Abstract
Extracellular vesicles are emerging as potent vehicles of intercellular communication. In this review, we focus on a subclass of extracellular vesicles called exosomes. Previously considered an unimportant catch-all, exosomes have recently been recognized for their role in various diseases and their potential for therapeutic use. We have examined the role of exosomes after human lung transplantation and have delineated the composition of circulating exosomes isolated from lung transplant recipients diagnosed with acute and chronic rejection, primary graft dysfunction, and respiratory viral infection. The presence of lung-associated self-antigens (K-alpha 1 Tubulin and collagen V) and mismatched donor HLA in exosomes isolated from lung transplant recipients signifies that these exosomes originated in the transplanted lungs, and therefore dramatically affect transplant biology and immune pathways. Exosomes released from transplanted organs also carry other proteins, costimulatory molecules, and nucleic acids. Therefore, these molecules may be used as biomarkers for allograft rejection and immunity.
Keywords: exosomes, transplantation, autoimmune response, alloimmune response
1. Introduction
Extracellular vesicles (EVs) are secreted by cells from multicellular and unicellular organisms. EVs are originated from plasma membranes either ectosomal or endosomal in origin. EVs are released from all type of cells undergoing stress including during transformation, activation, drug treatments, surgeries and infections (bacterial, viral, fungal, etc.). EVs are categorized according to size, and are sorted into groups such as ectosomes, oncosomes, shedding microvesicles (MVs), etc. But this classification varies according to different reports. In this review, we discuss MVs (100 to 1000 nm in size), apoptotic bodies (>1000 nm in size), exosomes (40 to 150 nm in size), and oncosomes (secreted by oncogenic cells; >1000 nm in size) [1, 2]. All the vesicles contain a subset of proteins, lipids, RNA and DNA corresponding to their origin from the parent cell. EVs are thought to be involved in intercellular communication by transferring information via small biological molecules (i.e, lipids, carbohydrates, proteins, small metabolites, and nucleic acids) [3, 4]. EVs are being explored for potential use for therapeutic applications, disease prognosis, and biomarker discovery for various diseases. [5]. The EV’s have been studied in cancer during the last two decades but study of EVs are limited in the field of transplantation. Since EVs are released by both immune and non-immune cells it is thought to play important roles in the regulation of immunity during disease, chronic illness, infections and solid organ transplants. In this review we will focus on allograft immunity following solid organ transplantation.
Transplantation is the last option for patients with end stage organ disease. Major organ transplants currently performed are kidney, heart, lung, liver, intestine and pancreas either alone or with kidney [6, 7]. Numbers of recipients seeking organ transplants are outnumbered as compared to availability of suitable donors. This varies for different organs.
A significant number of patients are on the waiting list for organ transplants which reflects the lack of organ donors. Success rate of the transplant depends on the organ transplanted donor/host compatibility and infections, etc. Transplants often undergo acute and chronic rejection which can be due to either cellular or humoral immune response or a combination of both. In addition, donor factors play a critical role especially during the early period following transplantation. Late allograft loss is most often due to chronic allograft damage resulting in progressive decline of graft function years after transplantation.
Although, in solid organ transplants, several mechanisms underlying acute or chronic rejection and leading to graft dysfunction have been reported which includes cellular immune response to mismatched donor human leucocyte antigens (HLA), development of donor specific antibody against mismatched HLA [8], as well as development of antibody tissue restricted self-antigens (SAgs) [9, 10]. Immune responses affecting mainly the small arteries and capillaries can result in cardiac allograft vasculopathy (CAV) [11–13] following heart transplantation, transplant glomerulopathy affecting glomerular basement membrane which histologically recognized as either by duplication, double contouring, or splitting [14, 15] following kidney transplantation. The mechanisms leading to chronic rejection of different organ transplants are currently unknown though both humoral and cellular immune mechanisms play an important role leading to chronic rejection.
Identification of biomarkers involved in allograft function (stable vs rejection) will assist in identifying transplant recipients who are at risk for developing acute or chronic rejection thereby will allow to develop strategies for early intervention to prevent further damage.
It is our contention that EVs, especially exosomes, play an important role in immune activation or suppression of allograft immunity. Presence of allo-antigens, cell specific antigens, peptides, and costimulatory molecules on the surface of exosomes, as well as the presence of nucleic acids, lipids, small RNAs and transcription factors inside the exosomes are released following transplantation making exosomes as one of the attractive targets towards identifying biomarkers associated with allograft immunity. In the current review we will explain the details of allograft immunity mediated by different kinds of EVs emphasizing exosomes released following transplantation during rejection process.
2.1. Microvesicles (MVs)
Description of MVs were first given by Chargaff and West in 1946 in the context with a factor which is perceptible in platelet free plasma and have potential to generate thrombin leading to blood coagulation. MVs were earlier referred as “platelet dust” due to their origin from platelets in plasma/serum [16, 17]. MVs are released by cells that measure between 100–1000 nm in size. MVs are released mostly during stress conditions by the budding/blebbing mechanism of the plasma membrane, and are secreted into the cellular milieu. MV’s are vesicles encapsulated by a phospholipid bilayer and their size overlaps with that of bacteria and insoluble immune complexes.
Vesicles secreted by the plasma membrane reflect intercellular communication through the exchange of cellular material, but the exact nature and origin of these MVs, as well as their secretion and ultimate fate, remain largely unknown.
2.2. Apoptotic Bodies
“Apoptotic body” term was used by Kerr in 1972 followed by Robert Horvitz et al. Apoptotic bodies are released from cells that undergo apoptosis, and their surface composition consists of phosphatidylserine [18, 19]. These apoptotic vesicles are also often referred to as apoptotic bodies. Apoptotic vesicles are reported to be larger than the other secreted vesicles, with their size ranging between 1–5 μm. Apoptotic bodies consist of parts of cells which are undergoing death. Apoptotic bodies consist of DNA (damaged and degraded DNA sequences), metabolites, remnants of cellular organelles. Annexin V and phosphatidyl serine and fragmented DNA are the markers of apoptotic bodies.
2.3. Oncosomes
EVs, from tumor cells are termed as oncosomes, they are large vesicles 1–10μm. Term oncosomes was first described by Janus Rak’s group in 2008 from tumors of the brain [20]. Oncosomes are reported to play a role in the tumor microenvironment by transporting bioactive molecules across tissue spaces and through the blood stream. Oncosomes are capable of carrying microRNA (miRNA), DNA, protein and metabolities. Oncosomes carry mutated DNA and RNA sequences of oncogenes and activated oncoproteins which can lead to horizontal transfer of biological material through intercellular trafficking [21–23]. Cellular uptake of oncogenic cargo through packaged oncosomes induces changes in phenotypes and behavioral pattern of naïve and healthy cells [24]. All the changes in healthy cells are dependent on the contents transferred through oncosomes. Horizontal transfer of mutated nucleic acids and proteins can also lead to resistance to existing therapies. Recent reports have demonstrated the effect on revival of dormant stages of resistant cells in hormone resistant breast cancers [25]. In summary, oncosomes play a critical role in intercellular communication between tumor cells and the tumor microenvironment [26–29].
2.4. Exosomes
Exosomes are membrane-bound vesicles measuring 40–150 nm in size. Exosomes are released by most cells, including mast cells, dendritic cells, B and T lymphocytes, neurons, adipocytes, endothelial cells, and epithelial cells [30–32]. Diseased, unhealthy cells have been noted to secrete more exosomes than healthier cells. The density of exosomes ranges from 1.13–1.19 g/mL, and they have been found in many types of bodily fluids, including blood, urine, ascites, breast milk, saliva, amniotic fluid, lymph fluid, and cerebrospinal fluid, from both healthy and unhealthy individuals [33–39]. Exosomes are cup-shaped vesicles, encapsulated by a lipid bilayer, and feature surface proteins, antigens, and specific markers (e.g, CD9, CD81, tetraspanins, Alix, CD63, tumor susceptibility gene 101, heat shock proteins, and specific markers) depending on the cells that release them [40, 41].
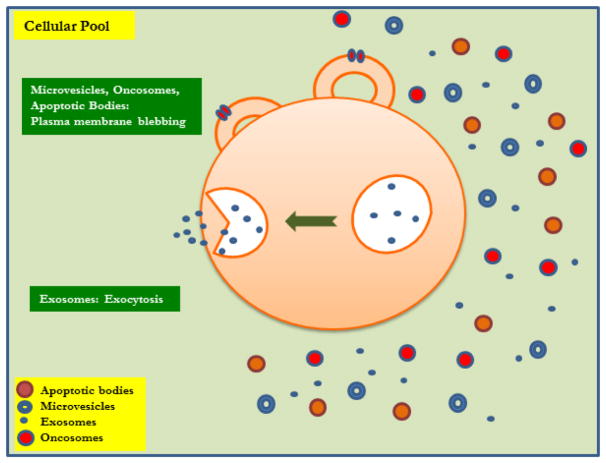
The composition of exosomes depends on their origin and the clinical condition of the individual (eg, cancer, infection, transplantation) [42]. According to a current exosome content database, Exocarta (Version 4), exosomes from various organisms and cell types have been characterized as containing 4563 proteins, 194 lipids, 1639 mRNAs, and 764 miRNA [43, 44]. Exosomes also contain biomolecules, including carbohydrates, proteins, lipids, nucleic acids (ie, DNA and RNA), and metabolites. The lipid and protein composition of exosomes have been studied, and researchers have found that exosomes originating from different types of cells have different lipid and protein compositions depending on the cells’ pathophysiological condition. These structural differences have been studied using Western blotting, fluorescence-activated cell sorting, electron microscopy, and mass spectrometry [45] (Figure 1).
Figure 1. Origin and distribution of Extracellular Vesicles.
Microvesicles, Oncosomes, Apoptotic Bodies and Exosomes.
All the different EV’s ie: MVs, apoptotic bodies, oncosomes and exosomes vary in their structure depending on the type of cytoskeletal proteins, cytoplasmic enzymes, cytokines, chemokines, cell specific antigens, cell signaling molecules, lipids and proteins present on the surface which is described in detail in Table 1. EV’s may be released from same or different cells but possess different functional aspects due to the difference in their architectural constituents and unique signaling molecules depending on the cells from which they are released. To study unique properties of different EV’s their difference in size and distribution have been made use of and differential centrifugation towards purifying EV’s have been employed by different research groups [46–50] (Table 1).
Table 1.
Extracellular Vesicles and their structures
| Exosomes | Microvesicles | Apoptotic bodies | Oncosomes | |
|---|---|---|---|---|
| Size range | Approximately 50–100 nm [30] | 100–1,000 nm (*100–400 nm in blood plasma) [88] | 1–5 μm [89] | 1–10 μm [86] |
| Mechanism of generation | By exocytosis of multivesicular bodies | By budding/blebbing of the plasma membrane | By release from blebs of cells undergoing apoptosis | By membrane blebbing mechanism |
| Isolation | Differential centrifugation and sucrose gradient ultracentrifugation 100,000–200,000g, vesicle density is 1.13–1.19 g/mL [90] size-based isolation techniques Immunoaffinity capture-based techniques Exosome precipitation(Total exosome isolation kit) Microfluidics-based isolation techniques [91] |
Differential centrifugation 18,000–20,000g [92] | Differential centrifugation 1000–2000g FACS based ApoBD [93] Established protocols are essentially lacking; most studies use co-culture with apoptotic cells instead of isolating apoptotic bodies |
Differential centrifugation 10000g [94] |
| Markers | CD9, CD81 CD63, TSPAN6, TSPAN8, CD151, CD37, CD53, Flotilin 1 and 2 [95] |
Annexin V binding, tissue factor and cell-specific markers [95, 96] CD9, CD81, CD82 |
Annexin V binding, DNA content | Oncogenic and tumor Suppressor protein mutated/oncogenic DNA [86] RNA/mi RNA |
| Cell adhesion molecules | Integrin, Lactadherin, ICAM [95] | Integrin, PECAM1, fibronectin [95] | ||
| Cell-type-specific proteins | MHC-I, MHC-II, APP, PMEL, TCR, FasL, CXCR4, HSPG, CD86, PrP, TFR, WNT [95] | MHC class I, LFA1, CD14 [95] |
2.4.1. Origin of EVs
The origin and biogenesis of EV’s are different that essentially distinguishes exosomes from MVs, oncosomes and apoptotic bodies. As early as 1980, researchers studying the process of reticulocyte maturation observed extra-vesicular secretion [51]. These extracellular vesicles are subcategorized based on their size, and they are endosomal in origin, due to the inward budding of endosomes. Some large EVs (ie, multivesicular bodies) have been shown to fuse with lysosomes or the plasma membrane, thereby releasing the contents needed for degradation into the extracellular space [52].
Exosomes, the smaller vesicles, are released from MV bodies and deliver functional RNAs (e.g, mRNA and miRNA) to other cells, which assist in intercellular communication [53–56]. The function of exosomes varies depending on their origin. Exosomes derived from antigen presenting cells can express major histocompatibility complex (MHC) class I and II molecules on the cell surface, which enables them to activate CD8β and CD4β T cells to induce specific immune responses [57–59].
2.4.2. Biology and Function of Exosomes
The secretion of exosomes into biological fluids contributes to their diverse physiological responses. Exosomes are found first to be involved in removing unnecessary proteins during the cell maturation process [60]. Exosomes can deliver diverse biological molecules, including proteins, lipids, RNAs (including long-coding, short-coding, and non-coding RNAs), DNA, and metabolites, depending on the cellular origin of the exosomes. Importantly, exosomes carry and present major histocompatibility peptide complexes to the exosomal surface, as well as inside the exosomes, and can therefore modulate immune responses [61]. Exosomes released by dendritic cells are currently being explored for their potential as therapeutic agents against infection and cancer [62]. EVs derived from cancer cells have been shown to promote angiogenesis and coagulation, to modulate immune responses, and to remodel surrounding parenchymal tissue, leading to tumor progression. Tumor cells secrete EVs that can be microvesicles, exosomes, or oncosomes [5].
Mechanisms by which exosomes influence various biological functions are not fully understood but the key features are; 1) direct contact of the antigens and markers present on the surface of exosomes to antigen presenting cells of the host leading to immune activation or deactivation, 2) fusion of exosomal and cell membrane or endocytosis through which biological materials present inside exosomes are released into the cells.
Exosomes are thought to play a critical role in both primary tumor growth and metastatic evolution. Exosomes orchestrate multiple systemic pathophysiological processes, such as coagulation, vascular leakiness, and reprogramming of stromal recipient cells, to support pre-metastatic niche formation and subsequent metastasis. Clinically, Exosomes have been useful as potential biomarkers. Further delineation of their progression and development of novel therapeutic targets may aid in prevention of cancer and metastasis [52, 63].
2.4.4. Source and Release of Exosomes
Exosomes are released by cells through reverse budding of the limiting membrane of late endosomes, via multivesicular bodies containing intraluminal vesicles [64]. Endosomal sorting complex is involved in the formation of intraluminal vesicles which requires transport (ESCRT) machinery ESCRT0, ESCRTI, ESCRTII and ESCRTIII. All the events in the reverse budding of MVs until the formation and release of exosomes occurs in the sequential pattern involving ESCRT complex [65, 66].
Release of exosomes from cells can be influenced by many circumstances, both physiological and pathological. Proteomic analyses reflect the presence of some common proteins in exosomes from all cell types [67, 68], but some contents (eg. nucleic acids, certain proteins, and metabolites) are unique to exosomes. These contents are secreted by various cells, depending on the external insult (eg, infection, interactions of host and pathogen cells, immune responses, physiological conditions, or disease state of the body) [69]. Exosomes are released into multiple biological fluids, including urine and breast milk. Unique signatures have been identified from vesicles that originate from different cells under various circumstances. Until recently, the origin of a particular biomarker found in urine, called aquaporin-2, has been a mystery. However, new reports [36, 70] have used mass spectroscopy to confirm that the presence of aquaporin-2 in urine signifies the presence of exosomes.
Ciprofloxacin, an antibacterial drug, has been shown to induce exosome release [71]. Cancer cells also release exosomes, which promote tumor growth, inhibits immune responses, and prompts complex communication between immune cells via nucleic acids (eg, DNA, RNA), proteins, and other metabolites in these exosomes [72]. According to a report by Mittra et al, patients diagnosed with blood cancer have fragmented dsDNA, which can integrate into the healthy cell genome via exosome-mediated transfer, leading to activation of the DNA damage repair pathway, followed by apoptosis [73]. Exosomes can contribute to cancer progression, leading to metastasis as well as activating DNA damage repair.
2.4.5. Exosomes in Allograft Immunity
Transplantation is the sole treatment option for many patients diagnosed with end-stage organ disease. However, long-term graft survival is hindered by rejection of the transplanted organ by immune and nonimmune processes. Acute and chronic rejection can occur after transplantation of any solid organ transplant, including kidney, heart, and lungs. Long-term immunosuppression has reduced the incidence of rejection. The mechanisms leading to rejection, especially chronic rejection, after solid organ transplant remain under investigation; however, both humoral and cellular immune processes are thought to play an essential role in the pathogenesis of rejection. During allograft rejection, the recipient’s immune system recognizes mismatched MHC antigens from the donor organ and activates T lymphocytes, a process known as allorecognition. This activation of T cells occurs through two distinct pathways: a direct pathway (ie, T cell stimulation by donor antigen presenting cells) and an indirect pathway (ie, T cell stimulation by self-antigen presenting cells) [74–80]. The recent discovery of T cell activation via exosomes is referred to as semi-direct pathway, but research on exosomes and their role in organ transplant is limited [81]. Exosomes released during the process of allograft rejection are currently being analyzed [82, 83]. The molecular and immune composition of exosomes isolated from urine varies, but exosomes isolated from urine can carry inter- and intracellular proteins as well as nucleic acid that reflect the state, stability and health of renal cells, physiological conditions, and pathological conditions [84–86]. Details of exosomes released during different organ transplants are provided in Table 2.
Table 2.
Characterization of Exosomes from Solid Organ Transplants
| Organ | Sample | Species | Condition | Exosome content | References |
|---|---|---|---|---|---|
| Lung | Serum, BALF | Human | Bronchiolitis obliterans syndrome (BOS)/acute rejection (AR) | Self-antigens (SAgs), Collagen V (Col-V) and Kα1 Tubulin (Kα1T), mismatched donor HLA,miR-92a [endothelial activation], miR-182 [inflammation], miR-142-5p [antibody-mediated rejection (AMR)/chronic rejection, and miR-155 [T cell activation] | [49] |
| Lung | serum | Human | BOS | Lung-associated SAgs, Ka1T and Col-V, MHC class II molecules, costimulatory molecules CD40, CD80, and CD86, and transcription factors class II MHC trans-activator, NF-kB, hypoxia-inducible factor 1-a, IL-1R – associated kinase 1, MyD88, and 20S proteasome | [97] |
| Lung | BALF | Human | AR | RNA and miRNA related to many immune pathways | [76] |
| Kidney | Urine | Human | Delayed graft function | Neutrophil gelatinase-associated lipocalin NGAL | [84] |
| Kidney | plasma | Human | AMR, cell-mediated rejection | mRNA transcripts of gene gp130, CCL4, TNFα, SH2D1B, CAV1, atypical chemokine receptor 1 | [98] |
| Kidney | Urine | Human | Slow graft function and vascular damage. | graft-specific HLA class I antigens, CD 133 | [99] |
| Kidney | Urine | Human | BK virus nephropathy in kidney transplant | bkv-miR-B1-5p and bkv-miR-B1-5p/miR-16 | [100] |
| Liver | serum | Human | Hepatocellular carcinoma after liver transplantation | miR-718 | [101] |
| Islet | Plasma | Mice | Human islet transplant to mice | Donor islet HLA, islet endocrine hormone markers insulin, glucagon, and somatostatin | [102] |
| Heart | Cell supernatant | Mice | Heterotopic (abdomen) vascularized cardiac transplantation | Donor HLA, miR-155 and contain heat-shock proteins | [74] |
| Heart | Serum | Mice | Syngeneic cardiac transplant rejection | miRNA involved in inflammation (ie, miR-10a, miR-155, and miR-142-5p) and fibrosis (ie, miR-21, miR-31, and miR-133b). SAgs, Col-V and Kα1T | [87] |
| Heart | Plasma | Human | AMR | Donor specfic HLA I, Troponin I, Complement C4 | [103] |
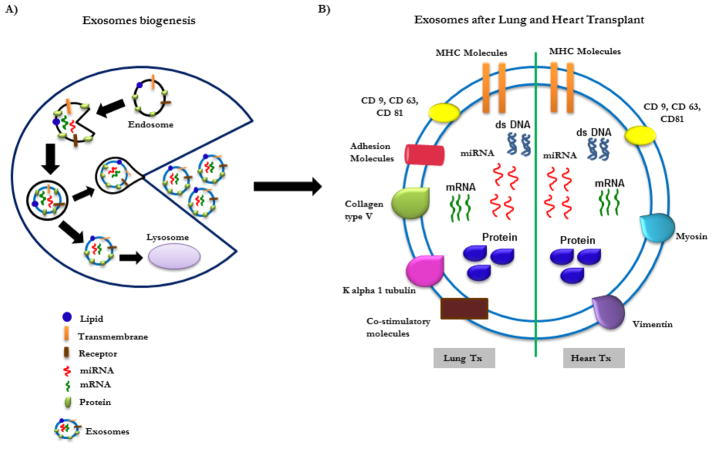
We have previously explored the role of exosomes after human lung transplantation and have demonstrated that exosomes contain not only mismatched donor HLA, but also contain the lung-associated SAGs, K-alpha 1 Tubulin (Kα1T) and collagen V (Col-V). Exosomes are released during allograft rejection, and Kα1T and Col-V are expressed on the surface of exosomes (Figure 2), suggesting that they can induce immune responses. More significantly, we have found that lung SAgs were not present in exosomes isolated from stable lung transplant recipients, suggesting that circulating exosomes with lung SAgs can be potential biomarkers for impending rejection. This also supports our conclusion that exosomes originate from the transplanted organ after immune insult or injury.
Figure 2. Exosomes biogenesis and composition.
A) Exosomes biogenesis. B) Composition of exosomes following heart and lung transplantation
We have also demonstrated that exosomes isolated from human lung transplant recipients undergoing rejection contain miRNAs known to induce inflammation, endothelial activation, antibody mediated chronic rejection, and Th17 differentiation [49]. This suggests that immune activation via the miRNA in exosomes can contribute to the pathogenesis of rejection. Recent studies have also shown that primary graft dysfunction, de novo donor-specific antibody development, and respiratory viral infections also induce circulating exosomes after lung transplant (unpublished research). Based on these findings, we propose that stress to the transplanted organs can release exosomes, and that persistence of these exosomes in the circulation can lead to immune activation and ultimately increase the risk of chronic rejection.
We recently found that induction of exosomes is not unique to lung transplant recipients diagnosed with rejection; instead, exosomes are also found in heart transplant recipients diagnosed with CAV [11–13] (akin to chronic rejection) and in renal transplant recipients diagnosed with transplant glomerulopathy (a condition that increases the risk of chronic rejection). In human heart transplant recipients with CAV, induction of circulating exosomes is correlated with development of antibodies to the cardiac SAgs, myosin and vimentin; in renal transplant recipients with transplant glomerulopathy, exosomes in the circulation are correlated with development of antibodies to the renal tissue SAgs, Col-IV and fibronectin. Our recent animal experiments have demonstrated that administration of antibodies to cardiac myosin immediately following syngeneic murine heterotopic cardiac transplantation resulted in graft failure within 8 days, and circulating exosomes with cardiac myosin and vimentin were present in those animals before graft failure occurred [87]. Further immunization with exosomes isolated from animals following cardiac graft failure can also lead to graft loss after syngeneic cardiac transplantation, and these animals also experience de novo development of antibodies to cardiac myosin and vimentin [87]. Both of these studies strongly demonstrate the importance of exosomes induced and released into circulation in the immunopathogenesis of allograft rejection.
3. Conclusions
As recently as a few decades ago, exosomes were treated as garbage bins, and their scope and potential role in carrying small molecules were underestimated. Exosome biology has now emerged as an exciting field for studies in cancer and, more recently, in transplantation. Our findings from lung transplant recipients clearly demonstrate the importance of circulating exosomes in the transplant immunity processes. The presence of the lung SAgs, Kα1T and Col-V, was noted primarily in the exosomes isolated from lung transplant recipients diagnosed with rejection or other clinical conditions thought to increase the risk for rejection (eg, severe primary graft dysfunction and respiratory viral infection). Our results in the animal model of cardiac transplantation also support the importance of exosomes in the allograft immune processes. Exosomes possess immunoregulatory molecules and also carry cell-derived antigens. Therefore, exosomes can be used for vaccination to induce immunity and or tolerance prior to transplantation. Exosomes with different cell surface antigens and factors within can also be employed as potential biomarkers for early detection of rejection following solid organ transplants and in other diseases of interest including cancers.
4. Future Directions
Exosomes are important not only in the field of cancer biology, but also in other pathological conditions such as infection and transplant immunology. The potential role of exosomes as biomarkers for lung allograft rejection as well as chronic rejection of other solid organs still needs to be validated. Unanswered questions include the presence of other factors, such as different antigens, costimulatory molecules, miRNA, and proteasomes, in the exosomes and their role in allograft immunity. Apart from the spread of small molecules during intercellular communication via exosomes, the fate of these exosomes, how they spread, and how they communicate with different cell types are other key questions that need further clarification. The role of exosomes in transplant tolerance is also of interest, and characterization of exosomes released during and after tolerance requires additional studies.
Highlights.
Different types of extracellular vesicles, their composition
Role for exosomes in allograft immunity
Exosomes with tissue associated self-antigens as biomarker for rejection
Acknowledgments
Funding sources
This work was supported by grants from the National Institutes of Health AI123034, HL056643, HL092514 (TM).
We would like to thank Clare Prendergast and Billie Glasscock for their assistance in preparing this manuscript.
Footnotes
Ethical statements
The authors declare no conflict of interest. All authors have reviewed and approved the manuscript and have contributed in a substantial and intellectual manner to the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 2.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 5.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 6.Scheuher C. A review of organ transplantation: Heart, lung, kidney, liver, and simultaneous liver-kidney. Crit Care Nurs Q. 2016;39(3):199–206. doi: 10.1097/CNQ.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 7.Kushner YB, Colvin RB. Organ Transplantation: A clinical guide. Cambridge University Press; Cambridge (UK): 2011. [Google Scholar]
- 8.Patel R, Terasaki PI. Significance of the positive crossmatch test in kidney transplantation. N Engl J Med. 1969;280(14):735–739. doi: 10.1056/NEJM196904032801401. [DOI] [PubMed] [Google Scholar]
- 9.Ramachandran S, Subramanian V, Mohanakumar T. Immune responses to self-antigens (autoimmunity) in allograft rejection. Clin Transpl. 2012:261–272. [PMC free article] [PubMed] [Google Scholar]
- 10.Bharat A, Mohanakumar T. Immune Responses to Tissue-Restricted Nonmajor Histocompatibility Complex Antigens in Allograft Rejection. Journal of Immunology Research. 2017;2017:8. doi: 10.1155/2017/6312514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nath DS, Basha HI, Mohanakumar T. Antihuman leukocyte antigen antibody-induced autoimmunity: role in chronic rejection. Current opinion in organ transplantation. 2010;15(1):16–20. doi: 10.1097/MOT.0b013e3283342780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiemann NE, Wellnhofer E, Knosalla C, Lehmkuhl HB, Stein J, Hetzer R, Meyer R. Prognostic impact of microvasculopathy on survival after heart transplantation: evidence from 9713 endomyocardial biopsies. Circulation. 2007;116(11):1274–1282. doi: 10.1161/CIRCULATIONAHA.106.647149. [DOI] [PubMed] [Google Scholar]
- 13.Rahmani M, Cruz RP, Granville DJ, McManus BM. Allograft vasculopathy versus atherosclerosis. Circ Res. 2006;99(8):801–815. doi: 10.1161/01.RES.0000246086.93555.f3. [DOI] [PubMed] [Google Scholar]
- 14.Porter KA, Andres GA, Calder MW, Dossetor JB, Hsu KC, Rendall JM, Seegal BC, Starzl TE. Human renal transplants. II. Immunofluorescent and immunoferritin studies. Lab Invest. 1968;18(2):158–171. [PubMed] [Google Scholar]
- 15.Najafian B, Fogo AB, Lusco MA, Alpers CE. AJKD Atlas of renal pathology: chronic antibody-mediated rejection. Am J Kidney Diseases. 2015;66(5):e41–e42. doi: 10.1053/j.ajkd.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166(1):189–197. [PubMed] [Google Scholar]
- 17.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13(3):269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 18.Kerr JF, Wyllie AH, Currie AR. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, caenorhabditis elegans. Dev Biol. 1977;56(1):110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 20.Rak J. Extracellular vesicles - biomarkers and effectors of the cellular interactome in cancer. Front Pharmacol. 2013;4:21. doi: 10.3389/fphar.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D’Asti E, Garnier D, Lee TH, Montermini L, Meehan B, Rak J. Oncogenic extracellular vesicles in brain tumor progression. Front Physiol. 2012;3:294. doi: 10.3389/fphys.2012.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minciacchi VR, Freeman MR, DiVisio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRclll by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 24.DiVizio D, Kim J, Hager MH, Morello M, Yang W, Lafargue CJ, True LD, Rubi MA, Adam RM, Beroukhim R, Demichelis F, Freeman MR. Oncosome formation in prostate cancer: association with a region of frequent chromosomal deletion in metastatic disease. Cancer Res. 2009;69(13):5601–55609. doi: 10.1158/0008-5472.CAN-08-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sansone P, Savini C, Kurelac I, Chang Q, Amato L, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L, Galkin A, Thakur B, Soplop N, Uryu K, Hishino A, Norton L, Bonafe M, Crissa M, Gasparre G, Lyden D, Bromberg J. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017;114(43):e9066–e9075. doi: 10.1073/pnas.1704862114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desrochers LM, Antonyak MA, Cerione RA. Extracellular vesicles: satellites of information transfer in cancer and stem cell biology. Dev Cell. 2016;37(4):301–309. doi: 10.1016/j.devcel.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fischer S, Cornils K, Speiseder T, Badbaran A, Reimer R, Indenbirken D, Grundhoff A, Brunswig-Spickenheier B, Alawi M, Lange C. Indication of horizontal DNA gene transfer by extracelluler vesicles. PLoS One. 2016;11(9):e0163665. doi: 10.1371/journal.pone.0163665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergsmedh A, Szeles A, Henriksson M, Bratt A, Folkman MJ, Spetz AL, Holmgren L. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci U S A. 2001;98(11):6407–6411. doi: 10.1073/pnas.101129998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TH, D’Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer - the emerging science of cellular debris. Semin Immunolpathol. 2011;33(5):455–467. doi: 10.1007/s00281-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 30.Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, ElAndaloussi S, Wood MJ, Vader P. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rashed MH, Bayraktar E, Helal KG, Abd-Ellah MF, Amero P, Chavez-Reyes A, Rodriguez-Aguayo C. Exosomes: from garbage bins to promising therapeutic targets. Int J Mol Sci. 2017;18:538. doi: 10.3390/ijms18030538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 33.Conde-Vancells J, Rodriguez-Suarez E, Gonzalez E, Berisa A, Gil D, Embade N, Valle M, Luka Z, Elortza F, Wagner C, Lu SC, Mato JM, Falcon-Perez M. Candidate biomarkers in exosome-like vesicles purified from rat and mouse urine samples. Proteomics Clin Appl. 2010;4:416–425. doi: 10.1002/prca.200900103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;5:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vella LJ, Greenwood DL, Cappai R, Scheerlinck JP, Hill AF. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet Immunol Immunopathol. 2008;124:385–393. doi: 10.1016/j.vetimm.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 38.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 39.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 41.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson SW, Nguyen J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J Control Release. 2016;228:179–190. doi: 10.1016/j.jconrel.2016.02.037. [DOI] [PubMed] [Google Scholar]
- 43.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 44.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suryadevara V, Govindkumari V. Exosomes and microparticles: The nanosized vesicles released from the cells that act as biomarkers for disease and treatment–riveting on lung diseases. Materials Today Proceedings. 2015;2:4626–4631. [Google Scholar]
- 46.Gallart-Palau X, Serra A, See Weng Wong A, Sandin S, Lai MKP, Chen CP, Kon OL, Sze SK. Extracellular vesicles are rapidly purified from human plasma by PRotein Organic Solvent PRecipitation (PROSPR) Sci Rep. 2015;5:14664. doi: 10.1038/srep14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corso G, Mager I, Lee Y, Gorgens A, Bultema J, Giebel B, Wood M, Nordin JZ, El Andaloussi S. Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Sci Rep. 2017;7:11561. doi: 10.1038/s41598-017-10646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Logozzi M, DeMilito A, Lugini L, Borghi M, Calabro L, Spada M, Perdicchio M, Marino ML, Federici C, Iessi E, Brambilla D, Venturi G, Lozupone F, Santinami M, Huber V, Maio M, Rivoltini L, Fais S. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLos One. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunasekaran M, Xu Z, Nayak DK, Sharma M, Hachem R, Walia R, Bremner RM, Smith MA, Mohanakumar T. Donor-derived exosomes with lung self-antigens in human lung allograft rejection. Am J Transpl. 2017;17(2):474–484. doi: 10.1111/ajt.13915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gardiner C, DiVizio D, Sahoo S, Thery C, Witwer KW, Wauben M, Hill AF. Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey. J Extracell Vesicles. 2016;5:32945. doi: 10.3402/jev.v5.32945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, Schmandt R, Lu KH, Mok SC. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;29:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 53.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valadi H, Eksstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature cell biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 55.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 56.Colombo M. Natural history and pathogenesis of hepatitis C virus related hepatocellular carcinoma. J Hepatol. 1999;31:25–30. doi: 10.1016/s0168-8278(99)80370-5. [DOI] [PubMed] [Google Scholar]
- 57.Théry C, Duban L, Segura E, Véron P, Lantz O, Amigorena S. Indirect activation of naïve CD4+ T cells by dendritic cell-derived exosomes. Nat Immunol. 2002;3:1156–1162. doi: 10.1038/ni854. [DOI] [PubMed] [Google Scholar]
- 58.Morelli AE, Larregina AT, Shufesky WJ, Sullivan ML, Stolz DB, Papworth GD, Zahorchak AF, Logar AJ, Wang Z, Watkins SC, Falo LD, Jr, Thomson AW. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3566. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- 59.De Toro J, Herschlik L, Waldner C, Mongini C. Emerging roles of exosomes in normal and pathological conditions: new insights for diagnosis and therapeutic applications. Front Immunol. 2015 May 4;6:203. doi: 10.3389/fimmu.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wen C, Seeger RC, Fabbri M, Wang L, Wayne AS, Jong AY. Biological roles and potential applications of immune cell-derived extracellular vesicles. J Extracell Vesicles. 2017;6:1400370. doi: 10.1080/20013078.2017.1400370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Greening GSDW, Xu R, Simpson RJ, Chen W. Exosomes and their roles in immune regulation and cancer. Semin Cell Dev Biol. 2015;40:72–81. doi: 10.1016/j.semcdb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Pitt JM, Kroemer G, Zitvogel L. Extracellular vesicles: masters of intercellular communication and potential clinical interventions. J Clin Invest. 2016;126:1139–1143. doi: 10.1172/JCI87316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Headley MB, Bins A, Nip A, Roberts EW, Looney MR, Gerard A, Krummel MF. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016;531:513–517. doi: 10.1038/nature16985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting or ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 65.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuffers S, Sem Wegner C, Stenmark H, Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10(7):925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 67.Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Möbius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 68.Buschow SI, van Balkom BW, Aalberts M, Heck AJ, Wauben M, Stoorvogel W. MHC class II-associated proteins in B-cell exosomes and potential functional implications for exosome biogenesis. Immunol Cell Biol. 2010;88:851–856. doi: 10.1038/icb.2010.64. [DOI] [PubMed] [Google Scholar]
- 69.Schorey JS, Cheng Y, Singh PP, Smith VL. Exosomes and other extracellular vesicles in host-pathogen interactions. EMBO Rep. 2015;16:24–43. doi: 10.15252/embr.201439363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanno K, Sasaki S, Hirata Y, Ishikawa S, Fushimi K, Nakanishi S, Bichet DG, Marumo F. Urinary excretion of aquaporin-2 in patients with diabetes insipidus. N Engl J Med. 1995;332:1540–1545. doi: 10.1056/NEJM199506083322303. [DOI] [PubMed] [Google Scholar]
- 71.Németh A, Orgovan N, Sódar BW, Osteikoetxea X, Pálóczi K, Szabó-Taylor KÉ, Vukman KV, Kittel Á, Turiák L, Wiener Z, Tóth S, Drahos L, Vékey K, Horvath R, Buzás EI. Antibiotic-induced release of small extracellular vesicles (exosomes) with surface-associated DNA. Scientific Reports. 2017;7:8202. doi: 10.1038/s41598-017-08392-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mittra I, Khare NK, Raghuram GV, Chaubal R, Khambatti F, Gupta D, Gaikwad A, Prasannan P, Singh A, Iyer A, Singh A, Upadhyay P, Nair NK, Mishra PK, Dutt A. Circulating nucleic acids damage DNA of healthy cells by integrating into their genomes. J Biosci. 2015;40:91–111. doi: 10.1007/s12038-015-9508-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Q, Rojas-Canales DM, Divito SJ, Shufesky WJ, Stolz DB, Erdos G, Sullivan ML, Gibson GA, Watkins SC, Larregina AT, Morelli AE. Donor dendritic cell-derived exosomes promote allograft-targeting immune response. J Clin Invest. 2016;126(8):2805–2820. doi: 10.1172/JCI84577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.D’Orsogna LJ, Roelen DL, van der Meer-Prins EM, van der Pol P, Franke-van Dijk ME, Eikmans M, Anholts J, Rossjohn J, McCluskey J, Mulder A, van Kooten C, Doxiadis II, Claas FH. Tissue specificity of cross-reactive allogeneic responses by EBV EBNA3A-specific memory T cells. Transplantation. 2011;91:494–500. doi: 10.1097/TP.0b013e318207944c. [DOI] [PubMed] [Google Scholar]
- 76.Gregson AL, Hoji A, Injean P, Poynter ST, Briones C, Palchevskiy V, Weigt SS, Shino MY, Derhovanessian A, Sayah D, Saggar R, Ross D, Ardehali A, Lynch JP, 3rd, Belperio JA. Altered exosomal RNA profiles in bronchoalveolar lavage from lung transplants with acute rejection. Am J Resp Crit Care Med. 2015;192:1490–1503. doi: 10.1164/rccm.201503-0558OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gonzalez-Nolasco B, Wang M, Prunevieille A, Benichou G. Emerging role of exosomes in allorecognition and allograft rejection. Curr Opin Organ Transplant. 2018;23:22–27. doi: 10.1097/MOT.0000000000000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gould DS, Auchincloss H. Direct and indirect recognition: the role of MHC antigens in graft rejection. Immunology Today. 1999;20:77. doi: 10.1016/s0167-5699(98)01394-2. [DOI] [PubMed] [Google Scholar]
- 79.Benichou G, Valujskikh A, Heeger PS. Contributions of direct and indirect T cell alloreactivity during allograft rejection in mice. J Immunol. 1999;162:352. [PubMed] [Google Scholar]
- 80.Marino J, Babiker-Mohamed MH, Crosby-Bertorini P, Paster JT, LeGuern C, Germana S, Abdi R, Uehara M, Kim JI, Markmann JF, Tocco G, Benichou G. Donor exosomes rather than passenger leukocytes initiate alloreactive T cell responses after transplantation. Sci Immunol. 2016;1:aaf8759. doi: 10.1126/sciimmunol.aaf8759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morelli AE, Bracamonte-Baran W, Burlingham WJ. Donor-derived exosomes: the trick behind the semidirect pathway of allorecognition. Curr Opin Organ Transplant. 2017;22:46–54. doi: 10.1097/MOT.0000000000000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morelli AE. Exosomes: From Cell Debris to Potential Biomarkers in Transplantation. Transplantation. 2017;101:2275–2276. doi: 10.1097/TP.0000000000001856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burlingham WJ. Exosomes: The missing link between microchimerism and acquired tolerance? Chimerism. 2014;5:63–67. doi: 10.1080/19381956.2015.1082026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alvarez S, Suazo C, Boltansky A, Ursu M, Carvajal D, Innocenti G, Vukusich A, Hurtado M, Villanueva S, Carreño JE, Rogelio A, Irarrazabal CE. Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplant Proc. 2013;45:3719–3723. doi: 10.1016/j.transproceed.2013.08.079. [DOI] [PubMed] [Google Scholar]
- 85.Park J, Lin HY, Assaker JP, Jeong S, Huang CH, Kurdi T, Lee K, Fraser K, Min C, Eskandari S, Routray S, Tannous B, Abdi R, Riella L, Chandraker A, Castro CM, Weissleder R, Lee H, Azzi JR. Integrated Kidney Exosome Analysis for the Detection of Kidney Transplant Rejection. ACS Nano. 2017;11:11041–11046. doi: 10.1021/acsnano.7b05083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sharma M, Liu W, Perincheri S, Gunasekaran M, Mohanakumar T. Exosomes expressing the self-antigens myosin and vimentin play an important role in syngeneic cardiac transplant rejection induced by antibodies to cardiac myosin. Amer J Transplant. 2018 doi: 10.1111/ajt.14650. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gyorgy B, Modos K, Pallinger E, Paloczi K, Pasztoi M, Misjak P, Deli MA, Sipos A, Szalai A, Voszka I, Polgar A, Toth K, Csete M, Nagy G, Gay S, Falus A, Kittel A, Buzas EI. Detection and isolation of cell-derived microparticles are comprised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117(4):e39–48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- 89.Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood. 2004;104(9):2761–2766. doi: 10.1182/blood-2003-10-3614. [DOI] [PubMed] [Google Scholar]
- 90.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In: Bonifacino JS, editor. Current Protocols in Cell Biology. 2006. pp. Unit 3–22. [DOI] [PubMed] [Google Scholar]
- 91.Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y. A comparative study of serum exosome isolation using differential ultracentrifucation and three commercial reagents. PLoS One. 2017;12(1):e0170628. doi: 10.1371/journal.pone.0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yuana Y, Bertina RM, Osanto S. Pre-analytical and analytical issues in the analysis of blood microparticles. Thromb Haemost. 2011;105(3):396–408. doi: 10.1160/TH10-09-0595. [DOI] [PubMed] [Google Scholar]
- 93.Atkin-Smith GK, Paone S, Zanker DJ, Duan M, Phan TK, Chen W, Hulett MD, Poon IK. Isolation of cell type-specific apoptotic bodies by fluorescence-activated cell sorting. Sci Rep. 2017;7:39846. doi: 10.1038/srep39846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morello M, Minciacchi VR, deCandia P, Yang J, Posadas E, Kim H, Griffiths D, Bhowmick N, Chkung LW, Gandellini P, Freeman MR, Demichelis D, DiVizio D. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12(22):3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.vanNiel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 96.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas El. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gunasekaran M, Sharma M, Hachem R, Bremner R, Smith MA, Mohanakumar T. Circulating exosomes with distinct properties during chronic lung allograft rejection. J Immunol. 2018;200(8):2535–2541. doi: 10.4049/jimmunol.1701587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang H, Huang E, Kahwaji J, Nast CC, Li P, Mirocha J, Thomas DL, Ge S, Vo AA, Jordan SC, Toyoda M. Plasma esocomes from HLA-sensitized kidney transplant recipients contain mRNA transcripts which predict development of antibody-mediated rejection. Transplantation. 2017;101(10):2419–2428. doi: 10.1097/TP.0000000000001834. [DOI] [PubMed] [Google Scholar]
- 99.Dimuccio V, Ranghino A, Pratico Barbato L, Fop F, Biancone L, Camussi G, Bussolati B. Urinary CD133+ extracellular vesicles are decreased in kidney transplanted patients with slow graft function and vascular damage. PLoS One. 2014;9(8):e104490. doi: 10.1371/journal.pone.0104490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kim MH, Lee YH, Seo JW, Moon H, Kim JS, Kin YG, Jeong KH, Moon JY, Lee TW, Ihm CG, Kim CD, Park JB, Chung BH, Kim YH, Lee SH. Urinary exosomal viral microRNA as a marker of BK virus nephropathy in kidney transplant recipients. PLoS One. 2017;12(12):e0190068. doi: 10.1371/journal.pone.0190068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, Shinden Y, Uguchi T, Eguchi H, Shirabe K, Ochiya T, Maehara Y, Mimori K. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocullular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–538. doi: 10.1038/bjc.2014.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vallabhajosyula P, Korutla L, Habertheuer A, Yu M, Rostami S, Yuan CX, Reddy S, Liu C, Korutla V, Koeberlein B, Trofe-Clark J, Rickels MR, Naji A. Tissue-specific exosome biomarkers for noninvasively monitoring immunologic rejection of transplanted tissue. J Clin Invest. 2017;127:1375–1391. doi: 10.1172/JCI87993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Korutla L, Habertheuer A, Hu R, Zielinski P, Reddy S, Naji A, Vallabhajosyula P. Characterization of circulating donor heart specific exosomes in clinical heart transplantation. J Heart and Lung Transplantation. 2018;37(4):S331. [Google Scholar]