SUMMARY
Platinum-based chemotherapeutics represent a mainstay of cancer therapy, but resistance limits their curative potential. Through a kinome RNAi screen, we identified microtubule-associated serine/threonine kinase 1 (MAST1) as a main driver of cisplatin resistance in human cancers. Mechanistically, cisplatin but no other DNA damaging agents inhibit the MAPK pathway by dissociating cRaf from MEK1, while MAST1 replaces cRaf to reactivate the MAPK pathway in a cRaf-independent manner. We show clinical evidence that expression of MAST1, both initial and cisplatin-induced, contributes to platinum resistance and worse clinical outcome. Targeting MAST1 with lestaurtinib, a recently identified MAST1 inhibitor, restores cisplatin sensitivity, leading to the synergistic attenuation of cancer cell proliferation and tumor growth in human cancer cells and patient-derived xenograft models.
Significance
Activation of proliferative signaling pathways following chemotherapy has been associated with therapeutic resistance. However, relatively little is known of its mechanism. Our findings provide insights into cancer cisplatin resistance by characterizing the role of MAST1 in driving cisplatin resistance and its mode of action in human cancers involving rewiring of the MEK signaling pathway. We offer a clinical strategy to enhance the susceptibility of cancers to cisplatin by identifying the small molecule kinase inhibitor lestaurtinib as a potent MAST1 inhibitor and cisplatin sensitizing agent. These findings not only highlight mechanisms by which MAST1 contributes to the development of cisplatin resistance via the MAPK pathway but also implicate MAST1 as a promising therapeutic target to battle cisplatin resistant cancers.
Summary
Jin et al. show that cisplatin dissociates cRaf from MEK1 to inhibit the MAPK pathway and identify MAST1 as a main cisplatin resistance driver that replaces cRaf to reactivate the MAPK pathway. They further show that inhibition of MAST1 by the multi-kinase inhibitor lestaurtinib restores cisplatin sensitivity.

INTRODUCTION
Platinum-based chemotherapy is employed for the treatment of a wide array of solid malignancies including head and neck, lung, and ovarian cancers (Galanski, 2006). Cisplatin and other similar platinum-based drugs lead to an initial therapeutic success, but many patients have tumors that are intrinsically resistant or develop resistance to cisplatin treatment (Giaccone, 2000; Koberle et al., 2010). Cisplatin exerts anticancer effects mainly by interacting with DNA to form mostly intrastrand crosslink adducts, which activates pro-apoptotic signal transduction pathways (Siddik, 2003). Cisplatin resistance likely occurs due to complex reasons, including increased drug efflux, drug breakdown, increased repair of damaged DNA, and increased activation of prosurvival pathways or inhibition of pathways that promote cell death (Siddik, 2003). Although a group of signaling effectors have been identified as predictive markers of cisplatin resistance including MRP2 (Liedert et al., 2003), ERCC1 (Metzger et al., 1998), ATPase7A/7B/11B (Aida et al., 2005; Moreno-Smith et al., 2013), ERBB2 (Fijolek et al., 2006), Bcl-2 (Han et al., 2003) and survivin (Ikeguchi and Kaibara, 2001), most of these studies lacked either an assessment of clinical correlation or an explanation for how these protein factors regulate pro-survival signals in the presence of cisplatin. Therefore, the detailed molecular mechanisms of platinum-based drug resistance still remain elusive.
Protein kinases are often involved in pro-survival signaling pathways (Datta et al., 1997; Persons et al., 2000). The serine/threonine kinase microtubule associated serine/threonine-protein kinase 1 (MAST1, a.k.a. SAST170) belongs to a family containing four members, MAST1-MAST4. MAST family members share approximately 49-64% sequence homology and contain four distinct domains including DUF1908, serine/threonine kinase domain, AGC-kinase C-terminal domain, and PDZ domain (Garland et al., 2008). MAST1 is reported to function as a scaffold protein to link the dystrophin/utrophin network with microfilaments via syntrophin (Lumeng et al., 1999). Recurrent rearrangement of the MAST1 gene has been observed in breast cancer cell lines and tissues (Robinson et al., 2011). Nonetheless, little is known about the biological role of MAST1 as a kinase and its role in human cancers.
Higher MEK1 expression in cancers is associated with platinum-based drug resistance and correlates with shortened progression free survival of patients. Activation of the MAPK family of proteins has been implicated in response to platinum-based chemotherapy. For instance, inhibition of MEK/ERK signaling augmented cisplatin sensitivity in human squamous cell carcinoma (Kong et al., 2015). Although the importance of MEK in cancer and its contribution to chemotherapy response is well studied, the detailed molecular mechanisms by which MEK is activated in response to platinum-based drug treatment, and how it consequently contributes to cisplatin response, is unclear. The goal of this study is to identify and characterize a critical kinase that drives cisplatin resistance in human cancers and evaluating its potential as a chemosensitizing therapeutic target for treatment of patients with cisplatin-resistant cancer.
RESULTS
MAST1 is important for cisplatin-resistant cancer cell proliferation and tumor growth
To gain insight into the role of protein kinase signaling in cancer chemoresistance, we performed kinome-wide RNAi screen using a lentiviral shRNA library targeting 781 human kinase genes and kinase-related genes represented by 4,518 shRNA constructs. The primary screen involved transducing cisplatin-resistant, KB-3-1cisR, or taxol-resistant, KB-3-1taxolR, human carcinoma cell lines and with a lentivirus pool containing shRNAs targeting each of the 781 individual genes, and treating with sublethal doses of cisplatin or taxol (Richert et al., 1985). From the primary screen using KB-3-1cisR, 567 of 781 genes were selected for ranking by excluding shRNAs with low infection efficiency and shRNA clones that alone induced more than 15% cell death (Figure 1A). The secondary screen evaluated the top 50 ranking candidate genes in four cisplatin-resistant cancer cell lines including head and neck squamous cell carcinoma (HNSCC) PCI-15A, lung cancer A549, and ovarian cancer A2780 in addition to KB-3-1 cells (Figures 1B, S1A, and S1B). Microtubule associated serine/threonine kinase 1 (MAST1) was identified as the third most effective target from the primary screen, and emerged as a lead hit from the secondary screen, as it sensitized cancer cells to cisplatin treatment across cancer types. MAST1 ranked 756 as an effective synthetic lethal target for taxol treatment, suggesting that targeting MAST1 selectively sensitizes cells to the chemotherapy agent cisplatin.
Figure 1. Targeting MAST1 sensitizes cisplatin treatment in vitro and in vivo.
(A) Primary screen testing 781 genes was carried out using sublethal dose (5 μg/ml) of cisplatin (left). Candidates with low virus infection efficiency (<25%; grey), and shRNAs that alone induced cell death (>15%; blue) were excluded (right). (B) Secondary screen used the top 50 candidates from the primary screen in 4 cisplatin-resistant cancer cell lines (KB-3-1cisR, A549cisR, A2780cisR and PCI-15AcisR) (left). 30 leads showing >10% cell death upon shRNA and cisplatin treatment are shown (right). (C-D) Cell viability and cisplatin sensitivity of KB-3-1cisR and A549cisR cells (C) and platinum-refractory (cisR) TKO-002 cells (D) with MAST1 knockdown. Cells were transduced with 3 different MAST1 shRNA clones followed by sublethal dose of cisplatin (5 μg/ml for KB-3-1cisR and 2 μg/ml for A549cisR) or vehicle treatment. Cisplatin IC50 was assessed after 72 hr. IP: immunoprecipitation. (E) Colony formation assays were performed using cancer cells with MAST1 knockdown and cisplatin treatment. (F-H) Effect of cisplatin treatment and MAST1 knockdown using two shRNA clones on tumor growth of KB-3-1cisR xenograft mice. Mice were treated with vehicle-control or cisplatin 3 days after xenograft and tumor size was monitored (F). Tumor weight (G) and MAST1 expression in tumor lysates are shown (H; top). Representative images of Ki-67 IHC staining in harvested tumors from each group are shown (H; bottom). Scale bars represent 10 mm for (F) and 50 μm for (H). Data are mean ± SD from three technical replicates of each sample for (C-E). Error bars represent SEM for (F) and SD for (G). p values were determined by two-tailed Student’s t test (ns: not significant; **: p < 0.01). See also Figure S1.
To confirm the screening results for MAST1, MAST1 stable knockdown cells were generated using individual shRNA clones. Targeting MAST1 with 3 different shRNA clones attenuated cell viability only in the presence of cisplatin in KB-3-1cisR and A549cisR cells (Figure 1C). Moreover, knockdown of MAST1 sensitized TKO-002 cell, which are derived from a patient with platinum-refractory small cell lung carcinoma (SCLC) (Owonikoko et al., 2016), to cisplatin treatment (Figure 1D). Sensitization was observed in cancer cells treated with carboplatin, another platinum-based compound, but not with taxol demonstrating that the synthetic lethal effect is specific to platinum (Figures S1C and S1D). In addition, MAST1 knockdown attenuated colony formation potential of cancer cell lines only in the presence of cisplatin (Figure 1E). We next functionally validated this in vivo. Xenograft tumors derived from KB-3-1cisR cells with MAST1 knockdown and treated with cisplatin showed dramatic decrease in tumor growth rate, tumor mass and tumor proliferation compared to tumors derived from cells with MAST1 knockdown but without cisplatin treatment or to tumors without MAST1 knockdown treated with cisplatin (Figures 1F-1H and S1E-S1F). These data suggest that MAST1 confers cisplatin resistance and targeting MAST1 sensitizes cancer cells to cisplatin by acting as a synthetic lethal partner.
MAST1 confers cisplatin resistance through cRaf independent MEK1 activation
We next explored whether the kinase activity of MAST1 is required to provide cisplatin resistance in cancer cells. Thus, we generated a kinase-dead form of MAST1 by mutating proton acceptor active residue aspartic acid (D) at 497 to alanine (A). D497A mutation in MAST1 abolished its kinase activity (Figure 2A). Overexpression of the WT but not the D497A mutant (DA) MAST1 conferred cisplatin resistance to cisplatin sensitive parental cells (Figure 2B). Consistently, shRNA resistant WT but not DA MAST1 rescued the cisplatin resistance attenuated due to MAST1 knockdown in cisplatin-resistant cells (Figure 2C). These data suggest that the kinase activity of MAST1 is required for cancer cells to acquire cisplatin-resistant pro-survival signals.
Figure 2. MAST1 directly phosphorylates MEK1 and activates anti-apoptotic signaling upon cisplatin treatment.
(A) MAST1 in vitro kinase assay using MAST1 wild type (WT) or kinase-dead mutant (DA; D497A). GST-MAST1 variants were enriched from 293T and kinase activity was assessed by ADP-Glo Kinase assay using myelin basic protein (MBP) as a substrate. (B) Cell viability of parental cancer cells with WT or DA MAST1 overexpression in the presence of cisplatin. (C) Cell viability of cisplatin-resistant (cisR) cells with endogenous MAST1 knockdown and expression of shRNA-resistant WT or DA MAST1. Relative viability was obtained by normalizing values to cisplatin untreated samples for (B) and (C). (D) Phospho-antibody array results using 1,318 antibodies in KB-3-1cisR lysates. Top seven protein factors whose phosphorylation states decreased in MAST1 knockdown and cisplatin treatment are shown. (E) Parental and cisR pairs of KB-3-1 and A549 cells with MAST1 knockdown and cisplatin treatment (5 μg/ml) were assayed for MEK1 phosphorylation by immunoblotting. (F) MAST1 in vitro kinase assay using recombinant inactive MEK1 (rMEK1 K79A) as a substrate. (G) Kinase activities of MEK1, cRaf and MAST1 in cells with MAST1 knockdown and cisplatin treatment. MEK1, cRaf and MAST1 were immunoprecipitated from KB-3-1 cells and kinase activities were determined by ADP-Glo kinase assay using recombinant inactive ERK2 or MEK1 as substrates. MEK inhibitor U0126 (10 μM) and Raf inhibitor L779450 (5 μM) were used as controls. (H) Western blot analysis of apoptosis-related factors. Cells with or without MAST1 shRNA were treated with cisplatin (5 μg/ml) for 24 hr. (I) Apoptosis assay using parental and cisR cells with or without MAST1 knockdown. Cells were treated with sublethal dose of cisplatin (2 μg/ml: parental, 5 μg/ml: cisR) for 48 hr and apoptotic cells were assayed by Annexin V staining. Error bars represent ± SD from three technical replicates. p values were determined by two-tailed Student’s t test (ns: not significant; *: 0.01 < p < 0.05; **: p < 0.01). See also Figure S2.
To investigate the mechanism of MAST1 as a kinase in cisplatin resistance, we performed high-throughput phosphorylation profiling with 1,318 site-specific antibodies from over 30 signaling pathways. We identified a spectrum of proteins whose phosphorylation levels were decreased in KB-3-1cisR cells only when MAST1 was knocked down and cells were treated with cisplatin (Figures 2D and S2A). Among these, a decrease in phosphorylation at S43 of cRaf, which is known to correspond to inhibition of cRaf activity, was observed. However, the greatest decrease was seen in phosphorylation of MEK1 at S221, which is involved in its activation. We next further investigated this by immunoblotting in diverse cell lines. Cisplatin uptake differed between parental and cisR cells (Figure S2B), therefore, the treatment conditions were adjusted throughout the study so that all cell lines could take up a similar amount of cisplatin. Immunoblotting confirmed that knockdown of MAST1 together with cisplatin treatment in cancer cells did not affect the phosphorylation levels of STAT3 or AKT (Figure S2C) but decreased the phosphorylation levels of MEK1 and its downstream ERK1/2 (Figures 2E and S2D). There was no change or slight increase in Ras and cRaf activity upon MAST1 knockdown and cisplatin treatment suggesting that the MEK activity decrease is not through its upstream Ras or Raf (Figure S2E). Furthermore, the decreased MEK/ERK activity was rescued by overexpression of WT but not DA MAST1 (Figure S2F). Moreover, MAST1 directly phosphorylates MEK1 at S221 in an in vitro MAST1 kinase assay (Figure 2F). Supporting this finding, the kinase activity of MEK1 was significantly decreased, but cRaf activity was not altered, by cisplatin treatment and MAST1 knockdown in KB-3-1 and KB-3-1cisR cells (Figures 2G and S2G). In line with our finding, targeting MEK1 by stable knockdown or its specific inhibitors AZD6244 and trametinib sensitized cancer cells to cisplatin, although MEK1 knockdown itself attenuated cell growth, which was not observed with MAST1 knockdown (Figures S2H and S2I). These results confirmed that MEK1 is a downstream effector of MAST1, which contributes to cisplatin resistance in cancer cells. Moreover, assessment of a group of apoptotic factors revealed that MEK1 and subsequent ERK inactivation upon MAST1 knockdown and cisplatin treatment resulted in the accumulation of pro-apoptotic factor BIM specifically (Figure 2H). Furthermore, MAST1 knockdown together with sub-lethal doses of cisplatin resulted in enhanced apoptotic cell death (Figure 2I). These data together suggest that MAST1 contributes to cisplatin resistance through a MEK1-mediated anti-apoptotic signaling pathway.
We further explored the molecular mechanism by which MAST1 controls MEK1 activation in the presence of cisplatin. Interestingly, cisplatin does not affect Ras or Raf activity but dissociates cRaf, the known upstream kinase of MEK, from MEK1 in diverse types of cancer cells (Figures 3A and S3A). The dissociation also occurred in cisR cells, although these cells required more cisplatin to mediate this effect (Figure S3B). Moreover, cisplatin dissociates cRaf from MEK1 in a dose-dependent manner in vitro using purified MEK1-cRaf complex isolated from KB-3-1 cells by MEK1 immunoprecipitation, suggesting that cisplatin directly induces dissociation of cRaf from MEK1 (Figure 3B). Biophysical assays including surface plasmon resonance (SPR), thermal shift, and microscale thermophoresis further demonstrated that cisplatin directly binds to MEK1 but not cRaf (Figures 3C and S3C). In addition, co-immunoprecipitation revealed that MAST1 forms a complex with cRaf-MEK1 and cisplatin dissociates cRaf from MEK1, but MAST1 remains within the complex (Figure 3D). In agreement with this, SPR using purified proteins showed that cisplatin binding to MEK1 weakens the interaction of MEK1 with cRaf, but not MAST1 (Figure 3E). SPR-based kinetic analyses of the binding between cisplatin and MEK1 using MEK1 cysteine mutants revealed that the C142 residue is responsible for cisplatin binding (Figures 3F and S3D). Cisplatin binding-deficient mutant MEK1 C142A was unable to separate cRaf from MEK1 in the presence of cisplatin in vitro (Figures 3F and S3E). Moreover, the C142A MEK1 neither separated cRaf from MEK1-cRaf complex nor attenuated MEK activity in cells in response to cisplatin, suggesting that cisplatin binds MEK1 and directly promotes the dissociation of cRaf (Figures 3G and S3F). MEK1 knockdown did not influence the MAST1 and cRaf interaction, suggesting that multiple binding sites exist in the complex, and MAST1 is coupled with cRaf not through MEK1 (Figure S3G). Next, we investigated whether cisplatin impacts the dependency of MEK1 on MAST1 or cRaf. Knockdown of cRaf decreased MEK1 phosphorylation in the absence but not the presence of cisplatin (Figure 3H). In contrast, MEK1 phosphorylation was decreased by MAST1 knockdown in the presence but not the absence of cisplatin (Figure 3H), and the decreased phosphorylation of MEK1 was restored by rescue expression of MAST1 in MAST1 knockdown cells with cisplatin (Figure 3I). Furthermore, in the presence of cisplatin, MAST1 WT but not MAST1 kinase-dead mutant DA induced MEK1 activation regardless of cRaf knockdown (Figure 3J). MEK1 activation status was also monitored by ERK phosphorylation (Figure S3H). These results suggest that cisplatin switches cRaf-dependent MEK1 activation to MAST1-dependence in cancer cells.
Figure 3. Cisplatin but no other DNA damaging agents dissociates cRaf from MEK1 and reactivates MEK1 through MAST1.
(A) Interaction between cRaf and MEK1 upon cisplatin treatment in diverse cancer types. Cells were treated with 5 μg/ml cisplatin for 24 hr prior to MEK1 immunoprecipitation. (B) Purified MEK1-cRaf complex from KB-3-1 cells were incubated with increasing concentrations of cisplatin (0-5 μg/ml) for 2 hr and applied to Western blotting. (C) Biacore SPR (left) and thermal shift assay (right) were performed using purified recombinant MEK1 or cRaf with increasing concentrations of cisplatin. (D) MAST1 interacts with cRaf and MEK1 in cancer cells. ov: overexpressed. (E) Dissociation constant (Kd) values for MEK1-cRaf or MEK1-MAST1 interaction in the presence and absence of cisplatin were determined by Biacore SPR analysis. (F) Interactions between MEK1 variants and cisplatin or cRaf in the absence and presence of cisplatin were determined by SPR and shown as Kd values. N.D. (not determined). (G) Binding affinity of WT and C142A MEK1 to cRaf in the presence of cisplatin. MEK1 shRNA-resistant flag-MEK1 WT and C142A were expressed in MEK1 knockdown KB-3-1 cells. Cells were treated with cisplatin (5 μg/ml) for the indicated time and MEK1-cRaf interaction was determined by flag pull down. (H) Effect of cRaf (top) or MAST1 (bottom) downregulation on MEK1 activation in the presence and absence of cisplatin. cRaf and MAST1 knockdown cells were treated with cisplatin for 48 hr and MEK1 activity was determined by p-MEK S217/S221 Western blotting. (I) Effect of MAST1 rescue expression on MEK1 activation in MAST1 knockdown cells with cisplatin treatment. (J) Effect of WT or DA MAST1 expression on MEK1 activation in cRaf knockdown cells in the presence or absence of cisplatin. (K) Purified MEK1-cRaf complex from KB-3-1 cells were incubated with 5 μg/ml cisplatin. Samples were collected at different time points and subjected to immunoblotting. Density analysis of relative amount of cRaf bound to MEK1 from three biological replicates are shown. (L) Accumulation of cisplatin-induced DNA damage and repair in KB-3-1 cells with or without MAST1 shRNA was determined by flow cytometry analysis of phospho-γH2AX (upper) and phospho-53BP1 (lower). For DNA repair, cells were treated with cisplatin (5 μg/ml) for 2 hr before cisplatin washed off and cells were incubated in fresh medium. (M) cRaf-MEK1 dissociation induced by DNA damaging agents and cytotoxic drugs. KB-3-1 cells were treated with different compounds as indicated, followed by MEK1 immunoprecipitation and immunoblotting. (N) Cells with or without MAST1 knockdown were treated with different concentrations of DNA damaging agents and cytotoxic drugs and IC50 values were obtained. Error bars represent ± SD from three biological (K) or technical (L) replicates. p values were determined by two-tailed Student’s t test (ns: not significant). See also Figure S3.
To further explore whether the cisplatin-mediated switch to MAST1 dependence is a result of a DNA damage response signaling, we examined the timing of the switch and the impact of MAST1 on cisplatin-DNA adduct and on DNA damage and repair in time course analyses. cRaf dissociated from MEK1 upon cisplatin treatment within one hr regardless of Raf activation such as BRaf V600E. However, cisplatin induced reactive oxygen species (ROS) and consequently activated Ras and the MEK/ERK cascade overshadowed the effect of dissociation on MEK/ERK suppression in the early hr of treatment, and this was abolished by antioxidant L-acetyl-cysteine (NAC) treatment. Whereas cisplatin-induced DNA damage was a later event, suggesting that MEK/ERK suppression is not a secondary effect of cisplatin treatment but occurs through cRaf-MEK1 dissociation (Figures 3K, S3I, and S3J). In addition, loss of MAST1 had no impact on DNA damage/repair and cisplatin adduct accumulation/removal (Figures 3L, S3K, and S3L). Moreover, MAST1 knockdown did not impact the Fanconi anemia DNA repair pathway and DNA damage response signaling (Figures S3M-S3O). Most importantly only cisplatin, but not other DNA damaging agents or the chemotherapy agent paclitaxel, induced dissociation of cRaf-MEK1 (Figure 3M). In line with this dissociation, knockdown of MAST1 only enhanced sensitivity to cisplatin but not to other DNA damaging agents or paclitaxel (Figures 3N and S3P). These data together suggest that MAST1 contributes to cisplatin resistant cell survival not by acting as a critical element of the DNA damage response but by activating MEK-mediated anti-apoptotic signaling.
We next investigated whether MEK1, as a downstream phosphorylation target of MAST1, contributes to MAST1-dependent pro-survival signals in response to cisplatin. Cancer cell lines with stable knockdown of MAST1 and forced expression of phospho-mimetic (S221D) or -deficient (S221A) mutant of MEK1 were generated and examined for proliferative potential in vitro and in vivo (Figure 4A). Silencing MAST1 using shRNA did not affect cell viability and apoptosis induction in the absence of cisplatin. But with cisplatin treatment, knockdown of MAST1 significantly decreased cell viability and enhanced apoptosis induction, whereas expression of S221D, but not S221A, MEK1 significantly rescued the phenotypes resulting from MAST1 knockdown (Figures 4B and 4C). Furthermore, S221D but not S221A MEK1 restored the attenuated tumor growth of MAST1 knockdown KB-3-1cisR xenografts treated with cisplatin (Figure 4D). These results suggest that MAST1 signals through MEK1 by phosphorylation at serine 221 to promote cisplatin-resistant cancer cell proliferation and tumor growth.
Figure 4. MAST1 induces cisplatin-resistant cancercell proliferation and tumor growth through MEK1 phosphorylation.
(A) MAST1 and flag-tagged S221A and S221D MEK1 expression were detected by immunoblotting in KB-3-1cisR, A549cisR and KB-3-1 cells. (B-C) Cell viability (B) and apoptosis (C) in MAST1 knockdown cells with the expression of the indicated MEK1 variants in the presence and absence of sublethal dose of cisplatin (2 μg/ml: parental, 5 μg/ml: cisR). (D) Tumor volume (left), tumor weight (middle), and Ki-67 expression (right) of KB-3-1cisR cell xenograft mice. Representative dissected tumors are shown on right top panel. KB-3-1cisR cells with MAST1 knockdown and MEK1 S221A or S221D expression were injected and cisplatin was administered by intraperitoneal injection when there were palpable tumors. Tumor volume and tumor weight were normalized to the corresponding cisplatin untreated group. Scale bars represent 5 mm for dissected tumors and 20 μm for Ki-67 IHC staining images. Data are mean ± SD from three technical replicates for (B) and (C). Error bars represent SEM for tumor growth and SD for tumor weight in panel (D). p values were determined by two-tailed Student’s t test (ns: not significant; *: 0.01 < p < 0.05; **: p < 0.01).
MAST1 expression correlates with cisplatin resistance of diverse cancer cell lines and primary tumor tissues
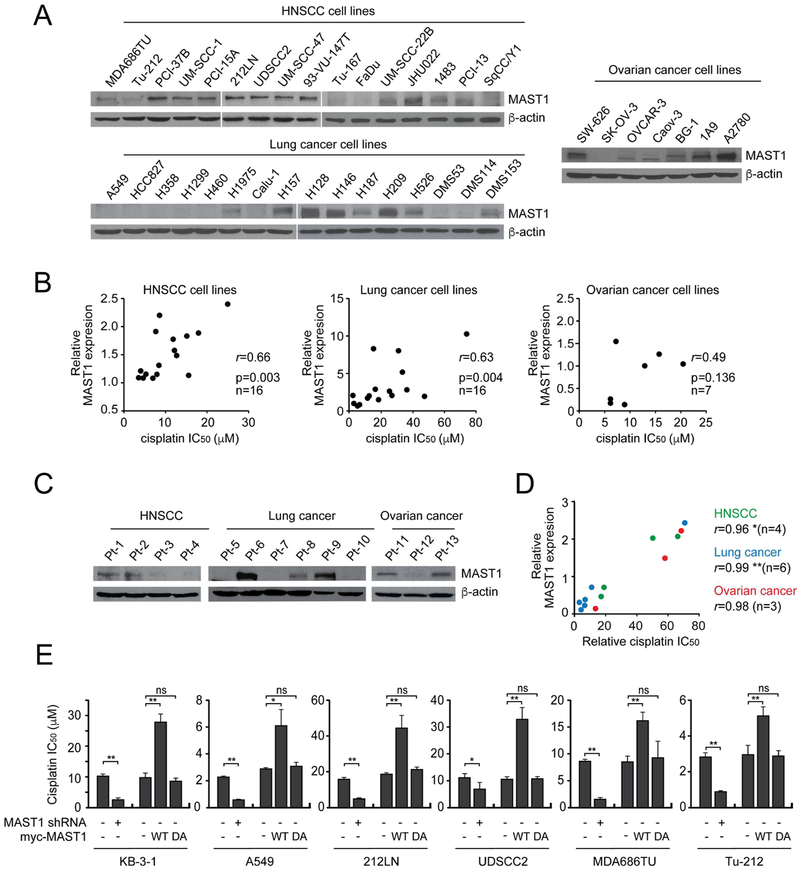
To investigate the expression of MAST1 and its relationship with cisplatin resistance in human cancer, MAST1 expression and cisplatin response were examined in 39 human cancer cell lines and 13 patient-derived xenograft (PDX) HNSCC, lung cancer, and ovarian cancers tissues (Geisinger et al., 1989; Lin et al., 2007). We found that MAST1 expression positively correlates with cisplatin IC50 (Figures 5A-5D and S4A-S4B). Knockdown of MAST1 by shRNA sensitized diverse cancer cells to cisplatin, while overexpression of WT but not DA MAST1 conferred cisplatin resistance to these cells (Figure 5E). We found that MAST1 expression levels, both protein and mRNA, were upregulated in cisplatin-resistant (cisR) cells that were chronically exposed to cisplatin, compared to parental cells (Figure S4C). Furthermore, we observed increased correlation between MAST1 and MEK phosphorylation in cisplatin treated cells compared to non-treated cells (Figure S4D).
Figure 5. MAST1 expression correlates with cisplatin resistance in cancer cell lines and patient-derived tumors.
(A) MAST1 expression in HNSCC, lung, and ovarian cancer cell lines. (B) Correlation between MAST1 expression and cisplatin IC50 in cancer cell lines shown in (A). (C) MAST1 expression in PDX tumors. (D) Correlation between MAST1 expression and cisplatin IC50 in PDX tumors shown in (C). r = Pearson’s correlation coefficient. (E) Effect of MAST1 knockdown or WT or DA MAST1 overexpression on cisplatin response in diverse cancer cell lines. Cisplatin response was determined using cisplatin IC50. Error bars represent SD from three technical replicates. p values were determined by two-tailed Student’s t test (ns: not significant; *: 0.01 < p < 0.05; **: p < 0.01). See also Figure S4.
To demonstrate the clinical relevance of our findings, we evaluated MAST1 and phospho-MEK levels in 97 HNSCC patient cases who received platinum-containing therapy or non-platinum based therapy such as radiation or surgery (Figures 6A, S5A-S5B, and Tables S1-S2). For cases of platinum-containing therapy, patients who showed no evidence of disease for over 2 years after chemotherapy with cisplatin and/or carboplatin were considered ‘Sensitive’ and patients with disease recurrence within 2 years were considered ‘Resistant’. To determine whether a high MAST1 level initially and after the chemotherapy confers cisplatin resistance, MAST1 and phospho-MEK levels were analyzed in tumor samples collected before and after platinum-containing chemotherapy. The ‘Resistant’ group showed significantly higher MAST1 and phospho-MEK1 levels compared to the ‘Sensitive’ group in both pre- and post-treatment tumor samples (Figures 6B and S5C). In support of our finding that cisplatin drives MAST1-mediated MEK activation, significant positive correlation between MAST1 and phospho-MEK levels were observed in post-platinum treatment samples, but not in samples collected before the treatment (Figure 6C). To determine whether MAST1 is induced during platinum treatment, we evaluated pre-and post-treatment samples. Analysis of pre- and post-treatment paired tumor samples obtained from individual patients showed that MAST1 staining increased after cisplatin treatment, whereas this increase was not observed in paired samples collected from cancer patients who received regimens other than platinum-based therapy (Figures 6D and S5D). Similar to paired tumors, the analysis of non-paired pre- and post-treatment tumors also demonstrated that MAST1 was higher after platinum-based therapy, but not after non-platinum-based therapy (Figure 6E). Furthermore, high MAST1 expression was associated with a worse clinical outcome only in cancer patients who received platinum-containing chemotherapy, but not in cancer patients who received non-platinum-based therapy (Figures 6F and 6G). Collectively, these results illustrate that initial MAST1 expression significantly correlates with platinum resistance (short term), and that MAST1 is further induced during platinum-based chemotherapy (long term), which together leads to platinum resistance and worse clinical outcome.
Figure 6. MAST1 and MEK activation is associated with cisplatin resistance and poor clinical outcome in human HNSCC.
(A) Schematic summary of HNSCC patients (n=97 cases) and samples (n=116 tumors) analyzed by IHC. Open circle: pre-therapy tumors; closed circle: post-therapy tumors. Tumor numbers of before or after CT or non-CT groups are indicated on the left. Case numbers for sensitive or resistant CT or non-CT treated patients are indicated at the bottom. CT: platinum-based chemotherapy. (B) MAST1 expression and MEK phosphorylation levels and drug response in specimens before (top) and after (bottom) platinum treatment. (C) Correlation between MAST1 and phospho-MEK in samples before (top) and after (bottom) platinum treatment. (D-E) Comparison of MAST1 status between pre- and post-therapy in paired (D) and non-paired (E) samples. Top: Platinum therapy; Bottom: non-platinum therapy. (F-G) Kaplan-Meier survival analysis of platinum-treated (F) and non-platinum-treated (G) patient groups. Patients were dichotomized by MAST1 expression level at median. Error bars represent ± SD for (B) and (E). p values were determined by Pearson correlation for (C), paired Student’s t test for (D), low-rank test for (F-G), and unpaired Student’s t test for the rest (ns: not significant; *: 0.01 < p < 0.05; **: p < 0.01). See also Figure S5.
Identification and characterization of lestaurtinib as a MAST1 inhibitor to target cisplatin-resistant cancer
Our finding that MAST1 is upregulated in cisplatin-resistant cancer and that attenuation of MAST1 sensitizes cancer cells to cisplatin treatment implicates MAST1 as a promising anti-cancer target to overcome cisplatin resistance. To our knowledge, there is no compound reported as a MAST1 inhibitor. We thus screened for a MAST1 inhibitor by testing the top 10 small molecules among 72 kinase inhibitors documented to bind to MAST1 (Figure 7A), according to the IUPHAR database, which provides binding reactivity of the 72 inhibitors against 456 kinases (Davis et al., 2011). 6 inhibitors out of 10 that showed a significant decrease in MAST1 activity were evaluated for their ability to sensitize cancer cells to cisplatin-induced cell death (Figure 7B). MAST1 inhibition efficacy strongly correlated with cisplatin response with r value of 0.82 (Figure 7C). Among these, lestaurtinib sensitized cells to cisplatin treatment the most using a concentration that attenuates cell viability by less than 20% when treated alone. Lestaurtinib effectively inhibited MAST1 in vitro and in cells (Figure 7D). We also showed that lestaurtinib inhibits MAST1 as an ATP competitive inhibitor (Figure 7E). To demonstrate the selectivity of lestaurtinib binding to MAST1, we generated a MAST1 L504D mutant. Leucine 504 in MAST1 is predicted to be critical for lestaurtinib binding based on the structure of lestaurtinib and PRK1, a kinase that has similar catalytic domain to MAST1 (Chamberlain et al., 2014). Cellular thermal shift assay showed that MAST1 L504D no longer binds to lestaurtinib (Figure 7F). Lestaurtinib attenuated the kinase activity of WT but not L504D MAST1 (Figure 7G). In line with the kinase activity, the lestaurtinib did not sensitize cells to cisplatin when harboring L504D MAST1, suggesting that lestaurtinib sensitizes cells to cisplatin by binding and inhibiting MAST1 kinase activity (Figure 7H). Overexpression of MAST1 enhanced cisplatin resistance and MEK activation but had no impact on cRaf activity in various cell lines with different cisplatin sensitivity, whereas 100 nM lestaurtinib was sufficient to fully inhibit both endogenous and overexpressed MAST1 in these cells suggesting that the drug capacity is high enough to inhibit any excessive MAST1 in cells (Figures 7I, S6A, and S6B). Lestaurtinib was not effective in cells overexpressing L504D MAST1 further supporting that the drug’s effect on cisplatin response and MEK activation is through MAST1 inhibition. Furthermore, lestaurtinib sensitizes cells to cisplatin treatment in diverse cell lines, while the effect was abolished in MAST1 knockdown cells in vitro and in vivo (Figures 7J, S6C, S6D, and 7K-7M, S6E-S6G, respectively). Lestaurtinib is a tyrosine kinase inhibitor known to inhibit FLT3, JAK2, Trk, and PRK1 (Shabbir and Stuart, 2010). Other known FLT3, Trk, or JAK2 inhibitors did not alter cisplatin sensitivity in cancer cells (Figure S6H). Furthermore, FLT3 and Trk were not expressed in the cancer cells we tested, and genetic inhibition of JAK2 did not alter cisplatin response (Figures S6I and S6J). These data together suggest that cisplatin sensitivity conferred by lestaurtinib occurs mainly through MAST1 inhibition, not through other kinases.
Figure 7. Validation of lestaurtinib as a MAST1 inhibitor.
(A) In vitro MAST1 kinas eassay using ten compounds potentially bind to MAST1. Compounds (10 μM) were incubated with purified GST-MAST1 and applied to the in vitro kinase assay. (B) Effect of top six MAST1 inhibitors from (A) on cisplatin sensitivity in KB-3-1 cells. A dose of inhibitor that did not affect viability when given alone (100 nM of lestaurtinib, dovitinib, staurosporine; 1 μM of sunitinib, SU14813, bosutinib) was used with increasing concentrations of cisplatin for 48 hr. (C) Correlation between MAST1 activity inhibition and cisplatin sensitivity of the top six MAST1 inhibitors. Red: lestaurtinib. (D) Effect of lestaurtinib on MAST1 activity in vitro (left) and in vivo in KB-3-1 cells (right). Purified GST-MAST1 variants were treated with increasing concentrations of lestaurtinib. (E) Effect of lestaurtinib on MAST1 activity at a range of ATP concentrations. (F) Cellular thermal shift assay using KB-3-1 cells harboring WT or L504D MAST1 treated without (−) or with (+) lestaurtinib. (G) Kinase activity of MAST1 WT or L504D treated with lestaurtinib. (H) cisplatin IC50 upon lestaurtinib treatment in KB-3-1 cells with endogenous MAST1 knockdown expressing MAST1 WT or L504D in KB-3-1 cells. (I) Effect of lestaurtinib and WT or L504D MAST1 overexpression on MEK and cRaf phosphorylation in KB-3-1 cells. (J) Cisplatin IC50 upon lestaurtinib treatment in cells with or without MAST1 knockdown in KB-3-1 cells. (K) Tumor growth of lestaurtinib and cisplatin treated mice carrying KB-3-1cisR cell xenografts. Error bars represent SEM. Representative tumors for each group are shown. Scale bar represents 5 mm. (L) Tumor weight (left) and Ki-67 IHC staining (right) at the experimental endpoint. Error bars represent SD and scale bars represent 50 μm. (M) MAST1 activities in tumor lysates are shown. Data are mean ± SD from three replicates of each sample except panels (K and L). p values were determined by two-tailed Student’s t test (ns: not significant; *: 0.01 < p < 0.05; **: p < 0.01). See also Figure S6.
Finally, we evaluated the efficacy of lestaurtinib in abrogating cisplatin resistance in vitro and in vivo. Treatment of lestaurtinib effectively sensitized multiple cisS and cisR cells to not only cisplatin (Figure S7A) but also carboplatin (Figure S7B). Furthermore, lestaurtinib and cisplatin synergistically attenuated cell viability in cells with combination index (CI) around 0.26 (Figure 8A, left). In KB-3-1 cells, lestaurtinib alone partially decreased cell viability and apparently this was not due to increased apoptosis. Similar results were obtained in colony formation assays (Figure 8A, middle). Consistent with the phenotype observed in MAST1 knockdown cells, inhibition of MAST1 by lestaurtinib in combination with cisplatin resulted in increased apoptotic cell death and decreased MEK activation (Figure 8A, right). Similar results were obtained in multiple cisS and cisR cells (Figure S6B). In addition, combination of lestaurtinib and cisplatin synergistically attenuated cell viability in primary platinum-refractory TKO-002 cells with CI value of 0.57 (Figure 8B). Consistent with MAST1 knockdown, targeting MAST1 with lestaurtinib only sensitized cells to cisplatin but not to other DNA damaging agents or chemotherapy agents, further supporting that the synergistic effect of MAST1 inhibition is specific to cisplatin (Figure S7C). Moreover, the effect of lestaurtinib was greater in cell lines and patient-derived tumors with higher MAST1 levels (Figures S7D and S7E). Lastly, we established patient-derived xenograft (PDX) models using diverse cancer patient tumors including HNSCC, lung cancer, and ovarian cancer. The dosage of 20 mg/kg lestaurtinib and 5 mg/kg cisplatin did not induce significant changes in body weight, histopathology, or hematopoietic properties (Figures S8A-S8C). The combination significantly reduced PDX tumor growth and tumor cell proliferation compared to single agent treatment (Figures 8C and 8D). Moreover, higher dose of lestaurtinib (30 mg/kg) achieved better efficacy in inhibiting MAST1 activity and tumor growth (Figure S8D). Lestaurtinib significantly attenuated MAST1 activity and MEK/ERK phosphorylation but not Ras or Raf activity in PDX tumors (Figures 8E, 8F, S8E, and S8F). Finally, to demonstrate whether the effect of MAST1 inhibitor lestaurtinib on the response to cisplatin therapy is mediated through MEK, the effect of a MEK inhibitor on enhancing cisplatin therapy was compared to that of lestaurtinib in vitro and in vivo in an additional PDX model of lung cancer. The combination of trametinib and cisplatin mimicked the lestaurtinib and cisplatin effect resulting in cisplatin sensitization and attenuated PDX tumor growth and proliferation, but lestaurtinib showed slightly greater synergistic effect than trametinib or AZD6244 (Figures S8G-S8K). These results together suggest that the MAST1-MEK1 signaling axis is a crucial pathway for cancer cisplatin resistance and lestaurtinib is a MAST1 inhibitor with promising anti-tumor effect in combination therapy with cisplatin in human cancers.
Figure 8. Lestaurtinib sensitizes cancer cells to cisplatin treatment in vitro and in vivo.
(A) Cell viability, colony formation, apoptosis induction, and MEK1 phosphorylation in KB-3-1 and 212LN with vehicle control, lestaurtinib, cisplatin and the combination. (B) Lestaurtinib effect on cell viability and cisplatin sensitivity of TKO-002 cells. Combination index (CI) values for synergistic effect are shown. (C) Effect of lestaurtinib, cisplatin and the combination effect on tumor growth of cisR HNSCC, lung cancer, and ovarian cancer PDX mice. Pt-8 tumor and Pt-11 tumor in Figure 5C were used for lung cancer PDX and ovarian cancer PDX, respectively. Error bars represent SEM. Scale bars for the dissected tumors represent 5 mm. (D) Ki-67 expression was determined by IHC staining. Scale bars represent 50 μm. (E) MAST1 activity was assessed by MAST1 in vitro kinase assay using MBP as a substrate. (F) MEK1 inhibition by lestaurtinib and cisplatin combination in PDX tumor lysates. (G) Proposed model for the role of MAST1 in cisplatin resistance in human cancer. Left: Cancer cells rely on cRaf-dependent MEK1 activation to promote tumor growth in the absence of cisplatin. Right: Cisplatin treatment dissociates cRaf from MEK1, while MAST1 phosphorylates MEK1 to activate the MAPK pathway in cRaf-independent manner, inhibiting BIM and providing a proliferative advantage to cancer cells. Targeting MAST1 by lestaurtinib restores cisplatin sensitivity to cells. Data are mean ± SD from three technical replicates of each sample except panel (C). p values were determined by two-tailed Student’s t test (*: 0.01 < p < 0.05; **: p < 0.01). See also Figures S7 and S8.
DISCUSSION
Here we identify MAST1 as a synthetic lethal partner of cisplatin that functions as a critical factor to program cisplatin-resistant pro-survival signaling in human cancers. Our findings suggest a mechanistic basis by which MAST1 controls cancer cells to evade cisplatin-induced cell death. Cells predominantly depend on Raf for MEK1 activation during active cell proliferation (Lavoie and Therrien, 2015). Consistently, we found that, although MAST1 is present in the cRaf-MEK1 complex, cancer cells depend on cRaf-dependent MEK1 activation to promote proliferation and tumor growth (Figure 8G, left). However, cisplatin treatment dissociates cRaf from MEK1, whereas MAST1 remains in complex with MEK1. MAST1 phosphorylates MEK1, leading to cRaf-independent activation of MEK1 and the downstream MAPK pathway including loss of the pro-apoptotic BIM. This MAST1-mediated MEK1 activation provides anti-apoptotic and proliferative protection to cancer cells treated with cisplatin, which, if sufficient to reverse the pro-apoptotic signaling induced by cisplatin, promotes cancer cell proliferation and tumor growth (Figure 8G, right). Our studies using clinical samples supports that upregulation of MAST1 positively correlates with non-response to platinum therapy. There may exist two complementary paths to MAST1-mediated platinum resistance, which are initially high levels of MAST1 and the ability to upregulate MAST1 in response to platinum.
Our study provides evidence that cisplatin inactivates the MAPK pathway by dissociating cRaf from MEK1, while MAST1 reactivates the MAPK pathway. Characterization of the DNA damage response upon MAST1 loss and demonstration of the specificity of the MAST1 effect for cisplatin but not broadly for DNA damaging drug reveals that targeting MAST1 may be impactful specifically for cisplatin therapy. The specific structure of cisplatin may disrupt the MEK1-cRaf complex resulting in MAST1 as the sole kinase remaining in the complex. Previous reports showed that cRaf binding sites in MEK1 consist of Ser221/217, whereas predicted cisplatin binding sites in MEK1 are Met56 and Cys104 (Caunt et al., 2015; Yamamoto et al., 2015). We revealed that cisplatin directly binds to MEK1 but not cRaf. We also found that MEK1 and cRaf dissociates in the presence of cisplatin, whereas the binding affinity of MEK1 and MAST1 is unaltered or slightly increased by cisplatin. It is possible that cisplatin binds to MEK1 and induces conformational changes that affect cRaf but not MAST1 binding. Since MAST1 remains within the complex while cRaf dissociates from MEK1 upon cisplatin treatment, cisplatin may not be involved in MAST1 recruitment or MAST1 binding to MEK1. Future mutational and structural studies are warranted to determine the structural mechanisms by which cisplatin affects cRaf and MAST1 function.
Since the dissociation of cRaf from MEK1 induces MAST1 to reactivate the MAPK pathway, any cellular events that affect cRaf-MEK1 binding may activate MAST1-dependent MEK1 activation. Indeed, our result showed that cRaf knockdown in the absence of cisplatin is sufficient to induce MAST1-dependent MEK1 activation, suggesting that MAST1 might represent an alternative upstream kinase of MEK1 that activates the MAPK pathway. In fact, similar “kinase switch” has been observed in cells. MEK1 can be activated by several alternative kinases such as MEKK1, Mos in oocytes, and Cot1 or MLK in melanoma bypassing Raf as one mechanism of resistance to BRaf inhibitors (Gardner et al., 1994; Johannessen et al., 2010; Marusiak et al., 2014; Verlhac et al., 2000). Although the MAPK pathway is implicated in cisplatin resistance, the detailed mechanism underlying its role is still unclear (Kong et al., 2015; Wang and Wu, 2014). Our findings reveal a mechanism by which a kinase MAST1-mediated cRaf-independent reactivation of the MAPK pathway contributes to the development of cisplatin resistance.
MAST1 appears to interact with syntrophin and links the dystrophin/utrophin network with microtubule filaments in cells. No disorders are reported to be associated with the MAST1 gene, and only a few studies reveal links between MAST family members and human cancer. For instance, MAST2 is known to bind and phosphorylate the tumor suppressor PTEN (Valiente et al., 2005). Robinson et al. demonstrated recurrent rearrangements involving MAST1 and MAST2 encoding genes in breast cancer, and overexpression of MAST1 or MAST2 gene fusions had proliferative effects (Robinson et al., 2011). Interestingly, two out of three identified MAST1 fusion genes lack its serine/threonine kinase domain-coding region, suggesting that a kinase-independent role of MAST1 may exist that contributes to proliferative potential in cancer. However, our data illustrate that kinase activity of MAST1 is required for cisplatin resistance.
From a clinical perspective, our findings support that MAST1 could serve as a predictive marker and as a promising therapeutic target to treat cancer patients in combination with platinum-based chemotherapy. We identified lestaurtinib as a potent MAST1 inhibitor with promising cisplatin sensitization potential. Although lestaurtinib was originally reported as a tyrosine kinase inhibitor that inhibits FLT3 as well as JAK2 and Trk (Shabbir and Stuart, 2010), it has been suggested that many multi-kinase inhibitors target both serine/threonine and tyrosine kinases (Bilanges et al., 2008; Dawson et al., 2010; Garcia-Manero et al., 2015). Consistent with this concept, we demonstrate that lestaurtinib effectively inhibits the serine/threonine kinase MAST1. Our findings that other FLT3, JAK2, and Trk inhibitors did not confer cisplatin sensitization and lestaurtinib had no effect on cisplatin response in cells deficient in MAST1 suggest that MAST1 represents the primary target through which lestaurtinib sensitizes cancer cells to cisplatin. Lestaurtinib is generally well tolerated in patients (Knapper et al., 2006) and is currently under clinical evaluation. We validated the efficacy of lestaurtinib in abolishing cisplatin resistance in patient-derived xenografts of diverse cancers, which suggests that our approach could be commonly applied to treat various types of cancers. MAST1-targeted therapy would be more beneficial to patients with advanced cancers or patients who received platinum-based therapy but recurred, in part, due to the induction of MAST1 during the treatment. Future pharmacokinetics and toxicity studies and clinical trials are warranted to evaluate and optimize dose and treatment conditions for the proposed anti-MAST1 therapy.
STAR Methods
CONTACT FOR REAGENT AND RESEOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sumin Kang (smkang@emory.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human studies
Approval to use human specimens was given by the Institutional Review Board of Emory University. All clinical samples were collected with informed consent under Health Insurance Portability and Accountability Act (HIPAA) approved protocols.
Animal studies
Animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of Emory University. Nude mice (athymic nu/nu, female, 4-6-week old, Envigo) or NSG mice (NOD scid gamma, female, 4-6-week old, Jackson Laboratory) were used for xenograft experiments.
Cell culture
H128 cells were cultured in Iscove Modified Dulbecco Medium (IMDM) with 20% FBS. DMS53, DMS114, and DMS153 cells were cultured in Waymouth medium with 10% FBS. SK-OV-3 cells were cultured in McCoy’s 5a medium with 10% FBS. SW-626 cells were cultured in Leibovitz’s L15 medium with 10% FBS. All HNSCC cell lines were cultured in Dulbecco Modified Eagle Medium (DMEM)/Ham’s F-12 50/50 mix medium with 10% FBS. KB-3-1, BG-1, Caov-3, and 293T cells were cultured in DMEM with 10% FBS. All the rest were cultured in RPMI 1640 medium with 10% FBS.
METHOD DETAILS
RNAi screens
Primary screen was performed using the human kinome shRNA library by transducing KB-3-1 cisplatin-resistant human carcinoma cells (KB-3-1cisR) with lentivirus pools targeting each gene individually. Cells were seeded into 6 replicates and infected with lentivirus in 96-well plates. 48 hr after infection, cells were treated with PBS, a sublethal dose of cisplatin, or puromycin (0.5 μg/ml), in duplicates. Cell viability was determined after 2 days of cisplatin treatment using CellTiter-Glo Luminescent Viability Assay (Promega). Gene candidates that induced more than 15% cell death by shRNA alone and/or had poor shRNA virus transduction efficacy assessed by puromycin selection were excluded. The 50 top ranking candidates from the primary screen were further confirmed in four cisplatin-resistant cancer cell lines PCI-15AcisR, A549cisR, A2780cisR, and KB-3-1cisR as performed in primary screen.
Virus production and infection for protein overexpression in cancer cells
Human MAST1 and MEK1 were myc or flag tagged by PCR and subcloned into pDEST27 and pLHCX-derived Gateway destination vectors. Selection was carried out with 300 μg/ml hygromycin for stable expression. MAST1 and MEK1 variants were generated by site-directed mutagenesis. Lentivirus carrying shRNA were generated by transfecting 293T cells with pLKO.1 vector encoding shRNA, pMD2.G, and psPAX2 and virus were collected. Cells were infected with lentivirus for 2 days and selected using puromycin (2 μg/ml).
Cell viability assay, colony formation assay, and apoptosis assay
Cells were treated with 5 μg/ml cisplatin for 48 hr for CisR cells and 24 hr for parental cells unless specifically indicated. Approximately 7000 cells/well were seeded in 96-well plates and treated with the indicated concentrations of cisplatin and lestaurtinib for 48 hr. Cell viability was measured using CellTiter-Glo Luminescent Viability Assay according to the manufacturer’s instructions. For colony forming assay, 200-500 cells were seeded in 35-mm dishes and treated with PBS, cisplatin, lestaurtinib or in combination for 24-48 hr. The colonies were stained with 0.5% crystal violet and counted by ImageJ software after 10 days. Apoptotic cells were assessed using FITC Annexin V Apoptosis Detection kit.
Kinase activity assays
MAST1 variants were precipitated from cell lysates with a glutathione-Sepharose 4B column or anti-MAST1 antibody. Eluted MAST1 was incubated with MBP or inactive MEK1 in kinase assay buffer (40 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 200 μM ATP, 0.1 mg/ml BSA) for 30 min at 30 °C. The activity of MAST1 was determined either by ADP-Glo Kinase Assay or Western blot using phospho-MEK S217/S221 antibody. cRaf and MEK1 kinase activities were measured by ADP-Glo kinase assay using recombinant inactive MEK1 and ERK2 as substrates, respectively.
Cisplatin or carboplatin sensitivity assay
To determine the IC50 of patient-derived xenograft (PDX) tumors, PDX tumors were digested with tissue dissociation buffer (0.1% collagenase, 0.01% hyaluronidase, 0.01% DNase I in HBSS) for 1 hr at 37°C to obtain single cell suspension. The enzyme digestion-released tumor cells were washed through a 70 mm strainer with HBSS and counted. 1~2 ×104 of live tumor cells for each PDX tumor were seeded in 96 well plates and treated with different concentrations of cisplatin or carboplatin. After 48~72 hr, the cell viability was determined by Cell Titer Glo assay and IC50 was calculated using Graphpad software with non-linear regression analysis. For cancer cell lines, approximately 0.5~1×104 of cells were seeded in 96 well plates and treated with different concentrations of cisplatin. After 48~72 hr, the cell viability was determined by Cell Titer Glo assay and IC50 was calculated using Graphpad software.
Phospho-Protein Profiling
Lysates obtained from KB-3-1cisR-pLKO.1 and KB-3-1cisR-pLKO.1-MAST1 shRNA cells treated with PBS or cisplatin were applied to the Phospho Explorer Antibody Array (Full Moon Biosystems), as previously described (Kang et al., 2010).
Surface Plasmon Resonance (SPR)
Biacore X100 system (GE Healthcare) was used to perform SPR experiments. 1 μM of recombinant MEK1 or RAF1 were coupled to CM5 sensor chip through amine chemistry at pH 5. For cisplatin-MEK1 or cisplatin-RAF1 binding analysis, a series dilution of cisplatin was prepared in 0.01 M HEPES pH 7.4, 0.15 M NaCl, 0.005% v/v Surfactant P20 and injected over MEK1 or RAF1 at 30 μl/min for a contact time of 180 s at 20 °C. For analysis of MAST1-MEK1 or RAF1-MEK1 interaction, single-cycle kinetic analysis was performed. Recombinant GST-MAST1 (0-100 nM) or GST-RAF1 (0-250 nM) at a series dilution were injected over the MEK1-coupled sensor chip in the absence or presence of 1 μM of cisplatin. The raw sensorgrams were blank-subtracted and analyzed using BIAevaluation Software (GE Healthcare).
Thermal Shift Assay
Thermal shift assay (Differential scanning fluorimetry) was performed using the Protein Thermal Shift Dye Kit. 1 μM of recombinant MEK1 or RAF1 was incubated with different concentrations of cisplatin. The fluorescence was recorded using Real-Time PCR Systems (Applied Biosystems) and data were analyzed using Prism Graphpad with Boltzmann model. The dissociation constant (Kd) was calculated using the equation below: Y=Bottom + ((Top-Bottom)*(1-((P-Kd-X+sqrt(((P+X+Kd)^2)-(4*P*X)))/(2*P))))
MicroScale Thermophoresis (MST)
A serial dilution of cisplatin was prepared (61 nM ~ 2 mM) in assay buffer (14 mM HEPES pH 7.5, 105 mM NaCl, 3.5 mM MgCl2, 0.04% Tween-20). 5 µl of each dilution step were mixed with 5 µl of the fluorescence-labeled recombinant MEK1 and filled in capillaries. The samples were analyzed on Monolith NT.115 Pico at 25°C, with 10% LED power and 80% Laser power. No sample aggregation or precipitation effects were observed in the normalized fluorescence.
Immunohistochemical staining
Formalin-fixed, paraffin-embedded tissue specimens from HNSCC patients were obtained from the Emory head and neck tissue bank. Approval to use human specimens was given by the Institutional Review Board (IRB) of Emory University. Clinical information for the patients was obtained from the pathology files at Emory University Hospital under the guidelines and with approval from the IRB of Emory University. All clinical samples were collected with informed consent under Health Insurance Portability and Accountability Act approved protocols. Tumors from HNSCC patients who received platinum or non-platinum-based therapy were used. IHC analyses of MAST1, phospho-MEK S217/S221, and Ki-67 were performed according to the protocol described previously (Li et al., 2013). Positive staining of MAST1 and phospho-MEK1 in the tumor cells was identified using IHC signal intensity, scored as 0 to 3+.
Xenograft studies
Animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee of Emory University. Athymic nude mice (athymic nu/nu, 6-week old, Envigo) were subcutaneously injected with 5 × 105 KB-3-1cisR cells with MAST1 knockdown and MEK1 variant expression. Cisplatin 5 mg/kg twice a week and lestaurtinib 20 mg/kg 5 times a week were administered by intraperitoneal (i.p.) injection and subcutaneous injection, respectively, from 3-7 days after xenograft for 16-28 days. For PDX mouse model, after informed consent and approval by the IRB of Emory, tumor tissues from a HNSCC patient and SCLC patients were biopsied and implanted into the hind flanks of 6-week old NOD scid mice (Charles River). Once the tumor size reached 1500 mm3, the tumor was excised and small pieces were implanted in the flank of 6-week old athymic nude mice. Ovarian cancer PDX donor mouse was obtained from Jackson Labs and serially transplanted to 6-week-old NOD scid gamma (NSG) mice. The mice were evenly divided into groups when the size of the tumor reached 100-150 mm3. Lestaurtinib was administered by subcutaneous injection of 20 or 30 mg/kg 5 times a week for 20-30 days. 40% polyethylene glycol 400, 10% povidone, and 2% benzyl alcohol in PBS was a diluent control. The combination group was given both cisplatin and lestaurtinib as described. Tumor growth was recorded by blind measurement of two perpendicular diameters of the tumors and tumor size was calculated using the formula 4π/3 × (width/2)2 × (length/2). Tumors were harvested at the experimental endpoint and tumor proliferation was determined by Ki-67 IHC staining. For all animal studies, animals were randomly chosen. Concealed allocation and blinding of outcome assessment were used. No statistical method was used to predetermine sample size.
Cellular thermal shift assay
Cellular thermal shift assay was performed as previously described (Gad et al., 2014; Martinez Molina et al., 2013). In brief, KB-3-1 cells with MAST1 knockdown were transfected with shRNA-resistant GST-fused MAST1 WT or L504D, and treated with DMSO or lestaurtinib (100 nM) for 24 hr. Cells were collected and resuspended in TBS. Multiple aliquots of cells were heated at 48, 50, 52, 54, 56, 58, 60, 62 and 64°C for 3 min. Cells were lysed, precipitates were removed, and the MAST1 in the soluble fraction was quantified by Western blot analyses.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis and graphical presentation was performed using Prism 6.0 (GraphPad). No statistical method was used to predetermine sample size. Data shown are from one representative experiment of multiple experiments. Data with error bars represent mean ± standard deviation (SD), except for xenograft tumor growth curves, which represent mean ± standard error of the mean (SEM). Statistical analysis of significance was based on paired two-tailed Student’s t test for Figure 6E and non-paired for all other figures. Statistical tests performed are based on a set of assumptions including normal distribution and homogeneity of variances. The variability within each group has been quantified with standard deviation and used for statistical comparison.
DATA AND SOFTWARE AVAILABILITY
GraphPad Prism 6 was used in this study for graphics and statistical analyses.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-beta-actin (clone AC -15) antibody |
Sigma-Aldrich | Cat#A1978; RRID: AB_476692 |
| Mouse monoclonal anti-MBP (clone F-6) antibody | Santa Cruz Biotechnology |
Cat#sc-271524; RRID: AB_10655672 |
| Mouse monoclonal anti-glutathione S-transferase (clone GST-2) antibody |
Sigma-Aldrich | Cat#G1160; RRID: AB_259845 |
| Mouse monoclonal anti-myc (clone 9B11) antibody | Cell Signaling Technology |
Cat#2276; RRID: AB_331783 |
| Rabbit monoclonal anti-phospho-MEK S217/S221 (clone 41G9) antibody |
Cell Signaling Technology |
Cat#9154; RRID: AB_2138017 |
| Rabbit polyclonal anti-phospho-MEK S221 (clone 166F8) antibody |
Cell Signaling Technology |
Cat#2338; RRID: AB_490903 |
| Mouse monoclonal anti-MEK1 (clone 61B12) antibody | Cell Signaling Technology |
Cat#2352; RRID: AB_10693788 |
| Rabbit monoclonal anti-phospho-c-Raf (S338) (clone 56A6) antibody |
Cell Signaling Technology |
Cat#9427; RRID: AB_10078092 |
| Rabbit polyclonal anti-c-Raf antibody | Cell Signaling Technology |
Cat#9422; RRID: AB_390808 |
| Mouse monoclonal anti-c-Raf (clone 53) antibody for immnuoprecipitation |
BD Biosciences | Cat#610152; RRID: AB_397553 |
| Rabbit polyclonal phospho-p44/42 MAPK (Erk1/2) (T202/Y204) (clone 20G11) antibody |
Cell Signaling Technology |
Cat#4376; RRID: AB_331772 |
| Rabbit monoclonal anti-p44/42 MAPK (Erk1/2) (clone 137F5) antibody |
Cell Signaling Technology |
Cat#4695; RRID: AB_390779 |
| Mouse monoclonal anti-p44/42 MAPK (Erk1/2) (clone 3A7) antibody |
Cell Signaling Technology |
Cat#9107; RRID: AB_2235073 |
| Rabbit monoclonal anti-BIM (clone C34C5) antibody | Cell Signaling Technology |
Cat#2933; RRID: AB_1030947 |
| Rabbit monoclonal anti-cleaved PARP (Asp214) (clone D64E10) XP antibody |
Cell Signaling Technology |
Cat#5625; RRID: AB_10699459 |
| Rabbit polyclonal anti-Bcl-2 antibody | Cell Signaling Technology |
Cat#2872; RRID: AB_10693462 |
| Mouse monoclonal anti-BID (clone 5C9) antibody | Santa Cruz Biotechnology |
Cat#sc-56025; RRID: AB_781628 |
| Mouse monoclonal anti-Mcl-1 (clone 22) antibody | Santa Cruz Biotechnology |
Cat#sc-12756; RRID: AB_627915 |
| Mouse monoclonal anti-Bcl-xL (clone H-5) antibody | Santa Cruz Biotechnology |
Cat#sc-8392; RRID: AB_626739 |
| Rabbit monoclonal anti-phospho-BAD (S112) (clone 40A9) antibody |
Cell Signaling Technology |
Cat#5284; RRID: AB_560884 |
| Rabbit monoclonal anti-BAD (clone D24A9) antibody | Cell Signaling Technology |
Cat#9239; RRID: AB_2062127 |
| Rabbit monoclonal anti-phospho-Histone gamma H2AX (S139) (clone 20E3) antibody |
Cell Signaling Technology |
Cat#9718; RRID: AB_2118009 |
| Rabbit polyclonal anti-phospho-53BP1 (S1778) antibody | Cell Signaling Technology |
Cat#2675; RRID: AB_490917 |
| Rabbit monoclonal anti-Cisplatin-modified DNA (clone CP9/19) antibody |
Abcam | Cat#ab103261; RRID: AB_10715243 |
| Mouse monoclonal anti-FLAG (clone M2) antibody | Sigma-Aldrich | Cat#F1804; RRID: AB_262044 |
| Rabbit polyclonal anti-FLAG antibody | Sigma-Aldrich | Cat#F7425; RRID: AB_439687 |
| Mouse monoclonal anti-phospho-STAT3 (Y705) (clone B-7) antibody |
Santa Cruz Biotechnology |
Cat#sc-8059; RRID: AB_628292 |
| Rabbit polyclonal anti-STAT3 (C-20) antibody | Santa Cruz Biotechnology |
Cat#sc-482; RRID: AB_632440 |
| Rabbit monoclonal anti-phospho-AKT (S473) (clone D9E) antibody |
Cell Signaling Technology |
Cat#4060; RRID: AB_2315049 |
| Mouse monoclonal anti-AKT (clone 2H10) antibody | Cell Signaling Technology |
Cat#2967; RRID: AB_331160 |
| Rabbit polyclonal FANCD2 antibody | Novus Biologicals | Cat#NB100–182; RRID: AB_10002867 |
| Rabbit polyclonal anti-phospho-ATR (S1989) antibody | GeneTex | Cat#GTX128145; RRID: AB_2687562 |
| Rabbit polyclonal anti-ATR antibody | Cell Signaling Technology |
Cat#2790; RRID: AB_2227860 |
| Rabbit monoclonal phospho-ATM (S1981) (clone EP1890Y) antibody |
Abcam | Cat#ab81292; RRID: AB_1640207 |
| Rabbit monoclonal anti-ATM (clone D2E2) antibody | Cell Signaling Technology |
Cat#2873; RRID: AB_2062659 |
| Rabbit monoclonal anti-phospho-Chk1 (S345) (clone 133D3) antibody |
Cell Signaling Technology |
Cat#2348; RRID: AB_331212 |
| Mouse monoclonal anti-Chk1 (clone 2G1D5) antibody | Cell Signaling Technology |
Cat#2360; RRID: AB_2080320 |
| Rabbit polyclonal anti-phospho-Chk2 (T68) antibody | Cell Signaling Technology |
Cat#2661; RRID: AB_331479 |
| Rabbit polyclonal anti-Chk2 antibody | Cell Signaling Technology |
Cat#2662; RRID: AB_2080793 |
| Rabbit polyclonal anti-FANCE antibody | Bethyl | Cat#A302–125A; RRID: AB_1720357 |
| Rabbit polyclonal anti-FANCG antibody | Novus Biologicals | Cat#NB100–2566; RRID: AB_921259 |
| Rabbit polyclonal anti-FANCI antibody | Abcam | Cat#ab15344; RRID: AB_443182 |
| Rabbit polyclonal anti-FANCA antibody | Bethyl | Cat#A301–980A; RRID: AB_1547945 |
| Mouse monoclonal anti-FANCF (clone D-2) antibody | Santa Cruz Biotechnology |
Cat#sc-271952; RRID: AB_10708556 |
| Mouse monoclonal anti-FANCL (clone C-4) antibody | Santa Cruz Biotechnology |
Cat#sc-137076; RRID: AB_2262590 |
| Rabbit polyclonal anti-FAMCM antibody | Bethyl | Cat#A302–637A; RRID: AB_10567252 |
| Mouse monoclonal anti-Histone H3 (clone 96C10) antibody |
Cell Signaling Technology |
Cat#3638; RRID: AB_1642229 |
| Rabbit monoclonal anti-pan-Trk (clone A7H6R) antibody | Cell Signaling Technology |
Cat#92991; RRID: N/A |
| Rabbit polyclonal anti-Flt-3/Flk-2 (C-20) antibody | Santa Cruz Biotechnology |
Cat#sc-479; RRID: AB_631052 |
| Rabbit monoclonal anti-Jak2 (clone D2E12) antibody | Cell Signaling Technology |
Cat#3230; RRID: AB_2128522 |
| Rabbit polyclonal anti-phospho BRaf (S445) antibody | Cell Signaling Technology |
Cat#2696; RRID: AB_390721 |
| Rabbit monoclonal anti-BRaf (clone 13) antibody | BD Biosciences | Cat#612374; RRID: AB_399736 |
| Rabbit polyclonal anti-MAST1 antibody | Novus Biological | Cat#NBP2–17228; RRID: N/A |
| Rabbit polyclonal anti-MAST1 antibody for IHC | Novus Biological | Cat#NBP1–81453; RRID: AB_11061334 |
| Rabbit monoclonal anti-Ki67 (clone EPR3610) antibody for IHC |
Abcam | Cat#ab92742; RRID: AB_10562976 |
| Rabbit polyclonal anti-phospho MEK (S218/S222) antibody for IHC |
Abcam | Cat#ab194754; RRID: N/A |
| Biological Samples | ||
| Human head and neck tumor tissues | This paper | N/A |
| Ovarian cancer patient-derived xenografts (PDX) | The Jackson Laboratory |
Cat#TM000335 |
| Head and neck cancer patient-derived xenografts (PDX) | This paper | N/A |
| Lung cancer patient-derived xenografts (PDX) | Owonikoko et al., 2016 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cisplatin | Sigma-Aldrich | Cat#P4394; CAS: 15663–27-1 |
| Carboplatin | Sigma-Aldrich | Cat#C2538; CAS: 41575–94-4 |
| FlexiTube GeneSolution GS5894 for Raf1 | Qiagen | Cat#1027416 |
| Negative Control siRNA | Qiagen | Cat#1027310 |
| Recombinant inactive MEK1 | SignalChem | Cat#M02–14BG |
| Recombinant active MEK1 | Thermo Fisher | Cat#PV3303 |
| Recombinant active MEK1 for MST | Proqinase | Cat#0550–0000-3 |
| Recombinant active c-Raf | SignalChem | Cat#R01–13G |
| Recombinant inactive ERK2 | SignalChem | Cat#M28–14G |
| Myelic Basic Protein | Sigma-Aldrich | Cat#M1891 |
| Mitomycin C | Sigma-Aldrich | Cat#M4287; CAS: 50–07-7 |
| Camptothecin | Selleckchem | Cat#S1288; CAS: 7689–03-4 |
| Doxorubicin | Sigma-Aldrich | Cat#D1515; CAS: 13192–04-6 |
| Paclitaxel | MilliporeSigma | Cat#580555; CAS: 25316–40-9 |
| Lestaurtinib | LC Laboratories | Cat#L-6307; CAS: 111358–88-4 |
| Dovitinib | LC Laboratories | Cat#D-3608; CAS: 405169–16-6 |
| Staurosporine | Sigma-Aldrich | Cat#M1323; CAS: 62996–74-1 |
| Midostaurin | LC Laboratories | Cat#P-7600; CAS: 120685–11-2 |
| Sunitinib | LC Laboratories | Cat#S-8877; CAS: 557795–19-4 |
| SU14813 | MedKoo | Cat#; CAS: 627908–92-3 |
| Bosutinib | LC Laboratories | Cat#B-1788; CAS: 380843–75-4 |
| Ruxolitinib | LC Laboratories | Cat#R-6600; CAS: 941678–49-5 |
| SB203580 | LC Laboratories | Cat#S-3400; CAS: 152121–47-6 |
| Ruboxistaurin | Sigma-Aldrich | Cat#SML0693; CAS: 169939–93-9 |
| MLN518 | Selleckchem | Cat#S1043; CAS: 387867–13-2 |
| AC220 | Selleckchem | Cat#S1526; CAS: 950769–58-1 |
| GNF5837 | Selleckchem | Cat#S7519; CAS: 1033769–28-6 |
| GW441756 | Selleckchem | Cat#S2891; CAS: 504433–23-2 |
| Fedratinib | Selleckchem | Cat#S2736; CAS: 936091–26-8 |
| AZD1480 | Selleckchem | Cat#S2162; CAS: 935666–88-9 |
| AZD6244 | Selleckchem | Cat#S1008; CAS: 606143–52-6 |
| Trametinib | Selleckchem | Cat#S2673; CAS: 871700–17-3 |
| Critical Commercial Assays | ||
| FITC Annexin V Apoptosis Detection Kit | BD Biosciences | Cat#556547 |
| CellTiter-Glo Luminescent Viability Assay | Promega | Cat#G7570 |
| ADP-Glo Kinase Assay | Promega | Cat#V6930 |
| Chromatin Extraction Kit | Abcam | Cat#ab117152 |
| Phospho Explorer Antibody Array | Full Moon Biosystems |
Cat#PEX100 |
| Ras Activation ELISA Assay Kit | EMD Millipore | Cat#17–497 |
| Amine Coupling Kit | GE Healthcare Life Sciences |
Cat#BR100050 |
| Sensor Chip CM5 | GE Healthcare Life Sciences |
Cat#BR100399 |
| Protein Thermal Shift Dye Kit | ThermoFisher | Cat#4461146 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems |
Cat#4368814 |
| Experimental Models: Cell Lines | ||
| Human: A549 cells | ATCC | Cat#CCL-185 |
| Human: A549 cisplatin resistant cells | This paper | N/A |
| Human: HCC827 cells | ATCC | Cat#CRL-2868 |
| Human: H358 cells | ATCC | Cat#CRL-5807 |
| Human: H1299 cells | ATCC | Cat#CRL-5803 |
| Human: H460 cells | ATCC | Cat#HTB-177 |
| Human: H1975 cells | ATCC | Cat#CRL-5908 |
| Human: Calu-1 cells | ATCC | Cat#CRL-5807 |
| Human: H157 cells | ATCC | Cat#CRL-5802 |
| Human: H128 cells | ATCC | Cat#HTB-120 |
| Human: H146 cells | ATCC | Cat#HTB-173 |
| Human: H187 cells | ATCC | Cat#CRL-5804 |
| Human: H209 cells | ATCC | Cat#HTB-172 |
| Human: H526 cells | ATCC | Cat#CRL-5811 |
| Human: DMS53 cells | ATCC | Cat#CRL-2062 |
| Human: DMS114 cells | ATCC | Cat#CRL-2066 |
| Human: DMS153 cells | ATCC | Cat#CRL-2064 |
| Human: SW-626 cells | ATCC | Cat#HTB-78 |
| Human: SK-OV-3 cells | ATCC | Cat#HTB-77 |
| Human: OVCAR-3 cells | ATCC | Cat#HTB-161 |
| Human: Caov-3 cells | ATCC | Cat#HTB-75 |
| Human: BG-1 cells | Geisinger et al. 1989 | RRID: CVCL_6570 |
| Human: 1A9 cells | Geisinger et al. 1989 | RRID: CVCL_H619 |
| Human: MDA686TU cells | Lin et al., 2007 | RRID: CVCL_6985 |
| Human: Tu-212 cells | Lin et al., 2007 | RRID: CVCL_4915 |
| Human: PCI-37B cells | Lin et al., 2007 | RRID: CVCL_C759 |
| Human: UM-SCC-1 cells | Lin et al., 2007 | RRID: CVCL_7707 |
| Human: PCI-15A cells | Lin et al., 2007 | RRID: CVCL_C184 |
| Human: PCI-15A cisplatin resistant cells | This paper | N/A |
| Human: 212LN cells | Lin et al., 2007 | RRID: CVCL_1T18 |
| Human: UDSCC2 cells | Lin et al., 2007 | RRID: CVCL_E325 |
| Human: UM-SCC-47 cells | Lin et al., 2007 | RRID: CVCL_7759 |
| Human: 93-VU-147T cells | Lin et al., 2007 | COSS2296308 |
| Human: Tu-167 cells | Lin et al., 2007 | RRID: CVCL_4912 |
| Human: FaDu cells | Lin et al., 2007 | RRID: CVCL_1218 |
| Human: UM-SCC-22B cells | Lin et al., 2007 | RRID: CVCL_7732 |
| Human: JHU022 cells | Lin et al., 2007 | RRID: CVCL_5991 |
| Human: 1483 cells | Lin et al., 2007 | RRID: CVCL_6980 |
| Human: PCI-13 cells | Lin et al., 2007 | RRID: CVCL_C182 |
| Human: SqCC/Y1 cells | Lin et al., 2007 | RRID: CVCL_0551 |
| Human: KB-3–1 cells | Richert et al., 1985 | RRID: CVCL_2088 |
| Human: KB-3–1 cisplatin resistant cells | Richert et al., 1985 | RRID: CVCL_IP04 |
| Human: A2780 cells | Sigma Aldrich | Cat#93112519 |
| Human: A2780 cisplatin resistant cells | Sigma Aldrich | Cat#93112517 |
| Human: MV-4–11 cells | ATCC | Cat#CRL-9591 |
| Human: HEL cells | ATCC | Cat#TIB-180 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Hsd:Athymic Nude-Foxn1nu | Envigo | Cat#069 |
| Mouse: NOD.CB17-Prkdcscid/NCrCrl | Charles Rivers | Cat#394; RRID: IMSR_CRL:394 |
| Mouse: NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ | The Jackson Laboratory |
Cat#JAX:005557; RRID: IMSR_JAX:005557 |
| Oligonucleotides | ||
| TRC Human Kinase shRNA Gene Family Library | Dharmacon | Cat#RHS4884 |
| shRNA targeting sequence: MAST1 #1: CCACTTCCTCTCCAAACACTT |
Dharmacon | Cat#RHS3979–9588955 |
| shRNA targeting sequence: MAST1 #2: CCACGGTCTACTTCTATGAAT |
Dharmacon | Cat#RHS3979–9588953 |
| shRNA targeting sequence: MAST1 #3: CGTGATGATGAATCACGTCTA |
Dharmacon | Cat#RHS3979–9588952 |
| shRNA targeting sequence: MEK1 #1: CTGATGCTGAGGAAGTGGATT |
Dharmacon | Cat#RHS3979–9570884 |
| shRNA targeting sequence: MEK1 #2: GATTACATAGTCAACGAGCCT |
Dharmacon | Cat#RHS3979–9570886 |
| shRNA targeting sequence: JAK2 #1: CAGTGTTAGATATGATGAGAA |
Dharmacon | Cat#RHS3979–9571807 |
| shRNA targeting sequence: JAK2 #2: GCTTTGTCTTTCGTGTCATTA |
Dharmacon | Cat#RHS3979–9571810 |
| Primer: MAST1 shRNA resistant silent-mutant Forward GTGGACGAGCTCCACTTCCTATCAAAACACTTCGG GAGCACC |
This paper | N/A |
| Primer: MAST1 shRNA resistant silent-mutant Reverse GGTGCTCCCGAAGTGTTTTGATAGGAAGTGGAGCT CGTCCAC |
This paper | N/A |
| Primer: MAST1 Forward TCTCTGGACCGCGCTTTCTA | This paper | N/A |
| Primer: MAST1 Reverse TGAGGCTTTTCCGATTACTGGT | This paper | N/A |
| Primer: FLT3 Forward GAATTCCCATGAAGCCCTGA | This paper | N/A |
| Primer: FLT3 Reverse CCCACTTTCCAATCACATCCA | This paper | N/A |
| Primer: JAK2 Forward TCTGGGGAGTATGTTGCAGAA | This paper | N/A |
| Primer: JAK2 Reverse AGACATGGTTGGGTGGATACC | This paper | N/A |
| Primer: TrkA Forward AACCTCACCATCGTGAAGAGT | This paper | N/A |
| Primer: TrkA Reverse TGAAGGAGAGATTCAGGCGAC | This paper | N/A |
| Primer: GAPDH Forward GACATCAAGAAGGTGGTGAA |
This paper | N/A |
| Primer: GAPDH Reverse TGTCATACCAGGAAATGAGC |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pLHCX | Clonetech | Cat#S1866 |
| Plasmid: Gateway pDEST27 | Invitrogen | Cat#11812013 |
| Plasmid: pLHCX-Gateway | This paper | N/A |
| Plasmid: pLHCX-myc-MAST1 WT | This paper | N/A |
| Plasmid: pLHCX-myc-MAST1 D497A | This paper | N/A |
| Plasmid: pDEST27-MAST1 WT | This paper | N/A |
| Plasmid: pDEST27-MAST1 D497A | This paper | N/A |
| Plasmid: pDEST27-MAST1 L504D | This paper | N/A |
| Plasmid: pLHCX-flag-MEK1 S221A | This paper | N/A |
| Plasmid: pLHCX-flag-MEK1 S221D | This paper | N/A |
| Software and Algorithms | ||
| GraphPad Prism software | GraphPad Software |
http://www.graphpad.com |
HIGHLIGHTS.
MAST1 is a synthetic lethal target for cisplatin therapy in cancer cells.
Cisplatin binds to MEK1 and dissociates cRaf but not MAST1 from MEK1.
MAST1 replaces cRaf to reactivate the MAPK pathway and confer cisplatin resistance.
Lestaurtinib is a promising MAST1 inhibitor.
ACKNOWLEDGEMENTS
We acknowledge Dr. Anthea Hammond for editorial assistance. We thank Thomas Schubert and Maximillian Plach at 2bind GmbH, Germany for performing microscale thermophoresis assay. ICP-mass spectrometry was performed at the Center for Applied Isotope Studies, University of Georgia. This work was supported in part by NIH grants R01 CA175316 (S.K.), R01 CA207768 (S.K.), F31 CA183365 (G.A.), DoD grant W81XWH-17-1-0186 (S.K.), Developmental Funds from the Winship Cancer Institute of Emory University (S.K.), Winship Cancer Institute #IRG-17-181-04 from the American Cancer Society (L.J.), and the Emory University Integrated Cellular Imaging Microscopy Core of the Winship Cancer Institute comprehensive cancer center grant, P30CA138292. G.A. is an NIH pre-doctoral fellow. J.K., F.R.K. and S.K. are Georgia Cancer Coalition Scholars. S. K is a Robbins Scholar and an American Cancer Society Basic Research Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.L., T.O., C.-K.Q., L.H.B. F.R.K., G.Z.C. and N.F.S. provided critical reagents. Z.L. and T.J.B. performed structural analyses. K.R.M. collected clinical samples and performed histopathological study. H.-B.K. generated kinome shRNA lentivirus. D.W. and G.Z. performed patient-derived xenograft. J.K. performed biostatistical study. X.W., C.E.S., and D.M.S. provided clinical information. L.J., J.C., C.P. D.L., J.F., R.L., L.S., and G.N.A. performed all other experiments. L.J. and S.K. designed the study and wrote the paper.
DECLARATION OF INTERESTS
There are no competing interests to declare.
REFERENCES
- Aida T, Takebayashi Y, Shimizu T, Okamura C, Higasimoto M, Kanzaki A, Nakayama K, Terada K, Sugiyama T, Miyazaki K, et al. (2005). Expression of copper-transporting P-type adenosine triphosphatase (ATP7B) as a prognostic factor in human endometrial carcinoma. Gynecologic oncology 97, 41–45. [DOI] [PubMed] [Google Scholar]
- Bilanges B, Torbett N, and Vanhaesebroeck B (2008). Killing two kinase families with one stone. Nature chemical biology 4, 648–649. [DOI] [PubMed] [Google Scholar]
- Caunt CJ, Sale MJ, Smith PD, and Cook SJ (2015). MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nature reviews Cancer 15, 577–592. [DOI] [PubMed] [Google Scholar]
- Chamberlain P, Delker S, Pagarigan B, Mahmoudi A, Jackson P, Abbasian M, Muir J, Raheja N, and Cathers B (2014). Crystal structures of PRK1 in complex with the clinical compounds lestaurtinib and tofacitinib reveal ligand induced conformational changes. PloS one 9, e103638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, and Greenberg ME (1997). Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell 91, 231–241. [DOI] [PubMed] [Google Scholar]
- Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, and Zarrinkar PP (2011). Comprehensive analysis of kinase inhibitor selectivity. Nature biotechnology 29, 1046–1051. [DOI] [PubMed] [Google Scholar]
- Dawson MA, Curry JE, Barber K, Beer PA, Graham B, Lyons JF, Richardson CJ, Scott MA, Smyth T, Squires MS, et al. (2010). AT9283, a potent inhibitor of the Aurora kinases and Jak2, has therapeutic potential in myeloproliferative disorders. British journal of haematology 150, 46–57. [DOI] [PubMed] [Google Scholar]
- Fijolek J, Wiatr E, Rowinska-Zakrzewska E, Giedronowicz D, Langfort R, Chabowski M, Orlowski T, and Roszkowski K (2006). p53 and HER2/neu expression in relation to chemotherapy response in patients with non-small cell lung cancer. The International journal of biological markers 21, 81–87. [DOI] [PubMed] [Google Scholar]
- Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Strom CE, Svensson LM, Schultz N, Lundback T, Einarsdottir BO, et al. (2014). MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 508, 215–221. [DOI] [PubMed] [Google Scholar]
- Galanski M (2006). Recent developments in the field of anticancer platinum complexes. Recent patents on anti-cancer drug discovery 1, 285–295. [DOI] [PubMed] [Google Scholar]
- Garcia-Manero G, Khoury HJ, Jabbour E, Lancet J, Winski SL, Cable L, Rush S, Maloney L, Hogeland G, Ptaszynski M, et al. (2015). A phase I study of oral ARRY-614, a p38 MAPK/Tie2 dual inhibitor, in patients with low or intermediate-1 risk myelodysplastic syndromes. Clinical cancer research : an official journal of the American Association for Cancer Research 21, 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner AM, Vaillancourt RR, Lange-Carter CA, and Johnson GL (1994). MEK-1 phosphorylation by MEK kinase, Raf, and mitogen-activated protein kinase: analysis of phosphopeptides and regulation of activity. Mol Biol Cell 5, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland P, Quraishe S, French P, and O’Connor V (2008). Expression of the MAST family of serine/threonine kinases. Brain research 1195, 12–19. [DOI] [PubMed] [Google Scholar]
- Geisinger KR, Kute TE, Pettenati MJ, Welander CE, Dennard Y, Collins LA, and Berens ME (1989). Characterization of a human ovarian carcinoma cell line with estrogen and progesterone receptors. Cancer 63, 280–288. [DOI] [PubMed] [Google Scholar]
- Giaccone G (2000). Clinical perspectives on platinum resistance. Drugs 59 Suppl 4, 9–17; discussion 37–18. [DOI] [PubMed] [Google Scholar]
- Han JY, Hong EK, Choi BG, Park JN, Kim KW, Kang JH, Jin JY, Park SY, Hong YS, and Lee KS (2003). Death receptor 5 and Bcl-2 protein expression as predictors of tumor response to gemcitabine and cisplatin in patients with advanced non-small-cell lung cancer. Medical oncology 20, 355–362. [DOI] [PubMed] [Google Scholar]
- Ikeguchi M, and Kaibara N (2001). Changes in survivin messenger RNA level during cisplatin treatment in gastric cancer. International journal of molecular medicine 8, 661–666. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. (2010). COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Elf S, Lythgoe K, Hitosugi T, Taunton J, Zhou W, Xiong L, Wang D, Muller S, Fan S, et al. (2010). p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. The Journal of clinical investigation 120, 1165–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapper S, Burnett AK, Littlewood T, Kell WJ, Agrawal S, Chopra R, Clark R, Levis MJ, and Small D (2006). A phase 2 trial of the FLT3 inhibitor lestaurtinib (CEP701) as first-line treatment for older patients with acute myeloid leukemia not considered fit for intensive chemotherapy. Blood 108, 3262–3270. [DOI] [PubMed] [Google Scholar]
- Koberle B, Tomicic MT, Usanova S, and Kaina B (2010). Cisplatin resistance: preclinical findings and clinical implications. Biochimica et biophysica acta 1806, 172–182. [DOI] [PubMed] [Google Scholar]
- Kong LR, Chua KN, Sim WJ, Ng HC, Bi C, Ho J, Nga ME, Pang YH, Ong WR, Soo RA, et al. (2015). MEK Inhibition Overcomes Cisplatin Resistance Conferred by SOS/MAPK Pathway Activation in Squamous Cell Carcinoma. Molecular cancer therapeutics 14, 1750–1760. [DOI] [PubMed] [Google Scholar]
- Lavoie H, and Therrien M (2015). Regulation of RAF protein kinases in ERK signalling.Nature reviews Molecular cell biology 16, 281–298. [DOI] [PubMed] [Google Scholar]
- Li D, Jin L, Alesi GN, Kim YM, Fan J, Seo JH, Wang D, Tucker M, Gu TL, Lee BH, et al. (2013). The prometastatic ribosomal S6 kinase 2-cAMP response element-binding protein (RSK2-CREB) signaling pathway up-regulates the actin-binding protein fascin-1 to promote tumor metastasis. The Journal of biological chemistry 288, 32528–32538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedert B, Materna V, Schadendorf D, Thomale J, and Lage H (2003). Overexpression of cMOAT (MRP2/ABCC2) is associated with decreased formation of platinum-DNA adducts and decreased G2-arrest in melanoma cells resistant to cisplatin. The Journal of investigative dermatology 121, 172–176. [DOI] [PubMed] [Google Scholar]
- Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, Ferris RL, and Lai SY (2007). Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head & neck 29, 163–188. [DOI] [PubMed] [Google Scholar]
- Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, and Chamberlain JS (1999). Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nature neuroscience 2, 611–617. [DOI] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, and Nordlund P (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. [DOI] [PubMed] [Google Scholar]
- Marusiak AA, Edwards ZC, Hugo W, Trotter EW, Girotti MR, Stephenson NL, Kong X, Gartside MG, Fawdar S, Hudson A, et al. (2014). Mixed lineage kinases activate MEK independently of RAF to mediate resistance to RAF inhibitors. Nature communications 5, 3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R, Leichman CG, Danenberg KD, Danenberg PV, Lenz HJ, Hayashi K, Groshen S, Salonga D, Cohen H, Laine L, et al. (1998). ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 16, 309–316. [DOI] [PubMed] [Google Scholar]
- Moreno-Smith M, Halder JB, Meltzer PS, Gonda TA, Mangala LS, Rupaimoole R, Lu C, Nagaraja AS, Gharpure KM, Kang Y, et al. (2013). ATP11B mediates platinum resistance in ovarian cancer. The Journal of clinical investigation 123, 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owonikoko TK, Zhang G, Kim HS, Stinson RM, Bechara R, Zhang C, Chen Z, Saba NF, Pakkala S, Pillai R, et al. (2016). Patient-derived xenografts faithfully replicated clinical outcome in a phase II co-clinical trial of arsenic trioxide in relapsed small cell lung cancer. Journal of translational medicine 14, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persons DL, Yazlovitskaya EM, and Pelling JC (2000). Effect of extracellular signal-regulated kinase on p53 accumulation in response to cisplatin. The Journal of biological chemistry 275, 35778–35785. [DOI] [PubMed] [Google Scholar]
- Richert N, Akiyama S, Shen D, Gottesman MM, and Pastan I (1985). Multiply drug-resistant human KB carcinoma cells have decreased amounts of a 75-kDa and a 72-kDa glycoprotein. Proceedings of the National Academy of Sciences of the United States of America 82, 2330–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. (2011). Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nature medicine 17, 1646–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir M, and Stuart R (2010). Lestaurtinib, a multitargeted tyrosine kinase inhibitor: from bench to bedside. Expert opinion on investigational drugs 19, 427–436. [DOI] [PubMed] [Google Scholar]
- Siddik ZH (2003). Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene 22, 7265–7279. [DOI] [PubMed] [Google Scholar]
- Valiente M, Andres-Pons A, Gomar B, Torres J, Gil A, Tapparel C, Antonarakis SE, and Pulido R (2005). Binding of PTEN to specific PDZ domains contributes to PTEN protein stability and phosphorylation by microtubule-associated serine/threonine kinases. The Journal of biological chemistry 280, 28936–28943. [DOI] [PubMed] [Google Scholar]
- Verlhac MH, Lefebvre C, Kubiak JZ, Umbhauer M, Rassinier P, Colledge W, and Maro B (2000). Mos activates MAP kinase in mouse oocytes through two opposite pathways. The EMBO journal 19, 6065–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, and Wu GS (2014). Role of autophagy in cisplatin resistance in ovarian cancer cells. The Journal of biological chemistry 289, 17163–17173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Tsigelny IF, Gotz AW, and Howell SB (2015). Cisplatin inhibits MEK1/2. Oncotarget 6, 23510–23522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.