Abstract
Comprehensive genomic cancer risk assessment (GCRA) helps patients, family members, and providers make informed choices about cancer screening, surgical and chemotherapeutic risk reduction, and genetically targeted cancer therapies. The increasing availability of multigene panel tests for clinical applications allows testing of well-defined high-risk genes, as well as moderate-risk genes, for which the penetrance and spectrum of cancer risk are less well characterized. Moderate-risk genes are defined as genes that, when altered by a pathogenic variant, confer a two to five-fold relative risk of cancer. Two such genes included on many comprehensive cancer panels are the DNA repair genes ATM and CHEK2, best known for moderately increased risk of breast cancer development. However, the impact of screening and preventative interventions and spectrum of cancer risk beyond breast cancer associated with ATM and/or CHEK2 variants remain less well characterized. We convened a large, multidisciplinary, cross-sectional panel of GCRA clinicians to review challenging, peer-submitted cases of patients identified with ATM or CHEK2 variants. This paper summarizes the inter-professional case discussion and recommendations generated during the session, the level of concordance with respect to recommendations between the academic and community clinician participants for each case, and potential barriers to implementing recommended care in various practice settings.
Keywords: Cancer genetics, ATM, CHEK2, moderate-risk gene, panel test, genomic cancer risk assessment (GCRA)
Introduction & Background
The clinical utility of genomic cancer risk assessment (GCRA) using single gene germline testing for cancer predisposition is well established [1–6]. Comprehensive GCRA helps patients, family members, and providers make informed choices about cancer screening, surgical and chemoprophylactic risk reduction, and genetically targeted cancer therapies. Historically, genetic testing evaluated for high-penetrance cancer predisposition genes such as BRCA1 and BRCA2. The advent of high-throughput next generation sequencing (NGS) and competitive marketing by multiple commercial vendors is driving down costs thereby increasing the availability of multigene panel tests for clinical applications. In addition to bundling well-defined high-risk genes, many panels also include a growing number of low and moderate-risk genes for which the penetrance and spectrum of cancer risk are less well characterized.
Moderate-risk genes are defined as genes that, when altered by a pathogenic variant, confer a two to five-fold relative risk (RR) of cancer [7]. Historically, the high cost of sequencing and a lower cancer incidence among carriers of moderate-risk pathogenic variants limited the identification of carriers for enrollment in retrospective and case-control studies. NGS and multigene panel testing are driving the identification of many more carriers of pathogenic variants in moderate-risk genes. The application of GCRA in carriers of moderate-risk genes is now common practice among many providers, however, prospective data and consensus management guidelines to help patients and providers make informed decisions are limited.
Among the moderate-risk genes appearing on commercial multigene cancer panels are the DNA repair genes ataxia-telangiectasia mutated (ATM) and checkpoint serine-threonine kinase 2 (CHEK2). ATM is located on chromosome 11 and encodes a serine threonine kinase that is activated in response to DNA double-strand breaks. ATM participates in DNA repair by phosphorylating downstream proteins involved in cell cycle checkpoint control, apoptosis, and DNA repair [8]. Pathogenic bi-allelic variants in ATM result in ataxia-telangiectasia (A-T), which is characterized by progressive cerebellar degeneration, oculocutaneous telangiectasia, immunological deficiency, radiosensitivity, and an increased risk of cancer [9–11]. Heterozygous carriers of pathogenic ATM variants do not display characteristic clinical features of autosomal recessive A-T, but do share an increased predisposition to cancer [10, 11]. The carrier frequency of ATM pathogenic variants is estimated at 0.5-1% in the general population [9]. Retrospective reviews undertaken to better quantify the relative risk of specific cancers in heterozygous carriers of pathogenic ATM variants demonstrate an increased risk of breast cancer (RR=2.8), but were unable to quantify postulated increased risk for colon, prostate, and pancreatic cancer[12, 13]. Consequently, the National Comprehensive Cancer Network (NCCN) consensus statements do not include recommendations for colorectal, prostate, or pancreatic cancer screening in patients with pathogenic ATM variants[14]. Further research is needed to better quantify the risk of cancer development and utility of increased screening in patients with inherited pathogenic ATM variants.
CHEK2 is located on chromosome 22 and also encodes a serine threonine kinase involved in the DNA damage response. CHEK2 is phosphorylated in response to double-strand DNA breaks, DNA alkylation, or replicative stress, and once phosphorylated, CHEK2 activates downstream targets including p53 to mediate cycle arrest and apoptosis [15, 16]. Germline CHEK2 sequence variants were first identified in Li-Fraumeni Syndrome families that do not carry TP53 pathogenic variants [15, 17]. While different studies have reported possible association with increased risk for breast, colon, and prostate cancer, precise relative risk estimates are limited to breast cancer; accordingly, at the time of the study, NCCN screening recommendations for CHEK2 carriers were limited to breast cancer screening [7, 18–20]. Notably, truncating and pathogenic missense variants in CHEK2 confer a RR of 3.0 (90% CI 2.6-3.5) and 1.58 (95% CI 1.42-1.75) respectively [21]. CHEK2 variant c. 1100delC (p.Thr367Metfs), a truncating variant, is the best-characterized CHEK2 pathogenic variant, seen in up to 1-2% of the general population [15, 17, 22].
An ever-increasing number of patients are being identified with pathogenic variants in moderate-risk genes, including, ATM and CHEK2. Recent work by Couch et al. evaluating pathogenic variants in 41,611 eligible consecutive white women with breast cancer referred for hereditary cancer genetic testing demonstrated that 6.18% of women harbored a pathogenic variant, excluding the high-risk genes BRCA1 and BRCA2 and low-risk founder CHEK2 variants c.470C>T (p.Ile157Thr) and c.1283C>T (p.Ser428Phe); among the most commonly mutated genes were ATM (1.06%) and CHEK2 (1.73%) [23]. Similarly, in a retrospective review of 337 patients who met NCCN criteria for HBOC assessment and underwent multigene panel evaluation, 25 (7.4%) patients had a pathogenic non-BRCA variant, and variants in CHEK2 and ATM each accounted for 15% of the total pathogenic variants identified [24]. Despite the inclusion of moderate-risk cancer genes on multigene panels and the frequency of pathogenic variants in these genes, there are currently only limited data to estimate the risk of cancer development in this patient population. There are few, if any, prospective studies of surveillance or risk reduction interventions to guide cancer risk management in these patients and management recommendations are often based on expert opinion. Further, genetic counselors and physicians often have limited clinical experience with the management of patients with pathogenic variants in moderate-risk genes. Tung et al. have proposed a general counseling framework for clinicians providing care to individuals with moderate-penetrance variants associated with an increased risk of cancer development [21].
Given the limited data regarding risk of cancer development and appropriate surveillance and risk reduction interventions in patients with ATM and CHEK2 variants, we hypothesized that there is heterogeneity among GCRA practitioners in the clinical management of ATM and CHEK2 patients. We convened a large, multidisciplinary, cross-sectional (academic and community oncology practices across the United States) panel of GCRA clinicians to review challenging, peer-submitted cases of patients identified with ATM or CHEK2 pathogenic variants. The review was integrated in a session at an annual cancer genomics conference, “New Frontiers in Diagnosis, Screening, and Management of Inherited Cancer Syndromes”, co-hosted by The University of Chicago and The City of Hope on April 8-9, 2016, in Chicago, IL (Supplement 1). Our aims were to share clinical experiences with reported variants in ATM and CHEK2 genes, identify provider knowledge gaps and areas of uncertainty, and survey whether patients get uniform access to recommended screening and prevention measures. This paper summarizes the inter-professional case discussion and recommendations generated during the session, the level of concordance with recommendations between the academic and community clinician participants for each case, and potential barriers to implementing recommended care in various practice settings.
Methods
Participants/Procedure
Participants included 104 conference attendees, comprised of physicians and allied health care professionals from diverse oncology practice settings across the United States (US) and internationally, including clinicians who participate in the nationwide, National Cancer Institute (NCI)-supported program, Clinical Cancer Genomics Community of Practice (CCGCoP), for GCRA training and practice support [25, 26]. Prior to the conference, participants were invited to submit cases from their clinical oncology practices with complex or challenging results, with a particular emphasis on pathogenic variants in ATM or CHEK2 yielded from clinical multigene panel tests.
The panel was comprised of five cancer genomics thought leaders representing different academic settings and disciplines. The session was co-moderated by a cancer genomics physician and a cancer risk genetic counselor. Presenters were invited to share each anonymized case, which included a brief clinical synopsis, pedigree, results of genetic testing, and specific management questions. The panel and participants discussed challenges related to each case and panelists generated recommendations in real-time that incorporated existing evidence from the literature, applied clinical judgment related to screening and risk management, and emphasized best practices in genetic counseling and patient and family communications. NCCN guidelines were cited when applicable, using the versions in effect at the time of the conference, NCCN Genetic/Familial High-Risk Assessment: Colorectal. Version 2.2015, and Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 2.2016 (detailed in Table 1). During the session, participants contributed to the oral case discussion and also documented their perceptions, interpretations, and feedback related to each case using a survey. Three cancer genomics clinician researchers documented key points and recommendations that were raised by the presenters or generated from panel discussion on each case.
Table 1.
NCCN Guidelines in publication at time of survey study (NCCN Genetic/Familial High-Risk Assessment: Colorectal. Version 2.2015), (NCCN Genetic/Familial High-Risk Assessment: Breast and Ovarian. Version 2.2016)
| Gene | Breast Cancer | Colon Cancer |
|---|---|---|
| ATM |
Screening: Annual mammogram and consider breast MRI starting at age 40 RRM: Consider based on family history |
Not addressed |
| CHEK2 |
Screening: Annual mammogram and consider breast MRI starting at age 40 RRM: Consider based on family history |
Not addressed |
Instrumentation
Participant demographic information including terminal degree, type of clinical practice, and geographic location of practice was collected on the conference registration form.
Case Submission Form
Case submissions were elicited from conference registrants one month prior to the conference via email, and included a de-identified pedigree, any relevant genetic testing results, and a brief description of specific challenges or questions related to the case.
Participant Feedback Survey
Participants completed a paper and pencil survey comprised of yes/no questions and open-ended prompts eliciting prior GCRA experiences, feedback on each case presented, and participant’s perceived learning value of the session (Supplement 2).
Data Analysis
Documentation outlining expert panel recommendations performed by three cancer genomics clinician researchers were compiled, compared for consistency, and summarized to reflect expert panel discussion and recommendations for each case. Quantitative and qualitative survey data from session participants were entered into Microsoft Excel 2013 spreadsheets and audited for accuracy [27]. Given the limited number of international participants and variation in screening practices by country, only participant data from clinicians practicing in the US was analyzed. Descriptive statistics were computed for demographic, yes/no, and rating scale items. Coding and thematic analyses of open-ended responses were conducted by two clinical researchers through a series of iterations, and frequencies of responses coded under each theme were tallied. Quantitative and qualitative outcomes were triangulated to increase the depth and validity of the findings [28]. Members of the expert panel and session moderators reviewed final panel and participant recommendations for relevance and accuracy.
Results
As summarized in Table 2, 104 (47%) of 219 clinicians who attended the conference participated in the case working session. Of these, 99 clinicians (95%) practiced within the US and were included in final data analysis. Eighty-one (82%) reported that they provide GCRA services as all or part of their practices, primarily in community hospital (52%) or private practice (29%) settings, and roughly half (51%) are active members of the CCGCoP.
Table 2.
Demographics of Participants in ATM/CHEK2 Case Working Session
| Participant Characteristics | N (%) |
|---|---|
| Terminal degree | |
| MD | 32 (32%) |
| RN/APN | 34 (34%) |
| GC | 25 (25%) |
| PhD | 1 (1%) |
| Other | 7 (7%) |
| Practice Setting | |
| Private Practice | 29 (29%) |
| Community Hospital | 51 (52%) |
| Academic Medical Institution | 11 (11%) |
| Other | 4 (4%) |
| Omitted | 4 (4%) |
| Participants currently delivering GCRA services as partor all of their practices | |
| Yes | 81 (82%) |
| No | 14 (14%) |
| Omitted | 4 (4%) |
| City of Hope Clinical Cancer Genomics Community of Practice members (CCGCoP) | |
| Yes | 50 (51%) |
| No | 48 (48%) |
| Omitted | 1 (1%) |
Of nine GCRA cases presented, the three ATM and two CHEK2 cases summarized below were selected to represent the key clinical challenges, discussion points, and recommendations generated during the case working session.
ATM
Cases one through three include patients with an inherited ATM variant.
Case 1: ATM Carrier Seeking Risk-Reducing Mastectomy
A 34-year-old unaffected female was referred for GCRA due to a family history of breast cancer and a known pathogenic ATM variant, c.1564_1565delGA (p.Glu522ILefs), that was previously identified in her sister, who was diagnosed with breast cancer at age 49. Family history (Figure 1) was also notable for breast cancer in the consultand’s mother at age 60 and maternal aunt at age 69, and for a maternal great-aunt with an unspecified female cancer as an adult. Her paternal family included cancer of unknown primary in a first cousin at age 50. Single-site testing for the pathogenic ATM variant, c.1564_1565delGA (p.Glu522ILefs), in the consultand was positive.
Fig. 1.
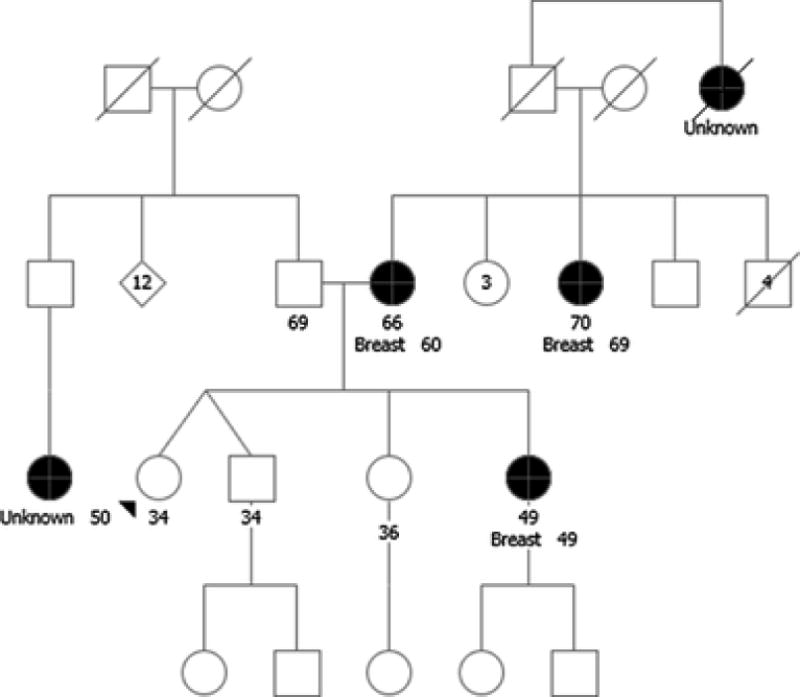
Case 1: ATM Carrier Seeking Risk-Reducing Mastectomy. Unknown, cancer of unknown type. Breast, breast cancer. Please see associated vignette for more details
Presenting Problem/Questions
1) What are the recommended breast cancer screening recommendations for the unaffected 34-year old consultand? 2) How does one address the question of risk reducing mastectomy (RRM) for this patient? 3) Do test results change management for the consultand given an empiric lifetime breast cancer risk estimate of 23% by the Claus model [29]?
Panel Recommendations
Screening for the consultand should include annual mammogram and breast MRI starting 10 years younger than the age of breast cancer diagnosis in the consultand’s sister [7]. With regard to breast cancer prevention, the panel advised that the consultand’s Gail score be calculated when she reaches 35 years of age, and tamoxifen may be considered if her 5-year risk of breast cancer development is greater than 1.67% [30]. RRM was not recommended given moderate lifetime breast cancer risk in carriers of a pathogenic ATM variant, however, the panel concurred that discussion of RRM may be appropriate depending on the consultand’s breast cancer risk-tolerance. The panel noted that risk management was not meaningfully altered by the ATM carrier status, given that MRI surveillance, in addition to mammogram, is recommended for women with a lifetime risk of breast cancer that exceeds 20% based on empiric risk models[29, 31]. The panel also emphasized the importance of testing other family members to identify other at-risk individuals and to determine on which side of the family the ATM variant segregated. The participants noted that, in their clinical practices, identification of ATM variants within families was often discordant with their expectations, (i.e., not segregating with the reported breast cancers). This reflects reported observations that the attributable risk and negative predictive value for ATM carriers is limited, emphasizing the importance of residual empiric risk based on family history. That is, unlike families with an inherited high-risk variant, testing negative for a known familial moderate-risk variant is not thought to reduce the individual’s risk to that of the general population.
Participant Feedback
Forty-seven (51%) participants noted that they had seen similar cases in their GCRA practices. Eighty-five (98%) fully or mostly concurred with the recommendations.
Case 2: Pancreatic Cancer Risk for ATM Carriers – True or Unrelated?
An 80-year-old female of Czech and Polish ancestry with a personal history of estrogen receptor positive (ER+), progesterone receptor positive (PR+), and Her2/neu negative invasive ductal carcinoma of the right breast diagnosed at age 65 was seen for GCRA. Family history (Figure 2) was significant for a brother who recently died from pancreatic cancer at age 81, and a sister who died of renal cell carcinoma at age 54. The proband’s son died of pancreatic cancer at age 53, and her 57-year-old daughter was diagnosed with early stage uterine cancer at age 52. A hereditary cancer panel evaluating 49 genes was ordered. Between the time of testing and result disclosure, the patient was diagnosed with metastatic pancreatic adenocarcinoma. Results from the panel revealed a pathogenic ATM variant, c.3850delA (p.Thr1284GInfs). Three of the patient’s adult children subsequently tested positive for the ATM variant and inquired regarding their own cancer screening recommendations.
Fig. 2.
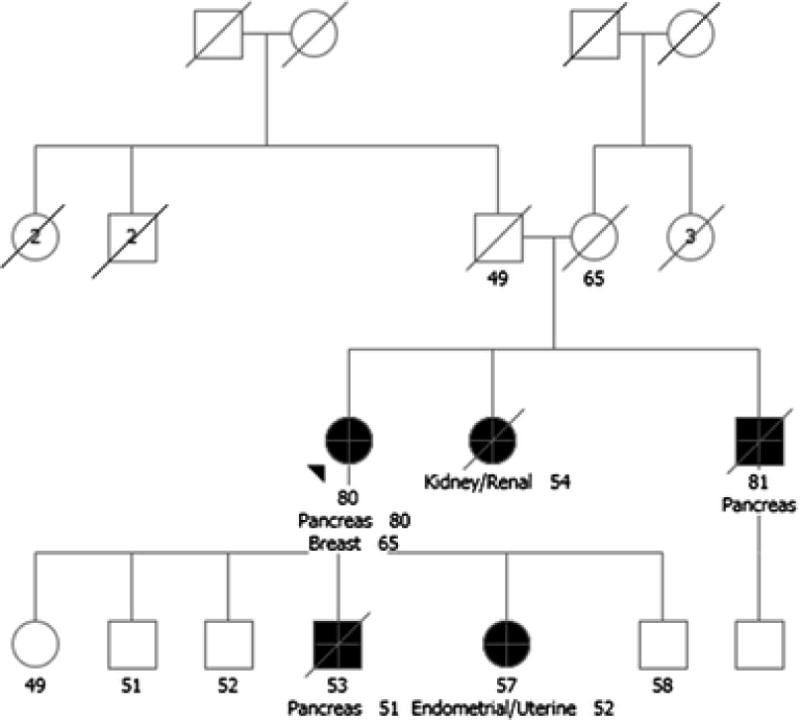
Case 2: Pancreatic Cancer Risk for ATM Carriers – True or Unrelated? Pancreas, pancreas cancer; Endometrial, endometrial cancer; Kidney, kidney cancer; Breast, breast cancer. Please see associated vignette for more details
Presenting Problem/Questions
How should one counsel regarding pancreatic cancer screening in patients with ann ATM variant, and in this family in particular?
How are recommendations influenced by ATM carrier status?
Panel Recommendations
Although there is evidence that pancreatic cancer risk is moderately increased (~5% lifetime) in ATM carriers, there are currently no NCCN guidelines for pancreatic cancer screening in ATM carriers [32]. Therefore, the panel advised that family history should guide the approach to pancreatic cancer screening for this family. A working definition of familial pancreatic cancer is a consultand with a pair of first-degree relatives affected by pancreatic cancer or a total of three family members affected by pancreatic cancer [33]. Given that the consultand’s children met criteria for familial pancreatic cancer, the panel recommended consideration of annual pancreatic cancer screening with alternating endoscopic ultrasound (EUS) and magnetic resonance cholangiopancreatography (MRCP) [34, 35]. These recommendations were independent of ATM carrier status. The panel also emphasized that screening may be considered in a consultand with an ATM variant and a single first-degree relative affected by pancreatic cancer depending on their risk-tolerance, after appropriate counseling regarding the risks and limitations of pancreatic cancer screening.
Participant Feedback
Twenty (21%) participants noted that they had seen similar cases in their GCRA practices. All participants agreed with the panel’s recommendations. Nineteen (25%) participants stated that their patients would not have access to all recommended pancreatic cancer screening due to lack of insurance coverage or availability of an EUS-qualified gastroenterologist. This raised the ethical issue of equity.
Case 3: Is radiation treatment contra-indicated in a carrier of homozygous ATM variants of uncertain significance?
A 48-year-old woman of Northern European ancestry, recently diagnosed with clinical stage II ER+, PR+, Her2/neu+ invasive breast cancer with nodal involvement, was referred for GCRA. Family history (Figure 3) included a diagnosis of melanoma at age 50 in the proband’s father and lung cancer in a paternal uncle, who was a smoker. Testing with an eight gene, breast cancer focused panel was performed. The proband was found to be homozygous for a variant of uncertain significance (VUS) in the ATM gene, c.4258C>T (p.Leu1420Phe), previously reported in both heterozygous and homozygous states. Per the report generated by the commercial genetic testing laboratory, this ATM VUS is not associated with A-T in the homozygous state. However, in a large case-control study of breast cancer patients, ATM c.4258C>T (p.Leu1420Phe) conferred a statistically significant increased risk of breast cancer when in the homozygous state (OR 5.31 CI=1.35-20.87) [36]. Increased breast cancer risk has not been observed in heterozygous carriers of this VUS [37].
Fig. 3.
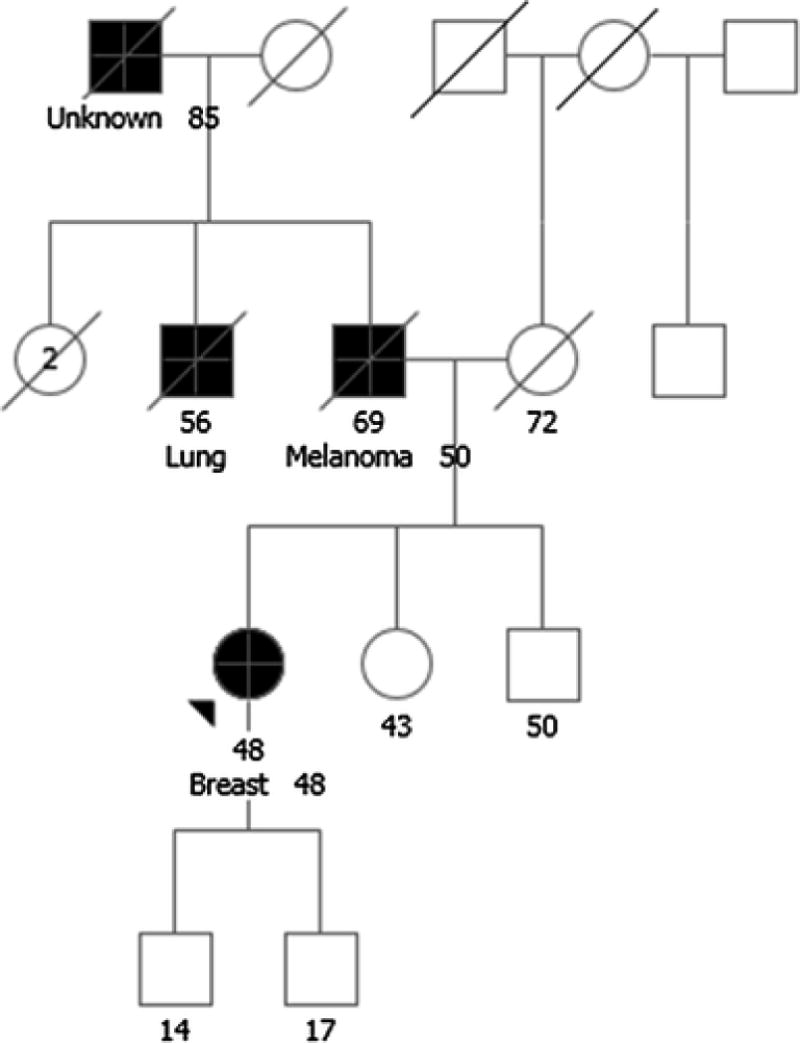
Case 3: Is radiation treatment contra-indicated in a carrier of homozygous ATM variants of uncertain significance? Lung, lung cancer; Breast, breast cancer. Please see associated vignette for more details.
Presenting Problem/Questions
1) Should the ATM VUS affect breast cancer treatment, specifically radiation therapy, in this proband? 2) What is the significance of this result for other family members?
Panel Recommendations
The panel advised that given the evidence that homozygous carriers of the ATM c.4258C>T (p.Leu1420Phe) VUS do not have a phenotype consistent with A-T, concern about radiosensitivity should not influence breast cancer treatment, and the proband should receive standard-of-care therapy for her malignancy. The panel also advised that there is insufficient evidence to recommend against radiation therapy for heterozygous ATM carriers if it is indicated based on cancer diagnosis; this information was not included in NCCN guidelines at the time of the conference, but has since been incorporated [38, 39]. With regard to other family members, genetic testing may also be considered to evaluate for homozygous carriers, specifically siblings who would each have a 1/4 chance of being homozygous for the ATM variant.. The panel emphasized that the proband should be encouraged to enroll in a research registry to aid in further characterizing the ATM VUS. Of note, a second case with a proband who is homozygous for the ATM c.4258C>T (p.Leu1420Phe) variant was submitted to the conference organizers from a separate clinical practice, reflecting the commonality of this alteration.
Participant Feedback
Seven (9%) participants noted that they had seen similar cases in their GCRA practices. All participants agreed with the panel’s recommendation. Participants who shared a similar experience expressed frustration with limited data in this context and emphasized the importance of encouraging patient participation in research.
CHEK2
Cases four and five represent challenges related to test results that revealed inherited pathogenic variants in the CHEK2 gene.
Case 4: Prioritizing Genetic Testing in Time Sensitive Situations
A 54-year-old Caucasian female with a personal history of ductal carcinoma in situ at age 44, treated with lumpectomy, and a new diagnosis of invasive ductal carcinoma at age 54, was referred for GCRA prior to cancer treatment. The proband’s mother was diagnosed with liver cancer at age 81; the proband had no knowledge of other cancers on either side of her family (Figure 4). Testing was initiated to evaluate BRCA1 and BRCA2, to ensure rapid results due to an upcoming surgical date, with reflex to a 21 gene breast/ovarian cancer panel. Findings included a pathogenic variant in CHEK2, c.1100delC (p.Thr367Metfs), a VUS in ATM, c.6067G>A (p.Gly2023Arg), and a VUS in BRIP1, c.577G>A (p.Val193Ile); since the time of original testing, the BRIP1 VUS was reclassified as benign[40].
Fig 4.
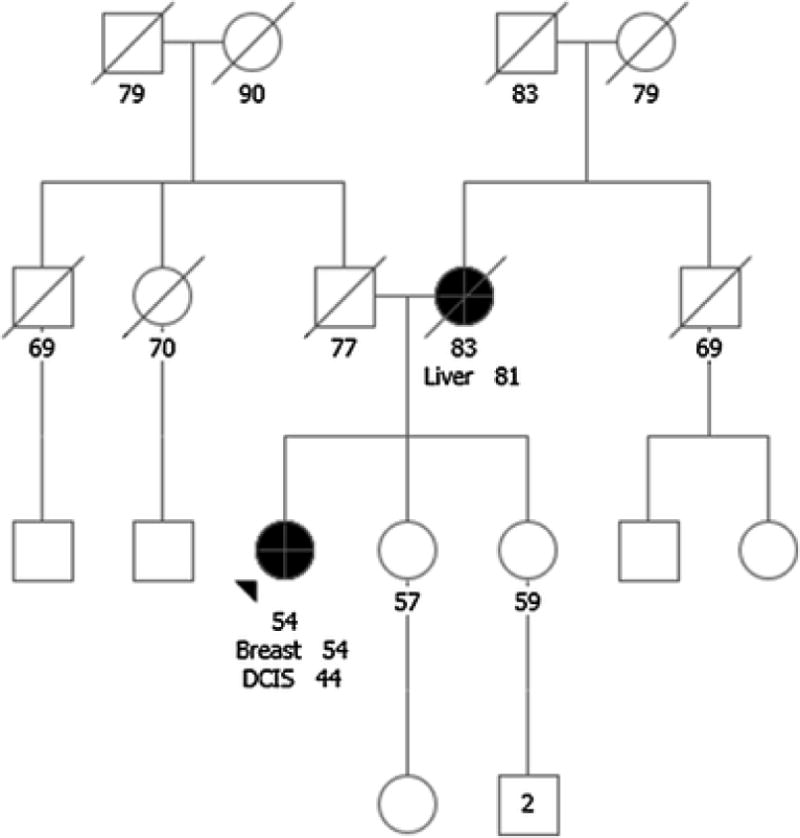
Case 4: Prioritizing Genetic Testing in Time Sensitive Situations. Breast, breast cancer; DCIS, ductal carcinoma in situ; Liver, liver cancer. Please see associated vignette for more details
Presenting problem/questions
1) What are the recommendations for breast cancer screening and management in CHEK2 carriers? 2) How should one approach selection of the appropriate gene panel in a time-limited situation? 3) Is there a potential for gene-gene interactions between the pathogenic CHEK2 variant and ATM VUS or BRIP1 VUS?
Panel Recommendations
The panel advised that, as per NCCN guidelines, evidence is insufficient to recommend consideration of RRM in CHEK2 carriers, and that management should be based on family history, treatment of the patient’s current cancer, and future cancer risk-tolerance given the limited data available to guide decision making. The panel reinforced NCCN guideline recommendations for annual mammogram and annual breast MRI in females with an inherited CHEK2 variant (Table 1). The panel also commented on the challenges of choosing the appropriate breast cancer specific gene panel, in particular, whether one should choose a high-risk breast cancer panel with reflex to moderate-risk genes if testing is initially ordered to guide surgical management recommendations. The panel concurred that a proximal surgery date may necessitate sending an abbreviated panel with rapid turnaround time to help decide between mastectomy and breast conserving surgeries. Regarding the question of potential significance of the findings of both a CHEK2 pathogenic variant and VUSs in ATM and BRIP1, the panel stated that there is no evidence for interaction of variants. The VUSs should be interpreted as uninformative and unless reclassified as pathogenic have no bearing on treatment or screening recommendations for the proband, nor should they be used to discern risk in family members. The panel emphasized that practices providing GCRA should have a follow-up model in place that performs regular review of the literature and updates carriers if a VUS is re-classified as pathogenic. Of note, two additional cases were submitted which discussed the question of gene-gene interaction, highlighting the frequency of identifying more than one variant on panel testing.
Participant Feedback
Thirty-four (45%) participants noted that they had seen similar cases in their GCRA practices. Fifty-seven (95%) fully or mostly concurred with the recommendations.
Case 5: Spectrum of Screening for CHEK2 Carriers
A 31-year-old female recently diagnosed with invasive breast cancer was referred for GCRA. The proband’s father was diagnosed with prostate cancer at age 43 and died of recurrence at age 65 (Figure 5). Her paternal aunt died of colon cancer at age 60 and a paternal uncle was diagnosed with prostate cancer at age 40. The patient was tested using a high-moderate-risk cancer panel, which detected a pathogenic CHEK2 variant, c.470C>T (p.Ile157Thr).
Fig. 5.
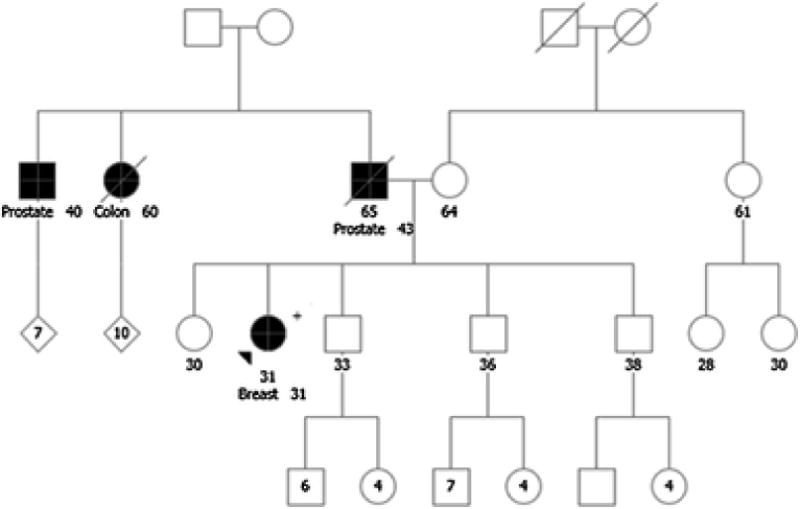
Case 5: Spectrum of Screening for CHEK2 Carriers. Colon, colorectal cancer; Prostate, prostate cancer; Breast, breast cancer. Please see associated vignette for more details
Presenting Problem/questions
1) What spectrum of cancers should one screen for in CHEK2 carriers? How does family history influence screening recommendations?
Expert Panel Recommendations
Pending completion of breast cancer treatment, the panel recommended that the proband undergo annual mammogram and breast MRI of remaining breast tissue. It was acknowledged that there is emerging evidence that breast cancer risk associated with the CHEK2 missense variant, c.470C>T (p.Ile157Thr) is less than that reported for the truncating variants; however, our current approach is to include MRI in screening recommendations for all carriers of a pathogenic CHEK2 variant, as we do not have sufficient data to make refined genotype/phenotype distinctions. For colonoscopy screening, the panel recommended colonoscopy every 5 years starting at age 40, though the association of colon cancer risk and this specific CHEK2 variant is not established. At the time of the study, specific recommendations for colon cancer screening in CHEK2 carriers were not included in NCCN guidelines, however, current guidelines are consistent with this recommendation (Table 1)[14, 41]. An association between CHEK2 variants and prostate cancer incidence are cited in the literature, but no screening guidelines are provided in NCCN [19, 20]. It was recommended that segregation analysis could be performed on the paternal uncle with prostate cancer to see whether the variant segregated with disease as this could be helpful for counseling the proband’s brothers; cancer site in a proband with a CHEK2 variant may influence the relative risk of specific cancers for his/her first-degree relatives [20]. Although the NCCN does not outline specific screening guidelines for prostate cancer in CHEK2 carriers, the panel recommended that annual PSA should be considered starting at age 40 based on family history. Finally, it is important to emphasize that mutations do not act in isolation and more research is needed to understand gene environment interactions in cancer development in CHEK2 carriers as well as other cancer predisposition genes.
Participant Feedback
Twenty-five (38%) participants noted that they had seen similar cases in their GCRA practices. Of the 12 (23%) participants who challenged the recommendations discussed, most questioned the merits of colon and prostate cancer screening in CHEK2 carriers. Participants indicated that their patients likely would not have access to recommended colonoscopies before age 50 in the absence of a significant family history of colorectal cancer.
Discussion
The increasing utilization of multigene panels evaluating high and moderate-risk genes has broadened the scope of GCRA while generating new challenges for clinical practitioners and affected families. This manuscript reviews recommendations for the management of patients with ATM and CHEK2 pathogenic variants, two frequently implicated moderate-risk genes included on many commercial testing panels, generated by a multidisciplinary, cross-sectional panel of GCRA clinicians at an annual genomic conference. Pathogenic variants in ATM and CHEK2 are associated with a moderately increased risk of breast cancer, RR 2.8 and 1.58-3.0, respectively [21]. However, the impact of screening and preventative interventions and spectrum of cancer risk beyond breast cancer associated with ATM and/or CHEK2 variants remain less well characterized. We reviewed 5 clinical cases involving ATM or CHEK2 encountered in the GCRA setting to help outline the current body of literature and also highlight expert recommendations for patient management. Key themes that surfaced during this discussion included: application of RRM in carriers of moderate-risk genes, accessibility of recommended screening and preventative services, and the influence of moderate-risk genes in the setting of a strong family history.
Application of RRM in carriers of moderate-risk genes
Discussion of the option of RRM is recommended in carriers of high-risk breast cancer genes such as BRCA1 or BRCA2 or individuals with a strong family history of breast cancer when genomic analysis does not reveal mutations in any of the known inherited cancer predisposition genes [7, 42–45]. This guideline is meant to balance the potential medical and psychological morbidities of RRM with up to a 90% decrease in the risk of developing breast cancer [46–49]. Important psychological considerations when pursuing RRM include the impact on a woman’s perceived quality of life, sexuality, and body image; investigations into whether there is a negative psychological impact on high-risk women following RRM are mixed [43]. Alternatively, the question of whether RRM, an invasive surgical intervention, is warranted as a preventative measure for a female carrier of a moderate-risk gene remains to be determined given concerns about over diagnosis and over treatment in healthy individuals. Specifically, NCCN guidelines outline that there is insufficient evidence to recommend RRM in carriers of ATM or CHEK2 variants based on associated relative risk of breast cancer [7]. However, panelists agreed that RRM may be discussed in ATM and CHEK2 carriers in the context of personal and family history, patient risk-tolerance, psychological and surgical morbidity, and alternative methods of risk reduction, such as chemoprevention and close surveillance. Ultimately, consideration of RRM is an individualized decision between the patient, her GCRA team, and her surgeon; and in the future, there are also likely to be increasing challenges by 3rd Party Payors. Prospective studies to define the risk reduction benefit of RRM in carriers of moderate-risk genes are not available to guide medical management. As such, more studies, with longer follow up, are needed to help GCRA practitioners educate patients when considering irreversible surgical intervention.
Accessibility of screening and preventative services
Performing heightened breast cancer screening measures such as breast MRI in carriers of variants in moderate-risk genes is outlined in current NCCN guidelines, although studies to support medical benefit and justify cost of intensive surveillance are lacking [7]. The panel participants stated that they observe heightened breast screening guidelines and shared the opinion that these guidelines facilitate obtaining insurance coverage for heightened screening although benefits are not as well defined. Additional surveillance studies advocated by experts, including pancreatic and colorectal cancer surveillance in ATM carriers and prostate cancer screening in CHEK2 carriers, raised a greater challenge for some panel participants. These interventions also lack a body of literature to support their implementation currently and are not recommended in NCCN. Barriers to these screening procedures include lack of insurance coverage and lack of access to trained specialists, particularly in the field of pancreatic cancer screening with EUS. This challenge raises the issue of equity of access to care in GCRA based on patient financial means or proximity to a specialized/major medical center. Over time, data from the Precision Medicine Initiative “All of Us” as well as ongoing registries and complementary consortia will help characterize the RR of additional cancers in ATM and CHEK2 carriers and lead to incorporation of additional cancer screening into national guidelines; although, one must use caution when interpreting registry data as this may not reflect true population risk estimates [50].. Until then, GCRA practitioners must consider cost effective ways to manage ATM and CHEK2 carriers, when national guidelines remain at the level of Expert Opinion.
Influence of moderate-risk genes in the setting of a strong family history
In some clinical situations, a patient undergoing GCRA may meet a level of risk that warrants heightened screening and chemoprophylaxis based on cancer risk models derived from family and personal history of breast cancer, prompting the question of whether genetic testing for moderate-risk genes adds value to the evaluation [29, 51, 52]. In this setting, genetic testing for moderate-risk genes may be warranted to help identify additional cancer risks. Further, identification of a moderate-risk gene may prompt cascade testing and segregation analysis to establish whether the variant is causative of the family cancer history, beneficial for patient counseling and future research. Unlike in the case of a high-risk gene, a family member who is a “true negative” and did not inherit a moderate-risk variant, does not eliminate his/her familial cancer risk. This interpretation of negative testing for moderate-risk genes emphasizes the importance of residual empiric risk based on family history. A pathogenic variant in a moderate-risk gene such as ATM or CHEK2 can only be interpreted as a part of the explanation in a family history of breast cancer.
The aforementioned themes highlight the need for additional research to better characterize cancer-specific relative risk, genotype-phenotype associations, influence of personal and family history, and impact of heightened surveillance and preventative measures in the evaluation and management of ATM and CHEK2 carriers. In the future, risk modifiers, such as the Polygenic Risk Scores, may also be incorporated to improve and better personalize risk stratification for ATM and CHEK2 mutation carriers[53]. Additional topics and challenges exist in the field of moderate-risk genes, which are beyond the scope of this manuscript. Research is ongoing to refine our understanding of moderate-risk genes and their utility in GCRA. Panel-based discussions and surveys of GCRA practitioners are incredibly useful as we aim to understand the current state of GCRA across the US, so that we may continue to refine our approach to the management of moderate-risk genes and welcome an era where comprehensive GCRA is widely available to the general public.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number R13CA206594-01 (PI: O. Olopade) and R25CA171998 (PIs: K. Blazer and J. Weitzel). A. West is supported by the National Cancer Institute of the National Institutes of Health under a Basic Medical Research Training in Oncology Award Number T32CA009566 (PI: O. Olopade). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: Dr. Olufunmilayo Olopade is co-founder of CancerIQ. All co-authors declare that they have no conflict of interest
Supplementary Material
Supplement 1. Cancer Genetics and Genomics Conference syllabus. Supplement 2. Participant survey
References
- 1.Hodgson SV. A Practical Guide to Human Cancer Genetics. Third. New York: Cambridge University Press; 2007. [Google Scholar]
- 2.Lindor NM, McMaster ML, Lindor CJ, Greene MH. Concise handbook of familial cancer susceptibility syndromes - second edition. J Natl Cancer Inst Monogr. 2008;38:1–93. doi: 10.1093/jncimonographs/lgn001. [DOI] [PubMed] [Google Scholar]
- 3.Offit K. Clinical Cancer Genetics: Risk Counseling and Management New York. New York: Wiley Liss; 1998. [Google Scholar]
- 4.Weitzel JN, Blazer KR, MacDonald DJ, Culver JO, Offit K. Genetics, genomics, and cancer risk assessment: State of the Art and Future Directions in the Era of Personalized Medicine. CA Cancer J Clin. 2011;61(5):327–59. doi: 10.3322/caac.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeMarco TA, Smith KL, Nusbaum RH, Peshkin BN, Schwartz MD, Isaacs C. Practical aspects of delivering hereditary cancer risk counseling. Semin Oncol. 2007;34(5):369–78. doi: 10.1053/j.seminoncol.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Hampel H, Sweet K, Westman JA, Offit K, Eng C. Referral for cancer genetics consultation: a review and compilation of risk assessment criteria. J Med Genet. 2004;41(2):81–91. doi: 10.1136/jmg.2003.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.National Comprehensive Cancer Network (NCCN) Genetic/Familial High-Risk Assessment: Breast and Ovarian Version 2. 2016 doi: 10.6004/jnccn.2016.0018. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Paull TT. Activation and regulation of ATM kinase activityin response to DNA double-strand breaks. Oncogene. 2007;26(56):7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 9.Cavaciuti E, Lauge A, Janin N, et al. Cancer risk according to type and location of ATM mutation in ataxia-telangiectasia families. Genes Chromosomes Cancer. 2005;42(1):1–9. doi: 10.1002/gcc.20101. [DOI] [PubMed] [Google Scholar]
- 10.Renwick A, Thompson D, Seal S, et al. ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet. 2006;38(8):873–5. doi: 10.1038/ng1837. [DOI] [PubMed] [Google Scholar]
- 11.Thompson D, Duedal S, Kirner J, et al. Cancer risks and mortality in heterozygous ATM mutation carriers. J Natl Cancer Inst. 2005;97(11):813–22. doi: 10.1093/jnci/dji141. [DOI] [PubMed] [Google Scholar]
- 12.Easton DF, Pharoah PD, Antoniou AC, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–57. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tung N, Lin NU, Kidd J, et al. Frequency of Germline Mutations in 25 Cancer Susceptibility Genes in a Sequential Series of Patients With Breast Cancer. J Clin Oncol. 2016;34(13):1460–8. doi: 10.1200/jco.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Comprehensive Cancer Network (NCCN) Genetic/Familial High-Risk Assessment: Colorectal Version 2. 2015 [Google Scholar]
- 15.Meijers-Heijboer H, van den Ouweland A, Klijn J, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*) 1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–9. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 16.Lee SB, Kim SH, Bell DW, et al. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res. 2001;61(22):8062–7. [PubMed] [Google Scholar]
- 17.Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71(2):432–8. doi: 10.1086/341943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suchy J, Cybulski C, Wokolorczyk D, et al. CHEK2 mutations and HNPCC-related colorectal cancer. Int J Cancer. 2010;126(12):3005–9. doi: 10.1002/ijc.25003. [DOI] [PubMed] [Google Scholar]
- 19.Cybulski C, Gorski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75(6):1131–5. doi: 10.1086/426403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gronwald J, Cybulski C, Piesiak W, et al. Cancer risks in first-degree relatives of CHEK2 mutation carriers: effects of mutation type and cancer site in proband. Br J Cancer. 2009;100(9):1508–12. doi: 10.1038/sj.bjc.6605038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tung N, Domchek SM, Stadler Z, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13(9):581–8. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell DW, Kim SH, Godwin AK, et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer. 2007;121(12):2661–7. doi: 10.1002/ijc.23026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couch FJ, Shimelis H, Hu C, et al. Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer. JAMA Oncol. 2017 doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor NS, Curcio LD, Blakemore CA, et al. Multigene Panel Testing Detects Equal Rates of Pathogenic BRCA1/2 Mutations and has a Higher Diagnostic Yield Compared to Limited BRCA1/2 Analysis Alone in Patients at Risk for Hereditary Breast Cancer. Ann Surg Oncol. 2015;22(10):3282–8. doi: 10.1245/s10434-015-4754-2. [DOI] [PubMed] [Google Scholar]
- 25.Blazer KR, MacDonald DJ, Ricker C, Sand S, Uman GC, Weitzel JN. Outcomes from intensive training in genetic cancer risk counseling for clinicians. Genet Med. 2005;7(1):40–7. doi: 10.109701/gim.0000151154.27612.49. [DOI] [PubMed] [Google Scholar]
- 26.Blazer KR, Macdonald DJ, Culver JO, et al. Personalized cancer genetics training for personalized medicine: improving community-based healthcare through a genetically literate workforce. Genet Med. 2011;13(9):832–40. doi: 10.1097/GIM.0b013e31821882b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Microsoft Excel. Redmond, WA: Microsoft; 2013. [Google Scholar]
- 28.Creswell JW. Mixed Methods Procedures Research design qualitative, quantitative, and mixed method approaches. 2nd. Thousand Oaks, Calif: Sage Publications; 2003. pp. 208–25. [Google Scholar]
- 29.Claus EB, Risch N, Thompson WD. Autosomal dominant inheritance of early-onset breast cancer. Implications for risk prediction. Cancer. 1994;73(3):643–51. doi: 10.1002/1097-0142(19940201)73:3<643::aid-cncr2820730323>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 30.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 31.National Comprehensive Cancer Network (NCCN) Breast Cancer Screening and Diagnosis Version 1. 2017 [Google Scholar]
- 32.Roberts NJ, Jiao Y, Yu J, et al. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov. 2012;2(1):41–6. doi: 10.1158/2159-8290.cd-11-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petersen GM. Familial pancreatic cancer. Semin Oncol. 2016;43(5):548–53. doi: 10.1053/j.seminoncol.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canto MI, Harinck F, Hruban RH, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62(3):339–47. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartsch DK, Slater EP, Carrato A, et al. Refinement of screening for familial pancreatic cancer. Gut. 2016;65(8):1314–21. doi: 10.1136/gutjnl-2015-311098. [DOI] [PubMed] [Google Scholar]
- 36.Fletcher O, Johnson N, dos Santos Silva I, et al. Missense variants in ATM in 26,101 breast cancer cases and 29,842 controls. Cancer Epidemiol Biomarkers Prev. 2010;19(9):2143–51. doi: 10.1158/1055-9965.epi-10-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bretsky P, Haiman CA, Gilad S, et al. The relationship between twenty missense ATM variants and breast cancer risk: the Multiethnic Cohort. Cancer Epidemiol Biomarkers Prev. 2003;12(8):733–8. [PubMed] [Google Scholar]
- 38.Bernstein JL, Haile RW, Stovall M, et al. Radiation exposure, the ATM Gene, and contralateral breast cancer in the women’s environmental cancer and radiation epidemiology study. J Natl Cancer Inst. 2010;102(7):475–83. doi: 10.1093/jnci/djq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.National Comprehensive Cancer Network (NCCN) Genetic/Familial High-Risk Assessment Breast and Ovarian Version 1. 2018 [Google Scholar]
- 40.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2015 Nov 17; doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Comprehensive Cancer Network (NCCN) Genetic/Familial High-Risk Assessment: Colorectal Version 3. 2017 doi: 10.6004/jnccn.2017.0176. [DOI] [PubMed] [Google Scholar]
- 42.Boughey JC, Attai DJ, Chen SL, et al. Contralateral Prophylactic Mastectomy (CPM) Consensus Statement from the American Society of Breast Surgeons: Data on CPM Outcomes and Risks. Ann Surg Oncol. 2016;23(10):3100–5. doi: 10.1245/s10434-016-5443-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunt KK, Euhus DM, Boughey JC, et al. Society of Surgical Oncology Breast Disease Working Group Statement on Prophylactic (Risk-Reducing) Mastectomy. Ann Surg Oncol. 2017;24(2):375–97. doi: 10.1245/s10434-016-5688-z. [DOI] [PubMed] [Google Scholar]
- 44.Giuliano AE, Boolbol S, Degnim A, Kuerer H, Leitch AM, Morrow M. Society of Surgical Oncology: position statement on prophylactic mastectomy. Approved by the Society of Surgical Oncology Executive Council, March 2007. Ann Surg Oncol. 2007;14(9):2425–7. doi: 10.1245/s10434-007-9447-z. [DOI] [PubMed] [Google Scholar]
- 45.Mai PL, Lagos VI, Palomares MR, Weitzel JN. Contralateral risk-reducing mastectomy in young breast cancer patients with and without genetic cancer risk assessment. Ann Surg Oncol. 2008;15(12):3415–21. doi: 10.1245/s10434-008-0160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22(6):1055–62. doi: 10.1200/jco.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 47.Brandberg Y, Sandelin K, Erikson S, et al. Psychological reactions, quality of life, and body image after bilateral prophylactic mastectomy in women at high risk for breast cancer: a prospective 1-year follow-up study. J Clin Oncol. 2008;26(24):3943–9. doi: 10.1200/jco.2007.13.9568. [DOI] [PubMed] [Google Scholar]
- 48.Hartmann LC, Schaid DJ, Woods JE, et al. Efficacy of bilateral prophylactic mastectomy in women with a family history of breast cancer. N Engl J Med. 1999;340(2):77–84. doi: 10.1056/nejm199901143400201. [DOI] [PubMed] [Google Scholar]
- 49.den Heijer M, Seynaeve C, Timman R, et al. Body image and psychological distress after prophylactic mastectomy and breast reconstruction in genetically predisposed women: a prospective long-term follow-up study. Eur J Cancer. 2012;48(9):1263–8. doi: 10.1016/j.ejca.2011.10.020. [DOI] [PubMed] [Google Scholar]
- 50.National Institutes of Health. All of Us Research Program. 2018 Jan 8; [Google Scholar]
- 51.Tyrer J, Duffy SW, Cuzick J. A breast cancer prediction model incorporating familial and personal risk factors. Stat Med. 2004;23(7):1111–30. doi: 10.1002/sim.1668. [DOI] [PubMed] [Google Scholar]
- 52.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81(24):1879–86. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 53.Chowdhury S, Dent T, Pashayan N, et al. Incorporating genomics into breast and prostate cancer screening: assessing the implications. Genet Med. 2013;15(6):423–32. doi: 10.1038/gim.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


