Abstract
Tuberculosis (TB) is the leading global infectious cause of death. Understanding TB transmission is critical to creating policies and monitoring the disease with the end goal of TB elimination. To our knowledge, there has been no systematic review of key transmission parameters for TB. We carried out a systematic review of the published literature to identify studies estimating either of the two key TB transmission parameters: the serial interval (SI) and the reproductive number. We identified five publications that estimated the SI and 56 publications that estimated the reproductive number. The SI estimates from four studies were: 0.57, 1.42, 1.44 and 1.65 years; the fifth paper presented age-specific estimates ranging from 20 to 30 years (for infants <1 year old) to <5 years (for adults). The reproductive number estimates ranged from 0.24 in the Netherlands (during 1933–2007) to 4.3 in China in 2012. We found a limited number of publications and many high TB burden settings were not represented. Certain features of TB dynamics, such as slow transmission, complicated parameter estimation, require novel methods. Additional efforts to estimate these parameters for TB are needed so that we can monitor and evaluate interventions designed to achieve TB elimination.
Key words: Reproductive number, serial interval, systematic review, tuberculosis
Introduction
Tuberculosis (TB), an airborne bacterial infection caused by the organism Mycobacterium tuberculosis (Mtb), has surpassed HIV/AIDS as the leading cause of death due to a single infectious organism worldwide [1]. It primarily attacks the lungs but can also infect other areas of the body [2, 3]. Those exposed to Mtb often develop latent TB infection (LTBI) and have a 5–10% lifetime risk of progressing to active TB [4, 5]. Worldwide, 2–3 billion people are infected with TB; an estimated 10.4 million people developed active TB disease in 2015 [4]. Major innovations in strategies and tools to monitor the success of new strategies are needed to achieve the World Health Organisation (WHO)’s ENDTB goals of reducing TB deaths by 95% and new cases by 90% by 2035 [4].
The reproductive number and serial interval (SI) are two key quantities in describing transmission of an infectious disease. The reproductive number is defined as the average number of secondary cases a primary infectious case will produce. In a totally susceptible population, it is referred to as the basic reproductive number (R0); it is referred to as the effective reproductive number (Re) if the population includes both susceptible and non-susceptible persons [6]. An Re > 1 indicates that the disease will continue to spread while an Re < 1 indicates that the disease will eventually die out. Although the reproductive number is usually defined as the average number of secondary cases, it is occasionally defined as the average number of secondary infections [7–10], a distinction that is important for a disease with a long incubation period (the time between infection and developing symptomatic disease) and/or only a fraction of infections progressing to disease. Depending on the setting, the reproductive number can be expressed as a function of parameters such as infection rate, contact rate, recovery rate, making it useful in determining whether or not a disease can spread through a population.
The serial interval (SI), defined as the time between disease symptom onset of a case and that of its infector [11], is a surrogate for the generation interval – an unobservable quantity defined as the time between the infection of a case and the time of infection of its infector [12]. The SI is an important quantity in the interpretation of infectious disease surveillance data, in the identification of outbreaks and in the optimisation of quarantine and contact tracing.
These two quantities have been used to inform control policies during outbreaks [13] by quantifying the transmission of infectious diseases such as influenza A (H1N1) [11, 12, 14], Severe Acute Respiratory Syndrome (SARS) [12, 15] and Ebola [16, 17], where progression to disease upon transmission occurs quickly. For example, Wallinga and Teunis [18] in 2004 demonstrated the impact of the first global alert against SARS on the change of the effective reproductive number.
TB has a slower transmission rate due to its much longer incubation period. Of the 5–10% of infections that develop into active (symptomatic and infectious) TB disease, it is thought that the majority occur within the first 2 years after infection [2, 5, 19], although active TB disease can develop decades after initial infection [20]. This is much longer than the aforementioned infectious diseases where cases show symptoms within days of infection. Although there is an increasing consensus that some transmission events may occur before the infector shows symptoms, many likely occur after the infector is symptomatic, therefore, the longer the incubation period is, the longer the SI (Fig. 1).
Fig. 1.
Important infectious disease intervals. The time between a and c is the serial interval; the time between b and c is the incubation period.
Development of TB disease can be caused by de novo infection, reactivation of the same bacterial strain as a previous infection [5, 21] or by infection with a bacterial strain different from the original infection (reinfection TB). This complicates estimation of the serial interval, unless molecular techniques are used to distinguish reinfection and reactivation [21]. To our knowledge, there has been no systematic review of methods to estimate the serial interval and reproductive number for TB. Therefore, in this paper we systematically review the literature to examine the methods applied to the estimation of TB transmission parameters and the estimates obtained from these methods. This compilation informs the gaps in our understanding of TB and identifies areas where further research is needed to develop methods to better understand TB transmission.
Methods
We conducted two searches in PubMed for publications in English – one for TB and serial interval; one for TB and reproductive number.
Tuberculosis and serial interval
(‘Tuberculosis’[MeSH] OR ‘Mycobacterium tuberculosis’[MeSH] OR ‘tuberculosis’[TI]) and (‘serial interval’[tiab] or ‘generation interval‘[tiab] or ‘serial distribution’ [tiab] or ‘secondary infections’ [tiab] or ‘secondary cases’ [tiab]).
TB and reproductive number
(‘Tuberculosis’[MeSH] OR ‘Mycobacterium tuberculosis’[MeSH] OR ‘tuberculosis’[TI] OR ‘pulmonary, tuberculosis [MeSH]’) and (‘reproductive number’[tiab] or ‘reproduction number’[tiab] or ‘reproductive rate’[tiab] or ‘reproduction rate’[tiab] or ‘reproduction ratio’[tiab] or ‘reproductive ratio’[tiab] or ‘reproduction value’[tiab] or ‘reproductive value’[tiab] or ‘R0’[tiab] or ‘secondary infections’[tiab] or ‘secondary cases’[tiab]).
Titles and abstracts of the publications referenced in the articles we found were reviewed for inclusion for either parameter. For the SI, as limited number of publications met our inclusion criteria, we also reviewed the titles and abstracts of publications that cited the serial interval articles that we included in a full-text review.
Two reviewers (two of YM, HEJ, LFW) independently screened all titles and abstracts, resolving discrepancies by consensus. Each publication was then independently reviewed by two reviewers (two of YM, HEJ, LFW) for inclusion. From the included articles, the same pairs of reviewers extracted the following details for all parameter estimates (if available): point estimates, confidence intervals, ranges, sample size and location/setting. We summarised the methods for analysis and aggregated those with similar estimation approaches.
Results
Serial interval
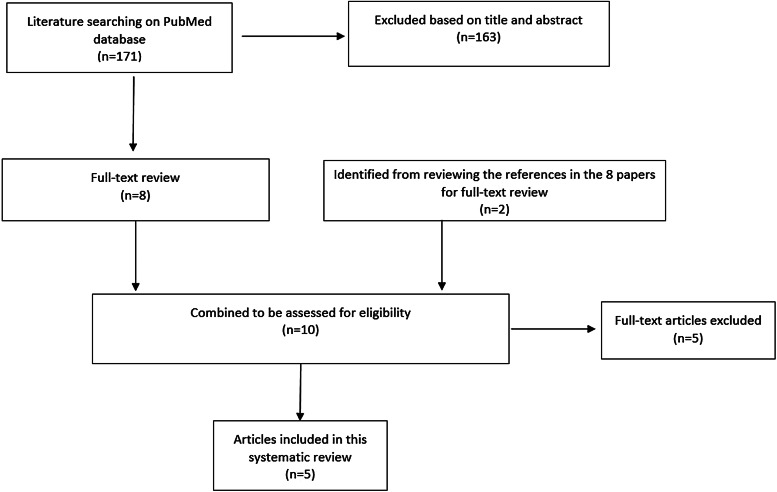
The serial interval query returned 171 articles (Fig. 2), of which 163 were excluded as they did not present any estimates. Leung et al. [22] reported the serial interval as the time from identification of primary case to secondary case as median 1.4 years (range: 0.4–5.2 years). This study used household transmission data from Hong Kong and focused on MDR- and XDR-TB. Vynnycky and Fine [23] analysed a population of white males in England and Wales in the 20th century using a mathematical compartmental model to estimate the SI as dependent on the age when infection occurred, distinguishing reinfection and reactivation in the model. In this model, the risk of developing disease was calibrated on incidence data. The estimates were presented as a frequency distribution. The most frequent time to develop disease was estimated at: between 20 and 30 years due to reinfection for those infected in the first year of life; between 10 and 14 years due to reinfection for those infected at age 10; <5 years due to recent infection for those infected at age 20 and those infected at age 40. ten Asbroek et al. [24] analysed genetic data for a Dutch sample from 1993 to 1996 to link infectors and infected people using DNA fingerprinting based on restriction fragment length polymorphism (RFLP) and estimated the serial interval at a geometric mean of 0.57 years (95% confidence interval (CI) 0.44–0.73). In this 4-year study, the probability of observing both the infector and the infected person depended on the time interval between isolates – the shorter this time interval was, the more likely that this couple was observed. Therefore, the observed serial intervals were weighted by the inverse of the difference between the length of the study period and the time between isolates of the infector and the infected person, allowing a rough correction for underrepresentation of longer SIs (Table 1).
Fig. 2.
Flow diagram of articles included in the search of estimates of the serial interval.
Table 1.
Estimates of the Serial Interval
| Name of first author [publication] | Objective | Method | Assumptions | Estimated serial interval |
|---|---|---|---|---|
| Leung [22] | Study household transmission of MDR-TB | Data on all MDR-TB in Hong Kong 1997–2006. Did contact investigations and DNA fingerprinting and linked index to secondary cases | No censoring in this estimate, not clear how long people were followed up for | 1.4 (0.4–5.2) years (median) |
| Vynnycky [23] | Demonstrate how the lifetime risk of disease, the incubation period and the serial interval changed | An age-dependent compartmental model | Assumed values for model input parameters such as the annual risk of infection | Estimated as dependent on age of infection and summarised as frequency distributions |
| ten Asbroek [24] | To determine the serial interval and incubation period of tuberculosis within 4 years of transmission | Descriptive approach on RFLP data (used to link infectors and infected people) | One source of infection for each infected cluster | 0.57 years (95% CI 0.44–0.73) |
| Borgdorff [25] | Same as [24] | Same as [24] | NA | Median: 1.44 years (95% CI 1.29–1.63) |
| Brooks-Pollock [26] | Estimate the relative contributions of household and community transmission, the serial interval and the immunity afforded by a previous TB infection | Descriptive approach for the serial interval | All members of the study cohort have been exposed to TB by living with someone with active disease | Mean serial interval: 3.5 years; median serial interval 1.65 years |
Two articles that cited the articles that met our inclusion criteria in the PubMed search reported estimates of the SI and were included for full-text review. Borgdorff et al. [25] used the same method on genetic data as [24] to estimate the median SI as 1.44 years (95% CI 1.29–1.63 years) for a Dutch sample from 1993–2007. Brooks-Pollock [26] in 2011 analysed cross-sectional household data for a sample in Lima, Peru from 1996 to 2002 and reported the time between the diagnosis of the infector and the infected person as an estimate for the SI with mean at 3.5 years and the median at 1.65 years.
Reproductive number
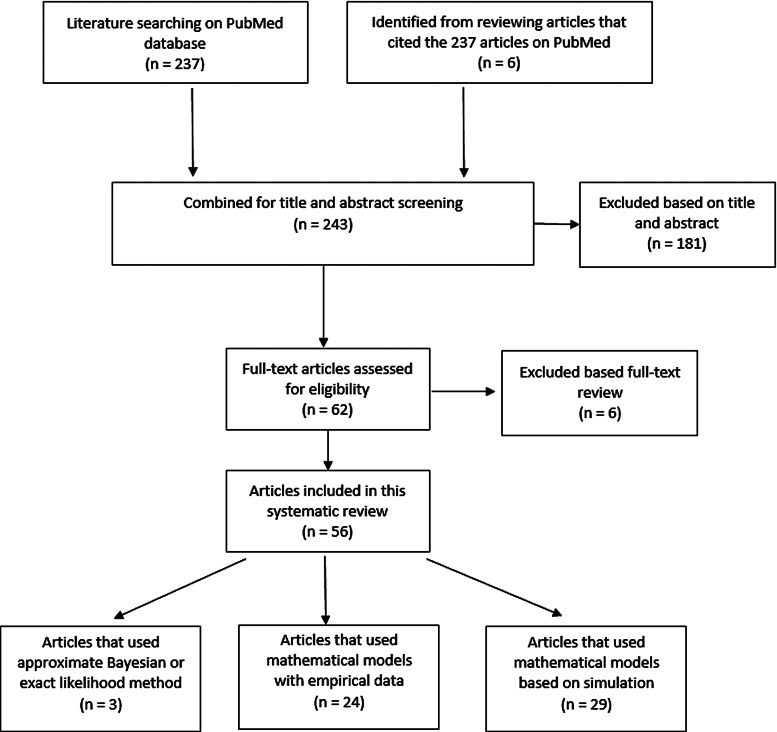
Two hundred and thirty-seven articles were identified for the reproductive number of TB. Additionally, six articles were included based on reviewing titles and abstracts of the articles that were referenced in the 237 articles, making the total number of articles 243. Fifty-six articles met our inclusion criteria and are described below. Three articles used either approximate Bayesian or exact likelihood methods, 24 articles used either a mathematical model fit with empirical data or a descriptive/regression approach on empirical data, and 29 articles used a simulation-based mathematical model (Fig. 3). Explicit estimates were extracted and summarised in Fig. 4. The estimates range from as low as 0.26 for the Netherlands in 1993–2007 to as high as 4.3 in China in 2012.
Fig. 3.
Flow diagram of articles included in the search of estimates of the reproductive number.
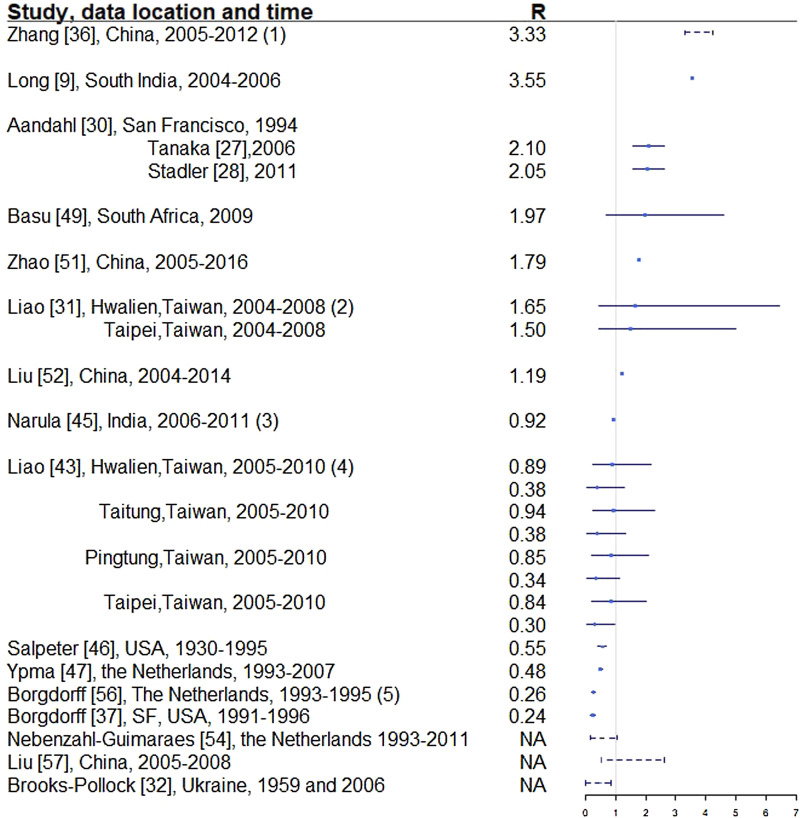
Fig. 4.
Reproductive number from studies with explicit R estimate from empirical data. Notes: (1) The range is for years 2005-2012, with the reproductive number estimated at 3.33, 3.72, 3.38, 3.97, 4.29, 3.32, 3.92 and 4.30, respectively. (2) For each location, the first R corresponds to drug-sensitive population and the second correspond to drug-resistant population. (3) R estimated for 35 states and union territories of India with estimates ranging from 0.72 to 0.98; 0.92 is the overall estimate for India. (4) For each location, the first R corresponds to drug-sensitive population and the second correspond to drug-resistant population. (5) Bordgorff in [27–29] estimated the reproductive number for the Netherlands from 1993 to 2007 at around 0.26 with lower bound of the 95% CI around 0.20 and upper bound around 0.32. (6) Broken lines indicate range; solid lines indicate 95% confidence interval. (7) Vynnycky and Fine [23] in 1998 estimated the basic reproductive number to decline from about 3 in 1900 to 2 in 1950 and to below 1 in about 1960 for England and Wales, which is not included in this graph.
Three articles (Table 2) used the same genetic RFLP data from TB diseased individuals during an outbreak in San Francisco in 1991–1992 [33]. They all estimated the effective reproductive number in a Bayesian framework. Tanaka et al. [30] used an approximated computation method to obtain an estimate of 3.4 (95% CI 1.4–79.7). Stadler [31] in 2013 used an exact likelihood method to obtain an estimate of 1.02 (95% CI 1.01–1.04) and claimed that the difference from the estimate in [30] was due to the lack of precision in the approximation of the posterior distribution in [30]. Aandahl et al. [32] in 2014 reconciled the two methods by specifying an informative prior for two parameters in [30] and improving the convergence performance of the Markov chain Monte Carlo (MCMC) sampler in [31]. The reconciled estimates were: 2.1 (95% CI 1.54–2.66) for the approximate method in [30] and 2.05 (95% CI 1.55–2.63) for the exact method in [31]. These papers used the same model but differed in the methods used to obtain the estimates. The assumptions of the model are listed in Table 2.
Table 2.
Estimates of the reproduction number using approximate Bayesian computation and exact likelihood methods (all methods used data from San Francisco on cases reported in 1994)
| Name of first author [publication] | Objective | Method | Assumptions | Estimated reproductive number (95% credible interval) |
|---|---|---|---|---|
| Tanaka [30] | Estimate TB transmission parameters: net transmission rate, doubling time and reproductive number | Approximate Bayesian computation | Constant supply of susceptible people; all genotypes are selectively neutral; mutation and transmission are independent; infinite alleles; epidemic spreads until N individuals are infected | 3.4 (1.4–79.7) |
| Stadler [31] | Estimate TB transmission parameters: net transmission rate and reproductive number | Exact likelihood | Constant birth-death rate; infinite alleles; epidemic started at a random time in the past; an isolate is sampled from an individual with probability P | 1.02 (1.01–1.04) |
| Aandahl [32] | Reconcile the different estimates in [30, 31] | Improved the method in [30] by specifying informative priors; improved convergence performance of the MCMC sampler in [31] | Fixed mutation rate; used Gaussian prior for the death/recovery rate |
Twenty-four articles analysed the reproductive number with empirical data (Table 3). Seventeen articles reported explicit estimates, with five estimating the effective reproductive number and 12 estimating the basic reproductive number. The majority of these articles used mathematical compartmental models with different variations in structure and parameterisation to address issues such as seasonality [43], the effect of age [46, 51] and HIV–TB co-epidemics [9].
Table 3.
Estimates of the reproductive number from mathematical models with empirical data
| Name of first author [publication] | Location, time of data | Type of data | Objective | Methods | Assumptions | Reproductive number type | Estimated reproductive number |
|---|---|---|---|---|---|---|---|
| Zhao [34] | China, 2005–2016 | CDC data | To investigate the impact of age on TB transmission | SEIR model with age structure; use least squares to get parameters that align with TB data in China; use Latin hypercube sampling to get CI | Although the susceptible compartment was stratified by age, the other compartments were not age-stratified thus assuming no difference in age for those compartments | Basic | 1.786 (95% CI 1.775–1.796) |
| Liu [35] | China, 2004–2014 | Annual TB case data | To use modelling to investigate the impact of different vaccination strategies (constant or pulse BCG) on TB transmission | Compartmental models with vaccination compartments | Assumptions made for all parameter values | Basic | 1.19 |
| Yang [36] | Shaanxi, China, 2004–2012 | Notifiable active TB cases by month | Study the seasonality impact on TB transmission dynamics | A seasonality TB compartmental model: subjects either entered latent or diseased compartment; contact rate, reactivation rate and disease-induced death rate are periodic continuous functions | Parameter values for recruitment rate, natural death rate, recovery rate | Basic | Dependent on parameter values |
| Nebenzahl-Guimaraes [28] | The Netherlands, 1993–2011 | Surveillance and RFLP data | Determine if mycobacterial lineages affect infection risk, clustering and disease progression among Mycobacterium tuberculosis cases | Descriptive and regression approach; DNA fingerprinting to link cases | All secondary cases captured in surveillance data; genetic matching accurately reflects transmission patterns | Effective | Range: 0.17–1.04 |
| Narula [37] | India, 2006–2011 | Quarterly reported data from Central TB Division | Estimate basic R0 for TB | Compartmental model with Bayesian melding technique to estimate parameters; Susceptible, latent, infected compartments instead of SIR | Some parameter values assumed with reference in the differential equations | Basic | 0.92, averaged for India overall with range 0.72–0.98 |
| Zhang [38] | China, 2005–2012 | Monthly case reporting data from CDC | Estimate effective R0 of TB by year | Compartmental model adding hospitalised compartment; Chi-square test for optimal parameters | An upper bound for number of initially susceptible people, natural death rate, initial number of latent individuals | Effective | Range from 3.318 to 4.302 from year 2005 to 2012 |
| Ypma [39] | The Netherlands, 1993–2007 | RFLP data | Explore the high heterogeneity in the number of secondary cases caused per infectious individual for TB | Model ‘superspreading’ parameter as a negative binomial distribution | Immigrants who have been in the country for less than 6 months at diagnosis are index cases themselves | Fingerprint reproductive number as a function of the effective reproductive number and the probability that the fingerprint of the infected person is different than its infector | 0.48 (95% CI 0.44–0.59) |
| Andrews [40] | Cape Town, South Africa, 2011 | Carbon dioxide data, public transit usage data from national survey | Estimate risk of TB transmission on 3 modes of public transit | Modified Wells-Riley model for airborne disease transmission | Duration of infectiousness of 1 year; used TB and HIV parameters from studies in the same area; natural history parameters from the literature | Basic | Dependent on duration of infectiousness and frequency of transit usage |
| Okuonghae [41] | Benin city, Nigeria, 2008 | Survey data | Assess how control strategies on addressing TB transmission parameters can minimise incidence | Compartmental model adding compartments of disease awareness level, identified infectiousness | Model parameter values such as recruitment rate, recovery rate from the literature | Basic, under treatment | Dependent on parameter values |
| Liao [42] | Taiwan, 2005–2010 | Monthly data from CDC | Estimate MDR-TB infection risk | Mathematical probabilistic two-strain model with compartments for drug-sensitive and drug-resistant subjects; dose–response model for relationship between R0 and total proportion of infected population | Some model parameter values from data, some from the literature; assumed 0.99 of people latently infected were drug sensitive and 0.01 were drug resistant | Basic |
Hwalien County: 0.89 (95% CI 0.23–2.17) for drug sensitive; 0.38 (95% CI 0.05–1.30) for multi-drug resistant; Taitung County: 0.94 (95% CI 0.24–2.28) for drug sensitive; 0.38 (95% CI 0.05–1.33) for multi-drug resistant; Pingtung County: 0.85 (95% CI 0.21–2.08) for drug sensitive; 0.34 (95% CI 0.04–1.13) for multi-drug resistant; Taipei City: 0.84 (95% CI 0.21–2.00) for drug sensitive; 0.30 (95% CI 0.04–0.97) for multi-drug resistant; |
| Liao [43] | Taiwan, 2004–2008; selected three areas with the highest incidence, one with the lowest incidence | Monthly disease burden TB data from Taiwan CDC | Examine TB population dynamics and assess potential infection risk | Compartmental model with susceptible, latently infected, infectious, non-infectious and recovered compartments; incorporated reactivation, relapse and reinfection | Some parameter values taken from the literature, some estimated from data | Basic, estimated as sum of fast, slow and relapse | Highest R0 total in Hwalien: 1.65 with 95th percentile range 0.45–6.45; Taipei lowest at 1.5 (0.45–4.98); Taitung: 1.72; Pingtung: 1.65 |
| Liu [44] | China, 2000–2008 | Data from the National Bureau of Statistics | Incorporate migration to study TB transmission | SEIR compartments for rural residents, migrant workers and urban population | Model parameters calculated from website data; migration rates | Basic | No explicit estimate |
| Borgdorff [29] | The Netherlands, 1993–2007 | RFLP data | Determine to what extent tuberculosis trends in the Netherlands depend on secular trend, immigration and recent transmission | DNA fingerprinting to link cases | All secondary cases captured in surveillance data; genetic matching accurately reflects transmission patterns | Basic | 0.24 (95% CI 0.21–0.26) |
| Liu [45] | China, Jan, 2005–Dec, 2008 | Monthly notification data from Ministry of Health | Develop a model incorporating seasonality and define basic reproduction ratio | Used periodic infection rate and reactivation rate to incorporate seasonality in the compartmental model; considered fast and slow progression | Parameters such as recruitment rate, natural death rate were assumed to be constants; some parameter values assumed and some taken from the literature | Basic | Dependent on parameter values with range 0.4–2.6 |
| Brooks-Pollock [46] | Ukraine, 1959 and 2006 | Mortality data | Explore the effect of age structure on TB infection and disease prevalence, basic reproductive number and impact of intervention | Basic SEIR mathematical model with assumptions about survivorship | A survivorship function which could be described in terms of age and life expectancy | Basic | Dependent on progression rate with range 0–0.85 |
| Basu [47] | KwaZulu-Natal, South Africa | Extensively drug-resistant TB data (XDR-TB) | Model XDR-TB transmission dynamics | Model XDR-TB incorporating the existing XDR detection rate and treatment system | Even mixing of air; range of key parameters in the model | Effective | 1.97, range 0.7–4.6; 1.23, range 0.4–3.1 when combining screening and therapy; 1.38, range 0.6–3.3 with South African strategic plan alone. |
| Furuya [7] | Japan, 2000–2005 | Exposure data | Quantify the risk of TB infection in an internet café where people without homes stayed overnight | Wells-Riley model to estimate the reproductive number | Patients stayed in a confined space for 150 days; some values in the Wells-Riley equation assumed, others from the literature | Estimated as a function of exposure period | Dependent on exposure period |
| Long [9] | Southern India, 2004–2006 | HIV-TB co-epidemics data | Model HIV-TB co-epidemics and explore hypothetical treatment effect | First model: susceptibility to either or both diseases compartments; second model: SII*SEI | A linear relationship between treatment levels and the associated parameters; model parameters from the literature | Basic | R = 3.55 when no active treatment for TB |
| Borgdorff [48] | The Netherlands, 1995–2002 | RFLP data | Assess progress towards TB elimination | DNA fingerprinting to link cases; survival analysis | All secondary cases captured in surveillance data; genetic matching accurately reflects transmission patterns | Basic | Dutch index cases: 0.23, non-Dutch index cases: 0.25 |
| Borgdorff [49] | San Francisco, USA, 1991–1996 | RFLP data | Determine tuberculosis transmission dynamics in San Francisco and its association with country of birth and ethnicity | Define effective reproductive number as a function of transmission index, which is a function of number of secondary cases and potential source cases in a given subgroup | Each cluster originates from a single source case in the database; either the first case of a cluster was its source case, or that the probability of being a source case declined exponentially over time by 0.77% per day | Effective, recent transmission | 0.24 (95% CI 0.17–0.31) |
| Davidow [50] | New York City, 1989–1993 | TB and AIDS surveillance data | Evaluate the importance of recent M. tuberculosis transmission | Estimated # of TB infectious cases 1 year ago and computed short-term R0; R0 = the average # of new infections caused by each case per year of infectiousness*the average duration of infectiousness*the probability of progressing to active TB within 1 year after infection | Some clinical assumptions; parameter values in equation taken from the literature or calculated from neighbourhood-specific data | Short-term | No explicit estimates; focused on percentage of TB cases due to infection 1 year ago |
| Vynnycky [51] | England and Wales, 1900 | Surveillance data; age and time-specific mortality rates | Describe transmission dynamics of all forms of pulmonary TB | Age-structured mathematical model with compartments for endogenous and exogenous diseases | General relationship between: first primary episode and age at infection, risk of exogenous disease and age at reinfection, endogenous disease and current age; risk of reinfection and first infection are identical; parameter values from the literature | Basic and net which is the same as effective | Net R at about 1 from 1900–1950; basic R0 declined from about 3 in 1900, reached 2 by 1950, and first fell below 1 in about 1960 |
| Salpeter [52] | USA, 1930–1995 | Case rates, correction for rates before 1975 | Estimate time delay from infection to disease and R | Estimate R as a function of case rate and the shape of the delay function | R and case rate constant with calendar time t; incidence rate of latent infection is independent of the age | Effective | 0.55, range 0.4–0.7 in sensitivity analysis |
| Borgdorff [27] | Netherlands, 1993–1995 | RFLP data | Quantifying transmission of TB between and within nationalities | Effective R0 estimated as a function of transmission index | Probability of a patient being the source of a cluster was proportional to the incidence rate of potential sources times the probability that a potential source would give rise to a cluster | Effective | 0.26, 95% CI (0.20–0.32); also estimated for different nationalities |
Two articles [7, 40] used the Wells–Riley model or a modified version of the model. In these models, the reproductive number was expressed as a function of infection risk, which was further expressed as proportionate to environmental factors such as the number of infectious people in a given space, per-person breathing rate and inversely proportionate to germ-free ventilation rate. One article derived the reproductive number as a function of the transmission index – defined as the ratio of the number of secondary cases to the sum of the number of source cases (infectors) and non-clustered cases where clusters are defined as groups of patients that had isolates with identical fingerprints [27]. The largest reproductive number (effective) was estimated in [38] using the Chinese Centre for Disease Control and Prevention (CDC) data from 2005 to 2012, where the annual reproductive number ranged between 3.33 and 4.30 for years 2005–2012 in China. The lowest (effective) reproductive number was estimated at 0.24 (95% CI 0.17–0.31) using RFLP data in San Francisco, USA from 1991 to 1996 [49]. Vynnycky and Fine [51] in 1998 used an age-structured mathematical model and estimated the effective reproductive number to be around 1 from 1900 to 1950 in England and Wales; the basic reproductive number was estimated to have declined from about 3 in 1900 to 2 by 1950, and first fell below 1 in about 1960. The assumptions of these models are listed in Table 3.
One article defined the reproductive number as the number of secondary infections caused by an infectious case [7]. As only a fraction of the infected people develops active disease, the estimated reproductive number was larger than those in the other papers. The median of the reproductive number in this article ranged from 14 to 45 as exposure time increased from 1 to 5 months.
Twenty-nine articles analysed the reproductive number through simulation based on a mathematical modelling framework (Table 4). These articles all used mathematical compartmental models with different variations to address issues such as reinfection [68], the interaction between HIV and TB [64], and drug-resistant and drug-sensitive TB [60]. The majority of them focused on studying the effect of these issues on TB transmission dynamics through simulations that were not based on a specific population. In this case, parameters for the model were based on estimates from studies performed in diverse settings or sampled over a range of feasible values. The analytical expression of the basic reproductive number was derived to study the disease-free equilibrium and endemic-persistent state of TB in these papers. Five articles [10, 60, 64, 73, 76] included drug-resistant TB cases as a compartment and four articles [58, 68, 72, 75] included HIV + TB cases as a compartment.
Table 4.
Estimates of the reproductive number from mathematical models based on simulation
| Name of first author [publication] | Objective | Methods, setting | Assumptions | Basic or effective R0 | Estimated R0 |
|---|---|---|---|---|---|
| Ren [53] | Develop SEIR model for imperfect treatment with age-dependent latency and relapse | SEIR model | TB infectious in latent period; age-dependence | Basic | Dependent on parameters |
| Jabbari [54] | To set up a model that can examine two TB strains (DS and DR) with multiple latent stages | Mathematical compartmental model with compartments for latency stages | The drug-sensitive strain will not play a role in the process of exogenous reinfection for the drug-resistant strain | Basic | Dependent on parameters |
| Okuonghae [55] | Study the effects of additional heterogeneities from the level of TB awareness on TB transmission dynamics and case detection rate | Expanding [34] by dividing both susceptible and latently infected compartment by level of TB awareness | Reasonable values and bounds for parameters such as transmission rate, recovery rate from the literature | Effective | Dependent on parameters such as active case finding rate and treatment rate |
| Liu [56] | Evaluate effect of treatment for TB | Compartmental model with treatment and two latent periods incorporated | Once the treatment of active TB cases is interrupted, there is no more treatment; specified model parameter values and their relationship with one another | Basic | Dependent on transmission coefficients |
| Silva [57] | Study optimal strategies for the controlling active TB infectious and persistent latent individuals | Compartmental model considering reinfection and post-exposure interventions with the addition of early latent and persistent latent compartments | Parameter values taken from the literature | Basic | Dependent on transmission coefficient |
| Hu [58] | Study the threshold dynamics of TB | Compartmental model with periodic functions for reactivation rate and infection rate; include additional compartment for treated people that do not return to the hospital for examination | NA | Basic | Dependent on transmission coefficient |
| Emvudu [59] | Address the problem of optimal control for TB transmission dynamics | Compartmental model with an additional compartment for loss to follow-up | Half of the parameter values were assumed; others taken from the Cameroon literature | Basic | Dependent on parameters such as transmission rate |
| Sergeev [60] | How drug-sensitive and drug-resistant strains mixed together can impacts long-term TB dynamics | Compartmental with the three compartments for both latent and infected: drug-resistant, drug-sensitive and mixed strains | Reasonable values for many parameters; few data exist to inform model parameters | Basic; estimated for drug-resistant, drug-sensitive and mixed strains | Dependent on model parameters |
| Roeger [61] | Model TB and HIV co-infection | Compartmental model for joint dynamics of TB and HIV and compute independent reproductive numbers for the two diseases | Probability of infection is the same for those treated with TB and those susceptible; assumed relationship among model parameters | Overall R0 as the max of R0 for TB and HIV | Dependent on model parameters |
| Gerberry [62] | Study the trade-off between BCG and detection, treatment of TB | Compartmental model with additional compartments for latently infected and unvaccinated, latently infected and vaccinated; establish thresholds for basic R0 | Throughout the duration of the vaccine's efficacy, latent TB completely undetectable | Basic | Dependent on model parameters |
| Bhunu [63] | Model HIV/AIDS and TB coinfection | Compartmental model for TB, HIV separately without intervention; full model with intervention | Parameter values from Central Statistics Office of Zimbabwe and literature; relationship amongst parameters in the model | Basic | Dependent on model parameters |
| Bhunu [8] | Model the effect of pre-exposure and post-exposure vaccines | Compartmental model with additional compartments for susceptible (vaccinated or not) and latent (history of vaccine or not) | Homogeneous mixing; recovered people would not develop disease from reinfection, but could be re-infected; parameter values taken from Central Statistics Office and literature | Basic | Dependent on model parameters |
| Sharomi [64] | Address the interaction between HIV and TB | TB-only, HIV-only and full model analysed with both susceptible and latent compartments divided according to TB and HIV status | Dually infected people could not transmit both diseases; some parameters taken from the literature, others assumed | Basic | Dependent on model parameters |
| McCluskey [65] | Address global stability of high dimensional TB model | Use Lyapunov function to demonstrate the stability of the endemic equilibria in mathematical models for TB: SEIR, SEIS and SIR; fast and slow progression incorporated | Basic | No explicit estimate | |
| Martcheva [66] | Address the issue of an infected person being subject to further contacts with infectious individuals—‘super infection’ | Subdivide the latent stage into one where the disease progresses and one where the disease development is on hold | Relationship among model parameters | Basic | No explicit estimate |
| Aparicio [67] | Express basic R0 as a function of cluster size | Divide individuals into either active clusters or otherwise | Homogeneous mixing | Basic | No explicit estimate; expressed as a function of household size |
| Feng [68] | Examine how exogenous reinfection changes the TB transmission dynamics | Include additional parameters in the mathematical model to model exogenous reinfection | Constant per capita removal rate to focus on the role of reinfection | Basic | No explicit estimate; analytical expression |
| Beatriz [69] | Assess the effects of heterogeneous infectivity | Divide infective period into k stages | Homogenous mixing; bilinear incidence rate | Basic | No explicit estimate; analytical expression |
| Castillo-Chavez [70] | Use an age-structure model to study the dynamics of TB | Use age-specific parameters in the compartmental model; transmission dynamics studied for with and without vaccine | Mixing between individuals is proportional to their age-dependent activity level; disease-induced death rate neglected | Net and basic | No explicit estimate; analytical expression |
| Lietman [71] | Test the hypothesis that exposure to TB leads to disappearance of leprosy | Add in leprosy compartment in the mathematical model | Cross-immunity is symmetric: same immunity for TB and leprosy | Basic | Dependent on R0 of leprosy and cross-protection rate |
| Sanchez [72] | Evaluate the effects of parameter estimation uncertainty on the value of R0 | Latin hypercube sampling used on parameters in the compartmental model in Blower [72] to evaluate uncertainty of R0 | Range for parameters in the compartmental model | Sum of R0 for fast, slow and relapse | Dependent on parameters in the model |
| Gumel [10] | Study the transmission dynamics of TB with multiple strains, in the presence of exogenous reinfection | Included drug-sensitive and resistant strains in the compartmental model; exogenous reinfection incorporated | Homogenous mixing | Effective R0 for the two strains | Dependent on parameters in the model |
| Singer [73] | Study the impact of different reinfection levels of latently infected individuals on TB transmission dynamics | Compartmental model for heterogeneous population: one group more susceptible to infection than the other | Parameter range uniformly distributed according to previous papers | Basic | No explicit estimate |
| Trauer [74] | Model TB transmission for highly endemic regions of the Asia-Pacific where HIV-coinfection is low | Compartmental models with compartments for immunisation, latency, reinfection, drug-resistance, etc. | Parameters fixed values according to papers and WHO | Basic | Dependent on parameters; computed as 8.34 for drug-susceptible and 5.84 for drug resistant at baseline |
| Dye [75] | To establish criteria for MDR-TB control | Compartmental models with compartments for drug-susceptible, drug-resistant, treatment failure, etc. | Parameters calculated from different populations | Basic | Dependent on parameters; best estimated of the model parameters yielded R0 = 1.6 (95% CI 1.02–2.67) |
| Blower [76] | Track the emergence and evolution of multiple strains of drug-resistant TB | Non-compartmental mathematical model | NA | Basic | Dependent on drug susceptibility of TB |
| Blower [77] | Model the transmission dynamics of TB | Compartmental models with latently infected, infectious, non-infectious, recovered compartments | Some model parameters assumed; some taken from references | Basic; defined as the sum of slow progression, recent transmission and relapse | Median of 4.47, range: 0.74–18.58 |
| Blower [78] | Understand, predict and control TB | Compartmental models with drug-sensitive and drug-resistant compartments | NA | Basic | Dependent on model parameters |
| Aparicio [67] | Evaluate homogeneous mixing and heterogeneous mixing models for TB | Three types of compartmental models: a standard incidence homogenous mixing mode; a heterogeneous mixing model; an age-structured model | Assumptions on model parameters | Basic | Dependent on model parameters |
Discussion
We found very few publications that reported estimates for the serial interval of TB. Estimates of the reproductive number were limited to seven countries, with the majority of the publications using mathematical compartmental models that did not base estimates on actual data. This indicates a need for a better understanding of these crucial parameters of TB transmission, which can help inform public health decisions in order to reach the WHO's End TB goals [4] of reducing TB deaths by 95% and incident cases by 90% by 2035.
Serial interval
We found only five articles that discussed the estimation of the SI for TB and presented explicit estimates. ten Asbroek [24] estimated the serial interval over 4 years as a geometric mean of 0.57 years (95% CI 0.44–0.73). Using the same method over a longer study period (15 years compared with 4 years in [24]), the estimated median was 1.44 years, which is comparable with the median serial interval of 1.65 years in [26] with a 6-year study period. This indicates that the study period could potentially bias the serial interval estimates, even though the method in [24] corrected for the underrepresentation of longer serial intervals. In contrast with other infectious diseases that progress much faster and have SIs measured in days, the SI of TB can be weeks, years and even decades [23]. This unique feature of TB makes it difficult to obtain an unbiased estimate of the SI as lengthy follow-up is required to observe the long period between presence of symptoms of the infector and the infected person. Additionally, uncertainty regarding the presence and impact of multiple infection events further complicates the observation of this interval. Currently, the most common way of monitoring TB is by looking at annual incidence rates in studies that are often no longer than 5 years [79, 80]. This creates two issues: right censoring as symptoms of the infected people can develop long after the end of studies, and interval censoring as the symptom onset time can fall during long intervals between two observed time points. Another issue is patients’ and doctors’ delay. Patients may not seek medical assistance immediately after symptoms develop and diagnosis may require lab-processing time which causes delay in establishing the diagnosis [24], creating a left censoring issue. Survival analysis techniques can be considered to address these issues but may need substantial modification. Further ambiguity exists due to the inconsistent availability of genetic typing of strains to link cases, and the further uncertainty about how to best link strains when genetic information is available, as such information may not account for mutation rate, or infection with multiple bacterial strains.
Reproductive number
The majority of the articles used mathematical compartmental models (a brief introduction can be found in the appendix) to describe the transmission dynamics of TB. These models have been widely used to understand the dynamics of infectious diseases including SARS, influenza and TB, and they either use empirical data to estimate the parameters in the model or are based on simulation.
The compartmental models using empirical data are distinguished from simulation-based models in two key ways. First, empirical models use data to estimate some of the model parameters, while others are taken directly from the literature or assumed. Simulation-based models do not use empirical data to parameterise the models. For example, in [42] where empirical data was used, the mortality rate due to drug susceptible TB was estimated from Taiwanese Centre of Disease Control data and the effective contact rate for TB was estimated based on the literature; in [41] where simulation was used, the recruitment rate was taken from the literature and awareness rate of TB was estimated from data.
A second distinction between models based on empirical data and simulation-based models is that the former often report explicit estimates of the reproductive number for a specific region, while the latter usually focus on studying the impact of a certain feature on TB transmission dynamics. For example, in [37] where empirical data were used, the reproductive number was reported for India overall and by regions; in [60] where a simulation-based approach was used, the impact of drug-sensitive and drug-susceptible strains mixed together on TB transmission dynamics was studied.
In developed countries, the reproductive number was sometimes estimated to be well below 1: for example, 0.55 in the USA from 1930 to 1995 [52] and 0.26 in the Netherlands from 1993 to 1995 [27]. In developing countries, the reproductive number was as high as 4.3 in China in 2012 [38] and 3.55 in Southern India from 2004 to 2006. In the Netherlands, the reproductive number has been consistently estimated at well below one, ranging from 0.24 [49] to 0.48 [39].
The same dataset in San Francisco, USA in 1991–1992 (published in 1994) was used to estimate the effective reproductive number in two separate studies [30, 31] that yielded disparate results. The estimates from these two papers were reconciled in [32] to an estimated effective reproductive number of approximately 2.1 by specifying an informative prior for two parameters in [30] and improving the convergence performance of the MCMC sampler in [31]. One can contrast this estimate with other estimates for the USA to see the range of values obtained. A study of the entire USA in [52] estimated the reproductive number to be 0.55 using case rates of active TB in USA from 1955 to 1994. As shown in [81], TB incidence in San Francisco peaked between 1991 and 1993, due to the TB/HIV co-epidemic, which is consistent with the higher estimated reproductive number (around 2.1) in [30–32]. When using TB case rates in the entire USA from 1955 to 1994 as in [52], the potential geographical and temporal heterogeneity in the estimates is not well represented, resulting in an estimated reproductive number of 0.55. We would expect a lower reproductive number, and in particular, a reproductive number below one, when using data from 1955 to 1994 because by 1955, effective antibiotics were in use and BCG had also been developed, both leading to a reduction in TB incidence across the USA. In addition, Borgdorff [49] reported an effective reproductive number of 0.24 using RFLP data in San Francisco from 1991 to 1996. In this paper, the ratio of secondary cases and source cases was used to estimate the reproductive number, which may be an oversimplified estimator of the reproductive number. Issues such as linking the secondary cases and the sources cases have not been addressed. These divergent results indicate the need for the use of whole genome sequencing (WGS), which can be used to effectively link source and secondary cases.
Similar to the more statistical analysis of the San Francisco and the entire USA data, we observe that mathematical models lead to inconsistent results, at least partially attributable to the varying assumptions they make in their structure and parameterisation. For example, even though both [38] and [42] used mathematical compartmental models with different variations for similar regions (China and Taiwan), they have quite different estimates: between 3.3 and 4.3 in China from 2005 to 2012 as compared with 0.9 for drug-sensitive TB, around 0.38 for multidrug-resistant TB (defined as a TB strain resistant to at least isoniazid and rifampicin) in Taiwan from 2005 to 2010. Both articles used incidence data from Chinese and Taiwanese CDC but formulated the compartments in the models differently. In [38], compartments ‘exposed’, ‘infectious and hospitalised’ and ‘infectious but not hospitalised’ were included; in [42], compartments ‘latent’, ‘infected’ were used for two sub-populations: drug-sensitive and multidrug-resistant. The model parameters were also differently specified: in [38], some parameters were assumed while others were estimated using minimum sum of square; in [42], some parameters were given a probabilistic distribution and estimated with a root-mean-squared error method while others were assumed. The difference between the estimated reproductive numbers produced from these two modelling exercises is striking, as the two regions and populations are quite comparable in terms of demographics, economic status and access to healthcare. One could similarly contrast the modelling approaches and estimates obtained in [34] and [43], two other studies from China and Taiwan from similar time periods that produced different estimates. The differing model structures, as well as the parameter estimates, including the recruitment rate, incidence rate, and mortality rate, likely drive these observed differences. It is difficult to say which model might be a more accurate reflection of reality.
The example above illustrates the challenges of interpreting and using mathematical models for estimation of the reproductive number. However, most estimates to date make use of this approach. One shortcoming of these models is that they require assumptions about parameter values that may be difficult to estimate, such as the transmission rate, the treatment rate and the recovery rate, which are often unobservable and not reliably estimated. As a result, most of the articles assume values for the parameters in the model based on evidence in the published literature, where it exists, sometimes without measures of uncertainty (e.g. standard errors). Model structure also varies substantially from study to study, with no generally agreed upon approach to model TB and estimate parameters. For example, in [35], compartments of different vaccine strategies were included in the model and in [38], a compartment of hospitalisation was included in the model. These models also often require assumptions about the parameters used to run the models, which are likely to differ by country and time period. Sometimes sufficient data are unavailable to parameterise a model and generalisations need to be made that may not always be appropriate. The majority of the existing publications use mathematical compartmental models, which are not often ideal for statistical inference and estimation due to strong assumptions for the model structure and parameters used to run the models. While these models have the flexibility of using different compartments to evaluate the impact of policies, they are not ideal for real-time analysis where the appropriate model structure and parameter values required fitting the model may not be clear. The complexity of the natural history of TB and important factors such as HIV and drug resistance complicate these models and require additional parameters for which the data are sometimes not available. We believe that it is important to develop, as a complementary approach to compartmental models, likelihood-based data-driven analytic tools. Ideally, these estimators can be used with datasets using minimal assumptions. In addition, as WGS data become more ubiquitous [82], it will be important to develop methods that use these data to estimate the reproductive number (Fig. 5).
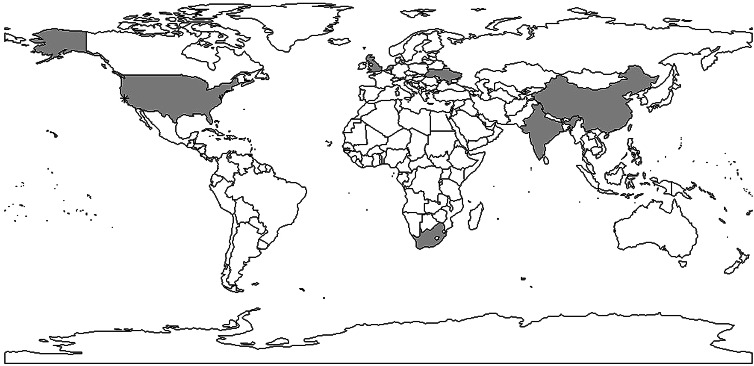
Fig. 5.
Shaded areas and stars indicate countries and cities with reproductive number estimates. Multiple estimates: China, Taiwan, USA, India; one estimate: Ukraine, the Netherlands, South Africa, the UK. *indicates San Francisco corresponding to data used in [30–32].
This review found that the reproductive number estimates for TB are very divergent – in reality, we would expect different results in different parts of the world, reflecting diversity in TB epidemics geographically. Therefore, it is important to have estimates from a wide range of settings. An ultimate goal of methods to estimate the reproductive number should be to use routinely collected data (including potentially WGS data) to be able to monitor the reproductive number in ‘real-time’ and evaluate interventions through this process.
Our review is subject to a number of limitations. It is possible that some useful papers could have been excluded due to our selection of search terms and our inclusion of reports in only English. These limitations are difficult to avoid in systematic reviews, in which the potential for increased yield from a wider search must be weighed against the increased feasibility of a tighter search. Additionally, our query was limited to searching in abstracts and titles, making it possible that we excluded articles where the keywords only appear in the text [25].
In conclusion, a limited number of studies have yielded explicit estimates for the serial interval and reproductive number of TB. When estimating the serial interval, it is difficult to observe the symptom onset of the infector and infected person with precision. Estimates of the reproductive number were limited geographically (Fig. 6) with estimates only available for seven countries. Settings with high TB burdens, especially high drug-resistant TB burdens such as the former Soviet Union [83] are not included in these papers. In addition, there was only one estimate from a high TB and high HIV burden country [47]. The lack of estimates could be because incidence and mortality rates are currently used to monitor TB control. These rates are not suitable for monitoring transmission; reductions in mortality could be attributed to improvements in treatment outcomes rather than any change in transmission and, due to the long incubation period of TB, changes in transmission could take years to impact incidence rates. In contrast, the reproductive number can provide a direct estimate of TB transmission itself. Most studies used mathematical models with various assumed model structures and parameters, making it difficult to compare the estimates and draw useful conclusions about the TB transmission dynamics by evaluating the reproductive number.
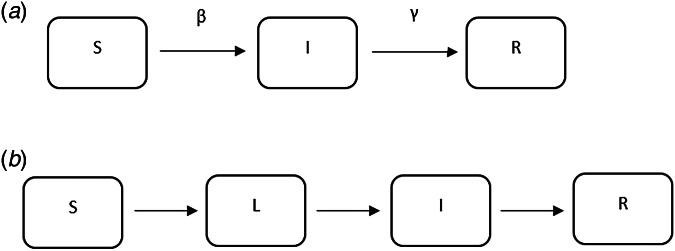
Fig. 6.
Examples of mathematical compartmental models.
The WHO End TB goals [4] include reducing TB deaths by 95% and incident cases by 90% by 2035. To achieve these goals, it is necessary to obtain improved estimates of the reproductive number and the SI as they can be used for monitoring and evaluating the effect of interventions on TB transmission. For example, the serial interval of TB can be used to determine how long one must monitor contacts of an infectious TB case to see if they will develop symptoms [84]. The effective reproductive number can be used to monitor the efficacy of interventions in reducing transmission. As interventions decrease transmission, estimates of the reproductive number should correspondingly decrease [41]; in particular, if the reproductive number can be maintained below one, the disease can potentially be eliminated.
The limited number of articles that we found and the lack of geographic representation, demonstrate a substantial gap in our understanding of these crucial parameters of TB transmission in diverse settings.
Acknowledgements
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number U19AI111276; and by the Providence/Boston Center for AIDS Research (P30AI042853). HEJ was supported by the U.S. National Institutes of Health (US NIH K01AI102944 award). LFW was supported by U.S. National Institutes of Health (R01GM122876). The content is solely the responsibility of the authors and does not necessarily represent the views of the U.S. National Institute of Allergy and Infectious Diseases or the U.S. National Institutes of Health.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Supplementary material
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268818001760.
click here to view supplementary material
Conflict of interest
None.
References
- 1.World Health Organization (2015) Global tuberculosis report 2015. World Health Organization. Available at http://www.who.int/iris/handle/10665/191102.
- 2.Center for Disease Control and Prevention (CDC) (2016) Basic TB facts, risk factors. [Internet]. Available at http://www.cdc.gov/tb/topic/basics.
- 3.American Lung Association (2016) Learn about tuberculosis. [Internet]. Available at http://www.lung.org/lung-health-and-diseases/lung-disease-lookup/tuberculosis/learn-about-tuberculosis.html.
- 4.World Health Organization (2016) Global tuberculosis report 2016. Available at http://www.searo.who.int/tb/documents/global-tuberculosis-report-2016/en/.
- 5.Horsburgh CR et al. (2004) Priorities for the treatment of latent tuberculosis infection in the United States. The New England Journal of Medicine 350, 2060–2067. [DOI] [PubMed] [Google Scholar]
- 6.Biggerstaff M et al. (2014) Estimates of the reproduction number for seasonal, pandemic, and zoonotic influenza: a systematic review of the literature. BMC Infectious Diseases 14, 480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuya H, Nagamine M and Watanabe T (2009) Use of a mathematical model to estimate tuberculosis transmission risk in an Internet café. Environmental Health and Preventive Medicine 14, 96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhunu CP et al. (2008) Modelling the effects of pre-exposure and post-exposure vaccines in tuberculosis control. Journal of Theoretical Biology 254, 633–649. [DOI] [PubMed] [Google Scholar]
- 9.Long EF and Brandeau ML (2009) Controlling co-epidemics: analysis of HIV and tuberculosis infection dynamics. Operations Research 56, 1366–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumel AB and Song B (2008) Existence of multiple-stable equilibria for a multi-drug-resistant model of Mycobacterium tuberculosis.pdf. Mathematical Biosciences & Engineering 5, 437–455. [DOI] [PubMed] [Google Scholar]
- 11.Boëlle PY et al. (2011) Transmission parameters of the A/H1N1 (2009) influenza virus pandemic: a review. Influenza and Other Respiratory Viruses 5, 306–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moser CB et al. (2015) The impact of prior information on estimates of disease transmissibility using Bayesian tools. PLoS ONE 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fraser C et al. (2009) Pandemic potential of a strain of influenza A(H1N1):early findings. Science (80-) 324, 1557–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White LF et al. (2009) Estimation of the reproductive number and the serial interval in early phase of the 2009 influenza A/H1N1 pandemic in the USA. Influenza and Other Respiratory Viruses 3, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riley S et al. (2003) Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science 300, 1961–1966. [DOI] [PubMed] [Google Scholar]
- 16.White LF and Pagano M (2008) A likelihood-based method for real-time estimation of the serial interval and reproductive number of an epidemic. Statistics in Medicine 27, 2999–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chowell G et al. (2004) The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. Journal of Theoretical Biology 229, 119–126. [DOI] [PubMed] [Google Scholar]
- 18.Wallinga J and Teunis P (2004) Different epidemic curves for severe acute respiratory syndrome reveal. American Journal of Epidemiology 160, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman-Adams H, Clark K and Juckett G (2014) Update on latent tuberculosis infection. American Family Physician Journal 89, 889–896. [PubMed] [Google Scholar]
- 20.Lillebaek T et al. (2002) Molecular evidence of endogenous reactivation of Mycobacterium tuberculosis after 33 years of latent infection. The Journal of Infectious Diseases 185, 401–404. [DOI] [PubMed] [Google Scholar]
- 21.Lambert ML et al. (2003) Recurrence in tuberculosis: relapse or reinfection? The Lancet Infectious Diseases 3, 282–287. [DOI] [PubMed] [Google Scholar]
- 22.Leung ECC et al. (2013) Transmission of multidrug-resistant and extensively drug-resistant tuberculosis in a metropolitan city. European Respiratory Society 41, 901–908. [DOI] [PubMed] [Google Scholar]
- 23.Vynnycky E and Fine PEM (2000) Lifetime risks, incubation period, and serial interval of tuberculosis. American Journal of Epidemiology 152, 247–263. [DOI] [PubMed] [Google Scholar]
- 24.ten Asbroek AH et al. (1999) Estimation of serial interval and incubation period of tuberculosis using DNA fingerprinting. International Journal of Tuberculosis and Lung Disease 3, 414–420. [PubMed] [Google Scholar]
- 25.Borgdorff MW et al. (2011) The incubation period distribution of tuberculosis estimated with a molecular epidemiological approach. International Journal of Epidemiology 40, 964–970. [DOI] [PubMed] [Google Scholar]
- 26.Brooks-Pollock E et al. (2011) Epidemiologic inference from the distribution of tuberculosis cases in households in Lima, Peru. The Journal of Infectious Diseases 203, 1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgdorff MW et al. (1998) Analysis of tuberculosis transmission between nationalities in the Netherlands in the period 1993–1995 using DNA fingerprinting. American Journal of Epidemiology 147, 187–195. [DOI] [PubMed] [Google Scholar]
- 28.Nebenzahl-Guimaraes H et al. (2015) Transmission and progression to disease of Mycobacterium tuberculosis phylogenetic lineages in the Netherlands. Journal of Clinical Microbiology 53, 3264–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgdorff MW et al. (2010) Progress towards tuberculosis elimination: secular trend, immigration and transmission. European Respiratory Society 36, 339–347. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka MM et al. (2006) Using approximate Bayesian computation to estimate tuberculosis transmission parameters from genotype data. Genetics 173, 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadler T (2011) Inferring epidemiological parameters on the basis of allele frequencies. Genetics 188, 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aandahl RZ et al. (2014) Exact vs. approximate computation: reconciling different estimates of Mycobacterium tuberculosis epidemiological parameters. Genetics 196, 1227–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Small PM et al. (1994) The epidemiology of tuberculosis in San Francisco a population-based study using conventional and molecular methods. The New England Journal of Medicine 330, 1703–1709. [DOI] [PubMed] [Google Scholar]
- 34.Zhao Y, Li M and Yuan S (2017) Analysis of transmission and control of tuberculosis in Mainland China, 2005–2016, based on the age-structure mathematical model. International Journal of Environmental Research and Public Health 14, 1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S et al. (2017) Mixed vaccination strategy for the control of tuberculosis: a case study in China. Mathematical Biosciences and Engineering 14, 695–708. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y et al. (2016) Seasonality impact on the transmission dynamics of tuberculosis. Computational and Mathematical Methods in Medicine 2016, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Narula P, Azad S and Lio P (2015) Bayesian melding approach to estimate the reproduction number for tuberculosis transmission in Indian States and Union Territories. Asia Pacific Journal of Public Health 27, 723–732. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Li Y and Zhang X (2015) Mathematical modeling of tuberculosis data of China. Journal of Theoretical Biology 365, 159–163. [DOI] [PubMed] [Google Scholar]
- 39.Ypma RJF et al. (2013) A sign of superspreading in tuberculosis: highly skewed distribution of genotypic cluster sizes. Epidemiology 24, 395–400. [DOI] [PubMed] [Google Scholar]
- 40.Andrews JR, Morrow C and Wood R (2013) Modeling the role of public transportation in sustaining tuberculosis transmission in South Africa. American Journal of Epidemiology 177, 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okuonghae D and Omosigho SE (2011) Analysis of a mathematical model for tuberculosis: what could be done to increase case detection. Journal of Theoretical Biology 269, 31–45. [DOI] [PubMed] [Google Scholar]
- 42.Liao CM and Lin YJ (2012) Assessing the transmission risk of multidrug-resistant Mycobacterium tuberculosis epidemics in regions of Taiwan. International Journal of Infectious Diseases 16, e739–e747. [DOI] [PubMed] [Google Scholar]
- 43.Liao C-M et al. (2012) A probabilistic transmission and population dynamic model to assess tuberculosis infection risk. Risk Analysis 32, 1420–1432. [DOI] [PubMed] [Google Scholar]
- 44.Liu L, Wu J and Zhao X-Q (2012) The impact of migrant workers on the tuberculosis transmission: general models and a case study for China. Mathematical Biosciences and Engineering 9, 785–807. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, Zhao XQ and Zhou Y (2010) A tuberculosis model with seasonality. Bulletin of Mathematical Biology 72, 931–952. [DOI] [PubMed] [Google Scholar]
- 46.Brooks-Pollock E, Cohen T and Murray M (2010) The impact of realistic age structure in simple models of tuberculosis transmission. PLoS ONE 5, 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Basu S et al. (2009) Averting epidemics of extensively drug-resistant tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 106, 7672–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borgdorff MW et al. (2005) Tuberculosis elimination in the Netherlands. Emerging Infectious Diseases journal 11, 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borgdorff MW et al. (2000) Transmission of tuberculosis in San Francisco and its association with immigration and ethnicity. International Journal of Tuberculosis and Lung Disease 4, 287–294. [PubMed] [Google Scholar]
- 50.Davidow AL, Aicabes P and Marmora M (2000) The contribution of recently acquired Mycobacterium tuberculosis infection to the New York City tuberculosis epidemic, 1989–1993. Epidemiology 11, 394–401. [DOI] [PubMed] [Google Scholar]
- 51.Vynnycky E and Fine PE (1998) The long-term dynamics of tuberculosis and other diseases with long serial intervals: implications of and for changing reproduction numbers. Epidemiology and Infection 121, 309–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salpeter EE and Salpeter SR (1998) Mathematical model for the epidemiology of tuberculosis, with estimates of the reproductive number and infection-delay function. American Journal of Epidemiology 142, 398–406. [DOI] [PubMed] [Google Scholar]
- 53.Ren S (2017) Global stability in a tuberculosis model of imperfect treatment with age-dependent latency and relapse. Mathematical Biosciences and Engineering 14, 1337–1360. [DOI] [PubMed] [Google Scholar]
- 54.Jabbari A et al. (2016) A two-strain TB model with multiple latent stages. Mathematical Biosciences and Engineering 13, 741–785. [DOI] [PubMed] [Google Scholar]
- 55.Okuonghae D and Ikhimwin BO (2016) Dynamics of a mathematical model for tuberculosis with variability in susceptibility and disease progressions due to difference in awareness level. Frontiers in Microbiology 6, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu L and Wang Y (2014) A mathematical study of a TB model with treatment interruptions and two latent periods. Computational and Mathematical Methods in Medicine 2014, Article ID 932186, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva CJ and Torres DFM (2013) Optimal control for a tuberculosis model with reinfection and post-exposure interventions. Mathematical Biosciences 244, 154–164. [DOI] [PubMed] [Google Scholar]
- 58.Hu X (2012) Threshold dynamics for a tuberculosis model with seasonality. Mathematical Biosciences and Engineering 9, 111–122. [DOI] [PubMed] [Google Scholar]
- 59.Emvudu Y, Demasse R and Djeudeu D (2011) Optimal control of the lost to follow up in a tuberculosis model. Computational and Mathematical Methods in Medicine 2011, Article ID 398476, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sergeev R, Colijn C and Cohen T (2011) Models to understand the population-level impact of mixed strain M. tuberculosis infections. Journal of Theoretical Biology 280, 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roeger L-IW, Feng Z and Castillo-Chavez C (2009) Modeling TB and HIV co-infections. Mathematical Biosciences and Engineering 6, 815–837. [DOI] [PubMed] [Google Scholar]
- 62.Gerberry DJ (2009) Trade-off between BCG vaccination and the ability to detect and treat latent tuberculosis. Journal of Theoretical Biology 261, 548–560. [DOI] [PubMed] [Google Scholar]
- 63.Bhunu CP, Garira W and Mukandavire Z (2009) Modeling HIV/AIDS and tuberculosis coinfection. Bulletin of Mathematical Biology 71, 1745–1780. [DOI] [PubMed] [Google Scholar]
- 64.Sharomi O and Podder C (2008) Mathematical analysis of the transmission dynamics of HIV/TB coinfection in the presence of treatment. Mathematical Biosciences and Engineering 5, 145–174. [DOI] [PubMed] [Google Scholar]
- 65.McCluskey CC (2006) Lyapunov functions for tuberculosis models with fast and slow progression. Mathematical Biosciences and Engineering 3, 603–614. [DOI] [PubMed] [Google Scholar]
- 66.Martcheva M and Thieme HR (2003) Progression age enhanced backward bifurcation in an epidemic model with super-infection. Journal of Mathematical Biology 46, 385–424. [DOI] [PubMed] [Google Scholar]
- 67.Aparicio JP, Capurro AF and Castillo-Chavez C (2000) Transmission and dynamics of tuberculosis on generalized households. Journal of Theoretical Biology 206, 327–341. [DOI] [PubMed] [Google Scholar]
- 68.Feng Z, Castillo-chavez C and Capurro AF (2000) A model for tuberculosis with exogenous reinfection. Theoretical Population Biology 57, 235–247. [DOI] [PubMed] [Google Scholar]
- 69.Beatriz M et al. (2000) The basic reproduction ratio for a model of directly transmitted infections considering the virus charge and the immunological response. Mathematical Medicine and Biology 17, 15–31. [PubMed] [Google Scholar]
- 70.Castillo-Chavez C and Feng Z (1998) Global stability of an age-structure model for TB and its applications to optimal vaccination strategies. Mathematical Biosciences 151, 135–154. [DOI] [PubMed] [Google Scholar]
- 71.Lietman T, Porco T and Blower S (1997) Leprosy and tuberculosis: the epidemiological consequences of cross- immunity. American Journal of Public Health 87, 1923–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sanchez MA and Blower SM (1997) Uncertainty and sensitivity analysis of the basic reproductive rate. American Journal of Epidemiology 145, 1127–1137. [DOI] [PubMed] [Google Scholar]
- 73.Singer BH and Kirschner DE (2004) Influence of backward bifurcation on interpretation of r(0) in a model of epidemic tuberculosis with reinfection. Mathematical Biosciences and Engineering 1, 81–93. [DOI] [PubMed] [Google Scholar]
- 74.Trauer JM, Denholm JT and McBryde ES (2014) Construction of a mathematical model for tuberculosis transmission in highly endemic regions of the Asia-pacific. Journal of Theoretical Biology 358, 74–84. [DOI] [PubMed] [Google Scholar]
- 75.Dye C and Williams BG (2000) Criteria for the control of drug-resistant tuberculosis. Proceedings of the National Academy of Sciences of the United States of America 97, 8180–8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blower SM and Chou T (2004) Modeling the emergence of the “hot zones”: tuberculosis and the amplification dynamics of drug resistance. Nature Medicine 10, 1111–1116. [DOI] [PubMed] [Google Scholar]
- 77.Blower S et al. (1995) The intrinsic transmission dynamics of tuberculosis epidemics. Nature Medicine 1, 815–821. [DOI] [PubMed] [Google Scholar]
- 78.Blower SM and Gerberding JL (1998) Understanding, predicting and controlling the emergence of drug-resistant tuberculosis: a theoretical framework. Journal of Molecular Medicine 76, 624–636. [DOI] [PubMed] [Google Scholar]
- 79.Morrison J, Pai M and Hopewell PC (2008) Tuberculosis and latent tuberculosis infection in close contacts of people with pulmonary tuberculosis in low-income and middle-income countries: a systematic review and meta-analysis. The Lancet Infectious Diseases 8, 359–368. [DOI] [PubMed] [Google Scholar]
- 80.Fox GJ et al. (2013) Contact investigation for tuberculosis: a systematic review and meta-analysis. European Respiratory Society 41, 140–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Higashi J. (2013). Report on Tuberculosis in San Francisco: 2012 [Internet]. Available at https://www.sfdph.org/dph/hc/HCCommPublHlth/Agendas/2013/2013/March/TB2012healthcommission.pdf.
- 82.Wyllie D et al. (2018) A quantitative evaluation of MIRU-VNTR typing against whole-genome sequencing for identifying Mycobacterium tuberculosis transmission: a prospective observational cohort study. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olson S, English R and Claiborne A. (2011). The New Profile of Drug-Resistant Tuberculosis in Russia [Internet]. Available at https://www.ncbi.nlm.nih.gov/books/NBK62461/.
- 84.Vink MA, Bootsma MCJ and Wallinga J (2014) Serial intervals of respiratory infectious diseases: a systematic review and analysis. American Journal of Epidemiology 180, 865–875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268818001760.
click here to view supplementary material