycine (Gly), or L-DOPA and NE (DA1m: n = 4 wells; DA1h: n = 3 wells; 200–400 cells/well; p > 0.05 for DA1m/h responses induced by DA compared with DA+SCH; p < 0.001 for DA1m/h responses induced by DA compared with DA+Halo, DA+Etic, 5-HT, His, Glu, GABA, Ado, ACh, Tyr, Oct, Gly and L-DOPA; comparing responses induced by NE with L-Dopa, p = 0.004 for DA1m and p <0.001 for DA1h). The insets show the normalized dose-dependent responses of DA1m- or DA1h-expressing cells to DA and NE application (n = 6 wells/group with 200–400 cells/well).
(E and F) Expression of GRABDA sensors in HEK293T cells. (E) Representative basal fluorescence Intensity (without DA) and responses to 100 μM DA. (F) Representative traces and group analysis of fluorescence changes in GRABDA-expressing cells in response to 100 μM DA followed by 10 μM Halo (DA1m: n = 18 cells from 4 cultures (18/4); DA1m-mut: n = 15/3; DA1h: n = 14/3; DA1h-mut: n = 14/3; p < 0.001 between DA1m and DA1m-mut; p < 0.001 between DA1h and DA1h-mut; p = 0.42 between DA1m and DA1h).
(G and H) Similar to (E and F), except that GRABDA sensors are expressed in cultured neurons (DA1m: n = 13/7; DA1m-mut: n = 14/5; DA1h: n = 16/4; DA1h-mut: n = 10/5; p < 0.001 between DA1m and DA1m-mut; p < 0.001 between DA1h and DA1h-mut; p = 0.88 between DA1m and DA1h).
Scale bars, 10 μm (E), 30 μm (G).
Values with error bars indicate mean ± SEM.
Students’ t-test performed; n.s., not significant; **, p < 0.01; ***, p < 0.001.
We next expressed GRABDA sensors in HEK293T cells and cultured neurons for further characterization. GRABDA sensors trafficked efficiently to the plasma membrane (Fig. 1E, G and S1F), and fluorescent signals were clearly distinguishable in subcellular compartments in neurons(Fig. S1F and G). Both DA1m and DA1h exhibited robust fluorescence increases to DA, which could be blocked by co-application of D2R antagonist haloperidol (Halo) (Sokoloff et al., 1990) (Fig. 1F and H). Mutant sensors (DA1m/h-mut) did not show detectable fluorescence increases to DA application (Fig. 1E-H).
GRABDA sensors exhibited similar or better photostability than EGFP or other cpEGFP-based sensors (Fig. S1C). Using a local perfusion system, both DA1m and DA1h showed rapid fluorescence increases (on rate) to DA application (Fig. S1D, E; 60 ± 10 ms for DA1m and 140 ± 20 ms for DA1h). The fluorescence decrease (off rate) in response to application of the antagonist Halo is slower in DA1h (2.5 ± 0.3 s) compared with DA1m (0.7 ± 0.06 s), consistent with the differences in EC50. For the specificity, bath application of DA elicited robust fluorescrence increase in GRABDA sensors expressing cells, which were completely blocked by co-application of D2R antagonists Halo or eticlopride (Etic), but not by the D1R antagonist SCH-23390 (SCH) (Fig. 1D). In addition, application of several other neurotransmitters did not elicit any detectable fluorescence changes, except for NE which drove modest fluorescence increase (Fig. 1D). Further characterization of the dose-dependent responses to DA and NE revealed a ~10-fold lower EC50 to DA than NE (Fig. 1D insets and Fig. S2A-D). Thus, both DA1h and DA1m are selective for DA over NE at physiological concentrations (DA: 10–100 nM; NE: 1–100 nM) (Bungay et al., 2003; Florin-Lechner et al., 1996; Kehr, 1992; Pacak et al., 1995; Schultz, 2007; Smith et al., 1992). Overall, both sensors show rapid and sensitive responses to physiological ranges of DA with little or no cross-talk to other neurotransmitters.
To test the coupling of GRABDA sensors to GPCR downstream pathways, we examined the coupling efficacies of GRABDA sensors to either G protein- or β-arrestin-dependent pathways (Beaulieu and Gainetdinov, 2011). Application of DA largely reduced the forskolin-induced cAMP increases in WT-D2R-expressing cells, but not in DA1h-expressing cells (Fig. S2E). Furthermore, co-expression of pertussis toxin (PTX) or GTPγS treatment did not alter the EC50 to DA for DA1m and DA1h sensors (Fig. S2F, G), suggesting the negligible coupling of GRABDA sensors to downstream G protein signaling. Next, we measured DA-induced internalization to indicate the coupling with β-arrestin pathway (Luttrell and Lefkowitz, 2002). Compared with WT-D2R that underwent rapid decrease of membrane fluorescence signal within 10 minutes, GRABDA sensors showed stable membrane fluorescence signals throughout 2-hour DA exposure (Fig. S2H, I). In addition, DA1h-expressing cells showed less β-arrestin-dependent reporter signal in the TANGO assay compared with WT-D2R-expressing cells (Fig. S2J). Collectively, these data suggest that GRABDA sensors do not engage the primary signaling pathways downstream of D2R.
Imaging DA dynamics in acute brain slices
To monitor endogenous DA release by GRABDA sensors, we virally expressed DA1m or DA1h by AAV into nucleus accumbens (NAc) of mice and prepared acute brain slices two weeks later. Strong fluorescence signals were detected in sensor-expressing slices, but not in uninjected control slices. Sensor fluorescence was in close proximity with tyrosine hydroxylase (TH) labeled dopaminergic fibers (Fig. 2A and Fig. S4A). Electrical stimulation of NAc core elicited transient fluorescence increases (Fig. 2A–E, Movie S1). The rising time constants of signals were fast (~0.1 s) for both DA1m and DA1h (Fig. 2F), whereas the decaying time constant of DA1h was slower (~17 s) than DA1m (~3 s) (Fig. 2F). Bath application of Halo abolished the evoked responses (Fig. 2G), verifying the signal specificity. Responses to repeated stimulus trains were stable over >30 min of recording (Fig. 2H).
Figure 2. Release of endogenous DA measured in acute mouse brain slices.
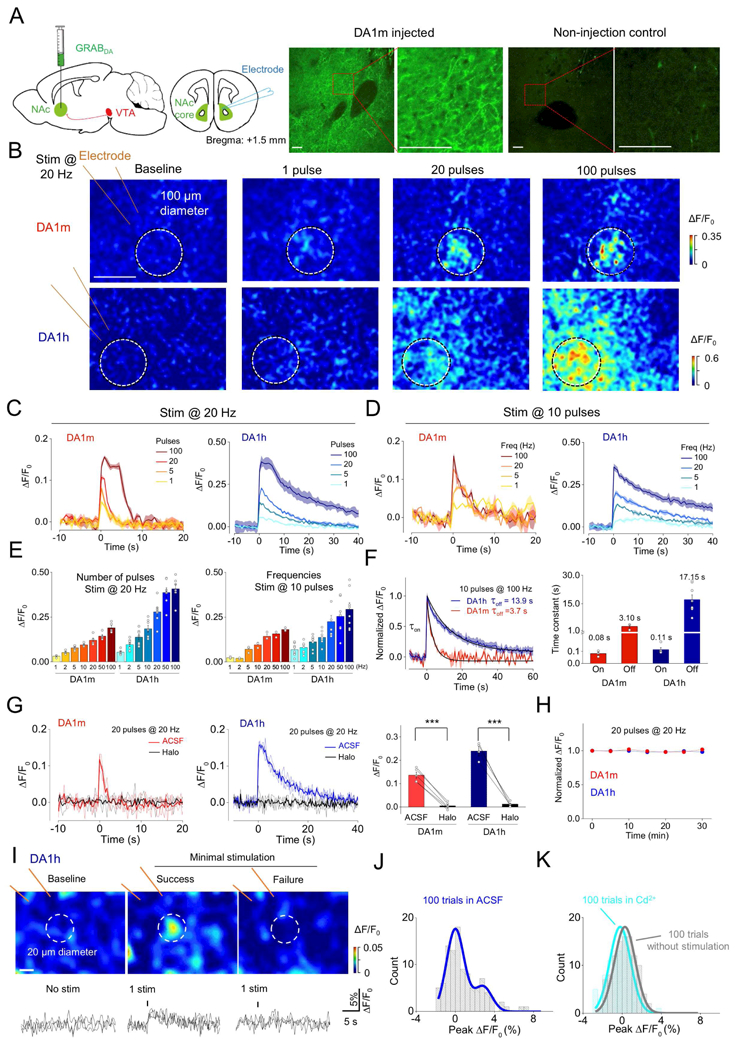
(A) Left two panels, schematic of the experimental protocol for expressing GRABDA sensors and imaging DA dynamics in mouse brain slices containing NAc. Right, the immunoreactive signals of GFP in NAc slices from either DA1m injected or non-injection control mice. Red squares in left panel indicate expanded region in right panel. Scale bars, 100 μm.
(B) Representative pseudocolor images of fluorescence responses in DA1m- or DA1h-expressing neurons following 20 Hz electrical stimulation containing the indicated pulse numbers. White circles represent the ROI selected for analysis. Scale bar, 100 μm.
(C-D) Representative fluorescence responses of DA1m- and DA1h-expressing neurons following 20 Hz electrical stimuli containing the indicated pulse numbers (C), or a 10 pulse train electrical stimulation at the indicated frequencies (D). Each trace is the average of 3 trials in one slice.
(E) Group analysis of the fluorescence response to electrical stimuli at different pulse numbers or frequencies (pulses: DA1m: n = 5 slices from 3 mice; DA1h: n = 7 slices from 4 mice; frequencies: DA1m: n = 3 slices from 2 mice; DA1h: n = 8 slices from 4 mice).
(F) Representative traces (left) and group analysis (right) of the normalized fluorescence changes and kinetics in DA1m- and DA1h-expressing neurons to 10 electrical pulses delivered at 100 Hz. The rising (on) and decaying (off) phases are fitted and summarized on the right. (DA1m: n = 3 slices from 2 mice; DA1h: n = 5–8 slices from 3 mice).
(G) Representative traces (left and middle, with 3 individual trials and the averaged trials) and group analysis (right) of DA1m- and DA1h-expressing neurons to 20 electrical pulses at 20 Hz, in control solution (ACSF) or solution containing 10 μM Halo (DA1m: n = 5 slices from 4 mice, p < 0.001 comparing ACSF with Halo; DA1h: n = 6 slices from 4 mice, p < 0.001 comparing ACSF with Halo).
(H) The fluorescence changes in DA1m- and DA1h-expressing neurons to multiple trains of electrical stimuli with an interval of 5 min. The fluorescence changes induced by the first train were used to normalize the data in each slice (DA1m: n = 3 slices form 2 mice; DA1h: n = 6 slices from 3 mice).
(I) Top: representative pseudocolor images of fluorescence responses during minimal stimulation (left, baseline; middle, success trial; right, failure trial). White circles represent ROI with ~20 μm diameter. Bottom, 3 exemplar trials for each condition, with the dashed lines indicating the stimulation. The data were processed with 3 × binning. Scale bar, 10 μm.
(J) The distribution of peak ΔF/F0 of DA1h-expressing neurons in 100 minimal stimulation trials in ACSF.
(K) Comparison of the distribution of peak ΔF/F0 in 100 trials without stimulation (gray) in ACSF and with stimulation in ACSF containing Cd2+ (light blue) from the same slice.
Values with error bars or shaded areas indicate mean ± SEM.
Student’s t-test performed; ***, p < 0.001.
To test whether these sensors could sensitively report DA release from single dopaminergic fibers, we conducted minimal stimulation experiments (Balaji and Ryan, 2007; Dobrunz and Stevens, 1999). We prepared acute slices of NAc expressing DA1h (Fig. S3A) and gradually turned down the stimulation strength until >50% of response failures occurred. We then repeated 100 trials at this stimulus strength in normal ACSF, followed by 100 trials in ACSF containing 200 μM Cd2+ (Swandulla and Armstrong, 1989) (Fig. 2I–K and Fig. S3B–E). Trials without simulation and stimulation trials in the presence of Cd2+ exhibited a single peak in the distribution of responses at ~0% ΔF/F0 (Fig. 2K and Fig. S3E, F). In contrast, responses in control ACSF showed a bimodal distribution, with an additional peak shifted by ~3% ΔF/F0 (Fig. 2J and Fig. S3B, G, H). This second peak represents the signal of dopamine release from putative activation of single fibers.
Imaging DA dynamics in Drosophila
To test the ability of GRABDA sensors to detect physiologically relevant DA dynamics in living animals, we started with Drosophila, because DA serves as a critical teaching signal in olfactory-associative learning in the fly brain (Burke et al., 2012; Davis, 1993; Heisenberg, 2003; Liu et al., 2012; Schwaerzel et al., 2003). Transgenic UAS-DA1m flies were generated and crossed with TH-GAL4 to express DA1m specifically in dopaminergic neurons (DANs). (Fig. 3A). Under two-photon imaging, the odor isoamyl acetate (IA) elicited time-locked fluorescence increases in the MB, most prominently in the β’ lobes (Movie S2), and the odor-evoked responses were blocked by Halo application. In contrast, no odor-evoked fluorescence response was observed in flies expressing DA1m-mut. When we expressed DA1m in Kenyon cells, which receive direct input from DANs (Aso et al., 2014; Mao and Davis, 2009; Tanaka et al., 2008), we observed odor-evoked responses in WT flies, but not in TH-deficient flies that lack DA synthesis, validating the specificity of the signals (Fig. 3B,C) (Cichewicz et al., 2017).
Figure 3. In vivo imaging of DA dynamics in the Drosophila brain.
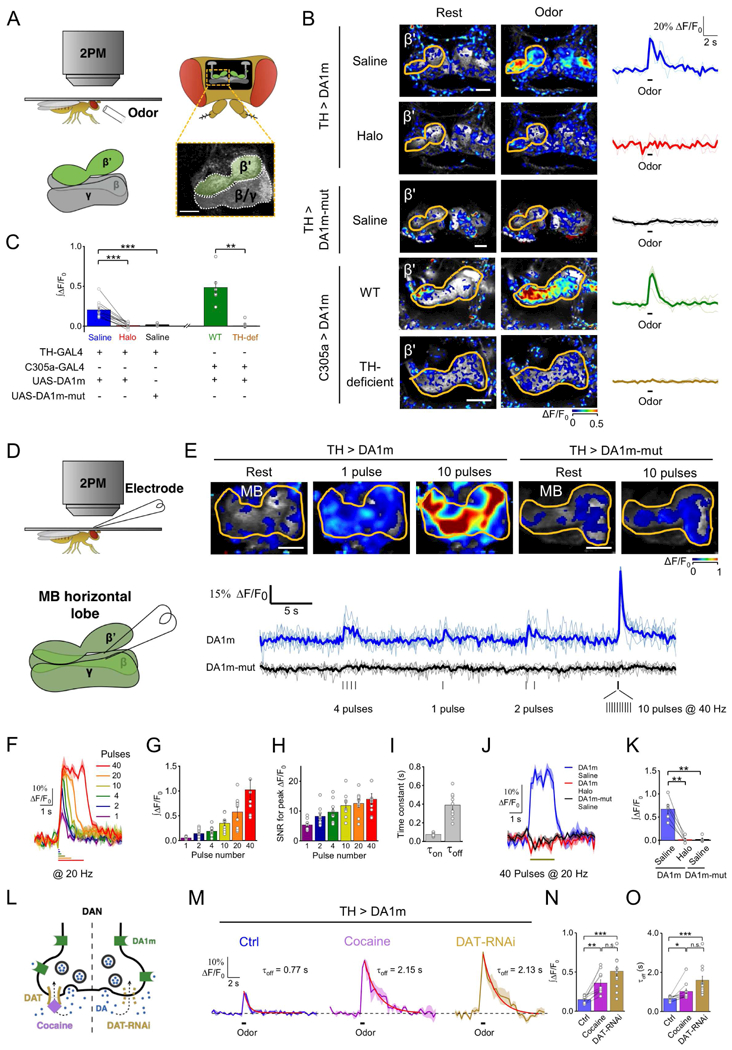
(A) Schematic for odor stimulation during two-photon microscopy in drosophila.
(B and C) Fluorescence changes of DA1m or DA1m-mut expressing flies to 1 sec odor stimulation. (B) Representative pseudocolor images, single trial traces (light) and averaged traces (bold) from one fly. (C) Group analysis (TH > DA1m: n = 12 flies; TH > DA1m-mut: n = 5 flies; C305a > DA1m WT flies: n = 6 flies; C305a > DA1m TH-deficient flies: n = 6 flies; p < 0.001 for TH > DA1m in saline compared with Halo; p < 0.001 for TH > DA1m compared with TH > DA1m-mut in saline; p = 0.002 for C305a > DA1m in WT flies compared with TH-deficient flies).
(D) Schematic depicting in vivo electrical stimulation in which an electrode was positioned near the DA1m-expressing DANs in order to evoke DA release.
(E) Top, representative pseudocolor images of TH > DA1m and TH > DA1m-mut flies in response to multiple trains of electrical pulses. Bottom, single-trial traces (light) and 6-trial averaged traces (bold) from one fly with indicated genotypes. Each vertical tick indicates 1-ms electrical pulse.
(F-I) Electrical stimulation of TH > DA1m flies. (F) Representative traces, (G) group analysis of integrated signal, (H) signal-to-noise ratios (SNR), and (I) kinetics of responses to electrical pulses (n = 9 flies/group).
(J-K) Fluorescence changes in TH > DA1m and TH > DA1m-mut flies in response to 40 pulses electrical stimuli (at 20 Hz), in normal saline or in saline containing 10 μM Halo (TH > DA1m: n = 5 flies; TH > DA1m-mutt: n = 5 flies; p = 0.004 for responses of TH > DA1m in saline compared with Halo; p = 0.007 for responses of TH > DA1m in saline compared with TH > DA1m-mut in saline). (J) Representative traces and (K) group analysis.
(L-O) Fluorescence changes in TH > DA1m flies in response to 1-s odor stimulation, in saline, saline containing 3 μM cocaine or when the DAT expression in DAN was impaired by DAT-RNAi. (L) Schematic of the experimental design. (M) Representative traces fitted with a single-exponential function (red traces), with the decay time constants shown. (N and O) The group analysis of integrals and the decay time constants (TH > DA1m: n = 10 flies; TH > DA1m, DAT-RNAi: n = 11 flies; between control and cocaine groups, p = 0.002 for integrals and p = 0.025 for decay time constants; between control and DAT-RNAi groups, p < 0.001 for both integrals and decay time constants; between cocaine and DAT-RNAi groups, p = 0.095 for integrals and p = 0.053 for decay time constants).
Averaged traces shaded with ± SEM are shown in (F), (J), and (M).
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Scale bars in (B) and (E), 25 μm.
To further characterize the sensitivity and kinetics of DA1m in vivo, we electrically stimulated MB DANs while imaging the MB horizontal lobes (Fig. 3D). We found that DA1m-, instead of DA1m-mut-, expressing DANs exhibited reproducible fluorescence increases in response to electrical stimulation, and single stimulus was sufficient to elicit a measurable fluorescence increase (Fig. responses were completely blocked by the Halo application (Fig. 3J, K and. 3I), and responses were completely blocked by the Halo application (Fig. 3J, K and S4G).
Within the MB γ lobe of DA1m-expressing flies (Fig. S5A), we observed that odor selectively elicited responses in the γ4 compartment (Fig. S5B1), while aversive electrical shock to the abdomen evoked responses in γ2 and γ3 compartments (Fig. S5B2). As a control, exogenous DA application caused overall fluorescence increases in all γ2–5 compartments (Fig. S5B3). These results provide direct evidence of compartmental DA dynamics during sensory processing in the MB, as postulated by indirect presynaptic Ca2+ imaging of DAN (Berry et al., 2012; Cohn et al., 2015).
To explore whether DA1m is sensitive and bright enough to report DA signals perceived by single neuron in vivo, we adopted MB296B-GAL4 to express DA1m in one DAN per hemisphere innervating MB γ2 compartment (Aso et al., 2014; Tanaka et al., 2008) (Fig. S5C). A brief aversive electrical shock to the fly’s abdomen elicited rapid, repeatable fluorescence rises in the γ2 compartment (Fig. S5C-E), suggesting DA1m’s feasibility for single cell imaging.
The DA transporter (DAT) is critical in regulating extracellular DA levels and serves as a primary target for drugs of abuse, including cocaine (Fig. 3L) (Bainton et al., 2000; Ritz et al., 1987). Indeed, cocaine application potentiated the odor-evoked responses in MB β’ lobes of TH > DA1m flies, and also prolonged the decay of signals. Knocking down the expression of DAT selectively in DANs phenocopied the effect of cocaine administration (Fig. 3M–O, Fig. S4H). Taken together, these data demonstrated that GRABDA sensors have sufficient sensitivity, kinetics, and specificity to report in vivo DA dynamics with sub-cellular spatial resolution, and sub-second temporal resolution in genetically-defined neurons of living flies.
Finally, we examined whether the ectopic expression of DA1m alters physiological properties of neurons. We observed no significant difference of odor-evoked Ca2+ signals in both DANs or Kenyon cells between the flies with or without expression of DA1m (Fig. S5F-H), suggesting that expression of the DA1m does not alter odor-evoked responses in neurons in the fly brain.
Imaging DA release in the intact zebrafish brain
Larval zebrafish has an optically transparent brain and can perform a wide range of behaviors, making them a powerful system to explore the structure and function of the vertebrate brain at cellular resolution. To test the feasibility of using GRABDA sensors in zebrafish larvae, we generated the transgenic line in which DA1m was expressed pan-neuronally throughout the brain, while TRPV1-TagRFP was expressed specifically in DANs to enable their chemogenetic activation by capsaicin (Fig. 4A). Exogenous DA application caused a fluorescence increase in DA1m-expressing neurons, that was blocked by co-application of the antagonist Halo (Fig. 4B–D). In the tectal neuropil downstream of DANs, repeated application of capsaicin caused a progressive fluorescence increase that was also blocked by Halo (Fig. 4E–H). Presentation of a threatening looming stimulus elicited time-locked fluorescence increase specifically in the neuropil of tectal neurons, but not in their cell bodies (Fig. 4I–L). In summary, DA1m is well suited to report in vivo DA dynamics in the brain of zebrafish larvae.
Figure 4. Monitoring in vivo DA release in transgenic zebrafish.
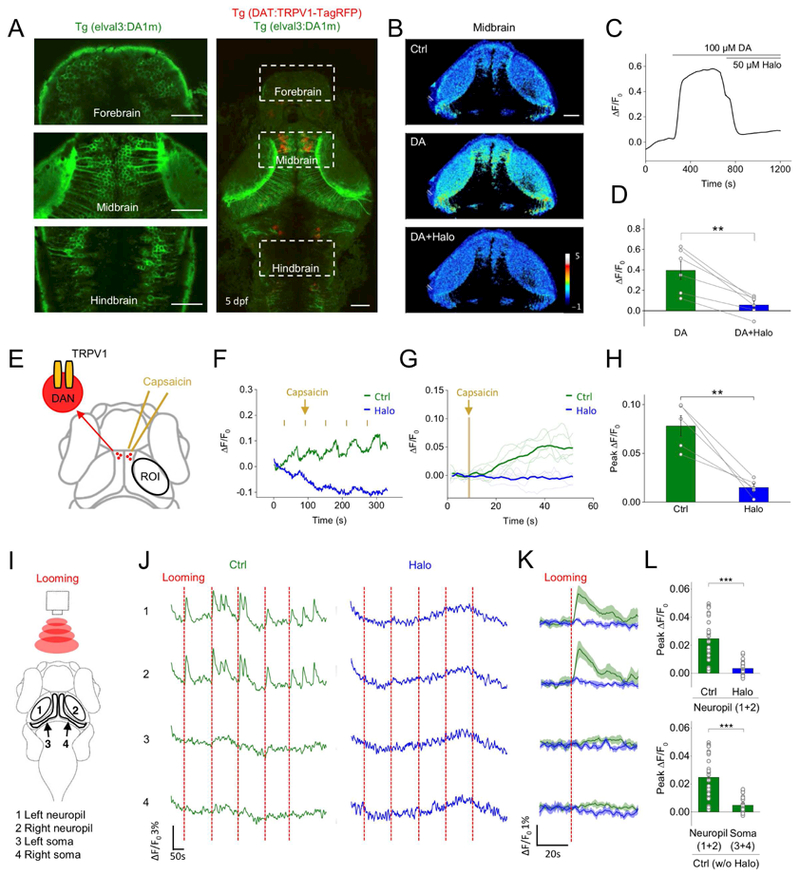
(A) Fluorescence images of a transgenic zebrafish larvae expressing DA1m (green) pan-neuronally and TRPV1-TagRFP (red) in DANs with expanded views of DA1m-expressing neurons in indicated brain regions (left).
(B-D) DA1m-expressing neurons to 100 μM DA followed by 50 μM Halo (n = 6 fishes; p = 0.002 between DA and DA+Halo). (B) Representative pseudocolor images, (C) traces and (D) group analysis.
(E) Schematic of chemogenetic activation of TRPV1-expressing DANs by capsaicin. The fluorescence signals in the tectal neurons (within ROI) were analyzed.
(F-H) Fluorescence changes of DA1m-expressing neurons to 5-trial capsaicin application, in control solution (green) or solution containing 50 μM Halo (blue). Representative traces encompassing 5 sequential stimulation trials and corresponding averaged traces from one fish are shown in F and G. (H) Group analysis (n = 5 fish; p = 0.006 between control and Halo).
(I) Schematic for visual stimulation in which red expanding dots were projected in front of the larva. The fluorescence responses in neuropil (1 and 2) and soma (3 and 4) regions of the optic tectum were analyzed, respectively.
(J-L) Fluorescence changes of DA1m-expressing neurons from each region during visual stimulation in control solution (green) or solution containing 50 μM Halo (blue). Vertical dashed red line, 3-s looming stimulation. Representative traces encompassing 5 sequential stimulation trials and the corresponding averaged traces from one fish are shown in J and K. Group analysis is shown in L (n = 30 trials from 3 fish for each condition; p < 0.001 in two panels).
Scale bars in (A) and (B), 50 μm.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; **, p < 0.01; ***, p < 0.001.
Combining optogenetics with GRABDA to measure the dynamics of DA in freely moving mice
To test the ability of GRABDA sensors to report DA dynamics in the mouse brain in vivo, we focused on DANs located in the substantia nigra pars compacta (SNc) that project to the dorsal striatum. We virally expressed DIO-C1V1 (Yizhar et al., 2011) in the SNc DANs of TH-Cre mice to permit their optogenetic activation. Coexpression of DA1m/DA1h-mut and tdTomato in the dorsal striatum allowed simultaneous monitoring of DA release and detection of movement-related artifacts (Fig. 5A and B). In freely moving mice, the ratio of DA1m to tdTomato was elevated upon the administration of methylphenidate, a known DAT blocker (Volkow et al., 1999), and was suppressed by subsequent administration of Etic, a D2R blocker (Fig. 5C). Transient fluctuations in the ratio are consistent with spontaneous DA release events during the animal movement (Balleine et al., 2007; da Silva et al., 2018; Howe and Dombeck, 2016). These transients were prolonged during methylphenidate application and their amplitudes were reduced by Etic administration. In contrast, mice expressing the DA1h-mut did not show observable fluorescence changes (Fig. 5C). Optogenetic activation of DANs in the SNc with C1V1 generated transient fluorescence increases in the dorsal striatum of DA1m-, but not DA1h-mut-, expressing mice (Fig. 5D), which could also be prolonged by methylphenidate and abolished by Etic (Fig. 5D–H).
Figure 5. Striatal DA dynamics measured in freely moving mice during optogenetic stimulation of the SNc.
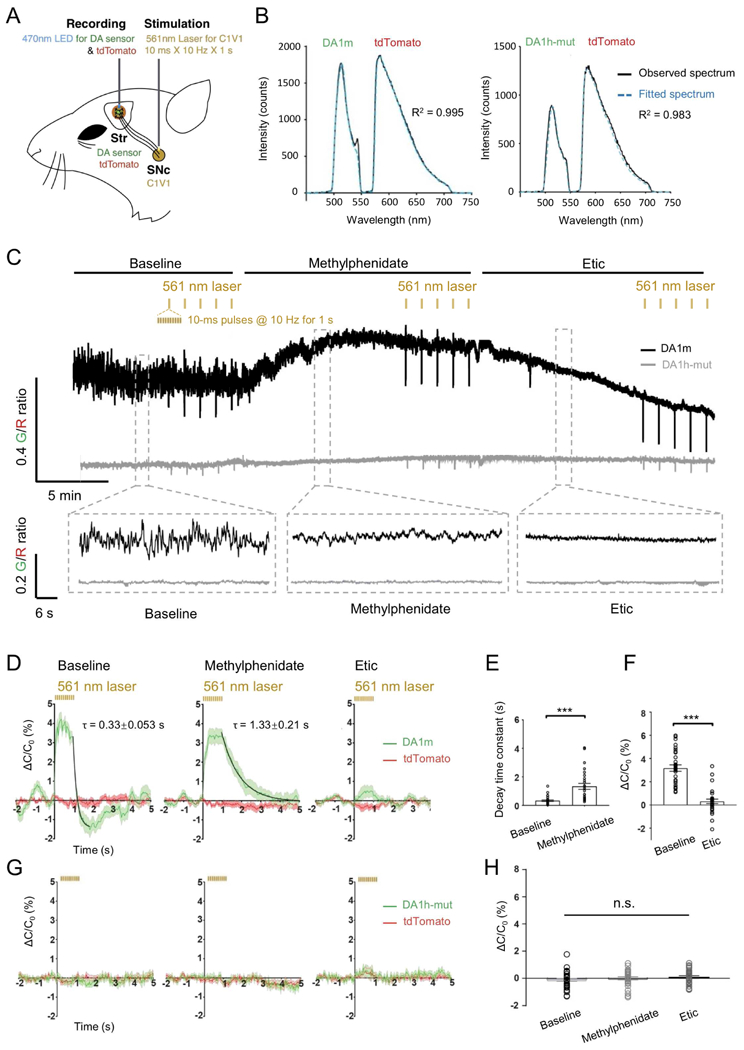
(A) Schematic depicting the dual-color optical recordings of DA1m-/DA1h-mut- and tdTomato-expressing neurons in the dorsal striatum during simultaneous optogenetic C1V1 stimulation of DANs in the SNc.
(B) Representative frames of the emission spectra of DA1m/DA1h-mut and tdTomato co-expressed in the dorsal striatum. Black traces show the measured spectrum; the blue dashed traces show the fitting curves generated by a linear unmixing algorithm.
(C) Representative traces showing the ratio of DA1m (black) or DA1h-mut (gray) to tdTomato coefficients in a freely moving mouse (top panel) and enlarged traces (bottom panel) in the baseline (left), 5 min after the i.p. injection of methylphenidate (10 mg/kg, middle), and 5 min after the i.p. injection of Etic (2 mg/kg, right). Black lines above indicate the time of compound administration. Yellow ticks indicate the time of optogenetic stimulation.
(D and G) Averaged fluorescence changes from DA1m/DA1h-mut (green) expressed in the dorsal striatum during optogenetic stimulation of DANs in the SNc by C1V1 under indicated conditions (n = 30 trials from 6 hemispheres of 3 mice for each condition). Baseline (left), after the i.p. injection of methylphenidate (middle), and after the i.p. injection of Etic (right). The off kinetics were fitted with a single-exponential function (black traces). ΔC/C0% represents the percent change of the fluorescence coefficient of each fluorophore (see methods for details). 5 data points (measured at 0.12 s, 0.32 s, 0.52 s, 0.72 s, and 0.92 s after the onset of the stimulation pulse train) were excluded to remove the stimulation artifacts.
(E) Comparison of the decay time constants of C1V1-evoked DA1m fluorescence responses between baseline group and methylphenidate group.
(F and H) Comparison of the magnitude of C1V1-evoked DA1m/DA1h-mut fluorescence changes between different groups.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; ***, p < 0.001.
Bi-directional modulation of DA dynamics in the NAc during Pavlovian conditioning
In addition to the nigrostriatal pathway, the dopaminergic projection from the ventral tegmental area (VTA) to the NAc regulates a variety of important functions including the reinforcement learning (Daw and Tobler, 2013; Glimcher, 2011; Gonzales et al., 2004). To test whether our sensor can detect behaviourally relevant changes in endogenous DA release, we first expressed DA1m or DA1h in the NAc of head-fixed, water-restricted mice and trained them to associate a brief auditory cue with an ensuing reward (a drop of water), or a punishment (a brief air puff to the face) (Fig. 6A). In every naïve mouse, reward or punishment delivery triggered respective increases or decreases in the fluorescence signals in the NAc. Over the course of training, mice selectively learned to associate the reward-predictive cue with delivery of reward, as the magnitude of the reward-evoked response decreased, while a response of similar sign gradually developed to the reward-predictive cue (Fig. 6B–H). In summary, both DA1m and DA1h sensors have high signal-to-noise ratio and temporal resolution to report the dynamic bi-directional changes in DA release over the course of Pavlovian conditioning.
Figure 6. Dopamine release in NAc measured during various training phases of an auditory Pavlovian conditioning task.
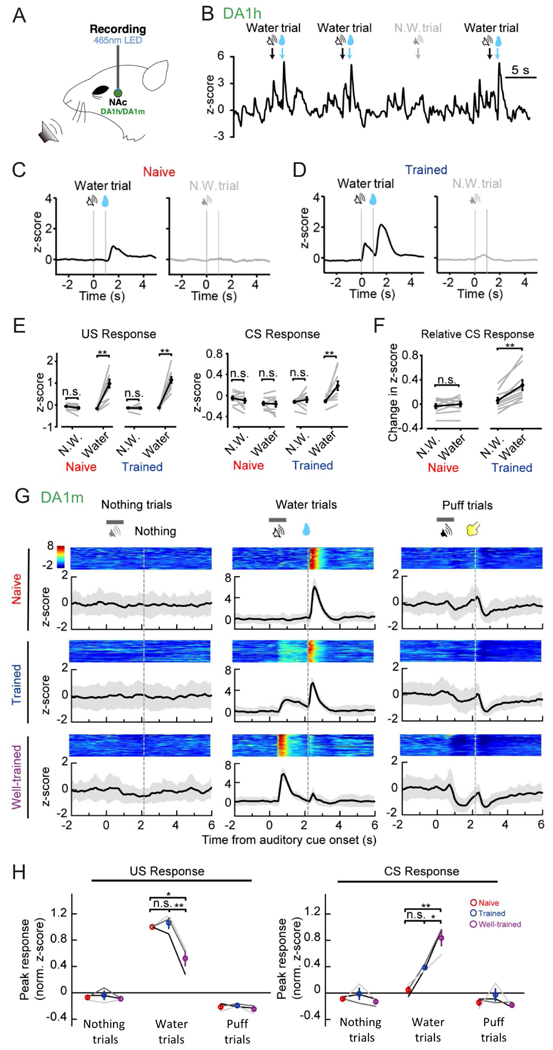
(A) Schematic for fiber photometry recording of GRABDA-expressing neurons from the NAc of a head-fixed mouse during auditory Pavlovian conditioning task.
(B) Exemplar trace of DA1h signals from a trained mouse, encompassing four sequential trials. The timings of cues (CS) or water reward (US) are indicated above.
(C and D) Exemplar time-aligned DA1h signals from a mouse in (C) naïve and (D) trained sessions. Note emergence of DA response to reward-predictive cue after training.
(E) Group analysis of DA1h responses to water (US, left) and cue (CS, right) of both naïve and trained mice (n = 9 mice; US response: naïve N.W.: p = 0.084; naïve water: p = 0.0020; trained N.W.: p = 0.56; trained water: p = 0.0020; CS response: naïve N.W.: p = 0.37; naïve water: p = 1.0000; trained N.W.: p = 0.043; trained water: p = 0.0020).
(F) Direct comparison of baseline-subtracted DA1h signals to cue (CS) (naïve: p = 0.43; trained: p = 0.0020).
(G) Exemplar time-aligned pseudocolor images and averaged traces (mean shaded with ± SD) from a mouse in naïve, trained and well-trained sessions.
(H) Group analysis of the normalized peak z-scores of DA1m signals to US and CS in different sessions. Each trace (coded with specific gray value) represents data from one animal (n = 3 mice; water trial US responses: p = 0.7638 between naive and trained, p = 0.0125 between naive and well-trained, p = 0.0080 between trained and well-trained; water trial CS responses: p = 0.1032 between naive and trained, p = 0.0067 between naive and well-trained, p = 0.0471 between trained and well-trained).
Values with error bars indicate mean ± SEM.
Signed rank test performed in (E) and (F); n.s., not significant; **, p < 0.01.
Post-hoc Tukey’s test was performed in (H); n.s., not significant; *, p < 0.05; **, p < 0.01.
Monitoring DA release in the NAc of mice during male mating behaviors
In contrast to the well-established involvement of DA in Pavlovian conditioning, DA dynamics during naturally rewarding social behaviors (Berridge and Robinson, 1998), such as courtship and mating, remain largely a matter of debate. Here, we took advantage of the high sensitivity and fast temporal resolution of DA1h to better understand DA dynamics during sexual behaviors. To confirm that DA1h can detect acute dopamine release in the NAc, we virally expressed Cre-dependent Chrimson-tdTomato in the DANs in the VTA using DAT-ires-Cre mice. We observed that optogenetic stimulation evoked time-locked fluorescence increases of DA1h (Fig. S6A–F). The DA1h signal peaks at the end of the 0.5 s stimulation and sharply decreases to pre-stimulation level by 2 s (Fig. S6G and H).
During the behavioral test, a sexually receptive C57BL/6J female was introduced into the home cage of the male mouse, and male quickly approached and investigated the female, and initiated mounting within the first minute (Fig. 7A and B). During sniffing female, the fluorescence of DA1h measured in the male’s NAc increased slightly in some animals. The fluorescence increases during mounting and intromission were highly reliable across all animals. During ejaculation, the fluorescence reached the largest response. Smaller but consistent fluorescence increases were also observed during penile grooming that typically occurred after intromission (Fig. 7C and D). The average fluorescence increases during all behaviors except sniffing female is significantly higher than values obtained from randomized controls (Fig. 7E and F). In animals that expressed DA1h-mutant in the NAc, we observed no significant increase of fluorescence during mounting, intromission or ejaculation, suggesting that the DA1h fluorescence increases during sexual behaviors were not due to movement artifacts (Fig. S6 I–L). These results indicate that DA is acutely released in the NAc during episodes of sexual behaviors and could encode information about specific features of courtship and mating.
Figure 7. Acute DA release in the NAc measured during male sexual behaviors.
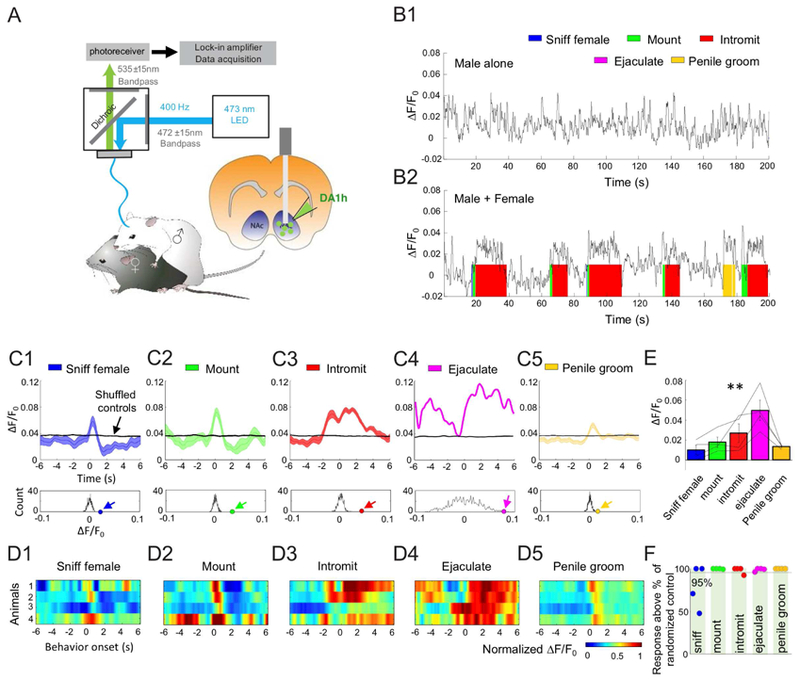
(A) Schematic depicting fiber photometry recording of DA1h-expressing neurons from the NAc of a male mouse during sexual behaviors.
(B1 and B2) Representative fluorescence changes (B1) before female introduction and (B2) during sexual behaviors. The colored shades indicate different behavioral events.
(C1–C5) Top, representative post-event histograms (PETHs, mean shaded with ± SEM) showing the DA1h signal aligned to onsets of various behavioral events from one mouse. Black lines show averaged PETHs of 1000× randomized controls. Bottom, the distributions of mean ΔF/F0 of randomized controls. Colored dots and arrows indicate the actual mean ΔF/F0 during the behaviors.
(D1–D5) Heat map showing the PETHs of all 4 animals during various behaviors. For each animal, ΔF/F0 is normalized with the maximum value during ejaculation.
(E) Group data summarizing the mean ΔF/F0 during various behaviors of all 4 animals. Error bar: ±SEM. One-way ANOVA with repeated measures. Among behaviors: F(3, 4) = 5.96. p = 0.01.
(F) Each dot indicates the mean ΔF/F0 value during one behavior of one animal in reference to the values of randomized controls. Most dots are at 100%, indicating that the mean ΔF/F0 is higher than 100% of the 1000× shuffled controls.
See also Fig. S6.
Discussion
Here we describe the development and characterization of a pair of novel genetically-encoded fluorescent sensors that enable specific, real-time detection of endogenous DA dynamics in several model systems, ex vivo and in vivo. In acute mouse brain slices, GRABDA sensors report stimulus-evoked DA release in mesolimbic pathway. In flies, GRABDA sensors detected odor-evoked DA release in the MB, and resolved DA release evoked by a single electrical stimulus. In transgenic zebrafish, GRABDA sensors reported chemogenetically evoked DA release in the optic tectum. In mice, combining optogenetic stimulation with GRABDA sensors enabled the simultaneous optical manipulation and detection of DA signals in vivo. Finally, GRABDA sensors revealed real-time DA dynamics in the NAc of behaving during Pavlovian conditioning or sexual behaviors.
Our GRABDA sensors exhibit several clear advantages over existing methods. First, GRABDA sensors are genetically-encoded by relatively small genes (~2 kb), making them amenable to transgenic approaches and viral packaging. Second, GRABDA sensors have high sensitivity to DA. In response to DA, DA1m and DA1h sensors achieve a maximal ΔF/F0 ~90% with ~130 nM and ~10 nM affinities, respectively. In contrast, conventional GPCR-based FRET probes for detecting neurotransmitters are usually limited to a maximum of ~5% under optimal conditions, and less than that in vivo (Vilardaga et al., 2003; Ziegler et al., 2011). Third, GRABDA sensors have high specificity for DA. Finally, GRABDA sensors have rapid response kinetics with a rise time of ≤ 100 ms (Fig. 1G, 3D and 4I). Although this response time of GRABDA sensors is slower than FSCV, it is sufficiently rapid for reporting most physiologically relevant DA dynamics and shares response kinetics similar to WT GPCRs (Lohse et al., 2008). A recently described fluorescent DA sensor (named dLight) utilizes a similar detection strategy to report DA signaling in vivo (Patriarchi et al., 2018). The applied dLight variants report in vivo DA dynamics in rodent brains with similar kinetics and signal-to-noise ratio as GRABDA sensors. However, the GRABDA sensors have been optimized for brightness, have more consistent sensitivity (EC50) to DA across different cell types, and have proven efficacy in multiple organisms in vivo.
We noted that the GRABDA sensors have negligible coupling with major GPCR downstream pathways. This is presumably due to the steric hindrance imposed by the bulky cpEGFP moiety that replaces parts of the ICL3, which is the critical position for G protein or arrestin to interact with the GPCR (Luttrell and Lefkowitz, 2002; Neves et al., 2002). Consistent with minimal coupling between GRABDA sensors and downstream signaling pathways, in vivo Ca2+ imaging experiments using the Ca2+ sensor jRCaMP1a revealed no measurable alteration in Ca2+ signaling in neurons of transgenic flies that overexpressing GRABDA sensors (Fig. S5F-H).
Using GRABDA sensors, we observed compartmentalized DA dynamics in the MB of flies, even down to single neuron level (Fig. S5). Thus, GRABDA sensors create new opportunities for exploring how specific compartments in the MB of the fly may exhibit distinct DA dynamics, which has been suggested by previous reports (Aso et al., 2014; Cognigni et al., 2017; Cohn et al., 2015; Mao and Davis, 2009). Experiments in flies also illustrate the power of GRABDA sensors to probe DAT function in vivo by directly measuring extracellular DA levels in real time. GRABDA sensors readily respond to visually induced DA release in the intact brain of the zebrafish larvae as well, providing a robust and convenient tool to examine DA dynamics in this classic vertebrate model system.
Our experiments in behaving male mice provide new insight into the dynamics of DA during sexual behaviors. Contrary to previous reports revealed by either microdialysis or FSCV measurement, which described slow changes of extracellular DA during sexual behaviors (Pfaus et al., 1990) or only transient increase during female introduction (Robinson et al., 2001), GRABDA sensors revealed a time-locked DA elevation aligned to various sexual behaviors, consistent with a model where DA encodes behavioral motivation, anticipation, or arousal. As recent breakthroughs in single-cell sequencing have highlighted a previously unrecognized molecular, morphological, physiological, and functional heterogeneity of DANs (Nair-Roberts et al., 2008; Ungless and Grace, 2012), we expected that the targeted expression of genetically-encoded GRABDA sensors with cell-type specificity could therefore provide a critical window into the coding strategy of dopaminergic transmission in complex behaviors.
We anticipate that future efforts will be able to further tune the affinity, enhance the selectivity, and increase the signal-to-noise ratio of the next-generation of GRABDA sensors by using the recently solved crystal structure of the D2R (Wang et al., 2018). Moreover, by adding a red fluorescent protein, GRABDA sensors can be readily transformed into ratiometric indicators, which could prove useful for more quantitative measurements of DA release across different experiments and preparations. Finally, a GPCR-based strategy was recently used to develop a genetically-encoded sensor (GACh) with high sensitivity and high selectivity for acetylcholine (ACh) (Miao Jing, 2018). Both sensors operate by coupling conformational changes in a GPCR induced by ligand binding to drive increases in cpEGFP florescence. Given the diverse ligand specificity of different GPCRs, a future goal will be to explore whether this principle can be expanded further to develop sensors for the entire range of known neurotransmitters and neuromodulators.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yulong Li(Yulongli@pku.edu.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Primary cultures
Rat cortical neurons were prepared from postnatal 0-day old (P0) Sprague-Dawley rat pups (male and female, random choice; Beijing Vital River). The cortical neurons were dissociated from the dissected rat brains in 0.25% Trypsin-EDTA (Gibco), and plated on 12-mm glass coverslips coated with poly-D-lysine (Sigma-Aldrich) in neurobasal medium (Gibco) containing 2% B-27 supplement (Gibco), 1% GlutaMax (Gibco), and 1% penicillin-streptomycin (Gibco). The neurons were cultured at 37°C in 5% CO 2.
Cell lines
HEK293T cell line (female, ATCC, CRL-3216) and Hela cell line (female, ATCC,CCL-2) were bought from ATCC and authenticated based on the morphology under microscope and the analysis of the growth curve. HEK293T cell lines stably expressing D2R or DA1h were estabilished in our laboratory. In brief, pPacific-D2R and pPacific-DA1h were constructed (pPacific was a self-constructed vector containing elements including 3’ TR, DNA encoding myc tag, 2A sequence, DNA encoding mcherry, DNA encoding puromycin and 5’ TR), and transfected with pCS7-PiggyBAC (S103P, S509G) (Yusa et al., 2011) into HEK293T cells, then the transfected cells were selected with 1 μg/ml puromycin. HEK293 cell line (female) stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion construct was a gift from Bryan L Roth (Kroeze et al., 2015). Cell lines were cultured in DMEM (Gibco) supplemented with 10% (v/v) FBS (Gibco) and 1% penicillin-streptomycin (Gibco) at 37°C in 5% CO 2.
Mice
P28−48 wild-type C57BL/6,TH-Cre mice and DAT-IRES-Cre mice were used to prepare the acute brain slices and in vivo fiber photometry experiments. Except in sexual behavior experiments the optical fibers were implanted on male mice, in other experiments fiber photometry recordings were performed in both male and female mice randomly, as the dopaminergic innervations mentioned in this paper are similar between them (Wu and Shah, 2011). All animals were maintained in the animal facilities and were family- or pair-housed in a temperature-controlled room with a 12-h/12-h light/dark cycle. All procedures for animal surgery and maintenance were performed using protocols that were approved by the Animal Care & Use Committees at Peking University, Chinese Academy of Sciences (CAS), New York University, University of California, San Francisco, and US National Institutes of Health, and were performed in accordance with the guidelines established by US National Institutes of Health guidelines.
Flies
To generate transgenic flies, the coding sequence of DA1m was integrated into the pUAST vector using Gibson Assembly (Gibson et al., 2009), which was then used in P-element-mediated random insertion. Transgenic Drosophila lines carrying DA1m on the chromosomes 2 (UAS-DA1m/cyo) and 3 (UAS-DA1m/TM2) with the strongest expression level after crossing with TH-GAL4 were used. The coding sequence of DA1m-mut was incorporated into pJFRC28 (Pfeiffer et al., 2012) (Addgene plasmid #36431) using Gibson Assembly, and this plasmid was used to generate transgenic flies using PhiC31-mediated site-directed integration into attp40. The embryo injections were performed at Core Facility of Drosophila Resource and Technology, Shanghai Institute of Biochemistry and Cell Biology, CAS. Transgenic flies were raised on conventional corn meal at 25°C, with ~70% humidity, under 12-h/12-h light/dark cycle. The fly lines used in this study: TH-GAL4, a gift and unpublished line generated by appending 2A-GAL4 to the last exon of TH, from Yi Rao, Peking University. C305a-GAL4 (BDSC: 30829) and 30y-GAL4 (BDSC: 30818), also gifts from Yi Rao. DTHFS+/−ple2/TM6B (Cichewicz et al., 2017), a gift from Jay Hirsh, University of Virginia. UAS-DAT-RNAi (TH01470.N) (Ni et al., 2011), from Tsinghua Fly center, Tsinghua University. UAS-jRCaMP1a (BDSC: 63792) (Dana et al., 2016), a gift from Chuan Zhou, Institute of Zoology, Chinese Academy of Sciences. MB296B-GAL4 (Aso et al., 2014), a gift from Hongtao Qin, Hunan University.
Adult Drosophila within 3 weeks after eclosion were used for imaging experiments. Both male and female were randomly used because dopaminergic projections to the MB are similar between them (Kimura et al., 2005). The flies corresponding to figures:
- Fig. 3A–C
- UAS-DA1m/cyo; TH-GAL4 (DANs)/TM6B
- UAS-DA1m-mut/+; TH-GAL4/+
- C305a-GAL4 (α’ and β’ Kenyon cells)/UAS-DA1m; DTHFS+/−ple2/+ (WT group)
- C305a-GAL4/UAS-DA1m; DTHFS+/−ple2 (TH-deficient group)
- Fig. 3D–K
- UAS-DA1m/cyo; TH-GAL4/TM6
- UAS-DA1m-mut/+; TH-GAL4/+
- Fig. 3L–O
- UAS-DA1m/cyo; TH-GAL4/TM6B
- UAS-DA1m/+; TH-GAL4/UAS-DAT-RNAi
- Fig. S5A–B
- UAS-DA1m/cyo; TH-GAL4/TM6B
- Fig. S5C–E
- MB296B-GAL4 > UAS-DA1m
- Fig. S5F–H
- TH-GAL4/ UAS-jRCaMP1a
- UAS-DA1m/+; TH-GAL4/UAS-jRCaMP1a
- 30y-GAL4/ UAS-jRCaMP1a
- UAS-DA1m/+; 30y-GAL4/UAS-jRCaMP1a
Zebrafish
The background strain is albino (slc45a2b4). To generate transgenic zebrafish, plasmids containing pTol2-elval3: DA1m (25 ng/μL) and Tol2 mRNA (25 ng/μL) were co-injected into fertilized eggs, and founders were screened three months later. Transgenic zebrafish adults and larvae were maintained at 28 °C on a 14-h/10-h light/dark cycle. Experiments were performed on 5 days-post-fertilization (dpf) larvae. Larval zebrafish do not have sex differentiation before 1-month post fertilization (Singleman and Holtzman, 2014)
METHOD DETAILS
Molecular biology
Plasmids were generated using Gibson Assembly. DNA fragments were generated using PCR amplification with primers (Thermo Fisher Scientific) with 30-bp overlap. The fragments were assembled using T5-exonuclease (New England Biolabs), Phusion DNA polymerase (Thermo Fisher Scientific), and Taq ligase (iCloning). All sequences were verified using Sanger sequencing (Sequencing platform in the School of Life Sciences of Peking University). DNA encoding the various DA receptor subtypes (D1R-D5R) was generated using PCR amplification of the full-length human GPCR cDNAs (hORFeome database 8.1). For characterization in HEK293T cells, the GRABDA constructs were cloned into the pDisplay vector (Invitrogen), with an IgK leader sequence inserted upstream of the coding region. The IRES-mCherry-CAAX gene was attached downstream of GRABDA and was used as a reference of membrane marker to calibrate the signal intensity. Site-directed mutagenesis of the N- and C-terminal linker sequences in cpEGFP was performed using primers containing randomized NNB codons (48 codons in total, encoding the 20 possible amino acids; Thermo Fisher Scientific). Site-directed mutagenesis of the D2R gene was performed using primers containing the target sites. For characterization in cultured neurons, the GRABDA constructs were cloned into the pAAV vector under the TRE promoter or the human synapsin promoter. The marker constructs RFP(mScarlet)-CAAX, EGFP-CAAX, KDELR1-EGFP, PSD95- RFP(mScarlet) and synaptophysin-RFP(mScarlet) were cloned into pEGFP-N3 vector. To assess the brightness of GRABDA sensors, D2R-EGFP chimera with EGFP inserted into the same insertion site within ICL3 was constructed as the reference. The pH sensitive fluorescent protein pHluorin (Miesenböck et al., 1998) was attached to the N terminus of D2R to measure the internalization of D2R in response to DA application. To generate stable cell lines expressing D2R or DA1h, a self-constructed vector containing elements including 3’ TR, DNA encoding myc tag, 2A sequence, DNA encoding mcherry,DNA encoding puromycin and 5’ TR was made. The DNA of D2R or DA1h was cloned into the pPacific vector using Gibson Assembly. Two mutations (S103P and S509G) were introduced to pCS7-PiggyBAC (VIEWSOLID BIOTECH) to generate a hyperactive piggyBac transposase (Yusa et al., 2011) for the construction of stable cell lines.
Expression of GRABDA in cultured cells and in vivo
The HEK293T cells and Hela cells were plated on 12-mm glass coverslips in 24-well plates and grown to ~50% confluence for transfection. Transfection was performed by incubating the cells with a mixture containing 1-μg DNA and 3-μg PEI for 6 h. Imaging was performed 24–48 h after transfection.
The cultured neurons were transfected 7–9 days later using the calcium phosphate transfection method. Transfection was performed by incubating the neurons with a mixture containing 125mM CaCl2, HBS (pH 7.04–7.12) and 2 μg DNA for 2 h. Then the DNA-Ca3(PO4)2 precipitate was removed from the culture by washing the coverslips with preheated HBS (pH 6.80). Imaging was performed 48–72 h after transfection.
For in vivo expression, mice with the age of P42–60 were first anesthetized by 2,2,2-Tribromoethanol (Avetin, 500 mg/kg, Sigma-Aldrich) through intraperitoneal injection, or by isoflurane (RWD Life Science), and then placed in a stereotaxic frame to inject AAVs with a microsyringe pump (Nanoliter 2000 Injector, WPI, or Nanoject II, Drummond Scientific). In Fig. 5, the AAVs of hSyn-DA1m/DA1h-mut and hSyn-tdTomato were injected in the dorsal striatum (AP: −0.5mm, ML: ±2.5mm, and DV: −2.2mm), and the AAV of Ef1a-DIO-C1V1-YFP was injected in the substantia nigra pars compacta (SNc) (AP: −3.1mm, ML: ±1.5mm, and DV: −4.0mm) in TH-Cre mice (B6.FVB(Cg)-Tg(TH-cre)FI172Gsat/Mmucd, MMRRC). In Fig. 6, 7 and S6I-L, the AAVs of hSyn-DA1m/DA1h/DA1h-mut were injected into NAc (Coordinates: (AP: −1.40 mm, ML: 1.00 mm, DV: 3.90 mm)) of wild-type C57/BL6 mice (Beijing Vital River Laboratory/Charles River Laboratories) unilaterally with ~300–500 nL per animal. In Fig. S6A–H, the AAV of hSyn-DA1h was injected into NAc (Coordinates: (AP: −1.40 mm, ML: 1.00 mm, DV: 3.90 mm)), and the AAV of hSyn-Flex-ChrimsonR-tdTomato was injected into the VTA (Coordinates: (AP: −3.28 mm, ML: 0.5 mm, DV: 4.0 mm)) of DAT-IRES-Cre mice (Jackson Laboratory).
Fluorescence imaging of cultured cells
Imaging was performed using an inverted Ti-E A1 confocal microscope (Nikon) and the Opera Phenix high content screening system (PerkinElmer). The Nikon confocal microscope was equipped with a 40×/1.35 NA oil immersion objective, a 488-nm laser and a 561-nm laser. During imaging, the cultured cells were bathed or perfused in a chamber with Tyrode’s solution containing (in mM): 150 NaCl, 4 KCl, 2 MgCl2, 2 CaCl2, 10 HEPES and 10 glucose (pH 7.4). Solutions containing the drug/compound of interest (DA (Sigma-Aldrich), Halo (Tocris), SCH 23390 (Tocris), Etic (Tocris), L-DOPA (Abcam), 5-HT (Tocris), histamine (Tocris), Glu (Sigma-Aldrich), GABA (Tocris), Ado (Tocris), ACh (Solarbio), NE (Tocris), Tyr (Sigma-Aldrich), Oct (Tocris)) were delivered via a custom-made perfusion system or via bath application. The chamber was fully cleaned with Tyrode’s solution and 75% ethanol between experiments. The GFP signals (e.g., the GRABDA sensors, the pHluorin-D2R, the iGluSnFR or EGFP-CAAX) were recorded using a 525/50-nm emission filter, and the RFP signals were collected using a 595/50-nm emission filter. The photostability was measured under 1-photon illumination (confocal microscopy) using a 488-nm laser with the laser power of ~350 μW and the intensity of ~1.75×102 W/cm2, and under 2-photon illumination using a 920-nm laser with the laser power of ~27.5 mW and the intensity of ~1.375×104 W/cm2. Photobleaching was applied to the entire sensor-expressing HEK293T cell with the area of ~200 μm2. The Opera Phenix high content screening system was equipped with a 60×/1.15 NA water immersion objective, a 488-nm laser, and a 561-nm laser. The GRABDA signals were collected using a 525/50-nm emission filter, and the mCherry signals were collected using a 600/30-nm emission filter. Where indicated, the culture medium was replaced with 100 μl of Tyrode’s solution containing various concentrations of the indicated drug/compound. The fluorescence intensities of the GRABDA sensors were calibrated using mCherry as the reference.
cAMP-dependent reporter gene assay
The assay was performed to compare forskolin-induced cAMP levels in WT-D2R- or GRABDA-expressing cells in response to DA application. The DA1h- or D2R-expressing cell line was transfected with the plasmid encoding the luciferase under the control of a cAMP response element. The culture medium was replaced to new culture medium before the experiment. DA was then applied to the cells to a final concentration of 100 μM. After 15 min, culture medium containing forskolin (Sigma-Aldrich) was applied to a final concentration of 10 μM to elevate the cAMP level. The same amount of culture medium was added to the control group. The cells were then cultured for 6 hours to enable the expression of the luciferase. Furimazine (NanoLuc® Luciferase Assay, Promega) was then applied to a final concentration of 5 μM and the luminescence was measured by VICTOR X5 multilabel plate reader (PerkinElmer).
GTP-γ-S binding assay
DA1m- or DA1h-IRES-mcherry-CAAX was expressed in HEK293T cells for over 24 hours in poly-D-lysine pre-coated 96-well plates and the medium was replaced by 100 μl of Tyrode’s solution before experiments. Cells were subsequently incubated with 50 μg/ml digitonin (Sigma-Aldrich) for 5 min to permeabilize the cell membrane and washed for 2 times with 100 μl Tyrode’s solution. Tyrode’s solution containing 100 μM GTP-γ-S (Sigma-Aldrich) was applied to the permeabilized cells in the experimental group and the cells were incubated for 10 min. The GRABDA signals and the membrane-localized RFP signals of both the experimental group and the control group were collected by Opera Phenix high content screening system (PerkinElmer) mentioned above, before and after, various concentration of DA (ranging from 0.01 nM to 1 μM) was applied.
Tango assay
DA of various concentration (ranging from 0.1 nM to 3.1 μM) was applied to D2R- or DA1h-expressing luciferase reporter cell lines stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion gene (Kroeze et al., 2015). The cells were then cultured for 12 hours to enable the expression of the luciferase. Furimazine (NanoLuc® Luciferase Assay, Promega) was then applied to a final concentration of 5 μM and the luminescence was measured by VICTOR X5 multilabel plate reader (PerkinElmer).
Fluorescence imaging of GRABDA in brain slices
Adeno-associated viruses (AAVs) carrying either DA1m or DA1h were injected into the nucleus accumbens (NAc) of mice and acute brain slices containing NAc were prepared two weeks later. Two weeks after the virus injection, the animals were anesthetized with IP injection of Avetin (250mg/kg) and then decapitated. The brains were removed immediately and placed directly in cold slicing buffer containing (in mM): 110 choline-Cl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 7 MgCl2, 25 glucose, and 2 CaCl2. The brains were then sectioned into 200-μm thick slices using a VT1200 vibratome (Leica), and the sectionswere transferred into the oxygenated Ringer’s buffer containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 1.3 MgCl2, 25 glucose, and 2 CaCl2. The slices were then allowed to recover in 34 °C for at least 40 min. For fluorescence imaging, the slices were transferred to an imaging chamber in an FV1000MPE two-photon microscope (Olympus) equipped with a 40×/0.80 NA water-immersion objective and a mode-locked Mai Tai Ti: Sapphire laser (Spectra-Physics) tuned to 920 nm for the excitation of GRABDA sensors and a 495~540-nm filter for signal collection. For electrical stimulation, a concentric electrode (model #CBAEC75, FHC) was positioned near the NAc core under the fluorescence guidance, and the imaging and stimulation were synchronized using an Arduino board with custom programs. The stimulationvoltage was set at 5–6 V except for minimal stimulation experiments, and the duration of each stimulation pulse was typically set at 2 ms (1 ms for minimal stimulation experiments).
For immunostaining of brain sections, GRABDA-expressing mice or non-injection control C57/BL6 mice were anesthetized with Avetin, and the heart was perfused with 0.9% NaCl followed with 4% paraformaldehyde (PFA). The brain was then removed and placed in 4% PFA for 4 hour, then cryoprotected in 30% (w/v) sucrose for 24 hour. The brain was embedded into tissue-freezing medium, and 50-μm-thick coronal sections were cut using a CM1900 cryostat (Leica). To label GRABDA and dopaminergic neurons in VTA and NAc, tissue sections were rinsed and then immunostained with chicken anti-GFP antibody (1:500, Abcam, Cat#ab13970) and rabbit anti-TH antibody (Millipore, Cat#ab152, 1:500 for VTA slices and 1:100 for NAc slices), followed by the Alexa-488-conjugated goat-anti-chicken (1:200, AAT-Bio, Cat#16687) and Alexa-555-conjugated goat-anti-rabbit (1:200, AAT-Bio, Cat#16690) secondary antibodies. The immunostained tissue sections were imaged using the same Nikon confocal microscope used in cell imaging.
Fluorescence imaging of transgenic flies
Adult Drosophila (within 3 weeks after eclosion) were used for imaging experiments. The fly was mounted on a customized chamber by tape, in a way the antenna and abdomen exposed to the air. A section of rectangular cuticle between the eyes, as well as air sacs and fat bodies were removed to expose the brain, which was bathed in saline, the so called adult hemolymph-like solution (AHLS): (in mM) 108 NaCl, 5 KCl, 5 HEPES, 5 Trehalose, 5 sucrose, 26 NaHCO3, 1 NaH2PO4, 2 CaCl2 and 1~2 MgCl2. The same Olympus two-photon microscope used for brain slices imaging was also used here. For DA1m imaging, 920-nm excitation laser and 495~540-nm filter were used. Fortwo-color imaging with DA1m and jRCaMP1a, 1000-nm excitation laser, 495~540-nm filter and 575~630-nm filter were used. For odor stimulation, the odorant isoamyl acetate (Sigma-Aldrich, Cat#306967) was firstly diluted by 200-fold in mineral oil in a bottle and secondly diluted by 5-fold in air, which was then delivered to the fly’s antenna at a rate of 1000 ml/min. Compounds such as Halo (Tocris) and cocaine (Qinghai Pharmaceuticals) were added directly to the AHLS to their final concentration, and the following experiments were performed 10 min after compound application. For electrical stimulation, a glass electrode (resistance ~0.2 M_) was placed in the region of the DANs in the MB and the stimulation voltage was set at 20~80 V. For body shock, two wires were attached to the abdomen of flies, and 60 V electrical pulses were delivered for 500 ms during stimulation. For DA perfusion, a patch of blood-brain-barrier of fly was carefully removed by tweezers, and AHLS containing 100 μM DA was delivered to the brain to exchange normal bath solution. Arduino was used to synchronized stimulation delivery and imaging with custom code. The sampling rates during odor stimulation, electrical stimulation, body shock and DA perfusion were 2.7 Hz, 12 Hz, 7 Hz and 0.5 Hz respectively.
Fluorescence imaging of zebrafish
All experiments were performed on 5 days-post-fertilization (dpf) larvae in 10% Hank’s solution containing (in mM): 140 NaCl, 5.4 KCl, 0.25 Na2HPO4, 0.44 KH2PO4, 1.3 CaCl2, 1.0 MgSO4, and 4.2 NaHCO3 (pH 7.2). Imaging of Tg (elval3: DA1m, DAT: TRPV1-TagRFP) larvae at 5-dpf was performed with an FV3000 inverted confocal microscope (Olympus) by using a 30× oil-immersion objective (1.05 NA, morphology imaging) or an FV1000 upright confocal microscope (Olympus) by using 40× water-immersion objective (0.8 NA, time-lapse imaging). After the larvae were paralyzed with α-bungarotoxin (100 μg/ml, Tocris), they were mounted dorsal side up in 1.5% low melting-point agarose (Sigma-Aldrich) and then immersed in an extracellular solution consisting of (in mM): 134 NaCl, 2.9 KCl, 4 CaCl2, 10 HEPES and 10 glucose (290 mOsmol/L, pH 7.8). To image the sensor expression pattern, images were acquired with a field of view consisting of 1,024×1,024 pixels with spatial resolution of 0.414×0.414×1 μm3 (x × y × z). For bath application of compounds, DA (100 μM in 1 mM ascorbic acid solution, Sigma-Aldrich) was added by pipette at ~4 min and Halo (50 μM in DMSO, Tocris) at ~12 min. These images were acquired with a view field of 640×640 pixels with spatial resolution of 0.497×0.497 μm2 (x × y) at ~1.5 Hz. For functional imaging, small anterior dissections initiated in ventricles were made, after that a glass pipette containing the TRPV1 agonist capsaicin (50 μM in absolute ethanol, Tocris) was advanced through the incision and placed near the cell bodies of the DANs. To activate the DANs, 5 pulses of puffs (9–10 psi, 100 ms) were delivered with 1-min interval. The larvae were bath in Halo (50 μM in DMSO, Tocris) for 10 min before imaging. For visual stimulation, red expanding dots were chosen in case of spectral interference. The visual stimulation was given for 3 s with ~2-min interval. These images were acquired with a field of view consisting of 800×800 pixels with spatial resolution of 0.397×0.397 μm2 (x × y) at ~ 1 Hz.
Fiber Photometry recording in freely moving mice
In all-optic experiments in Fig. 5, optical fiber probes (105 μm core/125 μm cladding) were implanted in the dorsal striatum and in SNc 4 weeks after the virus injection. Fiber photometry recording in the dorsal striatum was performed using a 50-μW 470-nm LED, and C1V1 in the SNc was stimulated using a 9.9-mW 561-nm laser. The measured emission spectra of DA1m and tdTomato were fitted using a linear unmixing algorithm (NIH, https://www.niehs.nih.gov/research/atniehs/labs/ln/pi/iv/tools/index.cfm). The coefficients of DA1m and tdTomato generated by the unmixing algorithm were used to represent the fluorescence intensities of DA1m and tdTomato, respectively (Meng et al., 2018). To evoke C1V1-mediated DA release in the dorsal lateral striatum, pulse trains (10-ms pulses at 10 Hz for 1 s) were delivered to the SNc using a 9.9-mW, 561-nm laser. In other experiments in Fig. 6, 7 and S6, an optic fiber (Thorlabs, FT200UMT, FT400UMT or BFH48–400) was attached to the implanted ferrule (Thorlabs, SF440–10) via a ceramic sleeve. A 400-Hz sinusoidal blue LED light (30 μW) (LED light: M470F1; LED driver: LEDD1B; both from Thorlabs) was bandpass filtered (passing band: 460–490 nm in Fig. 6; 472 ± 15 nm, Semrock, FF02–472/30–25 in Fig. 7 and S6) and delivered to the brain to excite GRABDA sensors. The emission light then traveled through the same optic fiber, was bandpass filtered (passing band: 500–550 nm in Fig. 6; 534 ± 25 nm, Semrock, FF01–535/50 in Fig. 7 and S6), detected by a Femtowatt Silicon Photoreceiver (Newport, 2151) and recorded using a real-time processor (RZ2 for Fig. 6; RZ5 for Fig. 7 and S6; TDT). The envelope of the 400-Hz signals that reflects the intensity of the fluorescence signals was extracted in real-time using a custom TDT program.
Behaviors
For the auditory conditioning task, mice were recovered for >3 days after surgery, and then water-restricted until reaching 85–90% of its original body weight and then prepared for behavior training. In the first Pavlovian task, the mice were trained on two frequency modulated pure tone auditory cues of 500 ms in duration, centered around 2.5 kHz and 11 kHz. For each mouse, one of the two tones was pseudo-randomly assigned to be the reward-predictive tone. Reward (water sweetened with 10% sucrose) was delivered through a water spout in front of the mouth following the reward-predictive cue with a variable 500–1500 ms delay. Rewarded and unrewarded trials were randomly interleaved with a variable inter-trial interval of 8–20 s. Mice experienced 200 trials (~100 rewards) per day in sessions lasting ~45 min.
In the subsequent Pavlovian conditioning task, the mice were trained on an auditory conditioning task, in which three pairs of auditory cues → outcomes pairs (or CS-US pairs; 8 kHz pure tone → 9 μl water; white noise → brief air puff to the face; and 2 kHz pure tone → no response) were delivered at random with a 10 −20 s randomized inter-trial interval. The duration and intensity of each auditory cue was 1 s and 70 dB, respectively. The respective outcomes were delivered 1 s after the end of each auditory cue. The behavioral setup consisted of a custom-built apparatus allowing head fixation of the mouse’s head to a Styrofoam rod (diameter: 15 cm). Rotation of the Styrofoam rod, which corresponds to the animal’s running speed, was detected using an optical rotatory encoder. Licking behavior was detected when the mouse’s tongue contacted the water delivery tube. Each lick signal was processed using an Arduino UNO board with custom code and sent digitally to the training program (written in MATLAB) via a serial port. Water delivery was precisely controlled using a stepping motor pump, and the air puff (15 psi, 25-ms duration) was controlled using a solenoid valve. Timing of the pump and valve was controlled using the same Arduino UNO board used for lick detection, which also provided synchronization between the training program and the TDT data acquisition system. During first two days of each training session, the outcomes were delivered without the prediction cues.
The sexual behaviors are defined following conventions in previous literature (Hull and Rodriguez-Manzo, 2009). DA1h was virally expressed in the NAc of male mice and an optical fiber was implanted to record the bulk fluorescence signal in freely moving animals To confirm that DA1h can detect acute dopamine release in the NAc, Cre-dependent Chrimson-tdTomato was virally expressed in the DANs in the VTA using DAT-ires-Cre mice. Optogenetic stimulation was done three weeks after viral injection. In details, sniffing female was defined as the male’s nose coming in close proximity to the female’s facial, body, and/or urogenital areas. “Mount” was defined as when the male posed his forelegs over the female’s back and with his hindfeet on the ground accompanying shallow pelvic thrusts. The mounting onset was defined as the moment at which the male tried to clasp female back. “Intromission” was defined as a deep rhythmic thrust following mounting. The onset of intromission was defined as the time at which the male performed the first deep thrusting toward the female with vaginal penetration. “Penile grooming” was defined when a male animal repeated grooming for his urogenital area after intromission and ejaculation. Ejaculation is detected when the male stopped thrusting and freeze for seconds. The putative ejaculation event was confirmed by the presence of vaginal copulatory plug.
QUANTIFICATION AND STATISTICAL ANALYSIS
Image data from cultured cells, acute brain slices and transgenic flies, were first processed with Image J software (NIH), traces were generated by Origin 9.1 and pseudocolor images were generated by custom-written MATLAB programs. The data of electrical stimulation experiment in acute brain slices and flies were first binned by 2× and averaged to generate representative traces. The signal to noise ratio (SNR) was calculated as the peak response divided by the standard error of the baseline fluorescence fluctuation. For zebrafish-relevant data, noise reduction by a custom-written MATLAB scripts was performed. For fiber photometry experiment in Fig. 6, baseline was first obtained by subtracting 2nd order exponential fitted data from the raw data after 10.17 Hz binning, and the fluorescence responses were indicated by Z score. The response to the CS was defined as the peak of the normalized PSTH between the CS onset and the US onset, and the response to US was calculated similarly using data from the US onset to data collected 2 s after the US onset. For fiber photometry experiment in Fig. 7 and S6, the signal baseline was first obtained by the MATLAB function “msbackadj” with a moving window of 25% of the total recording duration. The fluorescence responses were calculated as (Fraw–Fbaseline)/Fbaseline. To analyze event-evoked changes in DA release, we aligned each trial to the onset or offset of the behavior, and calculated the peri-stimulus time histogram (PSTH). To compare PSTH changes during different phases of the training, we used data from the 2nd day as naive, the 5–10th day as trained and >10th day as well-trained, and normalized the PSTH of each animal by water-evoked response during early training. The peak response during a behavior was calculated as the maximum ΔF/F0 during the behavior minus the average ΔF/F0 in the 2 s prior to the behavior onset.
Group differences were analyzed using the Student’s t-test, sign-rank test, One-Way ANOVA or post-hoc Tukey’s test. Except where indicated otherwise, all summary data presented as the mean ± SEM.
DATA AND SOFTWARE AVAILABILITY
The self-written MATLAB, Arduino and TDT programs will be provided upon request to the Lead Contact.
Supplementary Material
Movie S1: Single-trial fluorescence response of DA1h expressing neurons to a 50 pulses of electrical stimulation at 20Hz in acute NAc slice of mice, related to Fig.2.
Movie S2: Single-trial fluorescence response of TH > DA1m fly to 1-s odor stimulation, related to Fig.3.
Table S1: PCR primers, related to STAR Methods.
(A) The fluorescence and membrane trafficking of all five DRs with cpEGFP insertion. A membrane-localized RFP (RFP-CAAX) was co-expressed to indicate the plasma membrane and EGFP-CAAX was used as a control. Left, the fluorescence images of HEK293T cells expressing all five DR-based chimeras or EGFP-CAAX (green) and RFP-CAAX (red); scale bar, 10 μM; white bars in overlay panels, line-scanning region. Middle, the normalized line-scanning plots of the fluorescence signals in both green and red channels. Right, Pearson’s colocalization ratios of the DR-based chimeras or EGFP-CAAX according to RFP-CAAX (n = 30/2 for each protein; p < 0.001 comparing D2R with D3R, D4R and D5R; p = 0.001 between D2R and EGFP-CAAX; p = 0.006 between D2R and D1R).
(B) Relative brightness of DA1m and DA1h compared with D2R-EGFP chimeric construct in the absence and presence of 100 μM DA. (n = 4 wells/group with 100–200 cells/well).
(C) Photostability of DA1m and DA1h compared with other fluorescent probes. Top: Representative photobleaching curves of DA1m, DA1h, EGFP-CAAX and iGluSnFR expressed in HEK293T cells under confocal imaging (488 nm laser with the laser power of ~350 μW and the intensity of ~1.75×102 W/cm2 shown in left. The group data of decay time constants of each are shown in right (n = 10/3 of each group; p = 0.350 between DA1m and DA1h; p < 0.001 comparing DA1m with EGFP-CAAX and iGluSnFR). Bottom: Similar as top, except that two-photon laser (920nm laser withthe laser power of ~27.5 mW and the intensity of ~1.38×104 W/cm2) was used to test the photostability of DA1m, DA1h, EGFP-CAAX and GCaMP6s (n = 12/2 of each group; p = 0.251 between EGFP-CAAX and DA1m; p = 0.583 between EGFP-CAAX and DA1h; p = 0.537 between EGFP-CAAX and GCaMP6s; p = 0.051 between DA1m and DA1h; p = 0.678 between DA1m and GCaMP6s; p = 0.236 between DA1h and GCaMP6s).
(D) Schematic image showing the rapid local perfusion system. A glass pipette (black dashed lines) filled with DA or Halo was positioned close to the GRABDA-expressing cells, and fluorescence signals were measured using confocal line-scanning (red line).
(E) Left and middle: Representative fluorescence changes in GRABDA-expressing cells in response to the local perfusion (on rate: 100 μM DA in pipette with normal bath solution; off rate: 1 mM Halo in pipette with bath solution containing 10 μM DA for DA1m or 1 μM DA for DA1h). The traces are the average of 3 different ROIs on the scanning line, shaded with ± SEM and fitted with a single-exponential function (black traces, with the decay time constants shown). Right: Group data summarizing the response kinetics of GRABDA-expressing cells in response to DA (on) or Halo (off) (n = 8 cells/group; p = 0.0093 between on kinetics; p < 0.001 between off kinetics).
(F) Expression of GRABDA sensors in cultured neurons. Scale bars, 20 μm.
(G) Expression and localization of GRABDA sensors (green, G), subcellular markers (red, R) and the overlay (O) in the indicated subcellular compartments in cultured neurons. RFP-CAAX, PSD95-RFP and Synaptophysin-RFP were co-expressed as markers of the plasma membrane, dendritic spines, and presynaptic boutons, respectively. Arrow heads indicate synaptic boutons. Scale bars, 5 μm.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; **, p < 0.01; ***, p < 0.001.
(A and B) Normalized dose-dependent fluorescence responses to DA and NE of DA1m (A) or DA1h (B) expressing cultured neurons (DA1m+DA: n = 10/6; DA1m+NE: n = 12/5; DA1h+DA: n = 10/5; DA1h+NE: n = 11/8).
(C and D) Representative trace (C) and group analysis (D) of the fluorescence changes of DA1m-expressing neurons in response to the sequential application of the indicated compounds at 1 μM, including DA, L-Dopa, 5-HT, NE, His, ACh, DA(2nd) and DA+Halo (n = 12/12; p < 0.001 comparing responses to DA with L-Dopa, 5-HT, NE, His, ACh and DA+Halo).
(E) Fold of induction, which revealed the cAMP level, in the luciferase assay of D2R- (brown) or DA1h- (blue) expressing Nanoluc reporter cells bathed in control normal solution or solution containing 100 μM DA (n = 4 wells/group; p = 0.002 for D2R; p = 0.544 for DA1h).
(F) Normalized fluorescence changes in GRABDA-expressing cells in response to the application of DA, with or without the co-expression of pertussis toxin (PTX) (DA1m: n = 14/3; DA1m+PTX: n = 14/3; DA1h: n = 10/3; DA1h+PTX: n = 10/3; p = 0.680 comparing the EC50 of DA1m with DA1m+PTX; p = 0.810 comparing the EC50 of DA1h with DA1h +PTX).
(G) Fluorescence changes in GRABDA-expressing cells in response to the application of DA, with or without the pre-bathing of GTPγS (n = 3 well/group with 100–300 cell/well; p = 0.488 comparing the EC50 of DA1m with DA1m+GTPγS; p = 0.474 comparing the EC50 of DA1h with DA1h+GTPγS).
(H) Top: The representative trace of the total fluorescence changes in pHluorin-D2R-expressing Hela cells in response to the application of 100 μM DA followed by 10 μM Halo. Bottom: Exemplar traces of normalized fluorescence changes of DA1m- (red) and DA1h- (blue) expressing neurons during a 2-hour application of 100 μM DA.
(I) Quantification of relative internalization of pHluorin-D2R, DA1m and DA1h in response to the application of 100 μM DA (pHluorin-D2R: n = 12/2; DA1m: n = 20/12; DA1h: n = 14/6).
(L) Dose-dependent luminescence units in the tango assay of D2R- (brown) or DA1h- (blue) expressing cells in response to DA application (n = 3 well/group with 100–300 cell/well).
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; **, p < 0.01; 991 ***, p < 0.001.
(A) Schematic illustration of the DA1h virus injection, NAc slice preparation and minimal electrical stimulation in mice.
(B-D) Left, the fluorescence responses of DA1h-expressing neurons before (baseline) and after stimulation (stim) in a total of 100 trials under minimal stimulation condition (B), of which 31 trials were successes (C) and 69 trials were failures (D). The ΔF/F0 of 3 frames before stimulation and immediately after stimulation were averaged as “baseline” and “stim”.
Middle, the averaged fluorescence traces of all 100 trials (B), of 31 success trials (C), of 69 failure trials (D).
Right, the distribution of peak ΔF/F0 of all 100 trials (B, blue), with success trials highlighted in (C, red) and failure trials highlighted in (D, black).
(E) Similar as (B), except that ACSF containing 200 μM Cd2+ was bathed to the same slice.
(F) A different DA1h expressing slice where distributions of peak ΔF/F0 in 100 minimal stimulation trials in ACSF (gray) and in ACSF containing Cd2+ (light blue) were compared.
(G) Similar as (F), except the distributions of peak ΔF/F0 in ACSF (blue) and ACSF containing Cd2+ (light blue) were compared.
(H) Distribution of the group data under minimal stimulation, where peak ΔF/F0 from individual DA1h expressing slices was normalized and pooled together (~400 stimulation trials in total from 4 slices of 3 mice). Note, the normalization was done by first fitting for each set of data to identify the peak of failures as “zero” and peak of success as “one”.
Values with error bars indicate mean ± SEM.
(A) Top: representative immunoreactive signals of GFP (green) and TH (red) in NAc slices of DA1m injected mice (left) or non-injection control mice (right). Scale bar, 1mm.
Bottom: representative immunoreactive signals of GFP (green) and TH (red) in VTA slices of DA1m injected mice with (left) or without (right) the application of primary antibodies. Scale bar, 10 °m.
(B) Group analysis of the signal-to-noise ratio (SNR) of the fluorescence responses of DA1m- or DA1h-expressing neurons to a trial of electrical stimuli at different pulse numbers (left, n = 5 slices from 3 mice in DA1m; n = 7 slices from 4 mice in DA1h), or a trial of 10-pulse electrical stimuli with different frequencies (right, n = 3 slices from 2 mice in DA1m; n = 5–8 slices from 3 mice in DA1h).
(C) Three single trials (gray) and corresponding averaged trials (red in DA1m, blue in DA1h) of fluorescence responses of DA1m- or DA1h-expressing neurons to a train of 20 Hz electrical stimuli containing the indicated pulse numbers.
(D) Similar as in (C) except that a train of 10-pulse electrical stimuli at the indicated frequencies was applied.
(E-F) Fluorescence changes of TH > DA1m flies in response to electrical stimuli with indicated pulses at 20 Hz. Representative 3 single trial traces (light) with corresponding averaged traces (bold) from one fly are shown in (E). Group data of peak ΔF/F0 are summarized in (F) (n = 9 flies/group).
(G) Fluorescence changes of TH > DA1m and TH > DA1m-mut flies in response to 40-pulse electrical stimuli at 20 Hz, in normal saline or in saline containing 10 μM Halo. Representative 6 single trial traces (light) with corresponding averaged traces (bold) from one fly/group are shown.
(H) Fluorescence changes of TH > DA1m flies in response to 1-s odor stimulation in control condition (left), in the presence of the DAT blocker cocaine (3 °M) (middle), or in DAT-RNAi genetic background (right). Representative 2–3 single trial traces (light) with corresponding averaged traces (bold) from one fly/group are shown.
Values with error bars indicate mean ± SEM.
(A) Top panel, schematic illustration showing the TH > DA1m fly in vivo two-photon imaging. Bottom panels, the cartoon and the field of view indicate MB γ2–5 compartments in this focal plane.
(B1–B3) Fluorescence signals of TH > DA1m fly in response to 1-s odor, 500-ms body shock and 100 μM DA perfusion. Representative pseudocolor images and corresponding intensity profiles from one fly are shown in top and middle panels. Group data are summarized in bottom panels (n = 4 flies for odor; n = 6 flies for body shock; n = 3 flies for DA perfusion; for odor, comparing γ4 with γ2, γ3 and γ5, p = 0.011, 0.027 and 0.002; for body shock, comparing γ2 with γ3, γ4 and γ5, p = 0.211, 0.009 and 0.027; for DA perfusion, p = 0.967 between γ2 and γ3, p = 0.284 between γ2 and γ4; p = 0.305 between γ2 and γ5; p = 0.131 between γ3 and γ4; p = 0.282 between γ3 and γ5; p = 0.944 between γ4 and γ5).
(C) Schematic illustration and the field of view depicting the expression of DA1m in single DAN each hemisphere driven by MB296B-GAL4.
(D and E) Fluorescence signals of MB296B > DA1m fly in response to 1-s odor, 500-ms body shock and 100 μM DA perfusion. Representative pseudocolor images (averaged from 3-trials) and corresponding traces (light, 3 single trial traces; bold, 3-trial averaged traces) from one fly are shown in (D). The group analysis of peak ΔF/F0 are summarized in (E) (n = 5 flies for odor; n = 6 flies for body shock; n = 6 flies for DA perfusion; p < 0.001 between sham and body shock; p = 0.013 between sham and DA perfusion; p = 0.178 between body shock and DA perfusion).
(F) Schematic illustration of the MB β’ lobe.
(G and H) Fluorescence responses of jRCaMP1a- and DA1m-exprssing DANs (left) or Kenyon cells (right) to 1-s odor stimulation. Representative pseudocolor images and corresponding 3-trial-averaged traces shaded with ± SEM from one fly are shown in (G). Group data of the integrals of jRCaMP1a signals are summarized in (H) (TH > jRCaMP1a: n = 10 flies; TH > jRCaMP1a, DA1m: n = 11 flies; 30y > jRCaMP1a: 11 flies; 30y > jRCaMP1a, GRABDA1m: n = 12 flies; p = 0.503 between TH > jRCaMP1a and TH > jRCaMP1a, DA1m; p = 0.097 between 30y > jRCaMP1a and 30y > jRCaMP1a, DA1m).
Scale bars in (A), (C), and (G) are 25 μm.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Schematic diagram showing the experimental design used to record DA1h signals in the NAc (AAV hsyn-DA1h) while optogenetically activating the VTA DANs (AAV hsyn-Flex-ChrimsonR-tdTomato). 593-nm 20-ms yellow light was delivered at 20 Hz for 500 ms in each trial, and 60 trials were applied with 20-s interval to one animal.
(B-E) Representative fluorescence changes during optogenetic stimulations from one mouse. (B) Snapshot of 6-tral. (C) Heat map showing all 60-trial fluorescence signals aligned to the light onset. (D) Post-event histogram (PETH) showing the averaged fluorescence changes of 60-trial light stimulation (blue) and 1000× shuffled controls (black). Shades represent ±SEM. (E) The distributions of mean ΔF/F0 of shuffled controls. The colored dot and arrow indicate the actual mean ΔF/F0 during the light stimulation.
(F) Group data summarizing the mean ΔF/F0 increase during light stimulation (n = 4 animals). Error bar: ±SEM. Student t-test is performed to compare the mean ΔF/F0 of stimulation with that of shuffled controls, F(3) = 4.60, p = 0.019.
(G) Z score normalized PETHs of all 4 animals aligned to the light onset. The Z score at each time point is calculated using the -6 to 0 s prior to light onset as the baseline. Magenta line indicates the average latency to reach Z =2. Red line indicates the average latency to reach peak response.
(H) Group data summarizing the latency to reach significant increase (Z=2) and peak response after light stimulation.
(I) Schematic diagram showing the experimental design used to record DA1h-mutant signals in the NAc (AAV hsyn-DA1h-mut) of male mice during sexual behaviors.
(J1–J2) Representative fluorescence changes right before female introduction (J1) and during male sexual behaviors (J2). The shaded areas with colors indicate different behavioral events.
(K1–K5) Top: PETHs showing the fluorescence changes aligned to various behavioral events. Black lines show averaged PETHs of 1000× shuffled controls. Shades represent ±SEM. Bottom: The distributions of mean ΔF/F0 of shuffled controls. Colored dots and arrows indicate the actual mean ΔF/F0 during each behavior. Mean ΔF/F0 is calculated as averaged ΔF/F0 from 0–1s after the behavioral onset minus the baseline ΔF/F0. Baseline for sniff, mount, intromit and lick is defined as -12 to -2 s before the onset of each behavioral episode. Baseline for ejaculate is defined as 10 to 20 s after the onset of the behavior given that ejaculation is always preceded by mounting and intromission.
(L) Group data summarizing the mean ΔF/F0 during various behaviors of two animals. One-way ANOVA with repeated measures. Among behaviors: F(3, 4) = 0.63. p = 0.473.
Values with error bars indicate mean ± SEM.
Highlights.
GRABDA sensors are genetically-encoded dopamine (DA) sensors based on GPCR.
GRABDA enables fast, sensitive DA detection with molecular and cellular specificity.
Two separate constructs expand the effective dynamic range of DA detection.
Sensors report DA dynamics in vivo in multiple organisms & during complex behaviors.
Acknowledgements
This work was supported by National Basic Research Program of China (973 Program; grant 2015CB856402), the General Program of National Natural Science Foundation of China (project 31671118), the NIH BRAIN Initiative (NS103558), the Junior Thousand Talents Program of China, the grants from the Peking-Tsinghua Center for Life Sciences and the State Key Laboratory of Membrane Biology at Peking University School of Life Sciences to Y. Li; the NIH training grants F32 NS083369 and K99 MH110597 to S.F. Owen; the Junior Thousand Talents Program of China to S. Zhang; the grant from the Ministry of Science and Technology of the People’s Republic of China (2017YFA0505700) to M. Xu; the Uehara postdoctoral fellowship to T. Yamaguchi; the NIH R01MH101377, R21HD090563 and an Irma T. Hirschl Career Scientist Award to D. Lin; the Intramural Research Program of the NIH/NIEHS of the United States (1ZIAES103310) to G. Cui.
We thank Y. Rao for sharing the 2-photon microscope and X. Lei for the platform support of Opera Phenix high content screening system at PKU-CLS. We thank the Core Facilities at School of Life Sciences, Peking University for technical assistance. We thank R. Mooney, Y. Huang and L. Luo for valuable feedback of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Y.L. conceived and supervised the project. F.S., M.J., and J.Z. performed experiments related to sensor development, optimization and characterization in culture HEK cells, culture neurons, brain slices and transgenic flies, with the initial work from YC. L. and J. F., and help from H.W., Z.Y., F.L. and J.D. designed and performed experiments on transgenic fish. J.Z., Y.G., T.Y., W.P., S.O., L.W., S.Z., D.L., M.X., A.K., and G.C. designed and performed experiments in behaving mice. All authors contributed to data interpretation and data analysis. Y.L. wrote the manuscript with input from F.S., J.Z., M.J., D.L., S.O., M.X. and help from other authors.
Declaration of Interests
The authors declare competing financial interests. F.S., J.Z., M.J. and Y.L. have filed patent applications whose value might be affected by this publication.
References
- Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H , et al. (2014). The neuronal architecture of the mushroom body provides a logic for associative learning. Elife 3, e04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainton RJ, Tsai LT, Singh CM, Moore MS, Neckameyer WS, and Heberlein U (2000). Dopamine modulates acute responses to cocaine, nicotine and ethanol in Drosophila. Curr Biol 10, 187–194. [DOI] [PubMed] [Google Scholar]
- Balaji J, and Ryan TA (2007). Single-vesicle imaging reveals that synaptic vesicle exocytosis and endocytosis are coupled by a single stochastic mode. Proceedings of the National Academy of Sciences 104, 20576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleine BW, Delgado MR, and Hikosaka O (2007). The role of the dorsal striatum in reward and decision-making. J Neurosci 27, 8161–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnea G, Strapps W, Herrada G, Berman Y, Ong J, Kloss B, Axel R, and Lee KJ (2008). The genetic design of signaling cascades to record receptor activation. Proceedings of the National Academy of Sciences 105, 64–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, and Gainetdinov RR (2011). The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 63, 182–217. [DOI] [PubMed] [Google Scholar]
- Berridge KC, and Robinson TE (1998). What is the role of dopamine in reward: hedonic impact, reward learning, or incentive salience? Brain Res Brain Res Rev 28, 309–369. [DOI] [PubMed] [Google Scholar]
- Berry JA, Cervantes-Sandoval I, Nicholas EP, and Davis RL (2012). Dopamine is required for learning and forgetting in Drosophila. Neuron 74, 530–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungay PM, Newton-Vinson P, Isele W, Garris PA, and Justice JB (2003). Microdialysis of dopamine interpreted with quantitative model incorporating probe implantation trauma. Journal of Neurochemistry 86, 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke CJ, Huetteroth W, Owald D, Perisse E, Krashes MJ, Das G, Gohl D, Silies M, Certel S, and Waddell S (2012). Layered reward signalling through octopamine and dopamine in Drosophila. Nature 492, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chefer VI, Thompson AC, Zapata A, and Shippenberg TS (2009). Overview of Brain Microdialysis. Current Protocols in Neuroscience 47, 7.1.1–7.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichewicz K, Garren E, Adiele C, Aso Y, Wang Z, Wu M, Birman S, Rubin G, and Hirsh J (2017). A new brain dopamine - deficient Drosophila and its pharmacological and genetic rescue. Genes, Brain and Behavior 16, 394–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognigni P, Felsenberg J, and Waddell S (2017). Do the right thing: neural network mechanisms of memory formation, expression and update in Drosophila. Curr Opin Neurobiol 49, 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn R, Morantte I, and Ruta V (2015). Coordinated and compartmentalized neuromodulation shapes sensory processing in Drosophila. Cell 163, 1742–1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH Jr, Stein MA, Krasowski MD, Cox NJ, Olkon DM, Kieffer JE, and Leventhal BL (1995). Association of attention-deficit disorder and the dopamine transporter gene. American journal of human genetics 56, 993. [PMC free article] [PubMed] [Google Scholar]
- da Silva JA, Tecuapetla F, Paixao V, and Costa RM (2018). Dopamine neuron activity before action initiation gates and invigorates future movements. Nature 554, 244–248. [DOI] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, et al. (2016). Sensitive red protein calcium indicators for imaging neural activity. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL (1993). Mushroom bodies and Drosophila learning. Neuron 11, 1–14. [DOI] [PubMed] [Google Scholar]
- Daw ND, and Tobler PN (2013). Value Learning through Reinforcement: The Basics of Dopamine and Reinforcement Learning. In, pp. 283–298. [Google Scholar]
- Di Chiara G, and Imperato A (1988). Drugs abused by humans preferentially increase synaptic dopamine concentrations in the mesolimbic system of freely moving rats. Proc Natl Acad Sci U S A 85, 5274–5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrunz LE, and Stevens CF (1999). Response of hippocampal synapses to natural stimulation patterns. Neuron 22, 157–166. [DOI] [PubMed] [Google Scholar]
- Florin-Lechner SM, Druhan JP, Aston-Jones G, and Valentino RJ (1996). Enhanced norepinephrine release in prefrontal cortex with burst stimulation of the locus coeruleus. Brain Res 742, 89–97. [DOI] [PubMed] [Google Scholar]
- Glimcher PW (2011). Understanding dopamine and reinforcement learning: The dopamine reward prediction error hypothesis. Proceedings of the National Academy of Sciences 108, 15647–15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales RA, Job MO, and Doyon WM (2004). The role of mesolimbic dopamine in the development and maintenance of ethanol reinforcement, pp. 121–146. [DOI] [PubMed] [Google Scholar]
- Graybiel AM, Aosaki T, Flaherty AW, and Kimura M (1994). The basal ganglia and adaptive motor control. Science 265, 1826–1831. [DOI] [PubMed] [Google Scholar]
- Heisenberg M (2003). Mushroom body memoir: from maps to models. Nat Rev Neurosci 4, 266–275. [DOI] [PubMed] [Google Scholar]
- Holroyd CB, and Coles MGH (2002). The neural basis of human error processing: reinforcement learning, dopamine, and the error-related negativity. Psychol Rev 109, 679–709. [DOI] [PubMed] [Google Scholar]
- Howe MW, and Dombeck DA (2016). Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 535, 505–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howes OD, and Kapur S (2009). The dopamine hypothesis of schizophrenia: version III—the final common pathway. Schizophrenia bulletin 35, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki HK, de-Leon SB-T, Wong AM, Jagadish S, Ishimoto H, Barnea G, Kitamoto T, Axel R, and Anderson DJ (2012). Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signaling reveals appetite control of sugar sensing. Cell 148, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquins-Gerstl A, and Michael AC (2015). A review of the effects of FSCV and microdialysis measurements on dopamine release in the surrounding tissue. Analyst 140, 3696–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J (1992). Microdialysis/HPLC On-Line Dopamine and Metabolites, pp. 1–4. [Google Scholar]
- Kim MW, Wang W, Sanchez MI, Coukos R, Von Zastrow M, and Ting AY (2017). Time-gated detection of protein-protein interactions with transcriptional readout. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Ote M, Tazawa T, and Yamamoto D (2005). Fruitless specifies sexually dimorphic neural circuitry in the Drosophila brain. Nature 438, 229–233. [DOI] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang X-P, Lansu K, McCorvy JD, Giguère PM, Sciaky N, and Roth BL (2015). PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nature Structural &Amp; Molecular Biology 22, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Creed M, Jung K, Stefanelli T, Wendler DJ, Oh WC, Mignocchi NL, Lüscher C, and Kwon H-B (2017). Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nature methods 14, 495. [DOI] [PubMed] [Google Scholar]
- Liu C, Placais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, and Tanimoto H (2012). A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature 488, 512–516. [DOI] [PubMed] [Google Scholar]
- Lohse MJ, Hein P, Hoffmann C, Nikolaev VO, Vilardaga JP, and Bunemann M (2008). Kinetics of G-protein-coupled receptor signals in intact cells. Br J Pharmacol 153 Suppl 1, S125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotharius J, and Brundin P (2002). Pathogenesis of Parkinson’s disease: dopamine, vesicles and α-synuclein. Nature Reviews Neuroscience 3, 932. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, and Lefkowitz RJ (2002). The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci 115, 455–465. [DOI] [PubMed] [Google Scholar]
- Mao Z, and Davis RL (2009). Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front Neural Circuits 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Zhou J, Papaneri A, Peddada T, Xu K, and Cui G (2018). Spectrally Resolved Fiber Photometry for Multi-component Analysis of Brain Circuits. Neuron 98, 707–717 e704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Jing PZ, Guangfu Wang, et al. (2018). A genetically encoded acetylcholine indicator. Nature Biotechnology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenböck G, De Angelis DA, and Rothman JE (1998). Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192. [DOI] [PubMed] [Google Scholar]
- Missale C, Nash SR, Robinson SW, Jaber M, and Caron MG (1998). Dopamine receptors: from structure to function. Physiol Rev 78, 189–225. [DOI] [PubMed] [Google Scholar]
- Muller A, Joseph V, Slesinger PA, and Kleinfeld D (2014). Cell-based reporters reveal in vivo dynamics of dopamine and norepinephrine release in murine cortex. nAture methods 11, 1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair-Roberts RG, Chatelain-Badie SD, Benson E, White-Cooper H, Bolam JP, and Ungless MA (2008). Stereological estimates of dopaminergic, GABAergic and glutamatergic neurons in the ventral tegmental area, substantia nigra and retrorubral field in the rat. Neuroscience 152, 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neves SR, Ram PT, and Iyengar R (2002). G protein pathways, pp. 1636–1639. [DOI] [PubMed] [Google Scholar]
- Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat Methods 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieoullon A (2002). Dopamine and the regulation of cognition and attention. Progress in neurobiology 67, 53–83. [DOI] [PubMed] [Google Scholar]
- Pacak K, Palkovits M, Kopin IJ, and Goldstein DS (1995). Stress-induced norepinephrine release in the hypothalamic paraventricular nucleus and pituitary-adrenocortical and sympathoadrenal activity: in vivo microdialysis studies. Front Neuroendocrinol 16, 89–150. [DOI] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong W.-h., Folk RW, Broussard GJ, Liang R, Jang MJ, et al. (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. 4422, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaus JG, Damsma G, Nomikos GG, Wenkstern DG, Blaha CD, Phillips AG, and Fibiger HC (1990). Sexual behavior enhances central dopamine transmission in the male rat. Brain Research 530, 345–348. [DOI] [PubMed] [Google Scholar]
- Pfeiffer BD, Truman JW, and Rubin GM (2012). Using translational enhancers to increase transgene expression in Drosophila. Proceedings of the National Academy of Sciences 109, 6626–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz MC, Lamb R, and Kuhar M (1987). Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science 237, 1219–1223. [DOI] [PubMed] [Google Scholar]
- Robinson DL, Hermans A, Seipel AT, and Wightman RM (2008). Monitoring rapid chemical communication in the brain. Chem Rev 108, 2554–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DL, Phillips PEM, Budygin E.a., Trafton BJ, Garris P.a., and Wightman RM (2001). Sub-second changes in accumbal dopamine during sexual behavior in male rats. NeuroReport 12, 2549–2552. [DOI] [PubMed] [Google Scholar]
- Robinson DL, Venton BJ, Heien ML, and Wightman RM (2003). Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo. Clin Chem 49, 1763–1773. [DOI] [PubMed] [Google Scholar]
- Schultz W (2007). Behavioral dopamine signals, pp. 203–210. [DOI] [PubMed] [Google Scholar]
- Schultz W (2016). Dopamine reward prediction-error signalling: a two-component response. Nature Reviews Neuroscience 17, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaerzel M, Monastirioti M, Scholz H, Friggi-Grelin F, Birman S, and Heisenberg M (2003). Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in Drosophila. J Neurosci 23, 10495–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleman C, and Holtzman NG (2014). Growth and maturation in the zebrafish, Danio rerio: a staging tool for teaching and research. Zebrafish 11, 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AD, Olson RJ, and Justice JB (1992). Quantitative microdialysis of dopamine in the striatum: effect of circadian variation. Journal of Neuroscience Methods 44, 33–41. [DOI] [PubMed] [Google Scholar]
- Sokoloff P, Giros B, Martres M-P, Bouthenet M-L, and Schwartz J-C (1990). Molecular cloning and characterization of a novel dopamine receptor (D3) as a target for neuroleptics. Nature 347, 146–151. [DOI] [PubMed] [Google Scholar]
- Swandulla D, and Armstrong CM (1989). Calcium channel block by cadmium in chicken sensory neurons. Proceedings of the National Academy of Sciences 86, 1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka NK, Tanimoto H, and Ito K (2008). Neuronal assemblies of the Drosophila mushroom body. J Comp Neurol 508, 711–755. [DOI] [PubMed] [Google Scholar]
- Tidey JW, and Miczek KA (1996). Social defeat stress selectively alters mesocorticolimbic dopamine release: an in vivo microdialysis study. Brain Res 721, 140–149. [DOI] [PubMed] [Google Scholar]
- Ungless MA, and Grace AA (2012). Are you or aren’t you? Challenges associated with physiologically identifying dopamine neurons. Trends in neurosciences 35, 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga J-P, Bünemann M, Krasel C, Castro M, and Lohse MJ (2003). Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nature biotechnology 21, 807–812. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Fischman M, Foltin R, Abumrad NN, Gatley SJ, Logan J, Wong C, Gifford A, et al. (1999). Methylphenidate and cocaine have a similar in vivo potency to block dopamine transporters in the human brain. Life Sciences 65. [DOI] [PubMed] [Google Scholar]
- Wang S, Che T, Levit A, Shoichet BK, Wacker D, and Roth BL (2018). Structure of the D2 dopamine receptor bound to the atypical antipsychotic drug risperidone. Nature 2018, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RA (2004). Dopamine, learning and motivation. Nature reviews neuroscience 5, 483. [DOI] [PubMed] [Google Scholar]
- Wu MV, and Shah NM (2011). Control of masculinization of the brain and behavior. Curr Opin Neurobiol 21, 116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, Ogshea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, et al. (2011). Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K, Zhou L, Li MA, Bradley A, and Craig NL (2011). A hyperactive piggyBac transposase for mammalian applications. Proceedings of the National Academy of Sciences of the United States of America 108, 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler N, Batz J, Zabel U, Lohse MJ, and Hoffmann C (2011). FRET-based sensors for the human M1-, M3-, and M5-acetylcholine receptors. Bioorg Med Chem 19, 1048–1054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1: Single-trial fluorescence response of DA1h expressing neurons to a 50 pulses of electrical stimulation at 20Hz in acute NAc slice of mice, related to Fig.2.
Movie S2: Single-trial fluorescence response of TH > DA1m fly to 1-s odor stimulation, related to Fig.3.
Table S1: PCR primers, related to STAR Methods.
(A) The fluorescence and membrane trafficking of all five DRs with cpEGFP insertion. A membrane-localized RFP (RFP-CAAX) was co-expressed to indicate the plasma membrane and EGFP-CAAX was used as a control. Left, the fluorescence images of HEK293T cells expressing all five DR-based chimeras or EGFP-CAAX (green) and RFP-CAAX (red); scale bar, 10 μM; white bars in overlay panels, line-scanning region. Middle, the normalized line-scanning plots of the fluorescence signals in both green and red channels. Right, Pearson’s colocalization ratios of the DR-based chimeras or EGFP-CAAX according to RFP-CAAX (n = 30/2 for each protein; p < 0.001 comparing D2R with D3R, D4R and D5R; p = 0.001 between D2R and EGFP-CAAX; p = 0.006 between D2R and D1R).
(B) Relative brightness of DA1m and DA1h compared with D2R-EGFP chimeric construct in the absence and presence of 100 μM DA. (n = 4 wells/group with 100–200 cells/well).
(C) Photostability of DA1m and DA1h compared with other fluorescent probes. Top: Representative photobleaching curves of DA1m, DA1h, EGFP-CAAX and iGluSnFR expressed in HEK293T cells under confocal imaging (488 nm laser with the laser power of ~350 μW and the intensity of ~1.75×102 W/cm2 shown in left. The group data of decay time constants of each are shown in right (n = 10/3 of each group; p = 0.350 between DA1m and DA1h; p < 0.001 comparing DA1m with EGFP-CAAX and iGluSnFR). Bottom: Similar as top, except that two-photon laser (920nm laser withthe laser power of ~27.5 mW and the intensity of ~1.38×104 W/cm2) was used to test the photostability of DA1m, DA1h, EGFP-CAAX and GCaMP6s (n = 12/2 of each group; p = 0.251 between EGFP-CAAX and DA1m; p = 0.583 between EGFP-CAAX and DA1h; p = 0.537 between EGFP-CAAX and GCaMP6s; p = 0.051 between DA1m and DA1h; p = 0.678 between DA1m and GCaMP6s; p = 0.236 between DA1h and GCaMP6s).
(D) Schematic image showing the rapid local perfusion system. A glass pipette (black dashed lines) filled with DA or Halo was positioned close to the GRABDA-expressing cells, and fluorescence signals were measured using confocal line-scanning (red line).
(E) Left and middle: Representative fluorescence changes in GRABDA-expressing cells in response to the local perfusion (on rate: 100 μM DA in pipette with normal bath solution; off rate: 1 mM Halo in pipette with bath solution containing 10 μM DA for DA1m or 1 μM DA for DA1h). The traces are the average of 3 different ROIs on the scanning line, shaded with ± SEM and fitted with a single-exponential function (black traces, with the decay time constants shown). Right: Group data summarizing the response kinetics of GRABDA-expressing cells in response to DA (on) or Halo (off) (n = 8 cells/group; p = 0.0093 between on kinetics; p < 0.001 between off kinetics).
(F) Expression of GRABDA sensors in cultured neurons. Scale bars, 20 μm.
(G) Expression and localization of GRABDA sensors (green, G), subcellular markers (red, R) and the overlay (O) in the indicated subcellular compartments in cultured neurons. RFP-CAAX, PSD95-RFP and Synaptophysin-RFP were co-expressed as markers of the plasma membrane, dendritic spines, and presynaptic boutons, respectively. Arrow heads indicate synaptic boutons. Scale bars, 5 μm.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; **, p < 0.01; ***, p < 0.001.
(A and B) Normalized dose-dependent fluorescence responses to DA and NE of DA1m (A) or DA1h (B) expressing cultured neurons (DA1m+DA: n = 10/6; DA1m+NE: n = 12/5; DA1h+DA: n = 10/5; DA1h+NE: n = 11/8).
(C and D) Representative trace (C) and group analysis (D) of the fluorescence changes of DA1m-expressing neurons in response to the sequential application of the indicated compounds at 1 μM, including DA, L-Dopa, 5-HT, NE, His, ACh, DA(2nd) and DA+Halo (n = 12/12; p < 0.001 comparing responses to DA with L-Dopa, 5-HT, NE, His, ACh and DA+Halo).
(E) Fold of induction, which revealed the cAMP level, in the luciferase assay of D2R- (brown) or DA1h- (blue) expressing Nanoluc reporter cells bathed in control normal solution or solution containing 100 μM DA (n = 4 wells/group; p = 0.002 for D2R; p = 0.544 for DA1h).
(F) Normalized fluorescence changes in GRABDA-expressing cells in response to the application of DA, with or without the co-expression of pertussis toxin (PTX) (DA1m: n = 14/3; DA1m+PTX: n = 14/3; DA1h: n = 10/3; DA1h+PTX: n = 10/3; p = 0.680 comparing the EC50 of DA1m with DA1m+PTX; p = 0.810 comparing the EC50 of DA1h with DA1h +PTX).
(G) Fluorescence changes in GRABDA-expressing cells in response to the application of DA, with or without the pre-bathing of GTPγS (n = 3 well/group with 100–300 cell/well; p = 0.488 comparing the EC50 of DA1m with DA1m+GTPγS; p = 0.474 comparing the EC50 of DA1h with DA1h+GTPγS).
(H) Top: The representative trace of the total fluorescence changes in pHluorin-D2R-expressing Hela cells in response to the application of 100 μM DA followed by 10 μM Halo. Bottom: Exemplar traces of normalized fluorescence changes of DA1m- (red) and DA1h- (blue) expressing neurons during a 2-hour application of 100 μM DA.
(I) Quantification of relative internalization of pHluorin-D2R, DA1m and DA1h in response to the application of 100 μM DA (pHluorin-D2R: n = 12/2; DA1m: n = 20/12; DA1h: n = 14/6).
(L) Dose-dependent luminescence units in the tango assay of D2R- (brown) or DA1h- (blue) expressing cells in response to DA application (n = 3 well/group with 100–300 cell/well).
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; **, p < 0.01; 991 ***, p < 0.001.
(A) Schematic illustration of the DA1h virus injection, NAc slice preparation and minimal electrical stimulation in mice.
(B-D) Left, the fluorescence responses of DA1h-expressing neurons before (baseline) and after stimulation (stim) in a total of 100 trials under minimal stimulation condition (B), of which 31 trials were successes (C) and 69 trials were failures (D). The ΔF/F0 of 3 frames before stimulation and immediately after stimulation were averaged as “baseline” and “stim”.
Middle, the averaged fluorescence traces of all 100 trials (B), of 31 success trials (C), of 69 failure trials (D).
Right, the distribution of peak ΔF/F0 of all 100 trials (B, blue), with success trials highlighted in (C, red) and failure trials highlighted in (D, black).
(E) Similar as (B), except that ACSF containing 200 μM Cd2+ was bathed to the same slice.
(F) A different DA1h expressing slice where distributions of peak ΔF/F0 in 100 minimal stimulation trials in ACSF (gray) and in ACSF containing Cd2+ (light blue) were compared.
(G) Similar as (F), except the distributions of peak ΔF/F0 in ACSF (blue) and ACSF containing Cd2+ (light blue) were compared.
(H) Distribution of the group data under minimal stimulation, where peak ΔF/F0 from individual DA1h expressing slices was normalized and pooled together (~400 stimulation trials in total from 4 slices of 3 mice). Note, the normalization was done by first fitting for each set of data to identify the peak of failures as “zero” and peak of success as “one”.
Values with error bars indicate mean ± SEM.
(A) Top: representative immunoreactive signals of GFP (green) and TH (red) in NAc slices of DA1m injected mice (left) or non-injection control mice (right). Scale bar, 1mm.
Bottom: representative immunoreactive signals of GFP (green) and TH (red) in VTA slices of DA1m injected mice with (left) or without (right) the application of primary antibodies. Scale bar, 10 °m.
(B) Group analysis of the signal-to-noise ratio (SNR) of the fluorescence responses of DA1m- or DA1h-expressing neurons to a trial of electrical stimuli at different pulse numbers (left, n = 5 slices from 3 mice in DA1m; n = 7 slices from 4 mice in DA1h), or a trial of 10-pulse electrical stimuli with different frequencies (right, n = 3 slices from 2 mice in DA1m; n = 5–8 slices from 3 mice in DA1h).
(C) Three single trials (gray) and corresponding averaged trials (red in DA1m, blue in DA1h) of fluorescence responses of DA1m- or DA1h-expressing neurons to a train of 20 Hz electrical stimuli containing the indicated pulse numbers.
(D) Similar as in (C) except that a train of 10-pulse electrical stimuli at the indicated frequencies was applied.
(E-F) Fluorescence changes of TH > DA1m flies in response to electrical stimuli with indicated pulses at 20 Hz. Representative 3 single trial traces (light) with corresponding averaged traces (bold) from one fly are shown in (E). Group data of peak ΔF/F0 are summarized in (F) (n = 9 flies/group).
(G) Fluorescence changes of TH > DA1m and TH > DA1m-mut flies in response to 40-pulse electrical stimuli at 20 Hz, in normal saline or in saline containing 10 μM Halo. Representative 6 single trial traces (light) with corresponding averaged traces (bold) from one fly/group are shown.
(H) Fluorescence changes of TH > DA1m flies in response to 1-s odor stimulation in control condition (left), in the presence of the DAT blocker cocaine (3 °M) (middle), or in DAT-RNAi genetic background (right). Representative 2–3 single trial traces (light) with corresponding averaged traces (bold) from one fly/group are shown.
Values with error bars indicate mean ± SEM.
(A) Top panel, schematic illustration showing the TH > DA1m fly in vivo two-photon imaging. Bottom panels, the cartoon and the field of view indicate MB γ2–5 compartments in this focal plane.
(B1–B3) Fluorescence signals of TH > DA1m fly in response to 1-s odor, 500-ms body shock and 100 μM DA perfusion. Representative pseudocolor images and corresponding intensity profiles from one fly are shown in top and middle panels. Group data are summarized in bottom panels (n = 4 flies for odor; n = 6 flies for body shock; n = 3 flies for DA perfusion; for odor, comparing γ4 with γ2, γ3 and γ5, p = 0.011, 0.027 and 0.002; for body shock, comparing γ2 with γ3, γ4 and γ5, p = 0.211, 0.009 and 0.027; for DA perfusion, p = 0.967 between γ2 and γ3, p = 0.284 between γ2 and γ4; p = 0.305 between γ2 and γ5; p = 0.131 between γ3 and γ4; p = 0.282 between γ3 and γ5; p = 0.944 between γ4 and γ5).
(C) Schematic illustration and the field of view depicting the expression of DA1m in single DAN each hemisphere driven by MB296B-GAL4.
(D and E) Fluorescence signals of MB296B > DA1m fly in response to 1-s odor, 500-ms body shock and 100 μM DA perfusion. Representative pseudocolor images (averaged from 3-trials) and corresponding traces (light, 3 single trial traces; bold, 3-trial averaged traces) from one fly are shown in (D). The group analysis of peak ΔF/F0 are summarized in (E) (n = 5 flies for odor; n = 6 flies for body shock; n = 6 flies for DA perfusion; p < 0.001 between sham and body shock; p = 0.013 between sham and DA perfusion; p = 0.178 between body shock and DA perfusion).
(F) Schematic illustration of the MB β’ lobe.
(G and H) Fluorescence responses of jRCaMP1a- and DA1m-exprssing DANs (left) or Kenyon cells (right) to 1-s odor stimulation. Representative pseudocolor images and corresponding 3-trial-averaged traces shaded with ± SEM from one fly are shown in (G). Group data of the integrals of jRCaMP1a signals are summarized in (H) (TH > jRCaMP1a: n = 10 flies; TH > jRCaMP1a, DA1m: n = 11 flies; 30y > jRCaMP1a: 11 flies; 30y > jRCaMP1a, GRABDA1m: n = 12 flies; p = 0.503 between TH > jRCaMP1a and TH > jRCaMP1a, DA1m; p = 0.097 between 30y > jRCaMP1a and 30y > jRCaMP1a, DA1m).
Scale bars in (A), (C), and (G) are 25 μm.
Values with error bars indicate mean ± SEM.
Student’s t-test performed; n.s., not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
(A) Schematic diagram showing the experimental design used to record DA1h signals in the NAc (AAV hsyn-DA1h) while optogenetically activating the VTA DANs (AAV hsyn-Flex-ChrimsonR-tdTomato). 593-nm 20-ms yellow light was delivered at 20 Hz for 500 ms in each trial, and 60 trials were applied with 20-s interval to one animal.
(B-E) Representative fluorescence changes during optogenetic stimulations from one mouse. (B) Snapshot of 6-tral. (C) Heat map showing all 60-trial fluorescence signals aligned to the light onset. (D) Post-event histogram (PETH) showing the averaged fluorescence changes of 60-trial light stimulation (blue) and 1000× shuffled controls (black). Shades represent ±SEM. (E) The distributions of mean ΔF/F0 of shuffled controls. The colored dot and arrow indicate the actual mean ΔF/F0 during the light stimulation.
(F) Group data summarizing the mean ΔF/F0 increase during light stimulation (n = 4 animals). Error bar: ±SEM. Student t-test is performed to compare the mean ΔF/F0 of stimulation with that of shuffled controls, F(3) = 4.60, p = 0.019.
(G) Z score normalized PETHs of all 4 animals aligned to the light onset. The Z score at each time point is calculated using the -6 to 0 s prior to light onset as the baseline. Magenta line indicates the average latency to reach Z =2. Red line indicates the average latency to reach peak response.
(H) Group data summarizing the latency to reach significant increase (Z=2) and peak response after light stimulation.
(I) Schematic diagram showing the experimental design used to record DA1h-mutant signals in the NAc (AAV hsyn-DA1h-mut) of male mice during sexual behaviors.
(J1–J2) Representative fluorescence changes right before female introduction (J1) and during male sexual behaviors (J2). The shaded areas with colors indicate different behavioral events.
(K1–K5) Top: PETHs showing the fluorescence changes aligned to various behavioral events. Black lines show averaged PETHs of 1000× shuffled controls. Shades represent ±SEM. Bottom: The distributions of mean ΔF/F0 of shuffled controls. Colored dots and arrows indicate the actual mean ΔF/F0 during each behavior. Mean ΔF/F0 is calculated as averaged ΔF/F0 from 0–1s after the behavioral onset minus the baseline ΔF/F0. Baseline for sniff, mount, intromit and lick is defined as -12 to -2 s before the onset of each behavioral episode. Baseline for ejaculate is defined as 10 to 20 s after the onset of the behavior given that ejaculation is always preceded by mounting and intromission.
(L) Group data summarizing the mean ΔF/F0 during various behaviors of two animals. One-way ANOVA with repeated measures. Among behaviors: F(3, 4) = 0.63. p = 0.473.
Values with error bars indicate mean ± SEM.


